Abstract
Drug-related adverse events pose substantial risks to patients who consume post-market or Drug-related adverse events pose substantial risks to patients who consume post-market or investigational drugs. Early detection of adverse events benefits not only the drug regulators, but also the manufacturers for pharmacovigilance. Existing methods rely on patients’ “spontaneous” self-reports that attest problems. The increasing popularity of social media platforms like the Twitter presents us a new information source for finding potential adverse events. Given the high frequency of user updates, mining Twitter messages can lead us to real-time pharmacovigilance. In this paper, we describe an approach to find drug users and potential adverse events by analyzing the content of twitter messages utilizing Natural Language Processing (NLP) and to build Support Vector Machine (SVM) classifiers. Due to the size nature of the dataset (i.e., 2 billion Tweets), the experiments were conducted on a High Performance Computing (HPC) platform using MapReduce, which exhibits the trend of big data analytics. The results suggest that daily-life social networking data could help early detection of important patient safety issues.
General Terms: Algorithms, Experimentation, Theory
Keywords: Twitter mining, Big-data Analytic, High Performance Computing, MapReduce, Natural Language Processing, Public Health, Drug-related Adverse Events
1. INTRODUCTION
The United States Food and Drug Administration (FDA) defines an Adverse Event (AE) as “any undesirable experience associated with the use of a medical product”. In the arena of pharmacovigilance, identifying AEs in a timely manner plays a vital role as some of the AEs can be life-threatening.
In order to capture AEs and Adverse Drug Reactions (ADRs), various surveillance systems–spontaneous reporting systems (SRSs)—have been developed around the world. In U.S., the FDA’s Adverse Event Reporting System (AERS) [30] is a major SRS with more than four million reports. As of March 2012, the AERS data contains publicly available reports until September 2011. In European, the European Medicines Agency developed the EudraVigilince [12]; and the World Health Organization has an international pharmacovigilance system as well. Aforementioned SRSs (particularly AERS) contain many reported post-market drugs’ AEs. Although drug manufactures are required to report all known AEs, majority of the adverse events are detected by the physicians and patients, where reporting is voluntary [33]. Thus, the overall incidences of AEs are significantly underestimated. Moreover, before marketing a new drug, clinical trials have to be conducted to study the investigational new drugs (or unapproved use of a drug), which is a major avenue for discovering drug-related AEs. However, the current venues of capturing AEs in clinical trials are cumbersome.
The explosion of the social media websites like Twitter, Google+, and Facebook, poses a great potential for variety of research arenas from targeted marketing to contagious disease capture. Twitter, as a micro blogging plat-form, has enormous increasing number of users. On Twitter, users publish short messages using 140 or fewer characters to “tweet” about their opinion on various topics and to share information or to have conversations with the followers. Often, a Twitter user would share health-related information, such as “this warm weather + tamoxifen hot flushes is a nightmare!”, which indicates the drug use (“tamoxifen”) and associated side effects (“hot flushes”) of the user. Hence, we believe that Twitter can be a promising new data source for Internet-based real-time pharmacovigilance because of its message volume, updating frequency, and its public availability.
The goal of our research is to develop an analytic frame-work for extracting knowledge from Twitter messages that could indicate potential serious side effects caused by a drug of interest. The knowledge gained can be used to create a knowledge base for early detection of adverse events or identification of under-reported adverse events. Such framework is not only beneficial to government agencies such as the FDA for monitoring and regulating the drugs, but it also helps the pharmaceutical companies for pharmacovigilance and to provide decision support.
2. BACKGROUND
2.1 Clinical Studies, Investigational Drugs, and Adverse Events
Clinical studies are basic building blocks of developing a new drug and required to be completed before potential marketing of a drug. There are many regulations related to AEs for investigational drugs and can be categorized into three basic types. 1) Expected AEs, where some AEs are known to occur during the study design period and listed in the investigational brochure, the inform consent or as part of the general investigational plan. This type of AEs has to be recorded and reported but does not normally impose significant risks (i.e., excluded in reporting for AE Rate and Person Year Exposure etc.). 2) Serious AEs are the ones that may be fatal, medically significant, an anomaly, life-threatening, or may cause disability, hospitalization. This type of AEs have to be reported in 24 hours. 3) Other AEs that are not expected by the design of a study, but also considered as important and need to be reported. As these AEs occur, the Data Safety and Monitoring Boards (DSMB) and/or the sponsor of the study may suggest a change of the intervention (i.e. change in drug dose).
However, it is not easy to estimate expected AEs or capture them as they occur during the study period. Usually AEs are captured during a clinic visit by the health care provider based on the participant’s responses or the result of a test (i.e. lab, radiology etc.). Participant responses can be in form of patient diaries they complete in between visits and/or verbal responses during the office visit. There are challenges in both cases. In the case of patient diaries: it is very time consuming; patients are often not compliant; patient’s self-diagnosis or interpretation is required; and diaries can have complicated instructions that are not easy to follow for the patients. In the matter of reporting AEs during office visits: patients often either have difficult time remembering the potential AEs, its start and end date (if resolved) or intentionally hold back the information from the investigator to avoid being removed from the study. A number of studies have raise concerns related to the reliability and effectiveness of the current AE collection and reporting methods [25] [17] [21].
Various new data-driven analytical approaches have been proposed in literatures. Chee et. al. tried to find AEs from personal health messages posted in online health forums [5]. Kuhn et. al. have focused on extracting side effect information from medical literatures [18]. Friedman uses natural language processing to analyze electronic health records (EHR) to identify novel adverse drug events [13].
2.2 Twitter Mining
Numerous studies have been published on the topic of mining Twitter messages for health-related information. Cobb et. al. analyzed online social networks including Twitter to study how these platforms can facilitate smoking cessation [8]. Prier et. al. conducted an empirical study to explore tobacco-related tweets for identifying health-related topics [24]. Paul et. al. proposed an analysis model for mining public health topics from Twitter [23]. Both Culotta [1] and Aramaki et. al. [11] explored the avenue to detect influenza epidemics by analyzing Twitter messages. Moreover, various other analytic models have been proposed to mine Twitter messages for different information, range from predicting election voting results [28] to studying global mood patterns [14]. However, to our knowledge, there has been no study on mining Twitter messages for drug-related AEs.
3. METHODS
We start with a collection of over 2 billion Tweets collected (i.e., not specifically collected for this research) by Paul et. al. [23, 22] from May 2009 to October 2010, from which we try to identify potential adverse events caused by drugs of interest. The collected stream of Tweets was organized by a timeline. The raw Twitter messages were crawled using the Twitter’s user timeline API [29] that contains information about the specific Tweet and the user. We are only interested in and indexed the following four fields for each Tweet: 1) the Tweet id that uniquely identifies each Tweet; 2) the user identifier associated with each Tweet; 3) the timestamp of the Tweet; and 4) the Tweet text.
To mine Twitter messages for AEs, the process can be separated into two parts: 1) identifying potential users of the drug; 2) finding possible side effects mentioned in the users’ Twitter timeline that might be caused by the use of the drug concerned. Both processes involve building and training classification models based on features extracted from the users’ Twitter messages. Two-sets of features (i.e., textual and semantic features) are extracted from Twitter users’ timeline for both classification models. Textual features such as the bag-of-words (BoWs) model are derived based our analysis of the actual Twitter messages. Semantic features are derived from the Unified Medical Language System (UMLS) Metathesaurus [26] concept codes extracted from the Tweets using Metamap [2] developed at the National Library of Medicine (NLM).
To test our hypothesis, we selected five cancer drugs (see Table 1) to evaluate our method. The five drugs are chosen according to information in the ClinicalTrails.gov [31] based on the following criteria: 1) the drug is an investigational drug that is used in a cancer treatment study (i.e., categorized as targeting the condition “Cancers and Other Neoplasms” in ClinicalTrials.gov); 2) the study was started during May 2009 and October 2010. Our assumption is that patients enrolled in those studies might be given the drugs of interest during that period of time; and they may Tweet about the experience of their drug use. Table 1 lists the drugs of our interest, their synonyms, number of unique Tweets found for each drug, and number of unique users found that have tweeted about the drug.
Table 1.
Drugs of interest
| Drug name | Synonym(s) | # of tweets |
# of users |
|---|---|---|---|
| Avastin | Bevacizumab | 264 | 236 |
| Melphalan | ALKERAN | 23 | 15 |
| Rupatadin | Rupafin, Urtimed | 10 | 10 |
| Tamoxifen | Nolvadex | 147 | 124 |
| Taxotere | Docetaxel | 45 | 39 |
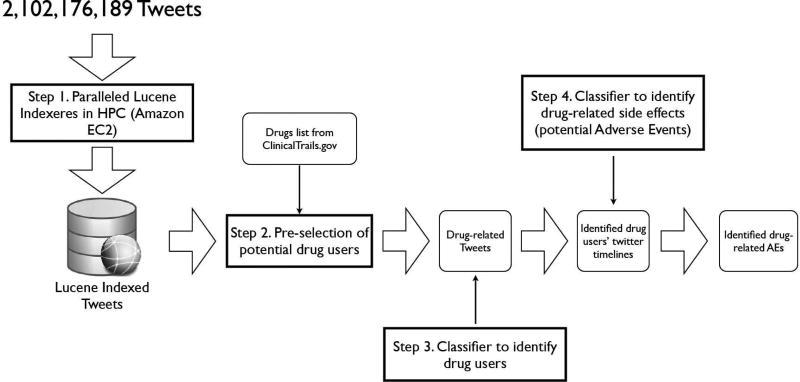
The data processing pipeline for the overall system is shown in Figure 1; and each step is explained below in detail.
Figure 1.
The data processing pipeline for the overall system.
3.1 Step 1 – Paralleled Lucene indexeres in a HPC platform
Searching and performing analysis over raw text files in such a massive volume (i.e., 2 billion Tweets in our study, and 1.5 terabytes storage space for both the text of the message and meta-data associated with the Tweet) is impractical. Rather, an efficient full text search capability is necessary to search over all 2B tweets containing a specific keyword (e.g., searching by a drug name, including its synonyms). Initially, we started with parsing the raw Twitter data and inserting the messages into a relational database (i.e., MySQL) that is capable of creating full text indexes. However, the performance of such attempt is undesirable (i.e., it took 38 hours to process 20 days’ Twitter messages). As another option, we used a specialized information retrieval library–Apache Lucene [27]. The use of Lucene resulted in significant performance improvements; particularly, it was possible to index 30 days tweets within 10 hours. However, considering that we have 18 months of Twitter data, it is still a computational intensive and time-consuming process. Fortunately, it is possible to parallelize the Lucene indexing process in a High Performance Computing (HPC) environment. In our case, we utilized the Amazon Elastic Compute Cloud (EC2) to run the Twitter indexers on 15 separate EC2 instances (i.e., High-Memory Double Extra Large Instance (m2.2×large), 34.2 GB of memory, and 13 EC2 Compute Units) in parallel, which we were able to parse and index all 2 billion Tweets within two days. The size of the Lucene indexes is 896 GB.
3.2 Step 2 – Pre-select potential drug users and extract users’ Twitter timeline
In this step, we searched over the 2B Twitter messages to find all Tweets that contained the drug name or one of its synonyms. We choose five cancer drugs (see Table 1) to evaluate our method. We also pre-processed the data to eliminate obvious outliners, such as re-Tweets (i.e., starts with ‘RT’), and Tweets that are not in English (i.e., we used the chromium-compact-language-detector [19] for language detection). After pre-processing, 239 users remain as potential drug users (i.e., this number seems to be low, but it is expected, since the drugs of interest we picked are drugs used in clinical trials that may not be on the market during that time frame) and their drug-related Tweets are collected. Since one user may have multiple Tweets about a drug of interest, we aggregate all these Tweets as one document to be processed by the classifier in the next step. In short, each document contains one or more Tweets related to the drug of interest tweeted by one user.
3.2.1 Step 3 – Building a classifier to identify drug users
In this step, we built a Support Vector Machine (SVM) to label a collection of Tweets (i.e., a document) posted by a Twitter user as whether the user herself or someone she knows has taken the drug of interest. For example, a document that contains “No more tamoxifen for me - finished 5 yrs of post cancer drug therapy.” is labeled as 1 indicating that the user is taking “tamoxifen”–a drug of our interest; while the user who tweeted “Please visit us at www.genglob.com For generic anti cancer drugs medicines alkeran, iressa, gefitinib, erlotinib, temonat, revlimid, Velcade,” should be classified as NOT a drug user, although she mentioned a drug of our interest–“alkeran”–in the Tweet. For cases where the Twitter user herself is not a drug user, but the Tweet indicate that a positive drug user she might know, we also labeled the document as positive. For example, “@whymommy so what does this mean for you?? What’s the game plan? I’m so sorry about Avastin”, where the context suggests that “@whymommy” is a user of “avastin”. To evaluate the performance of the classification model, the 239 collections of Twitter messages extracted from the previous step are labeled manually; where each document is reviewed by at least two domain experts to ensure the accuracy. 72 positive cases are found. This labeled dataset of the 239 documents is then used to train and test the classifier.
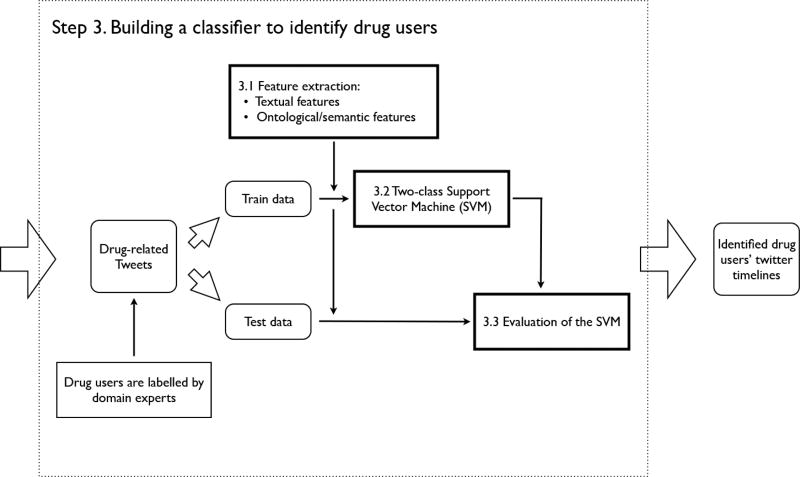
Figure 2 shows the detailed process of building and testing the classification model to identify drug users. It consists of three main parts, and the details are explained below.
Figure 2.
The detailed process of building and testing the classification model to identify drug users.
3.2.2 Step 3.1. Feature extraction
Two groups of features (i.e., 171 individual features) are extracted: 1) textual features that construct a specific meaning in the text; and 2) ontological/semantic features that express the existence of semantic properties. Seven textual features are considered based on our analysis of over 5, 000 drug-related Tweets (i.e., independent from our testing/training datasets for the SVM). Details about the choices of the textual features are discussed in the Discussion section. For semantic features, we use Metamap to discover UMLS Metathesaurs concepts expressed in the Twitter text. Feature values are then derived from the concept codes according to both the Semantic Type [32] and the more abstracted Semantic Group [20]. Before feeding the Twitter messages to Metamap, we pre-processed the text to eliminate elements that have no semantic meanings: 1) URLs; 2) user mentions; and 3) a list of words that will map to undesired concepts (e.g., the word ‘I’ will be mapped to “660 C0021966:I- (Iodides) [Inorganic Chemical]”, which is often incorrect within the context of Twitter messages). For each collection of twitter messages, Metamap generates a list of concept codes mapped from the free text, where each concept has: 1) a confidence score of the mapping; 2) a CUI code that uniquely identifies the concept; 3) the preferred name of the concept; and 4) the Semantic Type of the concept. Before deriving the feature values for each document, concepts with lower confidence score are dropped.
Further, semantic types in UMLS help to cluster the concepts into different categories. Therefore, the first set of semantic features we used is the count of concepts within each semantic type. The major semantic types are organisms, anatomical structures, manufactured object, substance, etc. However, for our purpose, we are mainly interested in semantic types that are related to drug (e.g., “Clinical Drug”, “Pharmacologic Substance”, etc.) and adverse effects (e.g., “Finding”, “Sign or Symptom”, “Disease or Syndrome”, etc.).
The current scope of the UMLS semantic types is quite broad (i.e., 135 semantic types and 54 relationships), allowing for the semantic categorization of a wide range of terminology in multiple domains. Therefore, in [20], McCray et. al. aggregated the UMLS semantic types into semantic groups to reduce conceptual complexity. In practice, the 135 semantic types are further abstracted into 15 groups, such as “Chemicals & Drugs”, “Disorders”, etc. Therefore, the second set of semantic features we used in this study is the count of concepts that fall into each semantic group.
Here is a summary of all the features we considered in this study.
Textual features that construct a specific meaning in the text:
Bag-of-words features that indicate an action or a state that the user has taken the drug
Number of hash-tags occurred in the document
Number of reply-tags occurred in the document
Number of words that indicate negation
Number of URLs
Number of pronouns
Number of occurrences of the drug name or its synonyms
Semantic features that express the existence of semantic properties (i.e., based on UMLS Concept Unique Identifiers (CUIs) extracted from the Tweets):
Number of CUIs in each Semantic Type
Number of CUIs in each Semantic Group
3.2.3 Step 3.2. Two-class Support Vector Machine (SVM)
Support Vector Machine (SVM) has been widely accepted as a effective technique for data classification. In a general classification problem, input data is split into training and testing sets. Each instance (i.e., sample) of the training set contains one class label (i.e., the target value) that indicates the category of the sample; and a vector of features (i.e., attributes, observed variables), which describes some characteristics of the instance. The goal of a SVM is to generate a prediction model based on training data that can accurately predict the class labels of the testing data given only the feature vectors of the testing data. Mathematically, given a training dataset (xi, yi), i = 1, …, l, where xi ∈ Rn is the feature vector of sample i and yi ∈ {+1, −1} is the class label of the same sample, the SVM [3, 9, 4] is to solve the following optimization problem,
| (1) |
subject to
The feature vectors (xi) are mapped into a higher (maybe infinite) dimensional space by the function ϕ(). SVM finds a linear separating hyperplane with the maximal margin in the higher dimensional space; C is the penalty parameter of the error term. Furthermore, SVM uses a kernel function (i.e., K(xi, xj) ≡ ϕ(xi)T ϕ(xj) to map the data into the higher feature space where a hyperplane can be drawn to do the separation. Often, the Gaussian Radial Basis Function (RBF) (i.e., K(xi, xj) = exp(−γ‖xi − xj‖2), γ > 0) is a reasonable first choice. RBF is a nonlinear mapping that can handle situations where the relation between the class labels and the features is nonlinear.
Features in the feature vectors are not necessarily all relevant to the target values (i.e., class labels). Feature selection is the process where a optimal subset of pertinent features for building more accurate and robust learning models. In this study, we found that the one-way Analysis of Variance (ANOVA) F-test [10] is simple, yet effective [7, 6]. Various standard techniques such as scaling, grid-based kernel parameter search, etc. [15] for building an efficient SVM have also been taken into account in this study.
3.2.4 Step 3.3. Evaluation of the SVM
Various classification metrics have been calculated to evaluate the SVM which include the accuracy, the precision, the recall, the Area Under the Curve (AUC) value, and the Receiver operating characteristic (ROC) curve.
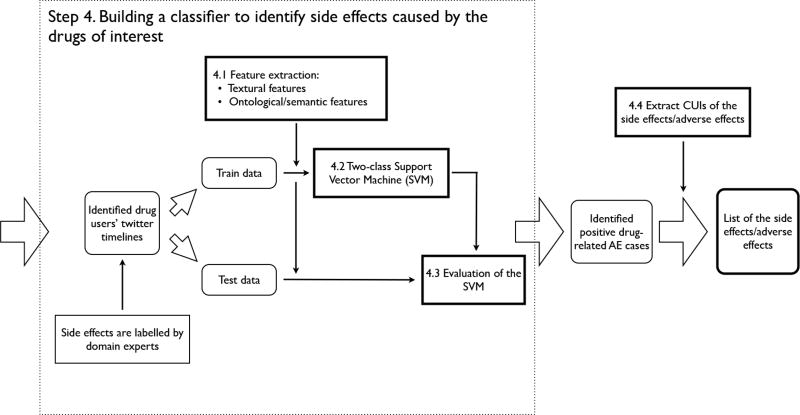
3.3 Step 4 – Building a classifier to identify side effects caused by the drugs of interest
The second part of this study is to build another classification model to identify side effects (i.e., adverse event indicators) caused by the use of the drugs. The overall process of this part as shown in Figure 3 is similar to Step 3 described above. We considered the same set of features (i.e., both textual and semantic features) as described in the previous section. However, there are three main differences.
Figure 3.
The process of building and testing the classification model to identify side effects caused by the drugs of interes.
First, the dataset of this classification model is the result of the previous step, where we only consider the positive cases identified by the previous model (i.e., 72 out of 239 cases are labeled as drug users). Similar to the process in the previous step, two domain experts evaluated all 72 positive cases and label each one as whether it can be considered as a report for an adverse event (i.e., side effects caused by the drug of interest). 27 out of 72 cases are labeled as positive.
Second, we consider more Tweets for each user. In the previous step, it is sufficient to only consider Tweets that contains the specific drug name or its synonyms. However, when mining for side effects, it is possible that the Twitter user complains about the harmful and undesired effects of the drug in a separate Tweet; and the Tweet can happen either before or after the mention of the drug. Therefore, in this classification model, we consider m Tweets before the drug-related Tweet plus n Tweets after it. In current implementation, m = 5 and n = 15, which gives us at least 21 (i.e., including the drug-related Tweet) to consider for each instance (i.e., some user mentioned the drug in multiple Tweets, therefore, it is possible to extract more than 21 Tweets from each user’s Twitter timeline to consider).
Third, from this model, we also extract a list of concept codes of the adverse effects for further analysis. One possible use of this list is to compare it with the reactions reported to the FDA’s Adverse Event Reporting System (AERS) [30] to find unreported or rare side effects.
4. RESULTS
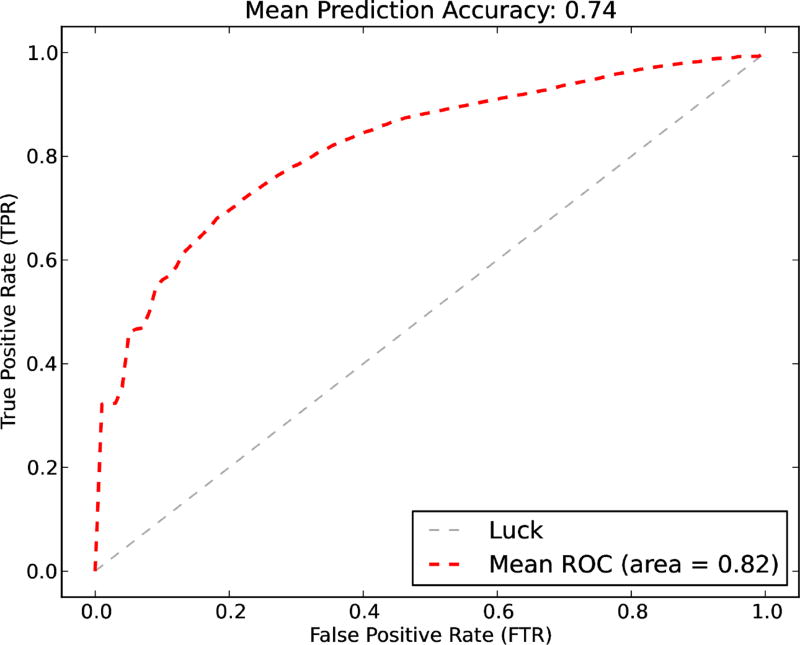
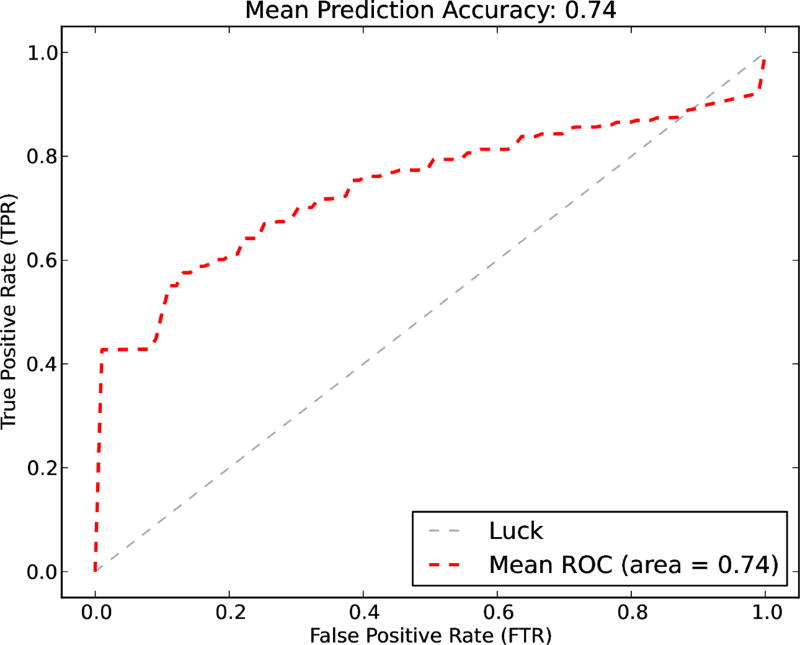
The results of both classification models are as the followings. For the drug user classifier, since there are only 72 positive cases out of 239 cases, the dataset is unbalanced. A naive SVM classifier cannot handle unbalanced dataset well. Therefore, we used down-sampling method to create a balanced dataset to improve SVM performance. To reduce bias, we bootstrap the down-sampling process 1000 times. In detail, at each iteration, we first randomly picked 72 negative cases to create a balanced dataset (i.e., 144 total cases, 72 positive plus 72 negative), then the samples are split into training and testing datasets (i.e., 2/3 is for training, 1/3 is left for testing). We trained a SVM classifier use the training test (i.e., including a f-score feature selection process), and measured the performance on the testing set. We drew the ROC curve using the mean values of the 1000 iterations. The prediction accuracy on average over the 1000 iterations is 0.74 and the mean AUC value is 0.82. Figure 4 shows the ROC curve of the model. The best features selected are: 1) from the textual feature set: the number of replay tags, the number of urls, and two word counts from the BoW features, which are the the word ‘got’ and the word ‘go’ (i.e., including counts of their variants); 2) from the semantic feature set: “Injury or Poisoning”, “Anatomical Abnormality”, and “Organ or Tissue Function”.
Figure 4.
Drug use identification - The mean prediction accuracy, the mean AUC value, and the ROC curve of the classifiers.
For the AE classification model, the same process applied. The prediction accuracy on average over the 1000 iterations is 0.74 and the mean AUC value is 0.74. Figure 5 shows the ROC curve of the model. In terms of best features, there is no textual feature selected; and the following semantic features give the best prediction results: “Sign or Symptom”, “Anatomical Abnormality”, “Intellectual Product”, “Human-caused Phenomenon or Process”, and “Behavior”.
Figure 5.
AE detection - The mean prediction accuracy, the mean AUC value, and the ROC curve of the classifiers.
5. DISCUSSION
The key factor of the classification performance are the features. In this study, we considered two groups of features extracted from the Twitter messages–textual and semantic features. Both classification models use the same feature extraction method, except in the drug user classification model we only considered the drug-related Tweets; while in the AE classification model we expanded the scope to consider Tweets around the drug-related Tweets (according to the timeline of the Tweets). Here, we first briefly discuss the reasons behind the choices of the textual features.
Bag-of-words (BOWs) features that indicate an action or a state that the user has taken the drug: The BOWs model is commonly in document classification, where it assumes that a text can be represented as an unordered collection of key words disregarding grammar. In our case, we carefully evaluated all the positive cases (i.e., we are not limited to the positive cases in the test dataset, which will lead to bias. Rather, we randomly picked another drug-related dataset and identified positive cases manually), and created a bag of 15 key words that might indicate the using of the drug. For example, a user tweeted that “Second Avastin injection down, two to go. It’s worse the second time around.”, where “injection” is a key word leads us to believe that the user is taking “Avastin”. We also considered the variants of the key words including the different part of speech (POS) of the same word (e.g., inject is the verb form of the noun injection), the different tenses of a verb (e.g., injected, inject, injecting), the singular or plural forms of a word (e.g., injects and inject), etc. The variants of the same word are considered as in one bag. A word count is calculated for each of the 15 key words present in a Tweet document [16].
Number of hashtags occurred in the document: Hash-tags are used in Twitter for marking keywords or topics in a Tweet, which often used by Twitter users as a way to categorize messages. The use of a hash-tag in a Tweet often indicates that the user wants to share a piece of information related to the drug, rather than the user has actually taken the drug. For example, there was a news break about the FDA revoking the approval of the breast cancer indication for “bevacizumab” (“Avastin®”, made by Genentech); and a user tweeted that “news: avastin may lose its FDA approval as breast cancer treatment. #avastin #breast-cancer”.
Number of user mentions occurred in the document: Mentions (i.e., starts with an ‘@’ sign followed by a Twitter user display name) are often used in Twitter by a user to reply other’s Twitter messages or indicate that the Tweet is intended for the users mentioned. Higher number of mentions occurred in a user’s Twitter timeline can indicate frequent discussions on a specific topic (e.g., “@HeyTammyBruce Where are the feminists on FDA story re: breakthrough breast cancer drug Avastin not being approved due to expense?”.
Number of words that indicate negation: A negation term is important feature to recognize, since it often indicates a negative intention. For example, “Still waiting for my first chemo. I will not be given the avastin as hoped, but that also means chemo will only take 5 months!”.
Number of Uniform resource locators (URL)s: Higher number of hyperlinks occurred in a user’s Twitter time-line often suggests that the user is merely showing information with his/her followers. For example, in “Avastinc an cause kidney damage http://bit.ly/9KWksC Higher doses and kidney cancer increase the risks; check for proteinuria w/ every cycle”, the user shares results of a “avastin” study reported by a news source; and the URL is used to link back to the original news article.
Number of pronouns: The existence of pronouns (e.g., I, she, he, etc.) in a Tweet is highly correlated to whether the user is a receiver of the drug. Further, pronouns can be an important factor to determine whether the drug user is the one who tweeted the message or it is merely someone the Twitter user knows. Although we do not use this information in this study, it is a valuable information for future research.
Number of occurrences of the drug name or its synonyms: Like the case with URLs, the fact that a user has a high number of Tweets that mentions the drug name hints that the user is trying to act as an information source (e.g., a Twitter user who gives constant update on breaking news) or the user could be a drug retailer, where the Twitter is used as a marketing platform.
The textual features are more tailored toward solving the first classification model to identify drug users. For finding potential adverse effects, the semantic features extracted using the UMLS concept codes fit the AE classification model better, since it contains features like number of concept codes that are “Finding”, “Sign or Symptom”, “Disease or Syndrome”.
The performance of both classification models are rather low. We think there are various reasons. First, the Twitter data is very noisy. People often use fragmented sentences or otherwise ungrammatical sequences and are filled with mis-spellings, odd abbreviations, and other non-word terms in their Tweets. The standard conventions for capitalization and punctuations are not strictly followed. Moreover, potential patients do not use standard medical terms, which often caused the Metamap to extract the wrong concept codes. Further, Metamap’s POS tagger is trained on medical documents that is more structured than Twitter messages. We intend to develop and train a social media specific POS tagger for biomedical information, which could potentially increase the classification accuracy.
6. CONCLUSION
In this research, we developed an analytic framework that combines natural language processing and machine learning methods to extract drug-related adverse events from the Twitter messages. Although the performance of both classification models is limited due to the high-level of noises existed in Twitter messages, this paper demonstrates the potential to meet our ultimate goal to automatically extract AE-related knowledges to support pharmacovigilance.
Acknowledgments
The work describe in this manuscript is supported by award UL1RR029884 through the NIH National Center for Research Resources and National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The work is also supported by the National Science Foundation EPSCoR Cyber infrastructure award #EPS-0918970. We thank Mark Dredze and Michael Paul [23, 22] from the Johns Hopkins University for sharing the raw Twitter data collected from May 2009 to October 2010.
Contributor Information
Jiang Bian, Division of Biomedical Informatics, University of Arkansas for Medical Sciences, Little Rock, AR.
Umit Topaloglu, Division of Biomedical Informatics, University of Arkansas for Medical Siences, Little Rock, AR.
Fan Yu, Research Systems, Information Technology, University of Arkansas for Medical Sciences, Little Rock, AR.
References
- 1.Aramaki E, Maskawa S, Morita M. Proceedings of the Conference on Empirical Methods in Natural Language Processing, EMNLP ’11. Stroudsburg, PA, USA: Association for Computational Linguistics; 2011. Twitter catches the flu: detecting influenza epidemics using twitter; pp. 1568–1576. [Google Scholar]
- 2.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 3.Boser BE, Guyon IM, Vapnik VN. Proceedings of the fifth annual workshop on Computational learning theory, COLT ’92. New York, NY, USA: ACM; 1992. A training algorithm for optimal margin classifiers; pp. 144–152. [Google Scholar]
- 4.Chang C-C, Lin C-J. Libsvm: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011 May;2(27):1–27. 27. [Google Scholar]
- 5.Chee BW, Berlin R, Schatz B. Predicting adverse drug events from personal health messages. AMIA Annu Symp Proc. 2011;2011:217–226. [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Lin C. Combining svms with various feature selection strategies. Feature Extraction. 2006:315–324. [Google Scholar]
- 7.Chen Y-W, Lin C-J. Combining SVMs with various feature selection strategies. In: Guyon I, Gunn S, Nikravesh M, Zadeh L, editors. Feature extraction, foundations and applications. Springer; 2006. [Google Scholar]
- 8.N. K. e. a. Cobb. Online social networks and smoking cessation: a scientific research agenda. J. Med. Internet Res. 2011;13:e119. doi: 10.2196/jmir.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes C, Vapnik V. Support-vector networks. Mach. Learn. 1995 Sep;20:273–297. [Google Scholar]
- 10.Richard G, Lomax, Hahs-Vaughn Debbie L, editors. S. C. A. S. Course. Statistical Concepts: A Second Course. 3. 2007. [Google Scholar]
- 11.Culotta A. Proceedings of the First Workshop on Social Media Analytics, SOMA ’10. New York, NY, USA: ACM; 2010. Towards detecting influenza epidemics by analyzing twitter messages; pp. 115–122. [Google Scholar]
- 12.European Medicines Agency. Eudravigilance – pharmacovigilance in eea. 2012 Mar; http://eudravigilance.ema.europa.eu/human/index.asp.
- 13.Friedman C. Proceedings of the 12th Conference on Artificial Intelligence in Medicine: Artificial Intelligence in Medicine, AIME ’09. Berlin, Heidelberg: Springer-Verlag; 2009. Discovering novel adverse drug events using natural language processing and mining of the electronic health record; pp. 1–5. [Google Scholar]
- 14.Golder SA, Macy MW. Diurnal and seasonal mood vary with work, sleep, and daylength across diverse cultures. Science. 2011 Sep;333(6051):1878–1881. doi: 10.1126/science.1202775. [DOI] [PubMed] [Google Scholar]
- 15.Hsu CW, Chang CC, Lin CJ. A practical guide to support vector classification. Department of Computer Science and Information Engineering, National Taiwan University; Taipei, Taiwan: 2003. [Google Scholar]
- 16.Iyengar A, Finin T, Joshi A. Content-based prediction of temporal boundaries for events in twitter. SocialCom/PASSAT. 2011:186–191. [Google Scholar]
- 17.Juergens JP, Szeinbach SL, Janssen MJ, Brown TR, Garner DD. An evaluation of interventions designed to stimulate physician reporting of adverse drug events. Top Hosp Pharm Manage. 1992 Jul;12(2):12–18. [PubMed] [Google Scholar]
- 18.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCandless M. chromium-compact-language-detector. 2012 Mar; http://code.google.com/p/chromium-compact-language-detector/
- 20.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Stud Health Technol Inform. 2001;84:216–220. [PMC free article] [PubMed] [Google Scholar]
- 21.Millman EA, Pronovost PJ, Makary MA, Wu AW. Patient-assisted incident reporting: including the patient in patient safety. J Patient Saf. 2011 Jun;7(2):106–108. doi: 10.1097/PTS.0b013e31821b3c5f. [DOI] [PubMed] [Google Scholar]
- 22.O’Connor B, Balasubramanyan R, Routledge BR, Smith NA. From tweets to polls: Linking text sentiment to public opinion time series; Proceedings of the Fourth International Conference on Weblogs and Social Media; 2010. [Google Scholar]
- 23.Paul MJ, Dredze M. You are what you tweet : Analyzing twitter for public health. Artificial Intelligence. 2011;38:265–272. [Google Scholar]
- 24.Prier KW, Smith MS, Giraud-Carrier C, Hanson CL. Proceedings of the 4th international conference on Social computing, behavioral-cultural modeling and prediction, SBP’11. Berlin, Heidelberg: Springer-Verlag; 2011. Identifying health-related topics on twitter: an exploration of tobacco-related tweets as a test topic; pp. 18–25. [Google Scholar]
- 25.Scharf O, Colevas AD. Adverse event reporting in publications compared with sponsor database for cancer clinical trials. J. Clin. Oncol. 2006 Aug;24(24):3933–3938. doi: 10.1200/JCO.2005.05.3959. [DOI] [PubMed] [Google Scholar]
- 26.Schuyler PL, Hole WT, Tuttle MS, Sherertz DD. The UMLS Metathesaurus: representing different views of biomedical concepts. Bull Med Libr Assoc. 1993 Apr;81:217–222. [PMC free article] [PubMed] [Google Scholar]
- 27.The Apache Software Foundation. Apache lucene core. 2012 Mar; [Google Scholar]
- 28.Tumasjan A, Sprenger TO, Sandner PG, Welpe IM. Predicting elections with twitter: What 140 characters reveal about political sentiment. In: Cohen WW, Gosling S, editors. ICWSM. The AAAI Press; 2010. [Google Scholar]
- 29.Twitter. Rest api resources. 2012 Mar; https://dev.twitter.com/docs/api.
- 30.U.S. Food and Drug Administration. Adverse event reporting system (aers) 2012 Mar; http://www.fda.gov/drugs/
- 31.U.S. National Institutes of Health. 2012 Mar; Clinicaltrails.gov. http://clinicaltrials.gov/
- 32.U.S. National Library of Medicine. Current semantic types. 2012 Mar; http://www.nlm.nih.gov/research/umls/META3_current_semantic_types.html.
- 33.Wang W, Haerian K, Salmasian H, Harpaz R, Chase H, Friedman C. A drug-adverse event extraction algorithm to support pharmacovigilance knowledge mining from PubMed citations. AMIA Annu Symp Proc. 2011;2011:1464–1470. [PMC free article] [PubMed] [Google Scholar]