Mycobacterial glucosylglycerate hydrolase is a highly conserved enzyme that is involved in recovery from nitrogen starvation. Here, the production, crystallization and structure solution of Mycobacterium hassiacum glucosylglycerate hydrolase using two-wavelength anomalous diffraction of selenomethionine-substituted crystals are presented.
Keywords: Mycobacterium hassiacum, GH63, glucosylglycerate hydrolase, nitrogen starvation, multiwavelength anomalous diffraction
Abstract
Glucosylglycerate hydrolase is highly conserved among rapidly growing mycobacteria and has been found to be involved in recovery from nitrogen starvation by promoting the rapid mobilization of the glucosylglycerate that accumulates under these conditions. Here, the production, crystallization and structure determination of glucosylglycerate hydrolase from Mycobacterium hassiacum using two-wavelength anomalous diffraction of selenomethionine-substituted crystals are described. The monoclinic (space group P21) crystals diffracted to ∼2.0 Å resolution at a synchrotron-radiation source and contained four molecules in the asymmetric unit, corresponding to a Matthews coefficient of 3.07 Å3 Da−1 and a solvent content of 59.9%. The quality of the experimental phases allowed the automated building of 1677 of the 1792 residues in the asymmetric unit.
1. Introduction
Nontuberculous mycobacteria (NTM), which include species other than the Mycobacterium tuberculosis complex and M. leprae, are receiving increasing attention. Many of these ubiquitous environmental bacteria are opportunistic pathogens that are capable of causing lymphadenopathy in children, pulmonary, skin and soft-tissue infections or even disseminated infections, particularly in susceptible individuals (Griffith et al., 2007 ▸; Piersimoni & Scarparo, 2008 ▸; Henkle & Winthrop, 2015 ▸). Since NTM are exposed to several environmental challenges, they display high resilience against stress conditions, including nutrient starvation, desiccation, pH and temperature variations, as well as antibiotic and disinfectant action (Falkinham, 2010 ▸). These characteristics allow them to colonize artificial environments such as domestic and hospital water-distribution systems, from where they easily access susceptible hosts. Moreover, NTM are naturally resistant to common antibiotics and some infections do not even respond to aggressive antitubercular treatments (Shahraki et al., 2015 ▸; Griffith et al., 2015 ▸; Nunes-Costa et al., 2016 ▸). NTM infections are thus becoming a considerable clinical challenge for which therapeutic solutions are scarce. No significant treatment advances for NTM infection in general have recently been accomplished and the increasing resistance of some mycobacterial species to the therapeutics currently in use reinforces the need for more active drug development (Nessar et al., 2012 ▸).
Glucosylglycerate hydrolase (EC 3.2.1.–) from M. hassiacum (MhGgH) catalyses the hydrolysis of glucosylglycerate (GG), which is accumulated during growth with limited nitrogen, into glucose and glycerate (Alarico et al., 2014 ▸). Mycobacteria seem to accumulate GG as part of a global response to nitrogen stress, so a mechanism relying on hydrolysis by MhGgH may allow the assimilation of this metabolite when nitrogen availability is restored and thus is likely to contribute to the rapid energy mobilization required for cell growth and division (Behrends et al., 2012 ▸; Nunes-Costa et al., 2017 ▸). This hydrolase is highly conserved among rapidly growing mycobacteria, reflecting its importance during evolution and its potential as a drug target for the treatment of NTM infections. On the basis of its amino-acid sequence, MhGgH is grouped into glycoside hydrolase family 63 (GH63) and it is likely to adopt the typical (α/α)6 architecture of this family. The GH63 family contains more than 1600 members, including a single enzyme sharing 36% amino-acid sequence identity with MhGgH that has been structurally characterized (Miyazaki et al., 2015 ▸). Here, we describe the expression, purification, crystallization and structure determination of MhGgH.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Production of MhGgH
The gene coding for MhGgH was amplified by PCR from a pET-30a-based construct (Alarico et al., 2014 ▸) and cloned into the NcoI/EcoRI sites of pETM11, giving the pETM11-MhGgH expression vector (see Table 1 ▸ for details). Escherichia coli BL21 (DE3) cells transformed with the pETM11-MhGgH plasmid were grown at 301 K in LB medium supplemented with 50 µg ml−1 kanamycin to an OD600 of 0.7–1.0. At this point, the temperature was decreased to 298 K and expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After overnight growth, the cells were harvested, resuspended in lysis buffer [20 mM Tris–HCl pH 8.0, 500 mM NaCl, 5 mM MgCl2, 1 µg ml−1 DNAse, 0.3 mg ml−1 lysozyme, 1 mM phenylmethylsulfonyl fluoride (PMSF)] and stirred gently on ice for 1 h. The protein extract was clarified by centrifugation, supplemented with 20 mM imidazole and loaded onto an immobilized metal-affinity column (Agarose Bead Technologies) loaded with nickel and pre-equilibrated in buffer A (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole). Bound MhGgH was eluted with 100 mM imidazole in buffer A. MhGgH-containing fractions were pooled and the His6 tag was removed by digestion with Tobacco etch virus (TEV) protease (1:5 molar ratio) at 277 K concomitantly with overnight dialysis against 20 mM Tris–HCl pH 8.0, 500 mM NaCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT) (dialysis buffer). Untagged MhGgH was separated from the His6 tag, tagged MhGgH and TEV protease by a second immobilized metal-affinity chromatography step using the same experimental conditions as described above. The MhGgH-containing column flowthrough was concentrated using a 10 kDa cutoff ultrafiltration device (Millipore) and loaded onto a HiPrep 26/60 Sephacryl S-200 HR size-exclusion column (GE Healthcare) using 20 mM Tris–HCl pH 8.0, 400 mM NaCl (storage buffer) as the mobile phase. Fractions containing MhGgH were pooled and concentrated using a 10 kDa cutoff ultrafiltration device (Millipore). The concentration of the purified protein was estimated by measuring its absorbance at 280 nm. The purified material was flash-frozen in liquid nitrogen and kept at 193 K until needed. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | M. hassiacum DSM 44199T (Tiago et al., 2012 ▸) |
| DNA source | pET-30a-MhGgH (Alarico et al., 2014 ▸) |
| Forward primer† | TATACGTCTCACATGCCGCACGACCCGAGTTTCAC |
| Reverse primer‡ | CGAATTCTTAGCCCAGCCAGTCGAGCACC |
| Cloning and expression vector | pETM11 |
| Expression host | MhGgH, E. coli BL21 (DE3); SeMet-MhGgH, E. coli B834 (DE3) |
| Complete amino-acid sequence of the construct produced§ | MKHHHHHHPMSDYDIPTTENLYFQGAMPHDPSFTPTQLAARAAYLLRGNDLGTMTTAAPLLYPHMWSWDAAFVAIGLAPLSVERAVVELDTLLSAQWRNGMIPHIVFANGVDGYFPGPARWATATLADNAPRNRLTSGITQPPVHAIAVQRILEHARTRGRSTRAVAEAFLDRRWGDLMRWHRWLAECRDRNERGRITLYHGWESGMDNSPRWDSAYANVVPGKLPEYQRADNVIITDPSQRPSDGEYDRYLWLLEEMKAVRYDDERLPSVMSFQVEDVFFSAIFSVACQVLAEIGEDYKRPHADVKDLYLWAERFRAGVVETTDQRTGAARDFDVLAEKWLVTETAAQFAPLLCGGLPHDRERALLKLLEGPRFCGHPDLKYGLIPSTSPVSRDFRPREYWRGPVWPVLTWLFSWCFARRGWAERARLLRQEGLRQASDGSFAEYYEPFTGEPLGSMQQSWTAAAVLDWLG |
| No. of amino acids | 448 |
| Theoretical molecular weight (Da) | 50897 |
The BsmBI recognition sequence is underlined.
The EcoRI recognition sequence is underlined.
The residues removed by TEV protease cleavage are italicized. The sequence of native MhGgH is underlined.
2.1.2. Production of selenomethionine-containing MhGgH
E. coli B834 (DE3) cells transformed with pETM11-MhGgH were grown in 25 ml LB medium overnight at 310 K. The cells were collected, washed thrice with sterile deionized water and used to inoculate 1 l of SelenoMethionine Medium (Molecular Dimensions). The culture was grown at 303 K to an OD600 of 0.7–0.8 before inducing expression with 0.5 mM IPTG. After overnight growth, the cells were harvested, resuspended in lysis buffer and incubated on ice with gentle agitation for 1 h. The protein extract was clarified by centrifugation, supplemented with 20 mM imidazole and 5 mM β-mercaptoethanol (β-ME), and loaded onto an immobilized metal-affinity column (Agarose Bead Technologies) loaded with nickel and pre-equilibrated with buffer A supplemented with 5 mM β-ME. Bound protein was eluted with 100 mM imidazole and 5 mM β-ME in buffer A. Fractions containing selenomethionine-labelled MhGgH (SeMet-MhGgH) were pooled and dialysed in the presence of TEV protease (1:5 molar ratio) against 50 volumes of dialysis buffer containing 400 mM NaCl for 2 h at room temperature followed by overnight incubation at 277 K. Untagged SeMet-MhGgH was separated from the cleaved affinity tag, noncleaved material and TEV protease by a second immobilized-metal affinity chromatography step under the same experimental conditions. The flowthrough containing the untagged protein was concentrated to 2.3 mg ml−1 in a 30 kDa cutoff ultrafiltration device (Millipore) with concomitant buffer exchange to storage buffer supplemented with 0.5 mM EDTA and 1 mM DTT.
2.2. Crystallization
Initial crystallization conditions were determined at 293 K with commercial sparse-matrix crystallization screens. Sitting-drop vapour-diffusion experiments were set up in 96-well CrystalQuick plates (Greiner Bio-One) using a Cartesian PixSys 4200 crystallization robot (Genomic Solutions) at the High Throughput Crystallization Laboratory (HTX Lab) of the EMBL Grenoble Outstation, Grenoble, France. The drops consisted of equal volumes (100 nl) of MhGgH (at 9.5 mg ml−1 in 20 mM Tris–HCl, 400 mM NaCl) and crystallization solution and were equilibrated against 88 µl reservoir solution. Three-dimensional MhGgH crystals appeared in condition No. 95 of the Morpheus sparse-matrix screen (Molecular Dimensions) after 1 d. The crystals were reproduced in-house using 24-well Cryschem M plates (Hampton Research) from drops composed of equal volumes (1 µl) of protein and precipitant solution equilibrated against 300 µl 0.1 M Tris–bicine pH 8.5, 0.1 M amino acids (glutamate, alanine, glycine, lysine and serine) and 24–34% GOL_P4K (glycerol, PEG 4000) as precipitant. Crystals were obtained after 1–2 d (Fig. 1 ▸ a). SeMet-MhGgH crystals were obtained using a drop ratio of 1:2 of protein solution (at 2.3 mg ml−1 in 20 mM Tris–HCl, 400 mM NaCl, 0.5 mM EDTA, 1 mM DTT) to precipitant equilibrated against a 300 µl reservoir of the crystallization condition identified for MhGgH but with 24–28% GOL_P4K. Two SeMet-MhGgH crystals were obtained after four months (Fig. 1 ▸ b). Crystallization information is summarized in Table 2 ▸.
Figure 1.
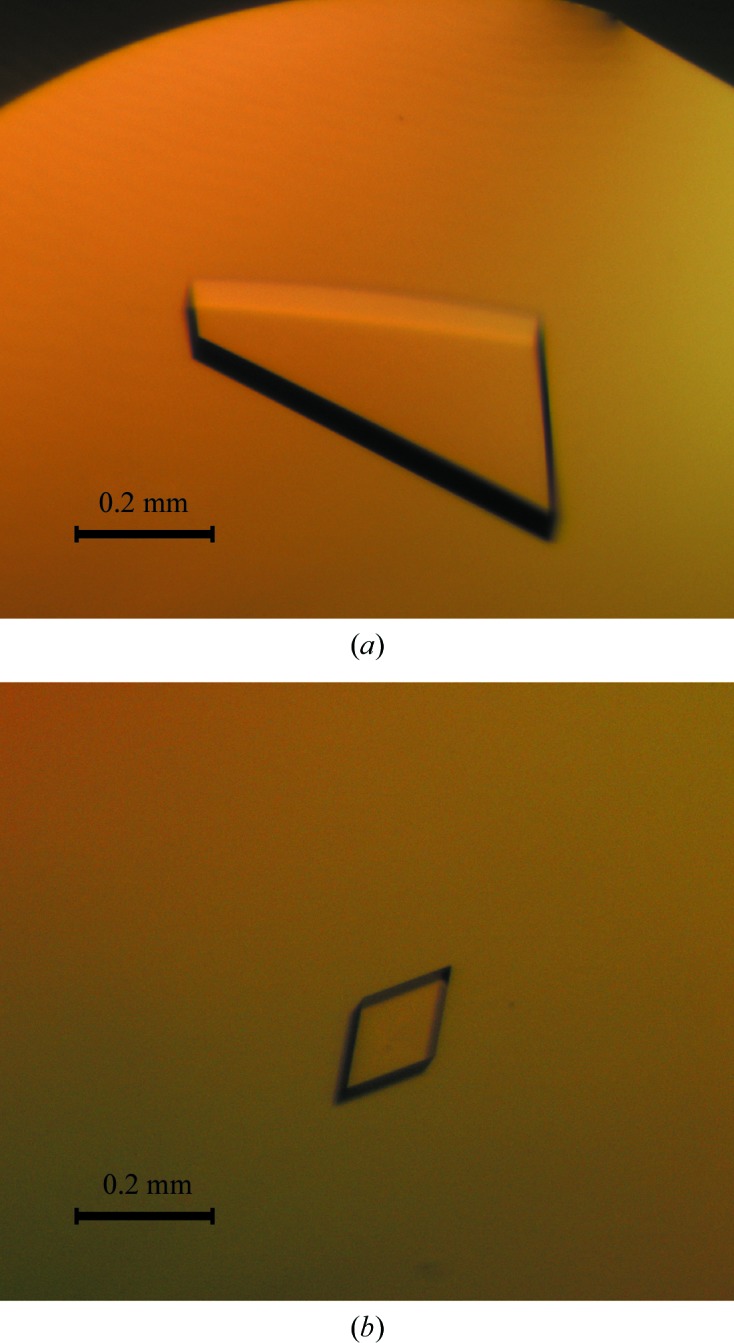
Crystals of MhGgH (a) and SeMet-MhGgH (b).
Table 2. Crystallization.
| Method | Sitting-drop vapour diffusion |
| Plate type | 96-well CrystalQuick plates (Greiner Bio-One) (screening); 24-well Cryschem M plates (Hampton Research) (production) |
| Temperature (K) | 293 |
| Protein concentration | MhGgH, 9.5 mg ml−1; SeMet-MhGgH, 2.3 mg ml−1 |
| Buffer composition of protein solution | MhGgH, 20 mM Tris–HCl pH 8.0, 400 mM NaCl; SeMet-MhGgH, 20 mM Tris–HCl pH 8.0, 400 mM NaCl, 0.5 mM EDTA, 1 mM DTT |
| Composition of reservoir solution | 0.1 M Tris–bicine pH 8.5, 0.1 M amino acids, 24–34% GOL_P4K |
| Volume and ratio of drop (protein:precipitant) | 0.1 µl, 1:1 (MhGgH screening); 2 µl, 1:1 (MhGgH production); 3 µl, 1:2 (SeMet-MhGgH) |
| Volume of reservoir | 88 µl (screening); 300 µl (production) |
2.3. Data collection and processing
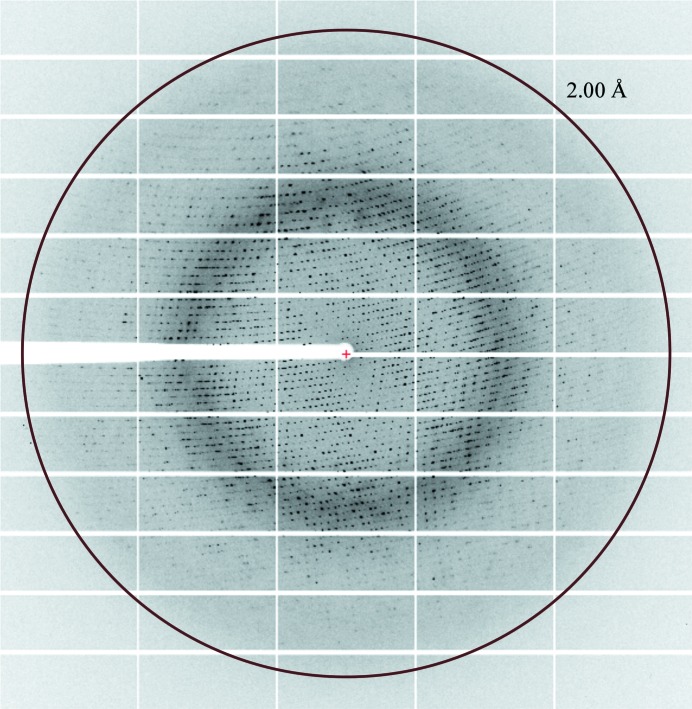
Diffraction data (Fig. 2 ▸) were collected from a single cryocooled (100 K) selenomethionine-containing MhGgH crystal that had previously been cryoprotected with crystallization solution containing 30% GOL_P4K and flash-cooled in liquid nitrogen on beamline ID29 (de Sanctis et al., 2012 ▸) of the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Two data sets were collected at wavelengths of 0.97909 and 0.97924 Å, corresponding to the peak and the inflection point of the K absorption edge of selenium, respectively. The data sets were automatically processed by the Grenoble Automatic Data procEssing (GrenADES) pipeline (Monaco et al., 2013 ▸) and scaled with XSCALE (Kabsch, 2010 ▸). X-ray diffraction data-collection and processing statistics are summarized in Table 3 ▸.
Figure 2.
X-ray diffraction pattern from a SeMet-MhGgH crystal. The circle corresponds to a resolution of 2.0 Å.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Peak | Inflection point | |
|---|---|---|
| Diffraction source | ID29, ESRF | |
| Wavelength (Å) | 0.97909 | 0.97924 |
| Temperature (K) | 100 | |
| Detector | PILATUS3 6M | |
| Crystal-to-detector distance (mm) | 344.2 | |
| Rotation range per image (°) | 0.1 | |
| Total rotation range (°) | 150 | |
| Exposure time per image (s) | 0.02 | |
| Space group | P21 | |
| a, b, c (Å) | 90.8, 86.1, 159.7 | 90.9, 86.3, 159.7 |
| α, β, γ (°) | 90.0, 93.0. 90.0 | 90.0, 93.0, 90.0 |
| Mosaicity (°) | 0.17 | 0.20 |
| Resolution range (Å) | 57.4–2.04 (2.07–2.04) | 58.6–2.06 (2.09–2.06) |
| Total No. of reflections | 428683 | 419987 |
| No. of unique reflections | 151619 | 148763 |
| Completeness (%) | 96.5 (82.1) | 97.4 (87.6) |
| Multiplicity | 2.8 (2.8) | 2.8 (2.7) |
| Anomalous completeness (%) | 68.3 (53.3) | 85.0 (71.9) |
| Anomalous multiplicity | 1.2 (1.7) | 1.3 (1.5) |
| Half-set correlation CC1/2 | 0.990 (0.659) | 0.990 (0.657) |
| Half-set anomalous correlation CCanom | 0.355 | 0.166 |
| 〈I/σ(I)〉 | 7.6 (1.6)† | 7.1 (1.8)‡ |
| R r.i.m. | 0.128 (0.776) | 0.129 (0.690) |
| Overall B factor from Wilson plot (Å2) | 20.4 | 25.4 |
〈I/σ(I)〉 falls below 2.0 at 2.15 Å resolution.
〈I/σ(I)〉 falls below 2.0 at 2.09 Å resolution.
3. Results and discussion
Recombinant MhGgH was expressed in E. coli and purified to homogeneity using a combination of immobilized metal-affinity and size-exclusion chromatography. The expressed construct encoded an N-terminal His tag, which was separated from the native MhGgH sequence by a TEV protease recognition site that allowed proteolytic tag removal. TEV protease cleavage resulted in an additional Gly-Ala dipeptide N-terminal to the native MhGgH amino-acid sequence (Table 1 ▸).
Initial crystallization conditions were identified using extensive sampling of commercial sparse-matrix crystallization screens at the EMBL Grenoble Outstation HTX Lab. The crystals were reproduced in-house, and optimized crystals obtained in 1–2 d diffracted X-rays to beyond 2.0 Å resolution at a synchrotron-radiation source. In order to solve the phase problem, selenomethionine-containing MhGgH was produced. It was found that selenomethionine-labelled MhGgH was considerably less soluble than the unlabelled protein under the same experimental conditions. Therefore, selenomethionine-labelled MhGgH could only be concentrated to a fourfold lower concentration than the unlabelled material. Under crystallization conditions similar to those for the wild-type protein, two SeMet-MhGgH crystals were obtained after four months (Table 2 ▸). The slower crystallogenesis of selenomethionine-labelled MhGgH is most likely to be owing to the lower sample concentration used in the crystallization assays.
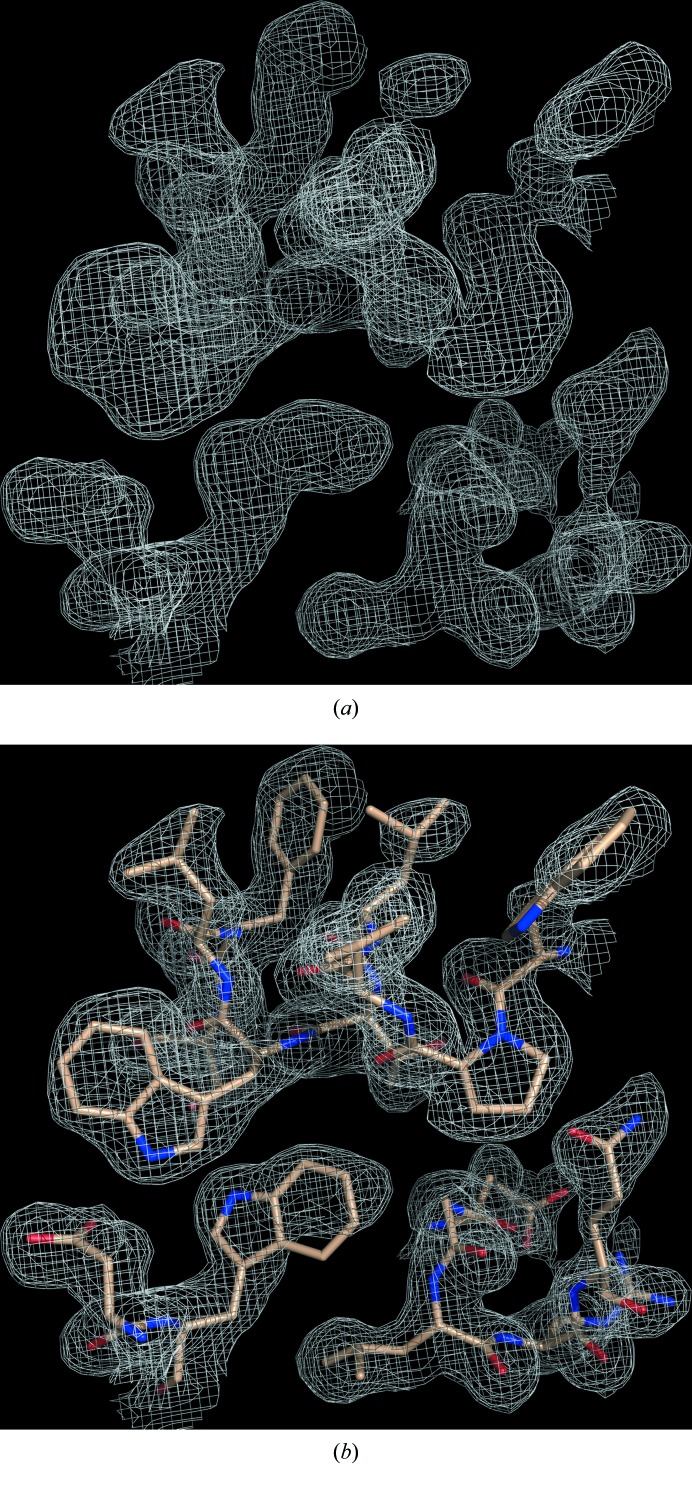
The diffraction from a SeMet-MhGgH crystal was measured on beamline ID29 at ESRF, Grenoble, France. The crystal belonged to the monoclinic space group P21 and diffracted to ∼2 Å resolution (Table 3 ▸). Analysis of the crystal content suggested the presence of four MhGgH molecules in the asymmetric unit, corresponding to a Matthews coefficient of 3.07 Å3 Da−1 and a solvent content of 59.9% (Winn et al., 2011 ▸). Two diffraction data sets were recorded from a single crystal at wavelengths corresponding to the peak and the inflection point of the K absorption edge of selenium (Table 3 ▸). The phase problem was solved by two-wavelength anomalous diffraction phasing with the SHELXC/SHELXD/SHELXE pipeline (Sheldrick, 2010 ▸) and the HKL2MAP GUI (Pape & Schneider, 2004 ▸). SHELXD identified 27 of the 36 (nine per molecule) Se atoms present in the asymmetric unit (CCall = 23.6%; CCweak = 17.8%), corresponding to SeMet residues 153, 232, 246 and 432 of all molecules, as well as SeMet residue 28 of molecules B and C, SeMet residue 39 of molecules A, B and C, and SeMet residues 75 and 181 of molecules A, B and D. Experimental phasing with density modification using SHELXE resulted in excellent electron-density maps (Fig. 3 ▸), with a final FOM of 0.652 and a pseudo-free CC of 71.3%. The autotracing function of SHELXE produced a 1411-residue poly-Ala model (CC for partial structure against native data = 36.1%). Subsequent automated model building with ARP/wARP (Langer et al., 2008 ▸) using the SHELXE-derived phases resulted in an initial model with 1677 (of 1792) residues built and sequenced. The MhGgH crystallographic model was further improved through alternative cycles of manual building with Coot (Emsley et al., 2010 ▸) and refinement with PHENIX (Adams et al., 2010 ▸) and is currently in the last stages of refinement.
Figure 3.
Experimental electron-density map for MhGgH. (a) Solvent-flattened electron-density map (grey mesh) from SHELXE (Sheldrick, 2010 ▸). (b) Electron-density map as in (a) with superposed protein model (sticks) automatically built by ARP/wARP (Langer et al., 2008 ▸).
Acknowledgments
We acknowledge the ESRF for the provision of synchrotron-radiation facilities and thank the staff for help with data collection. Transnational Access to the High Throughput Crystallization Laboratory of the European Molecular Biology Laboratory Grenoble Outstation was supported by the European Community Seventh Framework Program (FP7/2007–2013) Grant Protein Production Platform (PCUBE Agreement No. 227764).
Funding Statement
This work was funded by Fundação para a Ciência e a Tecnologia grants SFRH/BD/92955/2013, POCI-01-0145-FEDER-007274, UID/NEU/04539/2013, and SFRH/BPD/108299/2015. European Regional Development Fund grants POCI-01-0145-FEDER-007274, UID/NEU/04539/2013, and Norte-01-0145-FEDER-000012. COMPETE 2020 – Operational Programme for Competitiveness and Internationalization (POCI) grants POCI-01-0145-FEDER-007274 and UID/NEU/04539/2013. Norte Portugal Regional Operational Programme (NORTE 2020) grant Norte-01-0145-FEDER-000012.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Alarico, S., Costa, M., Sousa, M. S., Maranha, A., Lourenço, E. C., Faria, T. Q., Ventura, M. R. & Empadinhas, N. (2014). Sci. Rep. 4, 6766. [DOI] [PMC free article] [PubMed]
- Behrends, V., Williams, K. J., Jenkins, V. A., Robertson, B. D. & Bundy, J. G. (2012). J. Proteome Res. 11, 3888–3896. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Falkinham, J. O. (2010). Future Microbiol. 5, 951–960. [DOI] [PubMed]
- Griffith, D. E. et al. (2007). Am. J. Respir. Crit. Care Med. 175, 367–416. [DOI] [PubMed]
- Griffith, D. E., Brown-Elliott, B. A., Benwill, J. L. & Wallace, R. J. Jr (2015). Annals ATS, 12, 436–439. [DOI] [PubMed]
- Henkle, E. & Winthrop, K. L. (2015). Clin. Chest Med. 36, 91–99. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Miyazaki, T., Ichikawa, M., Iino, H., Nishikawa, A. & Tonozuka, T. (2015). J. Struct. Biol. 190, 21–30. [DOI] [PubMed]
- Monaco, S., Gordon, E., Bowler, M. W., Delagenière, S., Guijarro, M., Spruce, D., Svensson, O., McSweeney, S. M., McCarthy, A. A., Leonard, G. & Nanao, M. H. (2013). J. Appl. Cryst. 46, 804–810. [DOI] [PMC free article] [PubMed]
- Nessar, R., Cambau, E., Reyrat, J. M., Murray, A. & Gicquel, B. (2012). J. Antimicrob. Chemother. 67, 810–818. [DOI] [PubMed]
- Nunes-Costa, D., Alarico, S., Dalcolmo, M. P., Correia-Neves, M. & Empadinhas, N. (2016). Tuberculosis, 96, 107–119. [DOI] [PubMed]
- Nunes-Costa, D., Maranha, A., Costa, M., Alarico, S. & Empadinhas, N. (2017). Glycobiology, 27, 213–227. [DOI] [PubMed]
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst. 37, 843–844.
- Piersimoni, C. & Scarparo, C. (2008). Lancet Infect. Dis. 8, 323–334. [DOI] [PubMed]
- Sanctis, D. de et al. (2012). J. Synchrotron Rad. 19, 455–461.
- Shahraki, A. H., Heidarieh, P., Bostanabad, S. Z., Khosravi, A. D., Hashemzadeh, M., Khandan, S., Biranvand, M., Schraufnagel, D. E. & Mirsaeidi, M. (2015). Eur. J. Intern. Med. 26, 279–284. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2010). Acta Cryst. D66, 479–485. [DOI] [PMC free article] [PubMed]
- Tiago, I., Maranha, A., Mendes, V., Alarico, S., Moynihan, P. J., Clarke, A. J., Macedo-Ribeiro, S., Pereira, P. J. & Empadinhas, N. (2012). J. Bacteriol. 194, 7010–7011. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.