A mutation in a gene implicated in circadian clock function is associated with early flowering in a commercially significant chickpea cultivar.
Abstract
In climates that experience short growing seasons due to drought, heat, or end-of-season frost, early flowering is a highly desirable trait for chickpea (Cicer arietinum). In this study, we mapped, sequenced, and characterized Early flowering1 (Efl1), an ortholog of Arabidopsis (Arabidopsis thaliana) EARLY FLOWERING3 (ELF3) that confers early flowering in chickpea. In a recombinant inbred line population derived from a cross between CDC Frontier and ICCV 96029, this gene was mapped to the site of a quantitative trait locus on Ca5 that explained 59% of flowering time variation under short days. Sequencing of ELF3 in ICCV 96029 revealed an 11-bp deletion in the first exon that was predicted to result in a premature stop codon. The effect of this mutation was tested by transgenic complementation in the Arabidopsis elf3-1 mutant, with the CDC Frontier form of CaELF3a partially complementing elf3-1 while the ICCV 96029 form had no effect on flowering time. While induction of FLOWERING LOCUS T homologs was very early in ICCV 96029, an analysis of circadian clock function failed to show any clear loss of rhythm in the expression of clock genes in ICCV 96029 grown under continuous light, suggesting redundancy with other ELF3 homologs or possibly an alternative mode of action for this gene in chickpea. The 11-bp deletion was associated with early flowering in global chickpea germplasm but was not widely distributed, indicating that this mutation arose relatively recently.
Flowering time is a key factor in the geographical distribution of both native plant populations and cultivated crops (Jones et al., 2008). The two main environmental factors that regulate the transition from vegetative to reproductive development are photoperiod and vernalization, which, together, help ensure that flowering occurs when seasonal conditions are most likely to be conducive to seed set and maturation. The molecular pathways responsible for the regulation of flowering time in response to such environmental signals have been studied intensively in the model species Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), and more recently, there have been rapid advances in applying this knowledge to a wide range of important crops, including cereals, brassicas, and legumes, where the potential exists to further extend the range of cultivation through the selection of flowering gene variants (Osborn et al., 1997; Hecht et al., 2005; Higgins et al., 2010; Peng et al., 2015).
Domesticated chickpea (Cicer arietinum) is an important member of the legume family cultivated across more than 13.5 million ha globally, with major production centers in India, Australia, Pakistan, Turkey, Burma, Iran, Canada, and the United States (FAOSTAT, 2014; Gaur et al., 2015). Its wild progenitor, Cicer reticulatum, is a quantitative long day (LD) plant that exhibits responsiveness to vernalization (Smithson et al., 1985; Abbo et al., 2002). In its natural range within southeastern Turkey, C. reticulatum germinates in the autumn, flowers in the spring, and matures in the summer. The sensitivity of chickpea to ascochyta blight has confined the natural range of C. reticulatum to this limited region where winters are relatively dry and disease pressure is low (Abbo et al., 2002). Following the domestication of the wild progenitor, selection within domesticated chickpea for reduced vernalization responsiveness and increased photoperiod sensitivity is thought to have enabled the expansion of domestic chickpea cultivation into regions with damp winters and shorter growing seasons by enabling spring sowing, permitting the entire plant life cycle to be completed before the end of summer and thus avoiding conditions conducive to the spread of ascochyta blight (Kumar and Abbo, 2001). Domestication, together with the widespread adoption, early in the domestic crop’s history, of photoperiod-responsive, vernalization-insensitive genotypes suited to spring cropping, has left modern cultivated chickpea germplasm with greatly depleted genetic diversity (Kumar and Abbo, 2001; Abbo et al., 2003).
The resulting narrow genetic base presents difficulties for chickpea breeders now seeking to develop even earlier flowering cultivars that mature within the extremely short growing seasons encountered in many of the major chickpea growing regions. For example, in India, where spring cropping relies on residual winter rainfall, end-of-season drought during the pod-filling stage is a major constraint to chickpea production. In colder climates, such as those encountered in Saskatchewan, Canada, where 99% of Canada’s chickpeas are grown, commercial varieties of chickpea generally take around 110 to 130 d to mature, while the average frost-free period is just 110 to 120 d (Daba et al., 2016b). The identification of genes that confer early flowering to enable maturation within this short season would be highly desirable in order to mitigate the yearly risk of frost damage.
Currently, four major Mendelian loci controlling flowering time in chickpea have been identified through classical genetic analysis, with recessive alleles at these loci conferring early flowering. These have been named Early flowering1 (Efl1) to Efl4 (Gaur et al., 2015), with mutant alleles designated as efl1 to efl4. These were first identified in lines ICCV 2 (Efl1; Kumar and van Rheenen, 2000), ICC 5010 (Efl2; Or et al., 1999), BGD-132 (Efl3; Hegde, 2010), and ICC 16641 and ICC 16644 (Efl4; Gaur et al., 2015). In addition, six quantitative trait loci (QTLs) have been reported from chickpea linkage studies (Weller and Ortega, 2015). Several of these studies have involved ICCV 2, which is known to carry efl1, and each of these reported a major QTL on what is now known to be Ca5 (Cho et al., 2002; Vadez et al., 2012; Jamalabadi et al., 2013; Pushpavalli et al., 2015), while Efl2 has been shown to be associated with a QTL on Ca1 (Lichtenzveig et al., 2006). A recent genome-wide association study identified five further flowering time QTLs and put forward eight candidate genes for these QTLs, including an ortholog of the Arabidopsis gene PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1) on Ca4 that is proposed to be synonymous with the Efl1 locus (Upadhyaya et al., 2015).
One difficulty for the interpretation of previous genetic analyses of flowering time in chickpea has been the lack of common markers for comparison between different maps. As a result, there is considerable ambiguity surrounding the map positions of the chickpea Efl1 to Efl4 loci. In order to clarify the location, effect, and identity of genes that regulate flowering time, we identified QTLs and candidate genes for flowering time and photoperiod sensitivity in a recombinant inbred line (RIL) population derived from a cross between CDC Frontier, a photoperiod-sensitive Kabuli-type cultivar that was used recently to generate a whole-genome reference sequence (Varshney et al., 2013), and the extremely early-flowering, photoperiod-insensitive ICCV 96029, a Desi variety derived from ICCV 2, the efl1-type line. We present evidence that a major QTL conferring earliness in ICCV 96029 colocates with an ortholog of the key circadian gene ELF3 and that ICCV 96029 carries a mutation that impairs the function of this gene. We also show that the early-flowering phenotype of ICCV 96029 is associated with the elevated expression of several FLOWERING LOCUS T (FT) homologs.
RESULTS
QTL Analysis Reveals a Major Flowering Time Locus on Ca5
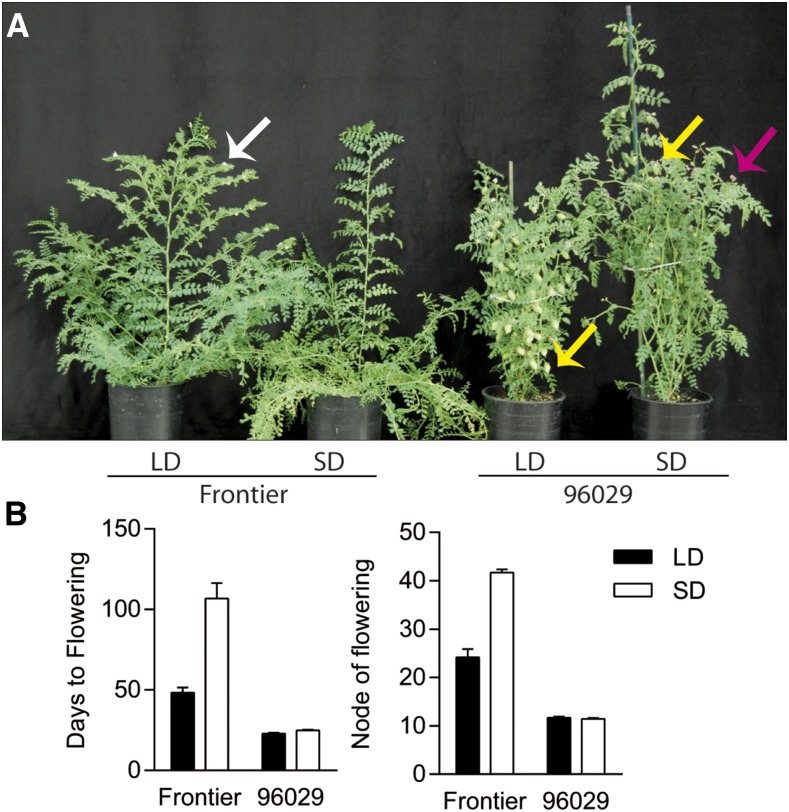
In order to characterize flowering behavior and photoperiod sensitivity of the biparental F8 mapping population CPR-01 (Tar’an et al., 2007; Deokar et al., 2014; Ali et al., 2016; Daba et al., 2016a), both RILs and parental lines CDC Frontier and ICCV 96029 were grown under short day (SD; 10 h of light/14 h of dark) and LD (16 h of light/8 h of dark) conditions. Figure 1 shows that CDC Frontier flowering time was highly photoperiod dependent, with days to flowering (DTF) significantly higher under SD as compared with LD (106.83 ± 9.49 versus 48.33 ± 3.23, respectively). By contrast, ICCV 96029 was largely photoperiod insensitive, with similar DTF in both SD and LD (24.88 ± 0.44 versus 22.89 ± 0.53, respectively). In both genotypes, the node of floral initiation was affected by photoperiod: SD-grown CDC Frontier flowered approximately 18 nodes later as compared with LD-grown CDC Frontier, while there was no difference in the node of floral initiation between LD- and SD-grown ICCV 96029 (Fig. 1B). CDC Frontier typically initiated abortive flower buds at nodes 16 to 20 prior to the formation of perfect flowers regardless of photoperiod, but the number of abortive buds was greater under SD. We observed that F1 populations derived from ICCV 96029 crosses were entirely late flowering under SD, whereas early flowering was recessive in the F2 populations derived from a range of crosses involving ICCV 96029 in our breeding program, consistent with previous reports of recessive early-flowering phenotypes in segregating populations derived from ICCV 96029 (Gumber and Sarvjeet, 1996; Or et al., 1999; Kumar and van Rheenen, 2000; Anbessa et al., 2006).
Figure 1.
ICCV 96029 is early flowering and shows reduced responsiveness to photoperiod compared with CDC Frontier. A, Comparison of CDC Frontier and ICCV 96029 phenotypes for plants grown under LD and SD. LD-grown CDC Frontier has initiated flowers (white arrow), while SD-grown CDC Frontier is vegetative. LD-grown ICCV 96029 has set strong seed and is beginning to dry off; SD-grown ICCV 96029 has produced weak pods and continues to flower (purple arrow). Pods are indicated by yellow arrows. Plants are 63 d from sowing. B, Effects of photoperiod on flowering behavior in CDC Frontier and ICCV 96029 plants. Values represent means ± se for n = 6 to 9 replicates.
We next examined the genetic basis for differences in flowering time and photoperiod responsiveness between these two parent lines using the CDC Frontier × ICCV 96029 CPR-01 RIL population (Deokar et al., 2014), identifying a total of eight QTLs for flowering time. These included four flowering time QTLs under LD (qdf-LD3, qdf-LD4, qdf-LD5, and qdf-LD8) that collectively explained approximately 75% of phenotypic variation under LD conditions and four flowering time QTLs under SD (qdf-SD3.1, qdf-SD3.2, qdf-SD4, and qdf-SD5) that explained approximately 81% of phenotypic variation under SD conditions (Table I). The most notable of these was a QTL on Ca5 (qdf-SD5) that accounted for approximately 59% of phenotypic variation in flowering time under SD (Table I). This is consistent with previous reports of a large flowering time QTL on Ca5 in crosses involving ICCV 2, one of the parents of ICCV 96029 (Jamalabadi et al., 2013; Pushpavalli et al., 2015).
Table I. A major QTL for flowering time under SD on Ca5.
Phenotypic variation explained (PVE; %) by QTLs is given for DTF under SD and LD in CPR-01. Sequences of flanking markers are shown in Supplemental Data Set S1.
| Photoperiod | QTL | Ca | Flanking Marker 1 | Flanking Marker 2 | Centimorgans | Log of the Odds | PVE | Total PVE |
|---|---|---|---|---|---|---|---|---|
| SD | qdf-SD3.1 | 3 | scaffold1313p59187 | scaffold498p29571 | 17.85–18.53 | 6.24 | 8.07 | |
| SD | qdf-SD3.2 | 3 | scaffold1871p14410 | scaffold1871p875930 | 35.65–36.85 | 2.98 | 3.60 | |
| SD | qdf-SD4 | 4 | cav1sc2.1p3082421 | KASP-CaGI-D627E | 29.93–30.48 | 7.80 | 10.51 | |
| SD | qdf-SD5 | 5 | cav1sc1.1p4774721 | cav1sc1.1p6476732 | 65.1–76.0 | 24.96 | 58.56 | 80.73 |
| LD | qdf-LD3 | 3 | scaffold28p2999987 | scaffold28p1702903 | 23.51–28.59 | 5.98 | 19.40 | |
| LD | qdf-LD4 | 4 | KASP-CaGI-D627E | scaffold405p948196 | 30.48–31.59 | 2.82 | 8.94 | |
| LD | qdf-LD5 | 5 | cav1sc1.1p4774721 | cav1sc1.1p6476732 | 65.1–76.0 | 3.56 | 11.17 | |
| LD | qdf-LD8 | 8 | scaffold267p297757 | scaffold937p67148 | 57.69–58.39 | 9.57 | 35.77 | 75.1 |
The early-flowering locus efl1 was first identified in a cross between ICCV 2 and JG 62 where flowering time distribution was bimodal, indicating that efl1 was the only major locus controlling differences in flowering time between these two lines (Kumar and van Rheenen, 2000). Consistent with this finding, we observed a Mendelian distribution of flowering time under SD within CPR-01, supporting the view that the efl1 locus carried by ICCV 96029 (Gaur et al., 2015) also is responsible for this large QTL (Supplemental Fig. S1). qdf-LD4 and qdf-SD4 had one flanking marker in common, while qdf-LD5 and qdf-SD5 shared the same markers, suggesting that these two loci confer earlier flowering under both LD and SD.
Physical Mapping Reveals Flowering Time Candidate Genes, Including an Ortholog of Arabidopsis EARLY FLOWERING3
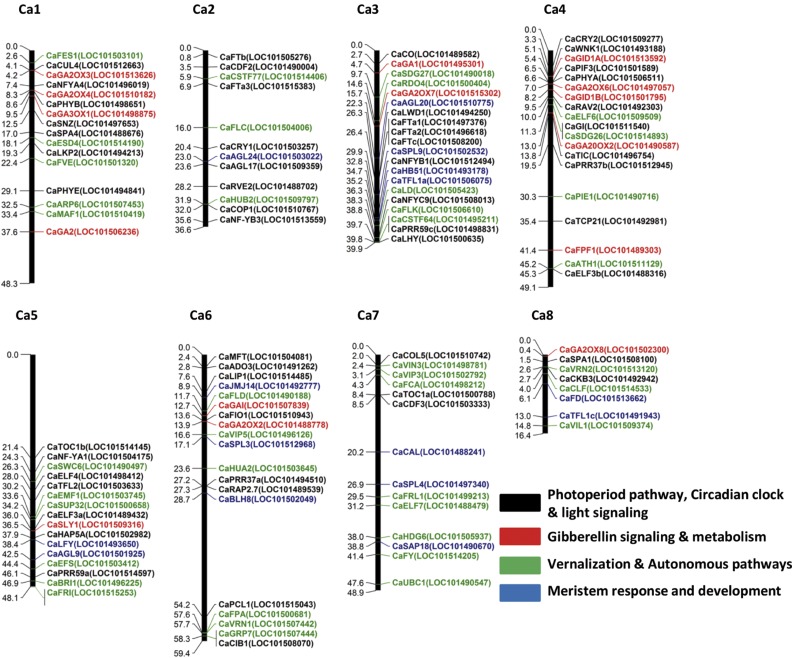
Comparative and functional genomic analyses have revealed that the functions of many Arabidopsis flowering time genes are conserved in legumes (Hecht et al., 2005; Kim et al., 2013; Weller and Ortega, 2015). Accordingly, we constructed a physical map with the positions of 138 chickpea orthologs of Arabidopsis flowering time genes, including those implicated in light signaling, the circadian clock, photoperiod sensing, and the autonomous pathways (Fig. 2). A total of 124 of these genes were located physically on chickpea pseudochromosomes Ca1 to Ca8, whereas the remaining 13 genes were located on 12 different unmapped scaffolds (Fig. 2; Supplemental Data Set S2). Based on their physical positions, we identified potential candidate genes for the flowering time QTLs identified in Table I. An ortholog of the LD flowering promoter GIGANTEA (GI; Ca_04359/XM_004496378) was found to colocalize within the confidence regions of qdf-SD4 and qdf-LD4, while an ortholog of the Arabidopsis gene TERMINAL FLOWER1 (TFL1; Ca_19775/XM_004515656) colocalized within the qdf-LD8 confidence region. The only flowering time candidate gene associated with the major qdf-SD5 and weaker qdf-LD5 QTLs on Ca5 in the 10.9 centimorgan between flanking markers cav1sc1.1p4774721 and cav1sc1.1p6476732 (35 and 36.5 Mb, respectively, on the CDC Frontier genome assembly Cav1.0; Varshney et al., 2013) was Ca_01620/XM_004501425, an ortholog of the Arabidopsis gene EARLY FLOWERING3 (ELF3), located at 36 Mb. This gene is known to be involved in gating light inputs to the circadian clock (McWatters et al., 2000; Covington et al., 2001; Carré, 2002), and mutations in ELF3 orthologs are associated with early flowering under SD and reduced photoperiod sensitivity in several crop species, including barley (Hordeum vulgare), lentil (Lens culinaris), and pea (Pisum sativum; Faure et al., 2012; Weller et al., 2012). Given this precedent for the conserved regulation of flowering by ELF3 orthologs under SD conditions across a diverse range of plant species, including legumes, the presence of an ELF3 ortholog at this QTL was highly suggestive.
Figure 2.
Physical maps of chickpea flowering time candidate genes. Chromosomes are represented as thick vertical lines with candidate gene positions indicated in Mb. Gene positions are based on the CDC Frontier genome assembly (ASM33114 version 1). A complete list of the 138 flowering time genes with their corresponding symbols and National Center for Biotechnology Information (NCBI) accession numbers can be found in Supplemental Data Set S2. Colors denote putative roles in the following pathways: black, photoperiod, circadian clock, and light signaling; red, GA signaling and metabolism; green, vernalization and autonomous pathways; blue, meristem response and development.
The CaELF3a Allele Derived from ICCV 96029 Is Associated with Early Flowering
The coding sequence (CDS) of 28 candidate genes within QTL intervals were sequenced in order to identify sequence variation between CDC Frontier and ICCV 96029 (Supplemental Data Set S3). The only two genes found to carry polymorphisms affecting the predicted protein sequence were orthologs of GI and ELF3. We found 10 single-nucleotide polymorphisms (SNPs) within the GI CDS, but all were synonymous except for two SNPs at nucleotide positions 1,881 (C/G) and 2,461 (G/C), which were predicted to result in the minor amino acid polymorphisms D627E and V821L, respectively. Alignment with GI amino acid sequences from selected angiosperms revealed that the D627E SNP was in a relatively conserved region and that the CDC Frontier GI allele was diverged at the Asp residue, whereas the ICCV 96029 Glu residue was conserved across all legume species examined (Supplemental Fig. S2). This SNP was used to create an allele-specific marker, and GI was genetically mapped to confirm its colocalization with the QTLs qdf-SD4 and qdf-LD4, supporting its status as a candidate gene for these QTLs (Supplemental Fig. S3). Cobos et al. (2007) reported previously a QTL in this region of Ca4 in a cross between CA 2156 and JG 62. Sequencing revealed the same SNP between these two parents, with the later-flowering CA 2156 matching the CDC Frontier genotype and the early-flowering JG 62 matching the ICCV 96029 genotype. This finding strengthens the idea that this SNP may be associated with flowering time.
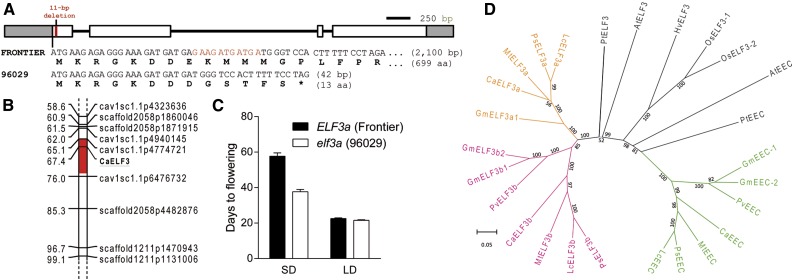
More significantly, the homolog of ELF3 in ICCV 96029 was found to carry an 11-bp deletion in the first exon. This deletion is predicted to result in six missense amino acids followed by a premature stop codon (Fig. 3A), although it is also possible that the presence of alternative start codons created by this deletion could simply modify the function or regulation of this gene without complete loss of function. Genotyping for this polymorphism in the RIL population revealed colocalization of the ELF3 deletion with the major SD flowering time QTL on Ca5, further supporting ELF3 as a strong positional candidate for Efl1 (Fig. 3B). Classification of the RIL population according to the CaELF3a genotype revealed that lines carrying the mutated form of caelf3a flowered slightly but significantly (P = 0.043) earlier under LD than those with the CaELF3a alleles derived from CDC Frontier (22.56 ± 0.38 versus 21.58 ± 0.29), while lines carrying the caelf3a mutation flowered 20 d earlier under SD than those carrying CaELF3a alleles (37.73 ± 1.23 versus 57.73 ± 1.85; Fig. 3C). Further classification of this population by GI genotype revealed a smaller, but clearly additive, effect of the GI locus on flowering time under SD conditions, with lines carrying the ICCV 96029 (putative wild-type) GI allele flowering earlier than those with the CDC Frontier GI allele (Supplemental Fig. S4). Although CDC Frontier is later flowering than ICCV 96029 under LD (Fig. 1), neither of these loci appears to strongly affect flowering time under LD, suggesting that this difference may be due to other unknown QTLs, potentially including qdf-LD8.
Figure 3.
ICCV 96029 carries an 11-bp deletion in an ELF3 homolog. A, Diagram of the chickpea ELF3a gene showing the location of the 11-bp deletion in the first exon of ICCV 96029 (red line). Exons are shown as boxes, with coding sequences in white and untranslated regions in gray. Nucleotides are shown commencing from the start codon, with the 11-bp polymorphism displayed in red. The premature stop codon in ICCV 96029 is indicated by the asterisk, and CDS and protein lengths of the two variants are indicated. aa, Amino acids. B, Location of the QTL controlling SD flowering on Ca5 in the CDC Frontier × ICCV 96029 mapping population CPR-01 (F8). The one-log of the odds confidence intervals around the QTL peak are indicated by red shading, with units in centimorgans. Marker sequences are provided in Supplemental Data Set S1. C, DTF in the CPR-01 mapping population under LD and SD conditions for plants carrying the CDC Frontier and ICCV 96029 forms of CaELF3a. Values represent means ± se for n = 35 (ELF3a) and n = 53 (elf3a). D, Phylogram of ELF3-like genes across several families. ELF3a, ELF3b, and EEC legume clades are indicated in pink, orange, and green, respectively. Bootstrap values obtained from 1,000 replicate trees are shown as percentages above each branch. Branches with bootstrap values less than 50% have been collapsed. The protein sequence alignment used to generate this phylogram is provided in Supplemental Data Set S4 along with accession numbers for each sequence. At, Arabidopsis thaliana; Ca, Cicer arietinum; Gm, Glycine max; Hv, Hordeum vulgare; Lc, Lens culinaris; Mt, Medicago truncatula; Os, Oryza sativa; Ps, Pisum sativum; Pt, Populus trichocarpa; Pv, Phaseolus vulgaris.
ELF3 is present as a single-copy gene in Arabidopsis, with the next most similar gene designated as ESSENCE OF ELF3 CONSENSUS (EEC; Liu et al., 2001). By contrast, a survey of ELF3-like genes in major crop legumes revealed that chickpea, pea, lentil, and Medicago spp. carry two copies of ELF3, forming two separate clades that we designated ELF3a and ELF3b (Fig. 3D), consistent with Rubenach et al. (2017). Common bean (Phaseolus vulgaris) databases lacked a representative in the ELF3a clade, while the tetraploid soybean (Glycine max) had only one copy in the ELF3a clade instead of the expected two. The chickpea gene corresponding to the candidate gene for Efl1 was located in the ELF3a clade (CaELF3a; NCBI locus XM_004501425) along with the previously characterized pea and lentil ELF3 homologs (Weller et al., 2012), while a second gene showing 47% homology to the CaELF3a protein sequence on Ca4 (XM_004498869) was named CaELF3b. Sequencing of the full CaELF3b genomic DNA (gDNA) in CDC Frontier and ICCV 96029 revealed no SNPs between these two parent lines. Genes in these two clades shared the same seven conserved domains described previously (Weller et al., 2012; Supplemental Data Set S4). A third ELF3-like clade also was present, but with the exception of PsEEC and GmEEC-1, members of this group showed greater homology to AtEEC than to AtELF3. The presence of a second ELF3 homolog in legumes raises the possibility that redundancy between CaELF3a and CaELF3b may reduce the impact of the 11-bp deletion on chickpea flowering time and photoperiod response.
The Function of the CaELF3a Protein Is Impaired by the 11-bp Deletion
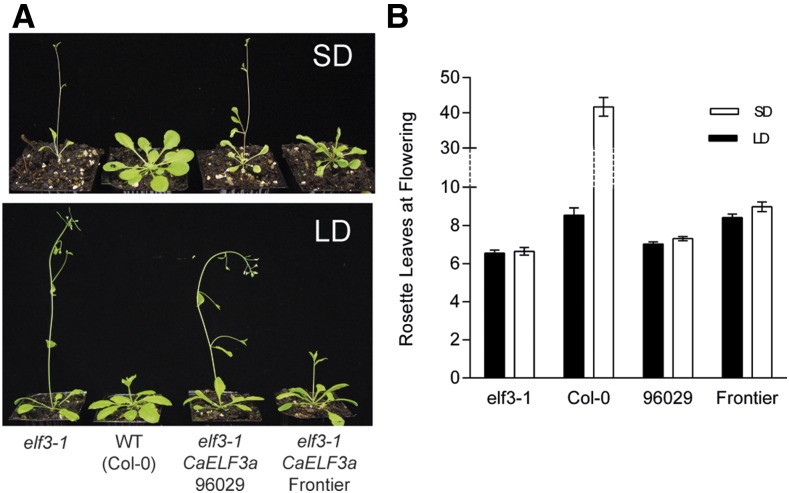
The association between the 11-bp deletion in CaELF3a and qdf-SD5.1 constitutes strong evidence that this mutation is responsible for a major proportion of the early-flowering phenotype in ICCV 96029. In order to confirm that the wild-type gene is functional and that the caelf3a mutation affects the functionality of the encoded protein, we performed a transgenic complementation assay using the Arabidopsis elf3-1 null mutant, which is photoperiod insensitive and flowers significantly earlier than the Columbia-0 wild type under both LD (6.55 ± 0.16 versus 8.54 ± 0.39 rosette leaves at flowering; P = 0.0002) and SD (6.64 ± 0.2 versus 41.7 ± 2.66 rosette leaves at flowering; P < 0.0001; Fig. 4; Supplemental Table S1). Flowering time was delayed significantly (P < 0.0001) in transgenic plants transformed with CaELF3a derived from CDC Frontier as compared with plants transformed with CaELF3a derived from ICCV 96029 under both LD (8.41 ± 0.18 versus 7.03 ± 0.11, respectively) and SD (8.97 ± 0.25 versus 7.31 ± 0.11, respectively). Under LD conditions, the CDC Frontier form of CaELF3a complemented elf3-1 (no significant difference when compared with the Columbia-0 wild type; P = 0.79), restoring later flowering (8.41 ± 0.18 rosette leaves), while the ICCV 96029 form had no significant effect on flowering time as compared with the elf3-1 control under LD (7.03 ± 0.11 rosette leaves; P = 0.085) and only a slight effect on flowering time as compared with the elf3-1 control under SD (7.31 ± 0.11 rosette leaves; P = 0.011; Fig. 4B; Supplemental Table S1). Transgenic Arabidopsis overexpressing the CDC Frontier CaELF3a variant was later flowering under SD (8.98 ± 0.25 rosette leaves) as compared with LD, but this difference was not statistically significant (P = 0.066), and the extreme late flowering observed in the wild type under LD was not restored (Fig. 4B; Supplemental Table S1). These results indicate that the 11-bp deletion in the ICCV 96029 allele affects the CaELF3 protein activity and support the idea that this gene is likely to be a key factor contributing to the early-flowering phenotype of ICCV 96029 grown under SD.
Figure 4.
The 11-bp deletion in CaELF3a affects the function of the protein in Arabidopsis. A, Degrees of complementation of flowering phenotypes of the wild-type (WT) Columbia-0 (Col-0) Arabidopsis elf3-1 mutant by CDC Frontier but not the ICCV 96029 form of 35S:CaELF3a under both 8-h SD and 16-h LD conditions. B, Rosette leaf number at flowering for the elf3-1 mutant, the wild type, and transgenic Arabidopsis lines grown under LD and SD. Because Columbia-0 wild-type lines were extremely late flowering under SD, a discontinuous y axis indicated with broken lines was used to enable the visualization of flowering time differences in other lines. For LD conditions, values represent means ± se for n = 91 and n = 66 for CDC Frontier and ICCV 96029, respectively. For SD conditions, values represent means ± se for n = 85 and n = 71 for CDC Frontier and ICCV 96029, respectively. For each photoperiod/transgene combination, eight to 11 independent transgenic lines were analyzed. Data are derived from T2 plants homozygous for the transgene. Control (wild type/elf3-1) values represent means ± se for n = 10 to 14.
The Expression of FT Genes Is Misregulated in a Line Carrying the caelf3a Mutation
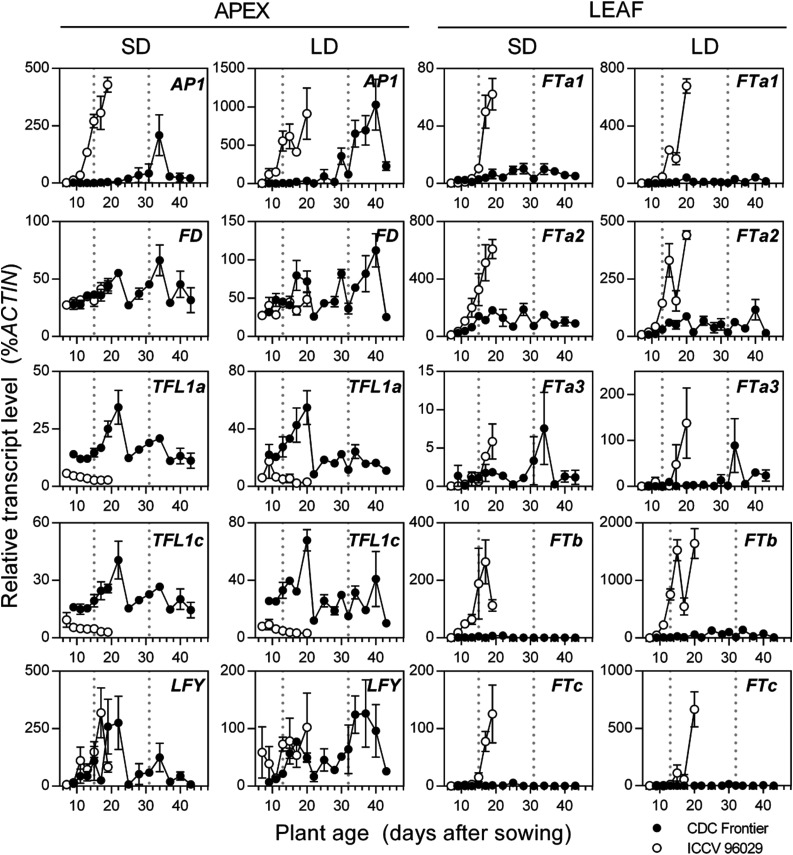
In order to understand the molecular basis for the differences observed in flowering time between parental lines, we next measured the expression of key genes involved in flowering time and meristem identity (Fig. 5). ICCV 96029 produced visible buds after 13 and 15 d under LD and SD, respectively, and flowered after 22 and 27 d under LD and SD, respectively. CDC Frontier had produced visible, abortive buds after 31 to 32 d in both LD and SD, prior to the eventual formation of flowers and anthesis, after 66 and 94 d under LD and SD, respectively. Consistent with this flowering behavior, we observed clear differences between the two lines in the expression of a homolog of APETALA1 (AP1), an inflorescence identity gene that serves as a developmental marker for inflorescence initiation in legumes (Hecht et al., 2011). Up-regulation of this gene above background levels occurred much earlier in ICCV 96029 than in CDC Frontier and occurred shortly prior to the observation of visible buds in both genotypes. TFL1 genes are known to inhibit flowering time promoters such as AP1 in Arabidopsis (Gustafson-Brown et al., 1994; Liljegren et al., 1999; Hanano and Goto, 2011). In ICCV 96029, the expression of both TFL1a and TFL1c was strongly repressed under LD and SD throughout plant development, reflecting the early flowering time observed in this line. Expression of these genes in CDC Frontier was at a higher level and increased throughout plant development before decreasing sharply at 22 d (LD) and 25 d (SD), possibly reflecting the transition to reproductive development. We did not observe any clear differences in the expression of CaFD or CaLFY between CDC Frontier and ICCV 96029, suggesting that the flowering time difference between CDC Frontier and ICCV 96029 is not associated with any difference in the regulation of these genes.
Figure 5.
The caelf3a mutation is associated with misregulation of flowering genes. Gene expression is shown in CDC Frontier and ICCV 96029 during development under LD and SD conditions. Relative transcript levels were determined in the second-uppermost fully expanded leaf for members of the FT family or dissected shoot apices for meristem identity genes. Values represent means ± se for n = 2 to 3 biological replicates, each consisting of pooled material from two plants. Developing floral buds were first visible in CDC Frontier at 31 and 32 d after sowing in SD and LD, respectively, and in ICCV 96029 at 15 and 13 d after sowing in SD and LD, respectively (broken lines).
In Arabidopsis, FT is a critical flowering promoter, and a role for FT homologs as integrators of internal and external flowering signals is widely conserved across a range of plant species (Pin and Nilsson, 2012). This gene family has been studied in several legume species (Kong et al., 2010; Hecht et al., 2011; Laurie et al., 2011; Yamashino et al., 2013; Zhai et al., 2014), where most species have at least five FT-like genes that may be divided into three distinct clades: FTa, FTb, and FTc (Hecht et al., 2011). We identified five members of this family in chickpea and observed clear differences in their expression between ICCV 96029 and CDC Frontier. In pea, FTb2 is the most strongly photoperiod-regulated FT gene, and its induction, which precedes the induction of any other FT gene, is closely correlated with the transition to reproductive development under inductive photoperiods (Hecht et al., 2011). Chickpea appears to have only a single FTb gene (CaFTb), and its induction at 9 d under LD and SD in ICCV 96029 was associated with the up-regulation of CaAP1 above background levels, also at 9 d (Fig. 5). All other CaFT genes were up-regulated in ICCV 96092 at a later stage, probably following the initiation of floral structures. Analysis of CDC Frontier expression data revealed a slight up-regulation of CaFTa1 and CaFTa2 expression under LD and SD at 19 to 20 d, along with an up-regulation of CaFTb at 25 d under LD, roughly corresponding to the gradual up-regulation of CaAP1 at 25 to 30 d; however, this up-regulation was very small compared with that seen in ICCV 96029. Given that the CDC Frontier buds observed throughout this time series were abortive, we considered the possibility that members of the chickpea FT family are not fully up-regulated until an abortion-inducing factor is overcome. However, additional measurements of FTb and FTa1 expression under SD continuing until flowering and anthesis revealed no major up-regulation associated with flowering (Supplemental Fig. S5). These clear differences between CDC Frontier and ICCV 96029 are consistent with the idea that CaELF3a regulates flowering via members of the CaFT family and that CaFTb is likely to be particularly important for the photoperiod-regulated induction of flowering.
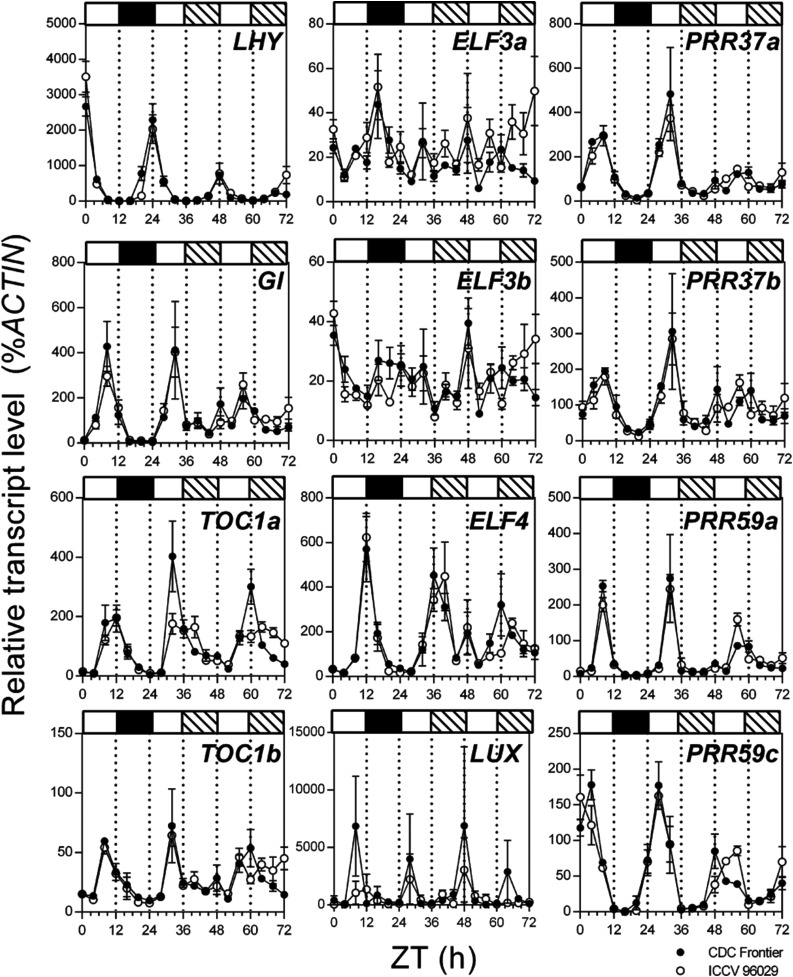
The caelf3a Mutation Has No Apparent Effect on the Rhythmic Expression of Clock Genes under Constant Light
In several species, mutations in the ELF3 gene result in a loss of rhythmic expression of clock genes when plants entrained to light/dark cycles are transferred to constant light (LL). In Arabidopsis, detailed studies point to a role for ELF3 in the generation and maintenance of circadian cycles in the absence of external light inputs. Therefore, we compared the regulation of several clock-related genes in ICCV 96029 and CDC Frontier grown under a 12-h photoperiod and transferred to LL. Unexpectedly, we did not observe any major phase shift or difference in amplitude of clock gene expression between these two lines (Fig. 6). Small changes in the expression peaks of CaELF4 and CaPRR59c during the LL period (particularly at Zeitgeber time 40–44) suggested the possibility of altered rhythms for these genes. However, most other genes, including LHY, GI, and several other members of the PSEUDO RESPONSE REGULATOR (PRR) family, showed very similar rhythms in ICCV 96029 and CDC Frontier. These results indicate that, even though CaELF3a has a large effect on flowering time, it is not essential for maintaining the rhythmicity of clock genes under LL conditions. One possible reason for this finding is that the mutation in ICCV 96029 CaELF3a may not result in a complete loss of function. Another possibility is that CaELF3b, the second copy of ELF3 in chickpea, may be partially compensating for the reduced, modified, or complete lack of function or regulation in CaELF3a derived from ICCV 96029.
Figure 6.
Rhythmic expression of circadian clock genes under LL are not affected by the caelf3a mutation. Plants were grown in growth chambers under a 12-h light/dark cycle at 22°C. Zeitgeber time (ZT) refers to the time since lights on of the last full entraining cycle, with ZT0 at 17 d after sowing. From ZT0, transcript levels were determined every 4 h in the uppermost fully expanded leaf of CDC Frontier and ICCV 96029 plants. At ZT24, plants were transferred to LL at 400 µmol m−2 s−1. Mean values ± se are shown for n = 2 to 3 biological replicates, each consisting of pooled material from two plants. Day and night periods are indicated by white and black bars, respectively, above the graphs, with subjective night periods during LL indicated by hatched bars.
The CaELF3a Deletion Arose Recently within Chickpea Germplasm
In view of the importance of introducing genes that confer early flowering to modern chickpea breeding germplasm, we sought to understand the origins and distribution of the caelf3a mutation in chickpea by screening a wide range of chickpea breeding material derived from around the world. Across 236 lines, we found 13 lines that carried the 11-bp mutation (Supplemental Data Set S5). Where pedigree records are available, these lines appear to be closely related to one another, and it is likely that they share the genetic region surrounding the Efl1 locus. ICCV 2, the immediate progenitor of ICCV 96029, was found to carry the caelf3a mutation, and given that these lines both carry the efl1 locus (Gaur et al., 2015), this is consistent with the view that Efl1 is equivalent to CaELF3a. These results indicate that the deletion mutation in CaELF3a appears to have arisen relatively recently within Indian germplasm and, thus, may have been selected within adapted material to further enhance an existing early-flowering phenotype.
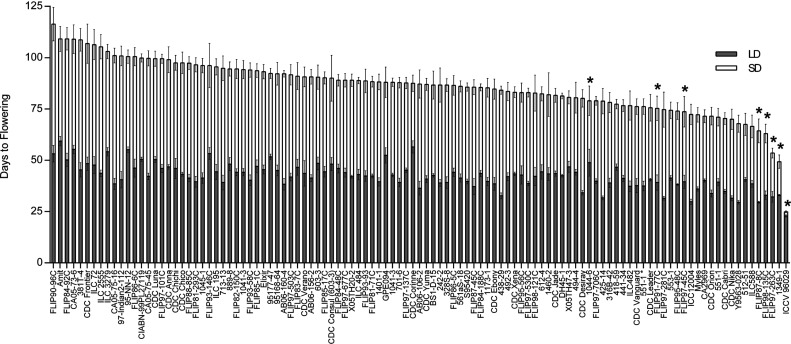
From this germplasm, a subset of 109 lines including eight lines carrying the elf3a mutation was phenotyped under SD and LD to evaluate flowering time. There was a small but significant reduction in DTF under LD for those lines carrying the 11-bp deletion (42.72 ± 0.57 versus 34.76 ± 2.74; P = 0.0004), along with a much more significant difference in DTF between the two groups under SD (88.13 ± 1.06 versus 60.38 ± 6.27 for ELF3a and elf3a groups, respectively; P < 0.0001). Figure 7 illustrates the association between the elf3a mutation and earlier flowering, with the five earliest flowering lines under SD carrying the elf3a mutation. Of these eight lines, ICCV 96029 flowered earliest by a large margin (24.5 d), indicating that, uniquely within the subset, this genotype carries one or more additional alleles besides caelf3a that strongly promote flowering under SD. This suggests that, although the 11-bp deletion in CaELF3a contributes significantly to the early-flowering phenotypes described in ICCV 2 and ICCV 96029, it is likely that a proportion of the ICCV 96029 phenotype is due to other genes with similar positive effects on flowering time (Gaur et al., 2015). To gain further insight into sequence variation at this locus, we resequenced the entire CaELF3a gDNA in these 109 lines. The results showed that the only CaELF3a sequence polymorphism present within this germplasm collection was the 11-bp deletion in the eight lines shown in Figure 7.
Figure 7.
The caelf3a mutation is associated with earlier flowering under SD in global chickpea germplasm in a survey of photoperiodic flowering in chickpea. Plants were grown in chambers under LD (16-h) and SD (10-h) photoperiod conditions for 12 h at 22°C and 12 h at 16°C. The eight lines carrying the 11-bp deletion in CaELF3a are indicated by asterisks. A full list of accessions carrying the 11-bp deletion in CaELF3a is provided in Supplemental Data Set S5. Values represent means ± se, n = 6.
DISCUSSION
A succession of evolutionary bottlenecks has resulted in domesticated chickpea germplasm with unusually limited genetic diversity (Abbo et al., 2003). The development of new varieties suited to diverse growing regions and the continued improvement of existing varieties depend on the identification and incorporation of allelic variation at key loci (Abbo et al., 2007). Currently, only four loci controlling the crucial trait of flowering time (Efl1–Efl4) have been identified in domesticated chickpea germplasm, and the genes responsible for these loci are unknown. Efl1, the first major flowering time locus described in chickpea, was initially defined in ICCV 2 (Kumar and van Rheenen, 2000), which has been used as a common parent in several crosses that report major QTLs on Ca5, suggesting a likely map position for Efl1 (Cho et al., 2002; Vadez et al., 2012; Jamalabadi et al., 2013; Pushpavalli et al., 2015). Allelism tests have provided strong genetic evidence that this early-flowering efl1 gene also is present in ICCV 96029, a direct descendent of ICCV 2 (Gaur et al., 2015). Consistent with this, our cross between ICCV 96029 and CDC Frontier revealed a major flowering time QTL on Ca5 that we conclude is equivalent to Efl1. We identified an ortholog of Arabidopsis ELF3 within the narrow confidence interval of this QTL and present evidence that this gene is responsible for the early-flowering Efl1 phenotype. Our findings are based on the discovery of a deletion in CaELF3a that is likely to be functionally significant, strong association of this mutation with phenotypes displaying recessive early flowering under SD, complementation of flowering phenotypes in the Arabidopsis elf3-1 mutant by CaELF3a, association between this mutation and early elevated expression of FT and AP1, and general early-flowering phenotypic similarity to other legume species carrying mutations in homologous ELF3 genes. Taken together, these results strongly implicate ELF3 as Efl1; however, we cannot definitively exclude the possibility that aspects of the Efl1 phenotype are due to other nearby genes.
The identification of flowering time QTLs in this study has revealed several genetic regions of interest for breeders seeking to develop chickpea cultivars with reduced photoperiod response and earlier flowering (Table I). A number of QTLs within these regions have been reported previously, including the QTLs on Ca4 (Cobos et al., 2007; Varshney et al., 2014) and Ca5 (Cho et al., 2002; Vadez et al., 2012; Jamalabadi et al., 2013; Pushpavalli et al., 2015). GI remains a potential candidate for qdf-SD4, with markers for CDC Frontier and ICCV 96029 alleles showing a strong association with flowering time under SD (Supplemental Fig. S4). In the model LD legume pea, the GI ortholog LATE1 regulates photoperiod sensitivity, with mutant alleles conferring late flowering under LD (Hecht et al., 2007), but we did not observe any significant association between the D627E SNP and flowering time under LD (Supplemental Fig. S4). Combined with fact that CDC Frontier appeared to carry the derived genotype for a relatively mild SNP, this raises the possibility that this polymorphism may represent a gain-of-function mutation resulting in increased photoperiod sensitivity and later flowering in CDC Frontier; further studies on this question are in progress. In addition, our physical map of chickpea flowering candidate genes (Fig. 2) will constitute a helpful guide to potential candidate genes controlling the other QTLs that have already been reported in other crosses.
Overall, our results lead us to conclude that the chickpea ortholog of ELF3 is responsible for the qdf-SD5 QTL on Ca5 and that this gene is equivalent to the previously described Efl1 locus. Efl1 has been associated previously with several QTLs on Ca3 (Cho et al., 2002; Jamalabadi et al., 2013), Ca4 (Upadhyaya et al., 2015), and Ca5 (Vadez et al., 2012; Pushpavalli et al., 2015). Changes to the naming convention and a lack of common markers between maps are likely to account for some of the reports of Efl1 on different chromosomes. For example, the strong QTL for DTF reported by Jamalabadi et al. (2013) in a RIL population derived from ILC 3279 × ICCV 2 was flanked by the markers TA117 and CaSTMS22, both of which are placed on Ca5 according to the current naming convention (Radhika et al., 2007; Hiremath et al., 2012; Varshney et al., 2013). Pushpavalli et al. (2015) provided recent evidence consistent with Efl1 as the candidate for qdf-SD5 when they reported a QTL explaining 25% to 27% of phenotypic variation in flowering time in an ICCV 2 × JG 11 mapping population, with flanking markers ICCM272-CaM0463 physically located at 34 to 44.8 Mb on Ca5 (CDC Frontier genome assembly Cav1.0; Varshney et al., 2013). This region closely corresponds to the location of the qdf-SD5 flanking markers in our population at 35 to 36.5 Mb and is consistent with the location of the CaSTMS22 marker reported by Jamalabadi et al. (2013) at 35.8 Mb on Ca5, close to the CaELF3a gene at 36 Mb. Given the relatively minor effect of CaELF3a on flowering time under LD in the majority of our accessions (Fig. 7), it is possible that the field conditions employed in these earlier studies failed to expose the more significant contribution of this region to DTF that we found associated with qdf-SD5 under controlled-environment SD. Recently, Upadhyaya et al. (2015) described a QTL on Ca4 as Efl1, implying that it is synonymous with Ca_14192/XM_012715013, a homolog of Arabidopsis PIE1. However, no data supporting the equivalence of this QTL with Efl1 were presented. The QTL was in close proximity to GI and the qdf-SD4 described in this study, but it is clearly distinct from the major qdf-SD5 QTL, and full-length sequencing of this gene in both CDC Frontier and ICCV 96029 revealed only two silent SNPs within the CDS, excluding it as a candidate for Efl1 in this population.
The elevated, early expression of members of the FT family in ICCV 96029 supports the idea that CaELF3a affects flowering time through the regulation of FT. However, our observation that the 11-bp deletion in CaELF3a was insufficient to induce the misregulation of circadian clock genes under LL was not anticipated given previous findings to the contrary in several other species (McWatters et al., 2000; Faure et al., 2012; Saito et al., 2012; Weller et al., 2012), although one study in Einkorn wheat (Triticum monococcum) by Alvarez et al. (2016) revealed relatively minor effects of an ELF3 mutation on circadian clock genes. One possibility consistent with this observation is that an alternative Met residue created by the 11-bp deletion could enable the transcription of a CaELF3a protein with incomplete loss of function. For example, the 11-bp deletion in CaELF3a could potentially create a start codon 23 bp into the CDS of mutated CaELF3a, conceivably resulting in the transcription of a protein with modified function or regulation lacking the first 11 residues of ELF3 domain I, which are highly conserved across legumes and members of the Rosaceae (Fig. 3A; Supplemental Data Set S4; Weller et al., 2012). Alternatively, it is possible that the second copy (CaELF3b) may play a complementary, additive role to CaELF3a in regulating light input into the circadian oscillator (Covington et al., 2001). Under this scenario, CaELF3b may be capable of partially fulfilling the role of CaELF3a, enabling the persistent rhythmic expression of clock genes under LL, while loss of the first copy could confer earlier flowering through either lower dosage effects or other unknown mechanisms. This view is consistent with a recent study by Rubenach et al. (2017) that showed that the pea ELF3b homolog PHOTOPERIOD partially compensates for the ELF3a homolog HR. A scenario involving ELF3 redundancy also is consistent with results in Figure 7 that indicate that, while the 11-bp deletion in CaELF3a is associated with early flowering under SD, it is not essential for conferring photoperiod sensitivity per se, raising the possibility that CaELF3b may be partially compensating. Alvarez et al. (2016) recently identified an ortholog of ELF3 as the earliness per se wheat locus Eps-Am1, which promotes flowering independently of photoperiod, and it is possible that CaELF3a functions in an analogous manner.
Several recent studies have highlighted a key role for ELF3 orthologs in the expansion of legume cultivation. For instance, natural variation at the soybean locus J, which was identified recently as an ortholog of ELF3, has been essential to the development of soybean cultivars adapted to low-latitude regions (Lu et al., 2017). In pea, a significant proportion of flowering time variation within global pea germplasm is controlled by the Arabidopsis ELF3 ortholog HR, with a single widespread functional variant conferring reduced photoperiod responsiveness (Weller et al., 2012). This variant was likely to have been selected during the expansion of legume cultivation. A similar functional variant that confers a spring growth habit has been reported in lentil cultivars commonly grown in short-season environments (Weller et al., 2012). We observed a similar association between ELF3a and early flowering under SD in global chickpea germplasm. However, our findings indicate a clear difference in the importance of this gene to domestication and spread between chickpea and other legume species. Fifteen distinct haplotypes of the pea HR/PsELF3 gene are present, with the hr mutation widespread within recently developed, domesticated germplasm and likely to have been essential to the domestication and spread of spring-cropped varieties (Weller et al., 2012). By contrast, we found just two CaELF3a haplotypes that differed only in whether they carried the 11-bp deletion, and the caelf3a haplotype represented a very small proportion of the total number of global chickpea accessions surveyed (Fig. 7). Together, these facts suggest that this mutation in CaELF3a arose following the shift from autumn to spring sowing in chickpea, and most likely as part of the relatively modern replacement of landraces by cultivars produced using modern breeding methods. ICCV 2, one of the parents of ICCV 96029, is the major early-flowering genetic variant to have enabled commercial cultivation in tropical regions of India and Burma, and it is likely that the efl1 mutation was a fundamental element in its widespread adoption. It was derived through the cross F3 [(K-850 × GW-5/7) × P-458] × F3 (L-550 × Gaumuchil)-2 in 1975/76 (ICRISAT, 1990), but records that may be used to identify the source of this mutation beyond these progenitors are restricted.
The identification of CaELF3a as a strong regulator of flowering time has practical applications, particularly for the development of varieties suited for SD environments, and we are currently introgressing this gene into other varieties to assess its effect on flowering time. ICCV 96029 is essentially photoperiod insensitive, whereas other lines carrying the efl1 mutation still show considerable response to daylength. This indicates that other factors contribute to the early flowering of this variety. Other candidate genes that may contribute to this early flowering time include GI on Ca4 and TFL1c on Ca8, and the QTLs identified in this study will be the subject of further investigation. The isolation and characterization of CaELF3a represents, to our knowledge, the first major chickpea flowering time locus for which a strong case has been built in support of the molecular identity of the gene. The exploitation of such early-flowering genes from cultivated germplasm in order to generate new varieties is helpful, but the introduction of variation from wild lines will be necessary to restore a measure of the lost genetic variation in chickpea and open up new regions for chickpea cultivation.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Phenotyping
CDC Frontier is a Kabuli-type chickpea (Cicer arietinum) cultivar developed at the University of Saskatchewan from a cross between FLIP 91-22C and ICC 14912 (Warkentin et al., 2005). Following its release in 2003, it was widely adopted due to its moderate resistance to ascochyta blight, caused by the fungus Ascochyta rabiei, and medium seed size. Under Saskatchewan field conditions, it matures relatively late as compared with newer cultivars. ICCV 96029 is a double-podded Desi cultivar with small brown seeds developed by the International Crop Research Institute for the Semi-Arid Tropics (ICRISAT), resulting from a cross between ICCV 2 and ICCV 93929. It is a transgressive segregant, flowering approximately 1 week earlier than both parents (Kumar and Rao, 2001). Released in 2000, it remains one of the world’s earliest flowering chickpea varieties (Kumar and Rao, 2001; Gaur et al., 2015).
QTL analysis was performed using the biparental mapping population CPR-01 (F8) of 92 RILs derived from a cross between these two parents, as described previously (Tar’an et al., 2007; Deokar et al., 2014; Ali et al., 2016; Daba et al., 2016a). Plants were grown in approximately 15-cm pots filled with Sunshine mix no. 4 medium (Sun Gro Horticulture Canada) in growth chambers for 12 h at 22°C and 12 h at 16°C. LD conditions consisted of a 16:8-h (light:dark) photoperiod under fluorescent cool-white lights at an irradiance of 350 to 400 µmol m−2 s−1, while SD conditions were 10:14-h light:dark. DTF was taken as the number of days from seedling emergence to the first fully open flower. The difference in DTF between SD and LD was used to determine photoperiod sensitivity.
QTL Analysis and Physical Mapping of Candidate Genes
A high-density genetic map consisting of 1,336 SNPs across the whole genome was generated previously for CPR-01 using a combination of Illumina 1,536 SNPs GoldenGate assay and Restriction-Site Associated DNA Sequencing genotyping-by-sequencing approaches (Deokar et al., 2014). These data were used for the QTL analysis (Table I) using the ICIM-ADD (composite interval mapping) method of QTL-IciMapping version 4.0.3.0 (Meng et al., 2015). We also identified 138 chickpea homologs of Arabidopsis (Arabidopsis thaliana) flowering genes (Fig. 2) using BLASTP searches against the Cicer arietinum NCBI RefSeq proteins of the CDC Frontier genome assembly (ASM33114 version 1; Varshney et al., 2013). BLASTP searches were performed with an e value cutoff of 1e-5 and query or subject coverage of at least 50%. CaELF3a and CaGI were mapped using Kompetitive Allele Specific chemistry (LGC Genomics).
Gene Expression Studies
Harvested tissue used in qRT-PCR experiments (Figs. 6 and 7) consisted of the second-uppermost fully expanded leaf or apical buds dissected to approximately 2 mm pooled from two plants. Samples were frozen in liquid nitrogen, and total RNA was extracted using the SV Total RNA Isolation System (Promega). RNA concentrations were determined using a NanoDrop 8000 (Thermo Scientific). Reverse transcription was performed in 20 µL with 1 µg of total RNA using the SensiFAST cDNA Synthesis Kit (Bioline). RT-negative (no enzyme) controls were included to monitor for gDNA contamination. The first-strand cDNA was diluted five times, and 2 µL was used in each real-time PCR. SYBR Green chemistry (SensiFAST; Bioline) was used to perform real-time PCR for 40 cycles in a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Two technical replicates and at least two biological replicates were performed for each sample. Primer details are included in Supplemental Table S2.
Construct Preparation and Arabidopsis Transformation
For the complementation study (Fig. 4), pCR8/GW/TOPO entry clones were generated for the Efl1 (CDC Frontier) and efl1 (ICCV 96029) alleles of CaELF3. The entry clones were then linearized with either XbaI or XhoI (New England Biolabs) and transferred by LR reaction (Invitrogen) into the pB2GW7 vector containing the cauliflower mosaic virus 35S promoter (Karimi et al., 2002). 35S:CaELF3a binary vectors were transformed into Agrobacterium tumefaciens GV3101 by electroporation. Arabidopsis (Columbia-0) was employed for this complementation assay, given the lack of a transformation system and ELF3 mutants in chickpea. In an earlier study, we used the same vectors and cloning methods to generate two constructs of 35S:PsELF3 (HR/hr), demonstrating that flowering in Arabidopsis transformed with 35S:hr is unaffected as compared with the elf3-1 mutant and that full complementation occurs where plants are transformed with 35S:HR (Weller et al., 2012). Thus, we have shown that the 35S promoter is able to direct sufficient expression of ELF3 to fully complement the elf3-1 mutation. Transformations were conducted using the method described by Martinez-Trujillo et al. (2004). Phosphinothricin-resistant T1 plants were selected, and T2 resistant transformants were grown under standard LD and SD conditions (16:8 h and 8:16 h, respectively) and scored for flowering time by counting the number of rosette leaves at flowering. T2 segregation ratios indicated the presence of a single insertion.
Phylogenetic and Statistical Analyses
For phylogenetic analyses, genes were identified by performing BLAST searches at NCBI (www.ncbi.nlm.nih.gov), Phytozome version 11.0 (www.phytozome.jgi.doe.gov), the pea (Pisum sativum) RNA-seq Gene Atlas (www.bios.dijon.inra.fr/FATAL/cgi/pscam.cgi), and KnowPulse (knowpulse.usask.ca) with reciprocal BLAST searches performed against Arabidopsis at TAIR (www.arabidopsis.org) to confirm gene identity. Full-length amino acid sequences were aligned using ClustalW (Supplemental Data Set S1; Thompson et al., 1994) and adjusted manually, where necessary, using GeneDoc (version 2.7.000; Nicholas and Nicholas, 1997; www.psc.edu/biomed/genedoc). These alignments were used to generate a neighbor-joining tree using MEGA version 6.0 (www.megasoftware.net). For statistical analysis of the data presented in Figures 4B and 7 as well as Supplemental Table S1, a two-tailed, unpaired Student’s t test was performed using GraphPad Prism (version 6.07; GraphPad Software).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in the supplemental figures, tables, and data sets.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Distribution of flowering time under SD for the RIL population CPR-01 derived from a cross between CDC Frontier and ICCV 96029.
Supplemental Figure S2. Alignment of partial GI amino acid sequences from selected angiosperms showing the location of the D627E SNP.
Supplemental Figure S3. Linkage map showing the position of the CaGI marker D627E on Ca4.
Supplemental Figure S4. DTF in the CPR-01 mapping population under LD and SD conditions.
Supplemental Figure S5. Postbud initiation expression of FTa1 and FTb in CDC Frontier grown under SD.
Supplemental Table S1. Statistical analyses for the transgenic complementation assay.
Supplemental Table S2. Primer details.
Supplemental Data Set S1. Sequences of flanking markers for QTLs shown in Table I and for markers shown in Figure 3B.
Supplemental Data Set S2. Chickpea flowering time candidate genes shown in Figure 2.
Supplemental Data Set S3. List of candidate genes sequenced in ICCV 96029 and CDC Frontier.
Supplemental Data Set S4. Alignment for Figure 3D.
Supplemental Data Set S5. Chickpea accessions carrying the 11-bp deletion in CaELF3a.
Acknowledgments
We thank Donna Lindsay, Parvaneh Hashemi, and Carmen Breitkreutz for assistance with plant husbandry and laboratory work; Raul Ortega for providing primers; Eldon Siemens, Adam Harrison, Dennis Wulf, and Brent Barlow for the use of the phytotron and field laboratory facilities; and Teresa Millán for providing seed.
Glossary
- LD
long days
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- SD
short days
- DTF
days to flowering
- CDS
coding sequence
- SNP
single-nucleotide polymorphism
- gDNA
genomic DNA
- LL
constant light
Footnotes
This research was supported by funding from the Saskatchewan Ministry of Agriculture through the Agriculture Development Fund and from Saskatchewan Pulse Growers.
Articles can be viewed without a subscription.
References
- Abbo S, Berger J, Turner NC (2003) Evolution of cultivated chickpea: four bottlenecks limit diversity and constrain adaptation. Funct Plant Biol 30: 1081–1087 [DOI] [PubMed] [Google Scholar]
- Abbo S, Lev-Yadun S, Galwey N (2002) Vernalization response of wild chickpea. New Phytol 154: 695–701 [DOI] [PubMed] [Google Scholar]
- Abbo S, Redden RJ, Yadav SS (2007) Utilization of wild relatives. In Yadav SS, Redden RJ, Chen W, Sharma B, eds, Chickpea Breeding and Management. CABI, Wallingford, UK, pp 338–354 [Google Scholar]
- Ali L, Deokar A, Caballo C, Tar’an B, Gil J, Chen W, Millan T, Rubio J (2016) Fine mapping for double podding gene in chickpea. Theor Appl Genet 129: 77–86 [DOI] [PubMed] [Google Scholar]
- Alvarez MA, Tranquilli G, Lewis S, Kippes N, Dubcovsky J (2016) Genetic and physical mapping of the earliness per se locus Eps-A (m) 1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct Integr Genomics 16: 365–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbessa Y, Warkentin T, Vandenberg A, Ball R (2006) Inheritance of time to flowering in chickpea in a short-season temperate environment. J Hered 97: 55–61 [DOI] [PubMed] [Google Scholar]
- Carré IA. (2002) ELF3: a circadian safeguard to buffer effects of light. Trends Plant Sci 7: 4–6 [DOI] [PubMed] [Google Scholar]
- Cho S, Kumar J, Shultz JL, Anupama K, Tefera F, Muehlbauer FJ (2002) Mapping genes for double podding and other morphological traits in chickpea. Euphytica 128: 285–292 [Google Scholar]
- Cobos M, Rubio J, Fernández-Romero M, Garza R, Moreno M, Millán T, Gil J (2007) Genetic analysis of seed size, yield and days to flowering in a chickpea recombinant inbred line population derived from a Kabuli × Desi cross. Ann Appl Biol 151: 33–42 [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daba K, Deokar A, Banniza S, Warkentin TD, Tar’an B (2016a) QTL mapping of early flowering and resistance to ascochyta blight in chickpea. Genome 59: 413–425 [DOI] [PubMed] [Google Scholar]
- Daba K, Tar’an B, Bueckert R, Warkentin TD (2016b) Effect of temperature and photoperiod on time to flowering in chickpea. Crop Sci 56: 200–208 [Google Scholar]
- Deokar AA, Ramsay L, Sharpe AG, Diapari M, Sindhu A, Bett K, Warkentin TD, Tar’an B (2014) Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC Genomics 15: 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2014) http://faostat.fao.org/faostat
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA (2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109: 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur PM, Samineni S, Tripathi S, Varshney RK, Gowda CLL (2015) Allelic relationships of flowering time genes in chickpea. Euphytica 203: 295–308 [Google Scholar]
- Gumber RK, Sarvjeet S (1996) Genetics of flowering time in chickpea: a preliminary report. Crop Improvement 23: 295–296 [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF (1994) Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76: 131–143 [DOI] [PubMed] [Google Scholar]
- Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrándiz C, Macknight R, Navarro C, Morin J, Vardy ME, Ellis N, Beltrán JP, Rameau C, et al. (2005) Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol 137: 1420–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJ, Weller JL (2007) Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 144: 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Laurie RE, Vander Schoor JK, Ridge S, Knowles CL, Liew LC, Sussmilch FC, Murfet IC, Macknight RC, Weller JL (2011) The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23: 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde VS. (2010) Genetics of flowering time in chickpea in a semi-arid environment. Plant Breed 129: 683–687 [Google Scholar]
- Higgins JA, Bailey PC, Laurie DA (2010) Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5: e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath PJ, Kumar A, Penmetsa RV, Farmer A, Schlueter JA, Chamarthi SK, Whaley AM, Carrasquilla-Garcia N, Gaur PM, Upadhyaya HD, et al. (2012) Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol J 10: 716–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICRISAT (1990) Chickpea Kabuli Variety ICCV 2. Plant Material Description 22, http://oar.icrisat.org/524/1/PMD_22.pdf
- Jamalabadi JG, Saidi A, Karami E, Kharkesh M, Talebi R (2013) Molecular mapping and characterization of genes governing time to flowering, seed weight, and plant height in an intraspecific genetic linkage map of chickpea (Cicer arietinum). Biochem Genet 51: 387–397 [DOI] [PubMed] [Google Scholar]
- Jones H, Leigh FJ, Mackay I, Bower MA, Smith LM, Charles MP, Jones G, Jones MK, Brown TA, Powell W (2008) Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol 25: 2211–2219 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim MY, Kang YJ, Lee T, Lee SH (2013) Divergence of flowering-related genes in three legume species. Plant Genome 6: doi/10.3835/plantgenome2013.3803.0008 [Google Scholar]
- Kong F, Liu B, Xia Z, Sato S, Kim BM, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, et al. (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154: 1220–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Abbo S (2001) Genetics of flowering time in chickpea and its bearing on productivity in semiarid environments. Adv Agron 72: 107–138 [Google Scholar]
- Kumar J, Rao BV (2001) Registration of ICCV 96029, a super early and double podded chickpea germplasm. Crop Sci 41: 605–606 [Google Scholar]
- Kumar J, van Rheenen HA (2000) A major gene for time of flowering in chickpea. J Hered 91: 67–68 [DOI] [PubMed] [Google Scholar]
- Laurie RE, Diwadkar P, Jaudal M, Zhang L, Hecht V, Wen J, Tadege M, Mysore KS, Putterill J, Weller JL, et al. (2011) The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol 156: 2207–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenzveig J, Bonfil DJ, Zhang HB, Shtienberg D, Abbo S (2006) Mapping quantitative trait loci in chickpea associated with time to flowering and resistance to Didymella rabiei the causal agent of Ascochyta blight. Theor Appl Genet 113: 1357–1369 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF (1999) Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zhao X, Hu Y, Liu S, Nan H, Li X, Fang C, Cao D, Shi X, Kong L, et al. (2017) Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet 49: 773–779 [DOI] [PubMed] [Google Scholar]
- Martinez-Trujillo M, Limones-Briones V, Cabrera-Ponce J, Herrera-Estrella L (2004) Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol Biol Rep 22: 63–70 [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3: 269–283 [Google Scholar]
- Nicholas KB, Nicholas HB (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. http://www.psc.edu/biomed/genedoc
- Or E, Hovav R, Abbo S (1999) A major gene for flowering time in chickpea. Crop Sci 39: 315–322 [Google Scholar]
- Osborn TC, Kole C, Parkin IA, Sharpe AG, Kuiper M, Lydiate DJ, Trick M (1997) Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana. Genetics 146: 1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Hu Z, Yang RC (2015) Genome-wide comparative analysis of flowering-related genes in Arabidopsis, wheat, and barley. Int J Plant Genomics 2015: 874361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Nilsson O (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ 35: 1742–1755 [DOI] [PubMed] [Google Scholar]
- Pushpavalli R, Krishnamurthy L, Thudi M, Gaur PM, Rao MV, Siddique KHM, Colmer TD, Turner NC, Varshney RK, Vadez V (2015) Two key genomic regions harbour QTLs for salinity tolerance in ICCV 2 × JG 11 derived chickpea (Cicer arietinum L.) recombinant inbred lines. BMC Plant Biol 15: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhika P, Gowda SJM, Kadoo NY, Mhase LB, Jamadagni BM, Sainani MN, Chandra S, Gupta VS (2007) Development of an integrated intraspecific map of chickpea (Cicer arietinum L.) using two recombinant inbred line populations. Theor Appl Genet 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Rubenach AJ, Hecht V, Vander Schoor JK, Liew LC, Aubert G, Burstin J, Weller JL (2017) EARLY FLOWERING3 redundancy fine-tunes photoperiod sensitivity. Plant Physiol 173: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, Matsubara K, Hori K, Yano M, Inoue H, et al. (2012) Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol 53: 717–728 [DOI] [PubMed] [Google Scholar]
- Smithson JB, Thompson JA, Summerfield RJ (1985) Chickpea. In Summerfield RJ, Roberts EJ, eds, Grain Legume Crops. Collins, London, pp 313–389 [Google Scholar]
- Tar’an B, Warkentin TD, Tullu A, Vandenberg A (2007) Genetic mapping of ascochyta blight resistance in chickpea (Cicer arietinum L.) using a simple sequence repeat linkage map. Genome 50: 26–34 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya HD, Bajaj D, Das S, Saxena MS, Badoni S, Kumar V, Tripathi S, Gowda CL, Sharma S, Tyagi AK, et al. (2015) A genome-scale integrated approach aids in genetic dissection of complex flowering time trait in chickpea. Plant Mol Biol 89: 403–420 [DOI] [PubMed] [Google Scholar]
- Vadez V, Krishnamurthy L, Thudi M, Anuradha C, Colmer TD, Turner NC, Siddique KHM, Gaur PM, Varshney RK (2012) Assessment of ICCV 2 × JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol Breed 30: 9–21 [Google Scholar]
- Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar’an B, et al. (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31: 240–246 [DOI] [PubMed] [Google Scholar]
- Varshney RK, Thudi M, Nayak SN, Gaur PM, Kashiwagi J, Krishnamurthy L, Jaganathan D, Koppolu J, Bohra A, Tripathi S, et al. (2014) Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor Appl Genet 127: 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkentin T, Banniza S, Vandenberg A (2005) CDC Frontier Kabuli chickpea. Can J Plant Sci 85: 909–910 [Google Scholar]
- Weller JL, Liew LC, Hecht VF, Rajandran V, Laurie RE, Ridge S, Wenden B, Vander Schoor JK, Jaminon O, Blassiau C, et al. (2012) A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Natl Acad Sci USA 109: 21158–21163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ortega R (2015) Genetic control of flowering time in legumes. Front Plant Sci 6: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Yamawaki S, Hagui E, Ueoka-Nakanishi H, Nakamichi N, Ito S, Mizuno T (2013) Clock-controlled and FLOWERING LOCUS T (FT)-dependent photoperiodic pathway in Lotus japonicus. I. Verification of the flowering-associated function of an FT homolog. Biosci Biotechnol Biochem 77: 747–753 [DOI] [PubMed] [Google Scholar]
- Zhai H, Lü S, Liang S, Wu H, Zhang X, Liu B, Kong F, Yuan X, Li J, Xia Z (2014) GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 9: e89030. [DOI] [PMC free article] [PubMed] [Google Scholar]