Abstract
Due to plummeting costs, whole genome sequencing of patients and cancers will soon become routine medical practice; however, we cannot currently predict how non-coding genotype affects cellular gene expression. Gene regulation research has recently been dominated by observational approaches that correlate chromatin state with regulatory function. These approaches are limited to the available genotypes and cannot scratch the surface of possible sequence combinations, and thus there is a need for perturbation-based approaches to better understand how DNA encodes gene regulatory functions. CRISPR/Cas9 genome editing has revolutionized our ability to alter genome sequence, and CRISPR/Cas9-based assays have already begun to contribute to new paradigms of gene regulation. We discuss the variety of arenas in which current and future CRISPR-based technologies will aid in developing predictive understanding of how genome sequence leads to gene regulatory function.
“Once a new technology rolls over you, if you’re not part of the steamroller, you’re part of the road.”
-Stewart Brand
CRISPR/Cas9 has hit the research community like a steamroller. Seemingly overnight, manipulation of the genome has transformed from a daunting task into a simple CRISPR/Cas9 targeting. While there has been much talk about the possible long-term implications of genome editing on humanity, from designer babies to genetic control of ecosystems, a more immediate revolution is underway in epigenetics research. CRISPR/Cas9 technology is providing a new perturbation-based approach to this field that has been dominated by observational research, promising to provide answers to questions that have been impossible to ask previously. In this article, we review the current status of CRISPR/Cas9 gene regulation research and what areas are most ripe for future insights from this disruptive technology.
The grand challenge in epigenetics
The advent of CRISPR/Cas9 follows on the heels of an equally momentous technological breakthrough: the decrease in the cost of DNA sequencing. The cost of human whole genome sequencing (WGS) has fallen from ~$10,000,000 in 2006 to $10,000 in 2011 to $1,000 currently [1]. In spite of this precipitous drop, whole exome sequencing (WES), sequencing only the 2% of the genome that codes for proteins, remains an order-of-magnitude cheaper. As a result, many large-scale sequencing projects have opted to perform WES [2]. Such large-scale WES has widened the list of disease-linked genes [2–4]; however, information on how diversity of non-coding DNA sequence in normal and cancerous genomes correlates with disease state has been slower because of the lack of data.
It seems that an inflection point where WGS becomes cheap enough to be the routine choice over WES is approaching, and a torrent of human WGS data with corresponding phenotypic data is not far off. Genomics England plans on sequencing 100,000 whole genomes by 2017 [5]; not to be outdone, the USA NIH’s Precision Medicine Initiative aims to sequence 1,000,000 genomes by 2020 [6]. Yet, making sense of this surge of data is not as straightforward as with WES data where the rules of missense and frameshift mutations simplify interpretation. We cannot currently predict how non-coding genotype affects molecular phenotypes such as nearby gene expression and are even worse at predicting how it affects organismal phenotypes such as disease susceptibility and cancer progression.
The approaching WGS revolution therefore creates an imperative to understand the rules by which DNA sequence encodes gene regulation. The vast majority of current effort in this arena is through correlative research. This is because observational epigenetic technologies such as ChIP-Seq to detect transcription factor binding sites, DNase-Seq to profile open chromatin, histone mark profiling to characterize chromatin states, and QTL studies to pair SNPs with differing molecular phenotypes are much more mature than those that perturb genome sequence.
While such observational research has vastly improved our understanding of epigenetics, there are inherent limitations to this approach. First, there are more combinations of a dozen nucleotides than there are people on earth, so even WGS and downstream molecular analysis of every human on the planet will not scratch the surface of possible genotypes. Second, no genotype exists in isolation, so it is not possible to control for the effects of differences in surrounding sequences. Current genetic association studies have power to detect disease links to single SNPs; however, detecting significantly disease-linked SNP combinations will require exponentially more patients which may not be possible. Third, genomes are shaped by evolution, which strongly confounds causal understanding of function. For example, redundancy in important regulatory regions such as promoters is selected for, so key motifs may be both causal to and dispensable for the formation of a particular promoter [7].
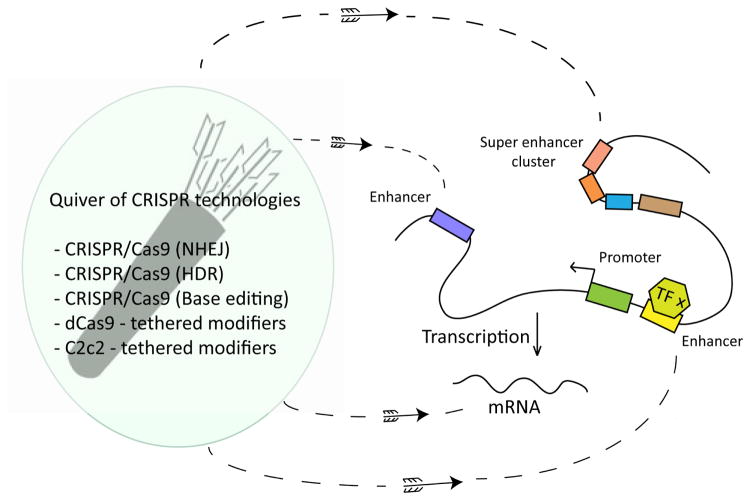
For all of these reasons, perturbational approaches to understanding genome sequence are sorely needed, and CRISPR/Cas9 has revolutionized our ability to alter genome sequence at just the right time (Figure 1). Below we describe how CRISPR/Cas9-based assays have begun to shed light on gene regulation and how it might be employed to do so in the future.
Figure 1. Applying CRISPR tools to gene regulation.
CRISPR provides a variety of tools to edit the genome. Applying each of these tools to enhance our understanding of gene regulation provides an exciting avenue for ongoing and future research.
Deciphering DNA codes in the non-coding genome
Given that each person has a never-before-seen genotype, if we are to interpret human genome sequencing data, we must be able to predict the full epigenetic consequences of an arbitrary DNA base change at every genomic position. Considering that full epigenetic consequences include binding of thousands of transcription factors in thousands of cell states, this is a daunting task.
The predominant approach used to date is to train a computational algorithm with genome-wide data for the epigenetic characteristic of interest in a particular cell state (e.g. DNase I hypersensitivity in lymphoid cells) and then test this algorithm’s ability to predict this epigenetic characteristic in held-out regions of the genome or alternate genotypes. It is assumed that this performance on held-out sequences equates to the algorithm’s general predictive power for novel sequences. One recent example of the state-of-the-art is DeepSea [8], which trains a deep learning algorithm on 690 TF binding profiles for 160 different TFs, 125 DHS profiles and 104 histone-mark profiles generated by the ENCODE and Roadmap consortia [9,10] and demonstrates impressive predictive power. However, algorithms like DeepSea are still far from the goal outlined above. They perform poorly or remain untested at the following tasks that will be required for bona fide gene regulatory codebreaking algorithms: predicting the magnitude of sequence-dependent changes in epigenetic characteristics; predicting cell type-specific epigenetic characteristics, especially of cell types without corresponding training data [11]; predicting the functional consequences of sequence changes on cells and organisms [8].
It is our belief that this approach of training algorithms on genome-wide epigenetic profiling data is approaching a limit to its accuracy unlikely to be overcome by more genomic data or more sophisticated algorithms. The genome is not large or random enough and there are not enough variant individuals to gather sufficient training data to predict epigenetic outcomes accurately. A solution is to alter genome sequence in high-throughput using CRISPR/Cas9, opening up a limitless trove of new training data to improve modeling accuracy.
Approaches that alter DNA sequence in high-throughput and assess epigenetic outcomes pre-date CRISPR. Libraries of DNA sequence variants have been used productively in reporter assays to assess the gene activation potential of each sequence [12–14] although the use of unintegrated episomal vectors likely alters the interpretation of such data. Such high-throughput library screening has cleverly been extended to improve prediction of RNA splicing [15] and protein translation [16], proving the benefits of training predictive models on perturbation-based data.
CRISPR/Cas9 has begun to be used in several ways to expand genotypic training data in a controlled fashion. The first approach employs CRISPR/Cas9 cleavage followed by non-homologous end-joining (NHEJ), an error-prone form of DNA damage repair that creates random indels and thus shuffles the local genotype at a precisely defined locus. Inducing CRISPR/Cas9-based NHEJ repair at an enhancer region in millions of cells followed by flow cytometric separation of cells based on reporter gene expression yields millions of variant genotypes paired with their gene regulatory phenotypic consequence. Deep sequencing of the enhancer region in such flow cytometrically separated populations has enabled the precise mapping of functionally important bases at a TF motif in that enhancer [17]. A similar approach was recently reported using zinc finger nuclease-dependent NHEJ [18].
The second approach employs CRISPR/Cas9 cleavage followed by homology-directed repair (HDR) to generate an array of local genotypes precisely determined by the library of input sequences. This approach was used to perform saturation replacement of two codons in the BRCA1 gene [19], an approach that could be extended to non-coding regions. Also recently, CRISPR/Cas9-based HDR was used to insert a library of 12,000 175-bp sequences into a defined genomic locus with minimal prior chromatin accessibility, and DNase I hypersensitivity analysis was used to assess how each of the DNA sequences encodes accessibility in this controlled context [11].
Comparing the NHEJ and HDR approaches reveals trade-offs. NHEJ can be performed efficiently at any locus in any cell type, can be multiplexed easily, and the number of resulting alleles is essentially limitless; however, the spectrum of CRISPR/Cas9-induced NHEJ mutations is dominated by short deletions and does not resemble the most common germline genotypic changes. HDR enables control of genotypes, yet it is more technically arduous, difficult to multiplex, and repair efficiency is lower and limited to certain cell types that undergo efficient HDR. Additionally, library oligonucleotide synthesis approaches currently allow a maximum of ~200 bp, limiting the breadth of what genomic HDR can address until synthesis improves [20]. Thus, both approaches will likely be useful in the future.
High-throughput in situ genome replacement assays should be paired in the future with a diverse set of downstream assays detecting TF binding, histone marking, and DNA methylation [21]. Importantly, testing the same sequences in different cell types will improve prediction of cell type-specific functions. An additional mode of CRISPR/Cas9-induced sequence change was recently developed by fusing Cas9 with a cytosine deaminase to enable spatially controlled C→T substitution in the genome [22]. By allowing predictable sequence change at each targeted locus, CRISPR/Cas9 base editing is ideally suited to multiplexed screens to assess the importance of SNPs (at least C→T SNPs) on gene expression and cellular phenotype. These approaches, by allowing controlled assessment of how an arbitrarily large collection of DNA sequences give rise to epigenetic phenotypes, will improve our ability to predict how genome sequence leads to gene regulatory phenotypes.
The elements of gene regulation
TFs bind in clusters known as promoters and enhancers, the chief functional units of gene regulation. Deciphering how such regulatory elements convey information is one of the key outstanding challenges in gene regulation research [23]. The workhorse assay to understanding the function of cis-regulatory sequence is the reporter assay. It can be scaled up to examine millions of putative regulatory sequences and can yield quantitative information on each sequence [13,24,25]. While valuable, this assay has several limitations. Technically, it is typically performed in episomal or randomly integrated contexts with short distances separating enhancers and promoters, possibly leading to inaccurate reflection of native activity. These flaws can all be solved by CRISPR/Cas9-based HDR. On a more fundamental level, however, it can only identify sequences that are sufficient to encode fully functional enhancers/promoters and thus ignores any sequence that is necessary but not sufficient for gene regulation. Since elements are tested individually, it does not yield any information about how gene regulatory regions combine to achieve target levels of gene expression. It also does not study regulatory regions in their native genomic context.
CRISPR/Cas9 has enabled a distinct approach in which regulatory elements are scanned by mutation to identify required regulatory elements. We developed the Multiplexed Editing Regulatory Assay (MERA), an assay in which a library of CRISPR guide RNAs (gRNAs) is constructed to tile a large swath of non-coding genomic space surrounding a gene of interest in an unbiased fashion [17]. The gene of interest is labeled with GFP, and the gRNA library is added such that each cell has a single focal mutation (on average affecting ~10 bp) in the surrounding genomic space. By detecting gRNAs enriched in cells that have partially or completely lost GFP, we identify which regions in the assayed genomic space are required for expression of the target gene. Applying MERA to four embryonic stem cell-specific genes, we found that gRNAs targeting the expected regions such as the GFP sequence, the gene body, promoter, and a subset of nearby enhancers were the most likely to induce loss of GFP expression. More interestingly, we also found that a number of unexpected non-coding regions were required for gene expression, including the promoters of neighboring genes and a set of regions with no known DNase I hypersensitivity or histone modifications associated with active chromatin, and we found that some annotated enhancers were dispensable for gene expression.
Other CRISPR/Cas9 non-coding region tiling screens have identified a class of temporarily required enhancers [26] and have identified distinct mechanisms of regulation in human and mouse at a conserved enhancer [27]. Additionally, a screen targeting most genomic p53 and ESR1 binding sites identified the individual sites most responsible for the oncogenic roles of these TFs in particular tumor cell line models [28]. Thus, CRISPR/Cas9 screens provide a new approach to systematically determine which non-coding regions surrounding a gene are required for expression.
These are early days for CRISPR/Cas9 mutagenesis screens, and many intriguing applications of this approach have yet to be explored. Replacing NHEJ mutation with deletion or base editing may alter sensitivity of the approach. Current screens have used blunt phenotypic readouts such as high, low, and absent expression or cell survival vs. death, so more subtle readouts are a must. So far, regulatory elements have been inactivated one at a time, and combinatorial CRISPR/Cas9 mutation will be useful to disentangle redundancies and dependencies among elements. Evidence from three-dimensional genome organization suggests that elements cluster in higher order structures [29], so expanding screens to larger genomic regions may hold additional surprises.
Super-enhancers/stretch enhancers (SEs), defined either by their length or amount of associated Mediator protein, have been shown to play an important role in controlling cell state [30–32]. It remains an open question whether SEs are qualitatively different than standard enhancers, and recently a spate of reports have used CRISPR/Cas9 to delete components of SEs to better understand their function [33–35]. Two of these reports suggest that SEs function hierarchically with vulnerable subparts whose mutation inactivates SE function [34,35], while one suggests that subparts within the SE act additively and thus indistinguishably from a collection of standard enhancers [33]. Interestingly, none of these results indicate that SEs act differently than standard enhancers. None report that SEs are particularly robust to mutation, which might be expected to result from the spatial concentration of enhancers. Such robustness could be queried using tiled CRISPR/Cas9 mutation screening. Additionally, HDR should be used to disrupt the physical co-localization of SE subparts to address whether their proximity is vital to their function. All in all, CRISPR-based perturbation promises to unlock the language by which gene regulatory elements instruct gene expression.
How best to “compress” gene regulatory function
One of the main promises of profiling epigenetic state is to compress information in the genome. For coding regions, such compression is straightforward and highly useful. Trinucleotide sequences can be represented by the amino acid they encode, compressing 64 possible trinucleotides into 21 amino acids. This compression is “lossy”, as codon decisions influence translation rate, mRNA folding, splicing and expression, which are ignored when coding regions are reduced to codons [36,37], yet it has proved immensely useful in interpreting genome function. Can we perform similar compression of regulatory DNA?
The most common regulatory DNA compression approaches divide the genome into “chromatin states” (e.g. H3K4Me1+H3K27Ac+ = active enhancer)[38], and these models show power in important genome interpretation tasks such as predicting which non-coding variants are likely to be deleterious. However, it is rarely asked how lossy these compression approaches are and which genomic features conserve the most predictive power. CRISPR/Cas9 provides an ideal tool for this question.
CRISPR/Cas9-based approaches are already finding that regulatory elements that surround the same gene and that share a chromatin state are quite heterogeneous in their effect on local gene expression [17,26,34]. This functional heterogeneity of elements predicted to be in an active chromatin state has been found consistently in reporter assay screens as well [39,40]. Additionally, there is some evidence that enhancers only function when paired to certain types of promoters [41–43], a pairing that appears to occur at the level of individual transcription factor binding sites that do not impart unique histone mark profiles.
Two observations about gene regulation cast doubt on whether chromatin states are the appropriate tool to bin regulatory element activities. First, cells utilize an array of different mechanisms to modulate RNA polymerase function above and beyond simple recruitment to promoters by coactivators [44], and this heterogeneity is not captured in chromatin states. Second, probably the chief evolutionary reason for dynamic gene regulation is to allow cells to respond to outside stimuli through signaling cascades, and thus it might make sense to account for each of these pathways separately to adequately predict how a cell will respond to a given perturbation. Nonetheless, given that there are three million enhancers each presumably evolved for a specific task [45], there may be no better way to “skin the cat” than chromatin states.
Studies that systematically address how replaceable a genetic element is with a different one of the same chromatin state are imperative. Using CRISPR/Cas9-based HDR, a genomic locus could be swapped out with thousands of replacement elements from elsewhere in the genome to compare function. Given 15 (or 100) possible compressed states, would an element’s chromatin state be the best predictor of its function or would features such as individual transcription factor binding sites or combinations thereof allow for more accuracy? Performing such assays in multiple native genomic loci with different modes of regulation, assaying for promoters, enhancers, and other elements, and performing multiplexed “slot machine” HDR to address pairing of elements will all help to address the most predictive features for genome abstraction.
One note of caution in this arena is that standard phenotypic readouts of gene regulatory elements such as their level of gene activation may overestimate the similarity of distinct elements. Work in which the Drosophila Snail promoter was replaced with distinct non-poised promoters showed only minimal effects in maximal gene activation but wide enough variation in the timing of transcriptional onset to disrupt the precisely coordinated process of mesoderm development [46]. Typical high-throughput cell line experiments ignore parameters such as timing and cell type-specificity [24] that may be crucial to the evolved function of regulatory elements. Thus, CRISPR/Cas9-based high-throughput screens must be paired with readouts that define function in holistic ways that accommodate stochasticity, timing, cell type-specificity and stimulus-response capabilities.
In the end, compressing genome information will always be a balance of information loss and savings in computational space and effort, and assessing the value of compression will depend on the breadth and sensitivity of functional assays. Yet, it is our opinion that the epigenetics field has devoted an overabundance of resources to histone mark-based compression without strong evidence that such assays hold the most predictive value. In situ, high-throughput locus replacement studies should enable much better estimation of which genomic features best summarize regulatory element function.
The causality of epigenetic marks
In spite of the vast efforts at mapping histone modifications and DNA methylation in different cell states, the importance of such epigenetic marks in causing epigenetic states as opposed to simply correlating with them is poorly understood. To address causality, focal manipulation of epigenetic state is required, and it has traditionally been tedious.
Perturbing histone modifying enzymes has pleiotropic effects on cells and organisms [47]. Thus, in order to pinpoint roles for single modifications at individual loci, researchers have attached histone modifying enzymes to DNA binding domains such as TALEs, zinc finger proteins, and more recently dCas9 [48]. To crudely summarize a vast body of literature, altering histone modifications can change function, for example inactivating an active gene, yet it provides predominantly short-term memory that is overridden by endogenous transcription factor binding activity.
One landmark study addressed the memory of targeted epigenetic modification by engineering small molecule-mediated recruitment of an epigenetic repressor (HP1α) to a defined genomic locus [49]. After short-term (7 day) small molecule-induced epigenetic silencing of an active locus, activity was quickly restored when the small molecule was removed; however, long-term (4.5 week) silencing was stably inherited. Short-term silencing correlated with H3K9Me3 deposition, while long-term silencing was correlated with DNA methylation, allowing the conclusion that the H3K9Me3 mark in this case is insufficient to propagate silencing.
Efforts to root out the causality of epigenetic marks have fallen short in several key areas in which further CRISPR/Cas9-based research could be beneficial. First, dynamic studies will be useful to determine the long-term effects of histone mark manipulation. How long-lasting are artificial histone mark changes and do they persist through cell division? Second, the chromatin code is highly combinatorial [50], yet manipulations to chromatin state have to date been individual. Technologies such as SunTag, in which a string of 10 antigens are attached to dCas9 [51], should allow locus-specific recruitment of combinations of chromatin modifiers. Third, it is clear that transcription factors interact with chromatin modifiers in complex ways, both inducing and responding to chromatin modifications. For example, Nrf1 binding is blocked by DNA methylation [52], but once bound, Nrf1 can open chromatin [53]. Deriving accurate mechanistic models for such interactions with feedback will be aided by combinatorial dCas9-based recruitment of transcription factors and chromatin modifiers, preferably with a dynamic component. One caveat to this line of work is that Cas9 by itself influences nucleosome positioning and adjacent transcription factor binding [54], so manipulating chromatin using Cas9 has side effects that must be controlled for. Altogether, epigenetic research lacks cause-and-effect mechanism, and CRISPR/Cas9 introduces a perfect tool set to begin such inquiry.
Contributions of RNA to epigenetic complexes
Many epigenetic modifying complexes such as the DNA methyltransferase machinery and Polycomb complex are known to associate with RNA [55,56]. It has even been proposed recently that the function of the transcription factor YY1 is modulated by local RNA [57]. However, the precise roles and sequence determinants for RNA in guiding these epigenetic complexes to targets in cis or in trans have not been verified. The newly discovered RNA-cleaving CRISPR enzyme C2c2, thus far only shown to work in bacteria, presents an intriguing tool to perturb interactions between specific RNAs and specific protein complexes [58]. Fusions between C2c2 and specific epigenetic complexes have the potential to unravel the local non-coding functions of RNA in gene regulation, which would provide a major leap forward in our understanding of gene regulation.
Conclusions
CRISPR/Cas9-based technologies will clearly play an outsized role in improving our understanding of gene regulation. This improved understanding should ultimately lead to better predictive interpretation of patient germline and cancer genome sequences, a crucial goal for the future of precision medicine. Could CRISPR also play a role in therapeutic intervention when gene regulation goes awry? Currently, it is difficult to control the outcome of CRISPR/Cas9-mediated genome editing in a high percentage of cells, even in vitro. As a result, seamless genome replacement is far off, and even introducing loss-of-function mutations may be dangerous, as chromosomal translocations are known to occur at detectable frequencies after targeting a population of cells [59]. Cas9 base editing [22] may offer a more attractive option for altering short regulatory regions in patient cells, although the APOBEC enzymes used in current versions of these strategies are potent oncogenic mutagens [60] that may necessarily introduce risk of off-target mutation. Nonetheless, the CRISPR field is young, and it is exciting to imagine a future in which predictive models of gene regulation can guide genome editing in patients to treat a panoply of diseases.
Acknowledgments
The authors acknowledge funding from The National Institutes of Health 1K01DK101684-01 and 1R01HG008363-01; the Human Frontier Science Program Young Investigator Grant RGY0084/2014; a Netherlands Organisation for Scientific Research VIDI; and a BWH Biomedical Research Institute Health and Technology Innovation Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.NHGRI. The Cost of Sequencing a Human Genome [Internet] 2016. [no volume] [Google Scholar]
- 2.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald TW, Gerety SS, Jones WD, van Kogelenberg M, King DA, McRae J, Morley KI, Parthiban V, Al-Turki S, Ambridge K, et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2014;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The 100,000 Genomes Project [Internet]. [date unknown], [no volume].
- 6.NIH awards $55 million to build million-person precision medicine study [Internet]. [date unknown], [no volume].
- 7.Spivakov M. Spurious transcription factor binding: non-functional or genetically redundant? BioEssays News Rev Mol Cell Dev Biol. 2014;36:798–806. doi: 10.1002/bies.201400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium EP. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto T, Sherwood RI, Kang DD, Rajagopal N, Barkal AA, Zeng H, Emons BJM, Srinivasan S, Jaakkola T, Gifford DK. A synergistic DNA logic predicts genome-wide chromatin accessibility. Genome Res. 2016 doi: 10.1101/gr.199778.115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–70. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Jr, Kinney JB, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–7. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patwardhan RP, Lee C, Litvin O, Young DL, Pe’er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberg AB, Patwardhan RP, Shendure J, Seelig G. Learning the Sequence Determinants of Alternative Splicing from Millions of Random Sequences. Cell. 2015;163:698–711. doi: 10.1016/j.cell.2015.09.054. [DOI] [PubMed] [Google Scholar]
- 16.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351:aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopal N, Srinivasan S, Kooshesh K, Guo Y, Edwards MD, Banerjee B, Syed T, Emons BJM, Gifford DK, Sherwood RI. High-throughput mapping of regulatory DNA. Nat Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vierstra J, Reik A, Chang K-H, Stehling-Sun S, Zhou Y, Hinkley SJ, Paschon DE, Zhang L, Psatha N, Bendana YR, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015;12:927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–3. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Cai Y, Carlson R, Chakravarti A, Cornish VW, Holt L, et al. The Genome Project-Write. Science. 2016;353:126–127. doi: 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]
- 21.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 22.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levo M, Segal E. In pursuit of design principles of regulatory sequences. Nat Rev Genet. 2014;15:453–468. doi: 10.1038/nrg3684. [DOI] [PubMed] [Google Scholar]
- 24.Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. Suboptimization of developmental enhancers. Science. 2015;350:325–328. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold CD, Gerlach D, Stelzer C, Boryń Ł, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–7. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 26.Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J, Maliskova L, Guan K-L, Shen Y, Ren B. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 2016;26:397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 30.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, et al. NISC Comparative Sequencing Program. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci US A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin HY, Willi M, Yoo KH, Zeng X, Wang C, Metser G, Hennighausen L. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat Genet. 2016;48:904–911. doi: 10.1038/ng.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan G-C, et al. Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev Cell. 2016;36:9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shabalina SA, Spiridonov NA, Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013;41:2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birnbaum RY, Patwardhan RP, Kim MJ, Findlay GM, Martin B, Zhao J, Bell RJA, Smith RP, Ku AA, Shendure J, et al. Systematic dissection of coding exons at single nucleotide resolution supports an additional role in cell-specific transcriptional regulation. PLoS Genet. 2014;10:e1004592. doi: 10.1371/journal.pgen.1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–11. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014;24:1595–1602. doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohler U, Wassarman DA. Promoting developmental transcription. Dev Camb Engl. 2010;137:15–26. doi: 10.1242/dev.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–44. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153:976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with “epigenetic” drugs: an update. Mol Oncol. 2012;6:657–682. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–60. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 51.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 53.Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–8. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barkal AA, Srinivasan S, Hashimoto T, Gifford DK, Sherwood RI. Cas9 Functionally Opens Chromatin. PLOS ONE. 2016;11:e0152683. doi: 10.1371/journal.pone.0152683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meller VH, Joshi SS, Deshpande N. Modulation of Chromatin by Noncoding RNA. Annu Rev Genet. 2015;49:673–695. doi: 10.1146/annurev-genet-112414-055205. [DOI] [PubMed] [Google Scholar]
- 57.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]