Abstract
Background
Published data on the relationship between matrix metalloproteinases (MMPs) polymorphisms and ovarian cancer risk have implicated inconclusive results. To evaluate the role of MMPs polymorphisms in ovarian cancer risk, a meta-analysis and systematic review were performed.
Methods
MMPs polymorphisms which could be quantitatively synthesized were involved in meta-analysis. Five comparison models (homozygote model, heterozygote model, dominant model, recessive model, additive model) were carried out, a subgroup analysis was performed to clarify heterogeneity source. The remaining polymorphisms which could not be quantitatively synthesized were involved in systematic review.
Results
10 articles with 20 studies were included in this paper. Among those studies, 8 studies involving MMP1 rs1799750 and MMP3 rs34093618 could be meta-analyzed and 12 studies involving 12 polymorphisms could not. Meta-analysis showed that no associations were found between MMP1 rs1799750 (homozygote model: OR = 0.93, 95%CI = 0.70–1.23, POR = 0.60; heterozygote model: OR = 1.09, 95%CI = 0.78–1.54, POR = 0.61; dominant model: OR = 1.02, 95%CI = 0.83–1.25, POR = 0.84; recessive model: OR = 0.95, 95%CI = 0.75–1.21, POR = 0.67; additive model: OR = 1.00, 95%CI = 0.85–1.17, POR = 0.99), MMP3 rs34093618 (homozygote model: OR = 1.25, 95%CI = 0.70–2.24, POR = 0.46; heterozygote model: OR = 1.08, 95%CI = 0.51–2.31, POR = 0.84; dominant model: OR = 0.97, 95%CI = 0.68–1.38, POR = 0.85; recessive model: OR = 1.12, 95%CI = 0.69–1.80, POR = 0.65; additive model: OR = 1.01, 95%CI = 0.79–1.31, POR = 0.91) and ovarian cancer. Furthermore, similar results were detected in subgroup analysis. The systematic review on 12 polymorphisms suggested that MMP2 C-735T, MMP7 A-181G, MMP8 rs11225395, MMP9 rs6094237, MMP12 rs2276109, MMP20 rs2292730, MMP20 rs12278250, MMP20 rs9787933 might have a potential effect on ovarian cancer risk.
Conclusions
In summary, polymorphisms of MMPs might not be associated with ovarian cancer risk. However, it is necessary to conduct more larger-scale, multicenter, and high-quality studies in the future.
Introduction
Ovarian cancer is main cause of death with gynecological tumors worldwide, and is often at an advanced stage by the time of diagnosis and has metastasized throughout the peritoneal cavity [1–2]. In 2013, there were an estimated 22,240 new cases and 14,030 new deaths [3]. Despite continuous advances in ovarian cancer research, diagnosis, and clinical treatment during the past 30 years [4], it has been still hard to find a cost-effective screening strategy to significantly increase the survival rate for early-stage ovarian cancer.
Genome-wide association studies (GWAS) concerning genetic aetiology of cancer have established more than 150 regions associated with various specific cancers, which expand the current understanding of carcinogenesis mechanisms [5]. Alterations in genetic sequence, such as single-nucleotide substitutions, lead to cancer formation by biologically regulating a handful of molecular activities [6].
Matrix metalloproteinases (MMPs), a family of more than 20 zinc-dependent enzymes known to degrade extracellular matrix and basement membrane components [7], are not only a prerequisite for multiple steps of cancer development but also play important roles in cancer invasion and metastasis [8]. MMPs are correlated with ovarian cancer, with the levels of MMP-2, MMP-7 and MMP-9 elevated in ovarian cancer patients [9–10]. At genetic level, a number of studies have been carried out to assess the association between polymorphisms of MMPs and ovarian cancer risk [11–27], but the conclusions have been still conflicted and even contradictory. For example, study by Ju [19] showed no associations existed between MMP1 rs1799750 and ovarian cancer in Korean, while study by Kanamori [11] showed 2G genotype of MMP1 rs1799750 might represent a risk factor for ovarian cancer in Japanese. Individual studies with a small sample size may result in incorrect conclusion. Therefore, a comprehensive meta-analysis and systematic review are necessary to precisely assess the relationships between MMPs polymorphisms and ovarian cancer risk.
Materials and methods
Search strategy
The databases Pubmed, Embase, Web of knowledge, were searched for all articles with the following search terms: (MMP OR MMPs OR matrix metalloproteinase OR matrix metalloproteinases) AND (polymorphism OR polymorphisms) AND (ovarian cancer OR ovarian carcinoma) up to search date: March 25, 2017. No limitation of publication language was defined for this search. Additional published data were identified by reviewing the bibliographical references listed in each retrieved article.
Inclusion criteria and exclusion criteria
All studies included in this meta-analysis were accorded with the following inclusion criteria: (a) study focused on the association between MMPs polymorphisms and ovarian cancer; (b) case-control design; (c) provided available frequency for each genotype in both cases and controls to calculate odds ratio (OR) and corresponding 95% confidence interval (95%CI). In addition, exclusion criteria were as follows: (a) reviews, editorials, comments or animal studies; (b) overlapped articles or studies with overlapping data.
Data extraction
Two investigators independently extracted the following data: first author’s name, year of publication, study country, ethnicity, source of controls, MMPs gene, polymorphisms, number of cases and controls, value of Hardy-Weinberg equilibrium (HWE). A consensus on the extracted items was reached by discussion between the two investigators.
Quality assessment
The quality of study was assessed according to the quality assessment criteria [28] (S1 Table), in which the quality scores ranged from 0 to 15. Studies with scores ≥9 were regarded as high quality.
Statistical analysis
In order to evaluate the association between MMPs polymorphisms and ovarian cancer risk, OR and 95% CI were summarized under five comparison models, including homozygote model, heterozygote model, dominant model, recessive model, additive model. The definition of comparison model was listed in S2 Table. The P value of the pooled ORs was considered significant if less than 0.05, which was examined by Z test. HWE in the control group was checked by chi-square test, deviation was considered with P<0.05. Heterogeneity assumption was checked by a chi-square-based Q statistic test and quantified by I2 value. If I2 value < 50% or P > 0.10, the fixed effect model was used [29]. Otherwise, random effect model was carried out [30], then a subgroup analysis by ethnicity was performed. Both funnel plot and Egger’s test were performed to test whether publication bias existed or not, bias was considered with P<0.05 in Egger’s test. The statistical analyses for the present study were completed by Review Manager software 5.1 (the Nordic Cochrane Center, Rigshospitalet, Copenhagen, Denmark) and Stata software 12.0 (StataCorp, College Station, TX, USA).
Results
Literature search and study characteristics
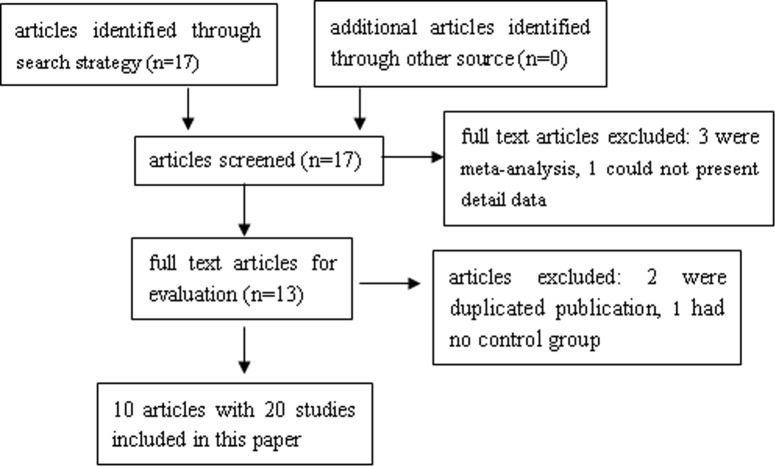
A total of 17 articles [11–27] were identified through search strategy. Reviewed on abstracts among these articles, 4 articles were excluded because 3 articles [25–27] were meta-analysis and 1 article [13] could not present detailed data. Then 13 full text articles were obtained for further evaluation, in which 3 articles were deleted for 2 articles [16, 21] were duplicated publication and 1 article [17] had no control group. Ultimately, 10 articles with 20 studies involving 14 polymorphisms were included in this paper. Among these studies, 8 studies with 2 polymorphisms [11, 12, 14, 15, 18, 19] (5 studies for MMP1 rs1799750, 3 studies for MMP3 rs34093618) involving 1019 ovarian cancer cases and 1609 controls could be quantitatively synthesized for meta-analysis. The remaining 12 studies with 12 polymorphisms [18, 20, 22, 23, 24] (12 polymorphisms including MMP2 C-1306T, MMP2 C-735T, MMP7 A-181G, MMP8 rs2155052, MMP8 rs11225395, MMP9 C-1562T, MMP9 rs6094237, MMP12 rs2276109, MMP13 rs17860523, MMP20 rs2292730, MMP20 rs12278250, MMP20 rs9787933) involving 2793 ovarian cancer cases and 3037 controls could not be quantitatively synthesized, thus the systematic review was performed. The flow diagram of study selection process was presented in Fig 1. The main characteristics of included articles or studies were listed in Table 1. The distributions of genotype in studies from meta-analysis and systematic review were in S3 Table and S4 Table.
Fig 1. Flow diagram of study selection process.
Table 1. Characteristics of studies included in the meta-analysis and systematic review.
| first author | year | contry | ethnicity | source of control | gene | polymorphisms | sample sizes (case/control) | HWE | quality score |
|---|---|---|---|---|---|---|---|---|---|
| Kanamori [11] | 1999 | Japan | East Asia | NA | MMP1 | rs1799750 | 163/150 | 0.009 | 5 |
| Biondi [12] | 2000 | Italy | Caucasian | NA | MMP1 | rs1799750 | 25/164 | 0.52 | 4 |
| MMP3 | rs34093618 | 25/164 | 0.217 | ||||||
| Wenham [14] | 2003 | USA | mixed | PB | MMP1 | rs1799750 | 311/387 | 0.264 | 12 |
| Smolarz [15] | 2003 | Poland | Caucasian | HB | MMP3 | rs34093618 | 118/110 | 0.587 | 8 |
| Li [18] | 2006 | China | East Asia | HB | MMP1 | rs1799750 | 122/151 | 0.002 | 9 |
| MMP3 | rs34093618 | 122/151 | 0.275 | ||||||
| MMP7 | A-181G | 138/160 | 0.714 | ||||||
| MMP9 | C-1562T | 138/160 | 0.263 | ||||||
| Ju [19] | 2007 | Korea | East Asia | HB | MMP1 | rs1799750 | 133/332 | 0.393 | 7 |
| Li [20] | 2008 | China | East Asia | PB | MMP2 | C-1306T | 246/324 | 0.862 | 10 |
| MMP2 | C-735T | 246/324 | 0.293 | ||||||
| Jia [22] | 2010 | China | East Asia | HB | MMP12 | rs2276109 | 300/300 | 0.746 | 12 |
| MMP13 | rs17860523 | 300/300 | 0.962 | ||||||
| Arechavaleta-Velasco [23] | 2014 | Mexico | mixed | NA | MMP8 | rs2155052 | 35/37 | 0.797 | 6 |
| MMP8 | rs11225395 | 35/37 | 0.013 | ||||||
| Wang [24] | 2015 | USA | mixed | HB | MMP9 | rs6094237 | 339/349 | 0.049 | 12 |
| MMP20 | rs2292730 | 339/349 | 0.01 | ||||||
| MMP20 | rs12278250 | 339/349 | 0.675 | ||||||
| MMP20 | rs9787933 | 339/349 | 0.59 |
NA, not available; HB, hospital based; PB, population based; MMP, matrix metalloproteinase; HWE, Hardy-Weinberg equilibrium
Meta-analysis and systematic review
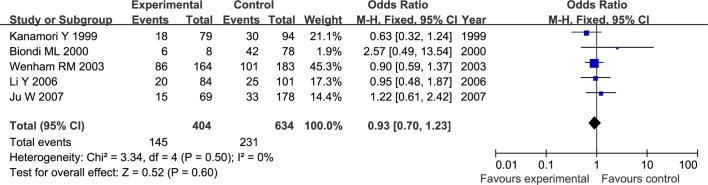
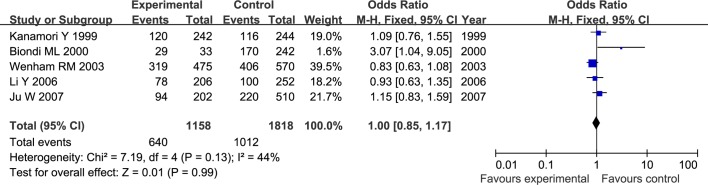
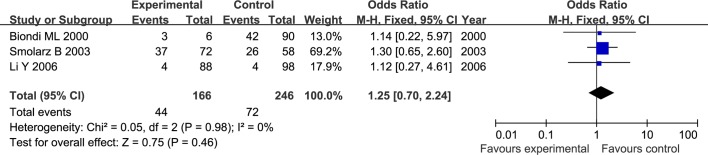
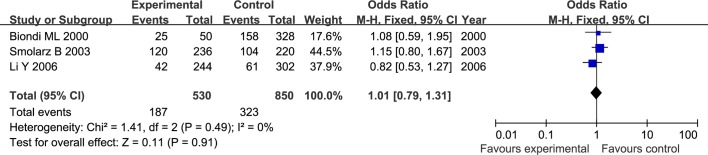
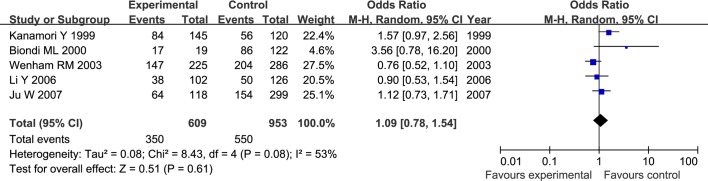
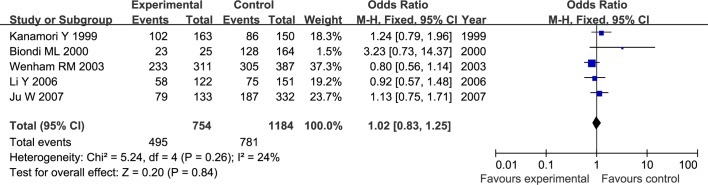
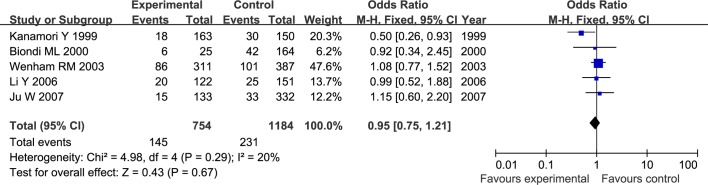
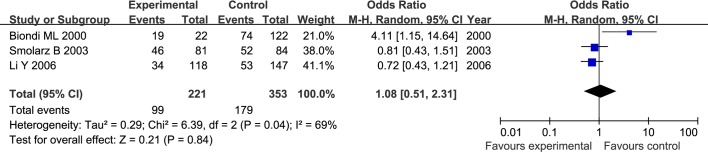
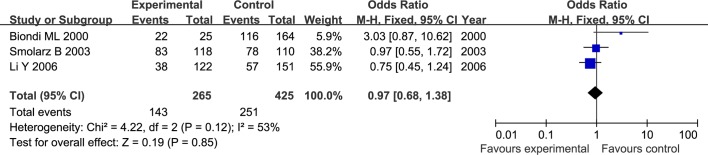
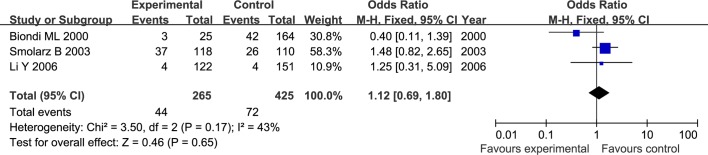
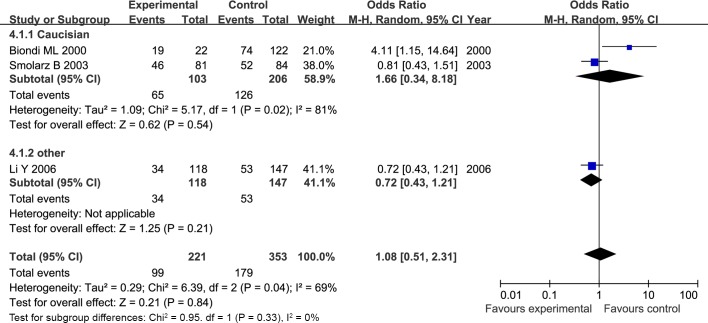
The results of meta-analysis for MMP1 rs1799750 and MMP3 rs34093618 polymorphisms were listed in Table 2. The forest plots for MMP1 rs1799750 were listed in Figs 2–6, and MMP3 rs34093618 were presented in Figs 7–11. On the whole, no significant association was found between MMP1 rs1799750 polymorphisms and ovarian cancer risk (homozygote model: OR = 0.93, 95%CI = 0.70–1.23, POR = 0.60; heterozygote model: OR = 1.09, 95%CI = 0.78–1.54, POR = 0.61; dominant model: OR = 1.02, 95%CI = 0.83–1.25, POR = 0.84; recessive model: OR = 0.95, 95%CI = 0.75–1.21, POR = 0.67; additive model: OR = 1.00, 95%CI = 0.85–1.17, POR = 0.99). For MMP3 rs34093618 polymorphism and ovarian cancer risk, overall, no significant association was found (homozygote model: OR = 1.25, 95%CI = 0.70–2.24, POR = 0.46; heterozygote model: OR = 1.08, 95%CI = 0.51–2.31, POR = 0.84; dominant model: OR = 0.97, 95%CI = 0.68–1.38, POR = 0.85; recessive model: OR = 1.12, 95%CI = 0.69–1.80, POR = 0.65; additive model: OR = 1.01, 95%CI = 0.79–1.31, POR = 0.91).
Table 2. Meta-analysis of association between MMPs polymorphism and ovarian cancer.
| comparison model | OR(95%CI) | PORa | I2 | Phetb |
|---|---|---|---|---|
| MMP1 rs1799750 | ||||
| 1G1G vs 2G2G | 0.93(0.70–1.23) | 0.60 | 0% | 0.50 |
| 1G2G vs 2G2G | 1.09(0.78–1.54) | 0.61 | 53% | 0.08 |
| 1G1G+1G2G vs 2G2G | 1.02(0.83–1.25) | 0.84 | 24% | 0.26 |
| 1G1G vs 1G2G+2G2G | 0.95(0.75–1.21) | 0.67 | 20% | 0.29 |
| 1G vs 2G | 1.00(0.85–1.17) | 0.99 | 44% | 0.13 |
| MMP3 rs34093618 | ||||
| 5A5A vs 6A6A | 1.25(0.70–2.24) | 0.46 | 0 | 0.98 |
| 5A6A vs 6A6A | 1.08(0.51–2.31) | 0.84 | 69 | 0.04 |
| 5A5A+5A6A vs 6A6A | 0.97(0.68–1.38) | 0.85 | 53 | 0.12 |
| 5A5A vs 5A6A+6A6A | 1.12(0.69–1.80) | 0.65 | 43 | 0.17 |
| 5A vs 6A | 1.01(0.79–1.31) | 0.91 | 0 | 0.49 |
a P value of the Z-test for odds ratio test
b P value of the Q-test for heterogeneity test.
Fig 2. Forest plot of MMP-1 rs1799750 and ovarian cancer risk (1G1G vs 2G2G).
Fig 6. Forest plot of MMP-1 rs1799750 and ovarian cancer risk (1G vs 2G).
Fig 7. Forest plot of MMP3 rs34093618 and ovarian cancer risk (5A5A vs 6A6A).
Fig 11. Forest plot of MMP3 rs34093618 and ovarian cancer risk (5A vs 6A).
Fig 3. Forest plot of MMP-1 rs1799750 and ovarian cancer risk (1G2G vs 2G2G).
Fig 4. Forest plot of MMP-1 rs1799750 and ovarian cancer risk (1G1G +1G2G vs 2G2G).
Fig 5. Forest plot of MMP-1 rs1799750 and ovarian cancer risk (1G1G vs 1G2G+2G2G).
Fig 8. Forest plot of MMP3 rs34093618 and ovarian cancer risk (5A6A vs 6A6A).
Fig 9. Forest plot of MMP3 rs34093618 and ovarian cancer risk (5A5A+5A6A vs 6A6A).
Fig 10. Forest plot of MMP3 rs34093618 and ovarian cancer risk (5A5A vs 5A6A + 6A6A).
The results of systematic review were presented in Table 3. Eight polymorphisms (MMP2 C-735T, MMP7 A-181G, MMP8 rs11225395, MMP9 rs6094237, MMP12 rs2276109, MMP20 rs2292730, MMP20 rs12278250, MMP20 rs9787933) were reported associated with ovarian cancer risk, while other polymorphisms could not be associated with ovarian cancer risk.
Table 3. Systematic review of association between MMPs polymorphisms and ovarian cancer.
| A | ||||||||||
| gene | polymorphisms | homozygote model | heterozygote model | dominant model | ||||||
| OR(95%CI) | PORa | OR(95%CI) | PORa | OR(95%CI) | PORa | |||||
| MMP7 | A-181G | NA | NA | NA | NA | NA | NA | |||
| MMP9 | C-1562T | 0.16(0.01, 3.46) | 0.25 | 0.20(0.01, 4.37) | 0.31 | 0.17(0.01, 3.57) | 0.25 | |||
| MMP2 | C-1306T | 3.86(0.45, 33.29) | 0.22 | 3.78(0.43, 33.3) | 0.23 | 3.84(0.45, 33.08) | 0.22 | |||
| MMP2 | C-735T | 1.12(0.52, 2.39) | 0.78 | 0.67(0.30, 1.47) | 0.32 | 0.93(0.44, 1.97) | 0.85 | |||
| MMP12 | rs2276109 | NA | NA | NA | NA | NA | NA | |||
| MMP13 | rs17860523 | 0.64(0.41, 1.02) | 0.06 | 0.84(0.56, 1.26) | 0.40 | 0.77(0.52, 1.12) | 0.17 | |||
| MMP8 | rs2155052 | NA | NA | NA | NA | NA | NA | |||
| MMP8 | rs11225395 | 0.38(0.08, 1.78) | 0.22 | 0.24(0.07, 0.79) | 0.02 | 0.26(0.08, 0.85) | 0.03 | |||
| MMP9 | rs6094237 | 2.00(1.28, 3.12) | 0.002 | 1.82(1.18, 2.980) | 0.007 | 1.90(1.27, 2.85) | 0.002 | |||
| MMP20 | rs2292730 | 0.53(0.34, 0.83) | 0.005 | 0.47(0.32, 0.70) | 0.0002 | 0.49(0.34, 0.72) | 0.0002 | |||
| MMP20 | rs12278250 | 0.81(0.18, 3.66) | 0.79 | 0.38(0.08, 1.77) | 0.22 | 0.73(0.16, 3.28) | 0.68 | |||
| MMP20 | rs9787933 | 1.46(0.32, 6.57) | 0.62 | 0.72(0.15, 3.37) | 0.68 | 1.29(0.29, 5.83) | 0.74 | |||
| B | ||||||||||
| gene | polymorphisms | recessive model | additive model | |||||||
| OR(95%CI) | PORa | OR(95%CI) | PORa | |||||||
| MMP7 | A-181G | 0.28(0.13, 0.63) | 0.002 | 0.30(0.14, 0.67) | 0.003 | |||||
| MMP9 | C-1562T | 0.76(0.42, 1.38) | 0.37 | 0.73(0.42, 1.26) | 0.25 | |||||
| MMP2 | C-1306T | 1.07(0.72, 1.58) | 0.74 | 1.11(0.78, 1.59) | 0.55 | |||||
| MMP2 | C-735T | 1.58(1.12, 2.23) | 0.009 | 1.36(1.02, 1.81) | 0.04 | |||||
| MMP12 | rs2276109 | 0.36(0.17, 0.73) | 0.005 | 0.37(0.18, 0.74) | 0.005 | |||||
| MMP13 | rs17860523 | 0.72(0.50, 1.04) | 0.08 | 0.80(0.61, 1.01) | 0.06 | |||||
| MMP8 | rs2155052 | 1.46(0.23, 9.28) | 0.69 | 1.94(0.34, 10.96) | 0.45 | |||||
| MMP8 | rs11225395 | 1.07(0.31, 3.69) | 0.92 | 0.63(0.33, 1.22) | 0.17 | |||||
| MMP9 | rs6094237 | 1.30(0.95, 1.77) | 0.10 | 1.37(1.10, 1.70) | 0.004 | |||||
| MMP20 | rs2292730 | 0.91(0.65, 1.28) | 0.60 | 0.76(0.62, 0.94) | 0.01 | |||||
| MMP20 | rs12278250 | 2.02(1.31, 3.11) | 0.001 | 1.79(1.20, 2.67) | 0.004 | |||||
| MMP20 | rs9787933 | 1.99(1.33, 2.97) | 0.0008 | 1.83(1.26, 2.66) | 0.002 | |||||
NA, not available
a P value of the Z-test for odds ratio test
Heterogeneity analysis and subgroup analysis
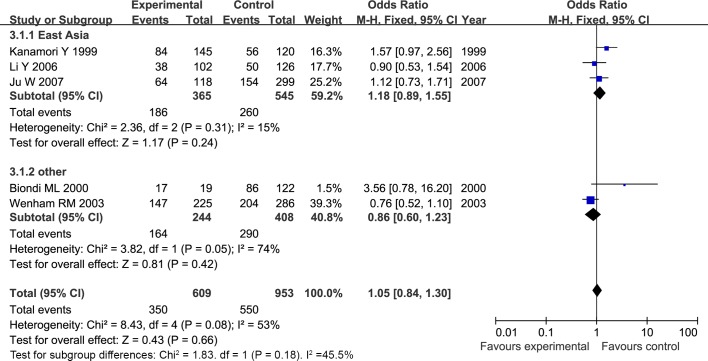
For both MMP1 rs1799750 and MMP3 rs34093618 polymorphism, there was obvious heterogeneity in heterozygote model (MMP1 rs1799750: I2 = 53%, Phet = 0.08; MMP3 rs34093618: I2 = 69%, Phet = 0.04). Then, a subgroup analysis by ethnicity was conducted to assess the source of heterogeneity. The forest plots of subgroup analysis for MMP1 rs1799750 and MMP3 rs34093618 were respectively presented in Figs 12 and 13. For MMP1 rs1799750, heterogeneity dramatically decreased when stratification analyses for Caucasian was conducted (I2 = 31%, Phet = 0.15), while MMP3 rs34093618 did not decreased (I2 = 81%, Phet = 0.02). No significant association was found between MMPs polymorphism and ovarian cancer in both two subgroup analysis.
Fig 12. Forest plot of MMP-1 rs1799750 and ovarian cancer risk stratified according to ethnicity (1G2G vs 2G2G).
Fig 13. Forest plot of MMP3 rs34093618 and ovarian cancer risk stratified according to ethnicity (5A6A vs 6A6A).
Publication bias analysis
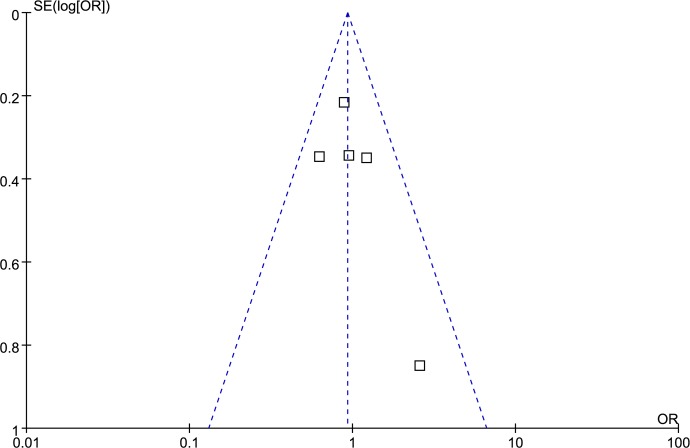
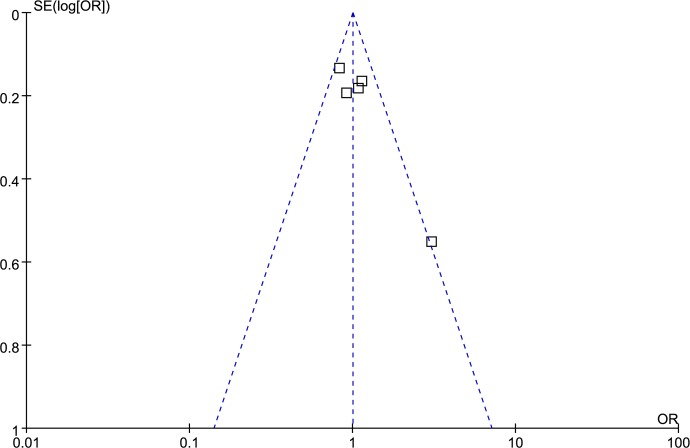
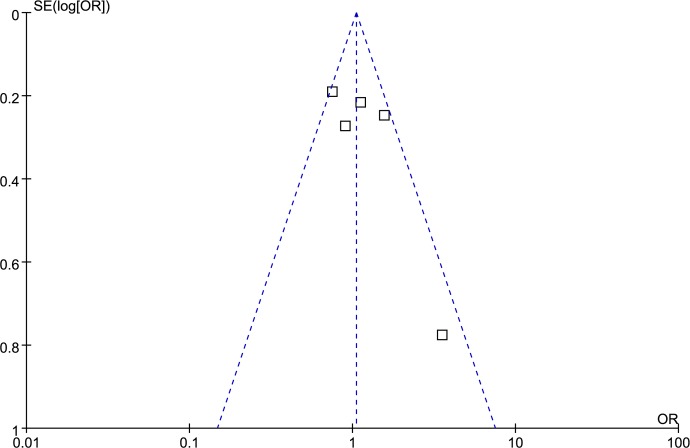
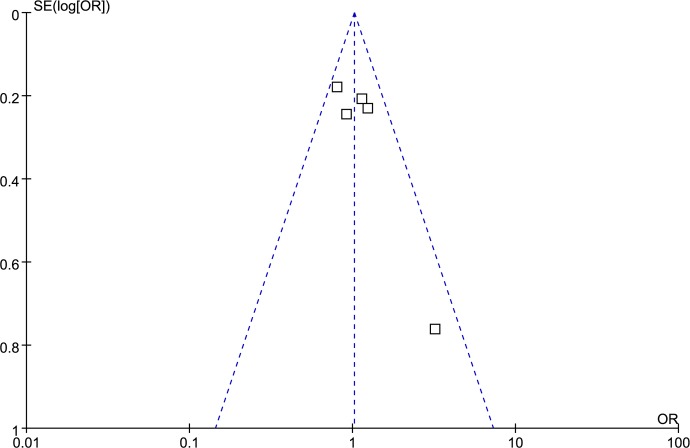
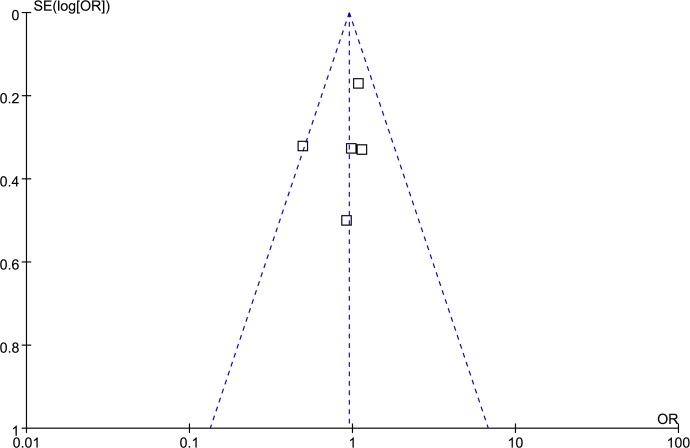
Funnel plot and Egger’s test were performed to access publication bias. Both funnel plots (Figs 14–18) and Egger’s test (homozygote model: P = 0.588; heterozygote model: P = 0.423; dominant model: P = 0.612; recessive model: P = 0.363; additive model: P = 0.534) suggested no evidence of publication bias in the meta-analysis of MMP1 rs1799750 polymorphism. For MMP3 rs34093618, publication bias analysis was not conducted for only 3 studies involved.
Fig 14. Funnel plot of MMP-1 rs1799750 and ovarian cancer risk (1G1G vs 2G2G).
Fig 18. Funnel plot of MMP-1 rs1799750 and ovarian cancer risk (1G vs 2G).
Fig 15. Funnel plot of MMP-1 rs1799750 and ovarian cancer risk (1G2G vs 2G2G).
Fig 16. Funnel plot of MMP-1 rs1799750 and ovarian cancer risk (1G1G +1G2G vs 2G2G).
Fig 17. Funnel plot of MMP-1 rs1799750 and ovarian cancer risk (1G1G vs 1G2G+2G2G).
Discussion
Study by Ju [19] showed no associations existed between MMP1 rs1799750 and ovarian cancer in Korean, while study by Kanamori [11] showed 2G genotype of MMP1 rs1799750 might represent a risk factor for ovarian cancer in Japanese. Therefore, a comprehensive meta-analysis and systematic review are necessary. As a powerful tool for summarizing the different studies, meta-analysis has been accepted as a significant tool to analyze cumulative data from limited study subjects [31].
This meta-analysis and systematic review, including 5 studies for MMP1 rs1799750 composed of 754 ovarian cancer cases 1184 and controls, 3 studies for MMP3 rs34093618 polymorphism composed of 265 cases and 425 controls, 12 studies for systematic review involving 2793 cases and 3037 controls, proved that MMP1 rs1799750 and MMP3 rs34093618 polymorphisms were not associated with ovarian cancer risk, in addition, subgroup analyses by ethnicity showed similar results. Although in systematic review eight polymorphisms, including MMP2 C-735T, MMP7 A-181G, MMP8 rs11225395, MMP9 rs6094237, MMP12 rs2276109, MMP20 rs2292730, MMP20 rs12278250, MMP20 rs9787933, might be associated with ovarian cancer risk, it was inconclusive results due to lack of relevant studies. Except eight above polymorphisms, it was revealed that other four polymorphisms in systematic review were not related with ovarian cancer risk.
The major strengths of our study were its comprehensive and systematic focus on the relationship between MMPs polymorphisms and ovarian cancer risk. Although a meta-analysis by Wang [32] has also investigated the relationship of MMP1 rs1799750 polymorphism with ovarian cancer (5 studies involving 754 cases and 1184 control) and produced similar results, our report identified 15 additional studies including 3058 cases and 3462 controls, which have not been included in report of Wang [32].
Also, some limitations still existed in our paper. First, control group was not uniformly defined, some controls were population-based while other controls were hospital-based. Second, significant heterogeneity was observed in a few comparison models. Although a subgroup analysis was performed to clarify sources, it was hard to find all potential sources. Third, departure from HWE was detected in some studies. Finally, there was a lack of a unified criterion for including studies, leading to failure to adjust them in age and lifestyle et al.
In summary, our reports showed that MMPs polymorphisms might not be associated with ovarian cancer risk. However, it is necessary to conduct more larger-scale, multicenter, and high-quality studies in the future.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Kaku T, Ogawa S, Kawano Y, Ohishi Y, Kobayashi H, Hirakawa T, et al. Histological classification of ovarian cancer. Med Electron Microsc. 2003; 36(1):9–17. doi: 10.1007/s007950300002 [DOI] [PubMed] [Google Scholar]
- 2.Al-Alem L, Curry TE Jr. Ovarian cancer: involvement of the matrix metalloproteinases. Reproduction. 2015; 150(2): R55–64. doi: 10.1530/REP-14-0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013; 63(1): 11–30. doi: 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 4.Ozols RF. Treatment goals in ovarian cancer. Int J Gynecol Cancer. 2005; 15suppl 1: 3–11. doi: 10.1111/j.1525-1438.2005.15351.x [DOI] [PubMed] [Google Scholar]
- 5.Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011; 130(1): 59–78. doi: 10.1007/s00439-011-1030-9 [DOI] [PubMed] [Google Scholar]
- 6.Geng P, Li J, Wang N, Ou J, Xie G, Liu C, et al. PSCA rs2294008 polymorphism with increased risk of cancer. PLoS One. 2015; 10(8): e0136269 doi: 10.1371/journal.pone.0136269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overall CM, Kleifeld O. Tumour microenvironment-opinion-validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006; 6(3): 227–239. doi: 10.1038/nrc1821 [DOI] [PubMed] [Google Scholar]
- 8.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002; 2(3): 161–174. doi: 10.1038/nrc745 [DOI] [PubMed] [Google Scholar]
- 9.Rasool M, Malik A, Basit Ashraf MA, Parveen G, Iqbal S, Ali I, et al. Evaluation of matrix metalloproteinases, cytokines and their potential role in the development of ovarian cancer. PLoS One. 2016; 11(11): e0167149 doi: 10.1371/journal.pone.0167149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu X, Li D, Zhang W, Zhou J, Tang B, Li L. Matrix metalloproteinase-9 expression correlates with prognosis and involved inovarian cancer cell invasion. Arch Gynecol Obstet. 2012; 286(6): 1537–1543. doi: 10.1007/s00404-012-2456-6 [DOI] [PubMed] [Google Scholar]
- 11.Kanamori Y, Matsushima M, Minaguchi T, Kobayashi K, Sagae S, Kudo R, et al. Correlation between expression of the matrix metalloproteinase-1 gene in ovarian cancers and an insertion/deletion polymorphism in its promoter region. Cancer Res. 1999; 59(17): 4225–4227 [PubMed] [Google Scholar]
- 12.Biondi ML, Turri O, Leviti S, Seminati R, Cecchini F, Bernini M, et al. MMP1 and MMP3 polymorphisms in promoter regions and cancer. Clin Chem. 2000; 46(12): 2023–2024 [PubMed] [Google Scholar]
- 13.Szyllo K, Smolarz B, Romanowicz-Makowska H, Niewiadomski M, Kozlowska E, Kulig A. The promoter polymorphism of the matrix metalloproteinase 3 (MMP-3) gene in women with ovarian cancer. J Exp Clin Cancer Res. 2002; 21(3): 357–361 [PubMed] [Google Scholar]
- 14.Wenham RM, Calingaert B, Ali S, McClean K, Whitaker R, Bentley R, et al. Matrix metalloproteinase-1 gene promoter polymorphism and risk of ovarian cancer. J Soc Gynecol Investig. 2003; 10(6): 381–387 [DOI] [PubMed] [Google Scholar]
- 15.Smolarz B, Szyłło K, Romanowicz-Makowska H, Niewiadomski M, Kozłowska E, Kulig A. PCR analysis of matrix metalloproteinase 3 (MMP-3) gene promoter polymorphism in ovarian cancer. Pol J Pathol. 2003; 54(4): 233–238 [PubMed] [Google Scholar]
- 16.Li Y, Wang Y, Kang S, Zhang JH, Guo W, Wang N, et al. Association of single nucleotide polymorphism in matrix metalloproteinases promoter with susceptibility to ovarian cancer. Zhonghua Fu Chan Ke Za Zhi. 2005; 40(7): 472–475 [PubMed] [Google Scholar]
- 17.Six L, Grimm C, Leodolter S, Tempfer C, Zeillinger R, Sliutz G, et al. A polymorphism in the matrix metalloproteinase-1 gene promoter is associated with the prognosis of patients with ovarian cancer. Gynecol Oncol. 2006; 100(3): 506–510. doi: 10.1016/j.ygyno.2005.08.049 [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Jin X, Kang S, Wang Y, Du H, Zhang J, et al. Polymorphisms in the promoter regions of the matrix metalloproteinases-1, -3, -7, and -9 and the risk of epithelial ovarian cancer in China. Gynecol Oncol. 2006; 101(1): 92–96. doi: 10.1016/j.ygyno.2005.09.058 [DOI] [PubMed] [Google Scholar]
- 19.Ju W, Kim JW, Park NH, Song YS, Kim SC, Kang SB, et al. Matrix metalloproteinase-1 promoter polymorphism and epithelial ovarian cancer: does ethnicity matter? J Obstet Gynaecol Res. 2007; 33(2): 155–160. doi: 10.1111/j.1447-0756.2007.00511.x [DOI] [PubMed] [Google Scholar]
- 20.Li XL, Kang S, Zhao XW, Zhang XJ, Zhou RM, Wang N, et al. Association of SNPs in the promoter of MMP-2 and TIMP-2 genes with epithelial ovarian cancer. Yi Chuan. 2008; 30(4): 455–462 [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Jia JH, Kang S, Zhang XJ, Zhao J, Wang N, et al. The functional polymorphisms on promoter region of matrix metalloproteinase-12, -13 genes may alter the risk of epithelial ovarian carcinoma in Chinese. Int J Gynecol Cancer. 2009; 19(1): 129–133. doi: 10.1111/IGC.0b013e31819a1d8e [DOI] [PubMed] [Google Scholar]
- 22.Jia J, Kang S, Zhao J, Zhang X, Wang N, Zhou R, et al. Association of functional polymorphisms on MMP-12 and MMP-13 gene promoter region with epithelial ovarian carcinoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010; 27(2): 209–213. doi: 10.3760/cma.j.issn.1003-9406.2010.02.020 [DOI] [PubMed] [Google Scholar]
- 23.Arechavaleta-Velasco F, Cuevas-Antonio R, Dominguez-Lopez P, Estrada-Moscoso I, Imani-Razavi FS, Zeferino-Toquero M, et al. Matrix metalloproteinase-8 promoter gene polymorphisms in Mexican women with ovarian cancer. Med Oncol. 2014; 31(8): 132 doi: 10.1007/s12032-014-0132-3 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Ye Y, Lin J, Meyer L, Wu X, Lu K, et al. Genetic variants in matrix metalloproteinase genes as disposition factors for ovarian cancer risk, survival, and clinical outcome. Mol Carcinog. 2015; 54(6): 430–439. doi: 10.1002/mc.22111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Kong B. Analysis of the association of matrix metalloproteinase-1 gene promoter (rs1799750) polymorphism and risk of ovarian cancer. Int J Gynecol Cancer. 2015; 25(6): 961–967. doi: 10.1097/IGC.0000000000000463 [DOI] [PubMed] [Google Scholar]
- 26.Chen SS, Song J, Tu XY, Zhao JH, Ye XQ. The association between MMP-12 82 A/G polymorphism and susceptibility to various malignant tumors: a meta-analysis. Int J Clin Exp Med. 2015; 8(7): 10845–10854. eCollection [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Sun J, Li G, Gu B, Wang X, Chi H,et al. Association between MMP-12-82A/G polymorphism and cancer risk: a meta-analysis. Int J Clin Exp Med. 2015; 8(8): 11896–11904. eCollection [PMC free article] [PubMed] [Google Scholar]
- 28.Thakkinstian A, McKay GJ, McEvoy M, Chakravarthy U, Chakrabarti S, Silvestri G, et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2011; 173(12): 1365–1379. doi: 10.1093/aje/kwr025 [DOI] [PubMed] [Google Scholar]
- 29.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst.1959; 22(4): 719–748 [PubMed] [Google Scholar]
- 30.Dersimonina R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986; 7(3): 177–188 [DOI] [PubMed] [Google Scholar]
- 31.Wu R, https://www.ncbi.nlm.nih.gov/pubmed/?term=Li%20B%5BAuthor%5D&cauthor=true&cauthor_uid=11318188LiB. A multiplicative-epistatic model for analyzing interspecific differences in outcrossing species. Biometrics. 1999; 55(2): 355–365 [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Kong B. Analysis of the association of matrix metalloproteinase-1 gene promoter (rs1799750) polymorphism and risk of ovarian cancer. Int J Gynecol Cancer. 2015; 25(6): 961–967. doi: 10.1097/IGC.0000000000000463 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.