BBM and other AIL transcription factors induce somatic embryogenesis in a dose- and context-dependent mechanism and through direct transcriptional regulation of major embryo identity genes.
Abstract
Somatic embryogenesis is an example of induced cellular totipotency, where embryos develop from vegetative cells rather than from gamete fusion. Somatic embryogenesis can be induced in vitro by exposing explants to growth regulators and/or stress treatments. The BABY BOOM (BBM) and LEAFY COTYLEDON1 (LEC1) and LEC2 transcription factors are key regulators of plant cell totipotency, as ectopic overexpression of either transcription factor induces somatic embryo formation from Arabidopsis (Arabidopsis thaliana) seedlings without exogenous growth regulators or stress treatments. Although LEC and BBM proteins regulate the same developmental process, it is not known whether they function in the same molecular pathway. We show that BBM transcriptionally regulates LEC1 and LEC2, as well as the two other LAFL genes, FUSCA3 (FUS3) and ABSCISIC ACID INSENSITIVE3 (ABI3). LEC2 and ABI3 quantitatively regulate BBM-mediated somatic embryogenesis, while FUS3 and LEC1 are essential for this process. BBM-mediated somatic embryogenesis is dose and context dependent, and the context-dependent phenotypes are associated with differential LAFL expression. We also uncover functional redundancy for somatic embryogenesis among other Arabidopsis BBM-like proteins and show that one of these proteins, PLETHORA2, also regulates LAFL gene expression. Our data place BBM upstream of other major regulators of plant embryo identity and totipotency.
Plant cells show a high degree of developmental plasticity and can be induced readily to regenerate new tissues or organs (pluripotency) and even embryos (totipotency) from in vitro-cultured explants. Somatic embryogenesis is a type of plant totipotency in which embryos are induced to form on vegetative explants, usually in response to exogenous hormones, especially auxins, and/or stress treatments (Fehér, 2015). Somatic embryogenesis is used extensively as a clonal propagation tool in biotechnology applications (Lelu-Walter et al., 2013; Sharma et al., 2013b; Park and Paek, 2014), and while the tissue culture requirements for somatic embryo induction are well known (Gaj, 2004), only a few of the molecular components that drive this process have been described (Elhiti et al., 2013).
A number of plant transcription factors have been identified that can convert somatic cells into embryogenic, totipotent cells. One of these transcription factors, Brassica napus BABY BOOM (BBM), encodes an AINTEGUMENTA-LIKE (AIL) APETALA2/ethylene-responsive element-binding factor (AP2/ERF; Boutilier et al., 2002). In Arabidopsis (Arabidopsis thaliana), AIL genes form a small, eight-member clade within the AP2/ERF transcription factor family, which, in addition to BBM, comprises AINTEGUMENTA (ANT), AIL1, PLETHORA1 (PLT1), PLT2, AIL6/PLT3, CHOTTO1 (CHO1)/EMBRYOMAKER (EMK)/AIL5/PLT5, and PLT7. AIL genes are expressed in dividing tissues, including root, shoot, and floral meristems, where they act in a redundant manner to maintain a meristematic state (Horstman et al., 2014). Single AIL knockout mutants show no or only minor defects, but double or triple mutants have stronger phenotypes related to reduced cell proliferation or altered cell identity, including smaller floral organs with partial loss of identity (Krizek, 2009, 2015), embryo arrest (Galinha et al., 2007), and impaired root and shoot meristem maintenance (Aida et al., 2004; Galinha et al., 2007; Mudunkothge and Krizek, 2012). BBM is expressed in the embryo and root meristem, where it regulates cell identity and growth together with other AIL proteins (Aida et al., 2004; Galinha et al., 2007). In line with their loss-of-function phenotypes, overexpression of Arabidopsis AIL transcription factors induces pluripotency, totipotency, and/or cell proliferation, with different functions being assigned to specific AIL proteins (Krizek, 1999; Nole-Wilson et al., 2005; Galinha et al., 2007; Tsuwamoto et al., 2010; Krizek and Eaddy, 2012). However, unlike other AIL genes, the genetic pathways in which BBM functions have not been well characterized (Horstman et al., 2014), and it is not known how this single protein controls both pluripotent and totipotent growth.
The overexpression of native or heterologous BBM genes also induces regeneration in other species (Morcillo et al., 2007; Deng et al., 2009; El Ouakfaoui et al., 2010; Heidmann et al., 2011; Lutz et al., 2011; Bandupriya et al., 2014; Yang et al., 2014; Florez et al., 2015; Lowe et al., 2016) and, therefore, is used as a biotechnology tool to improve plant transformation in model and crop species (Deng et al., 2009; Heidmann et al., 2011; Lutz et al., 2011; Florez et al., 2015; Lowe et al., 2016).
Other transcription factors also induce somatic embryogenesis when expressed ectopically in seedlings, including LEAFY COTYLEDON1 (LEC1), which encodes subunit B of a nuclear factor Y protein (NF-YB), and the B3 domain protein LEC2 (Lotan et al., 1998; Stone et al., 2001). LEC1 and LEC2, together with LEC1-LIKE (L1L) and the B3 domain proteins ABSCISIC ACID-INSENSITIVE3 (ABI3) and FUSCA3 (FUS3), are collectively referred to as the LAFL network (for LEC1/L1L, ABI3, FUS3, and LEC2; Jia et al., 2014). LAFL proteins function throughout embryogenesis, where they redundantly regulate early developmental processes such as embryo identity and later processes such as embryo maturation (storage product accumulation and desiccation tolerance) and dormancy (Jia et al., 2013). Although FUS3 and ABI3 do not induce somatic embryogenesis when overexpressed, they do confer embryo traits to seedlings (Parcy et al., 1994; Parcy and Giraudat, 1997; Gazzarrini et al., 2004). LEC1 and LEC2 directly regulate the expression of seed maturation and auxin response and biosynthesis genes (Lotan et al., 1998; Braybrook et al., 2006), and both of these functions could play a role in inducing a totipotent state (Braybrook and Harada, 2008). Moreover, LEC2 directly activates the MADS box transcription factor gene AGAMOUS-LIKE15 (AGL15), which enhances somatic embryogenesis from immature zygotic embryos when overexpressed (Harding et al., 2003; Braybrook et al., 2006). LAFL gene expression is controlled by the chromatin remodeler PICKLE (PKL) and by B3 domain-containing VIVIPAROUS1/ABI3-LIKE (VAL)/HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE (HSI) transcription factors. Mutations in PKL or VAL genes lead to increased LAFL gene expression and maintain embryo identity in seedlings (Ogas et al., 1999; Rider et al., 2003; Henderson et al., 2004; Suzuki et al., 2007).
While both BBM and LEC promote totipotency, it is not known whether they function in the same developmental pathways. To gain insight into the signaling pathways regulated by BBM, we performed a global analysis of BBM DNA-binding sites in somatic embryo tissue (Horstman et al., 2015). Here, we show that BBM induces cell totipotency during seed germination through transcriptional activation of the LAFL network and that BBM induces somatic embryogenesis in a dose- and context-dependent manner.
RESULTS
BBM Binds and Transcriptionally Activates LAFL Genes
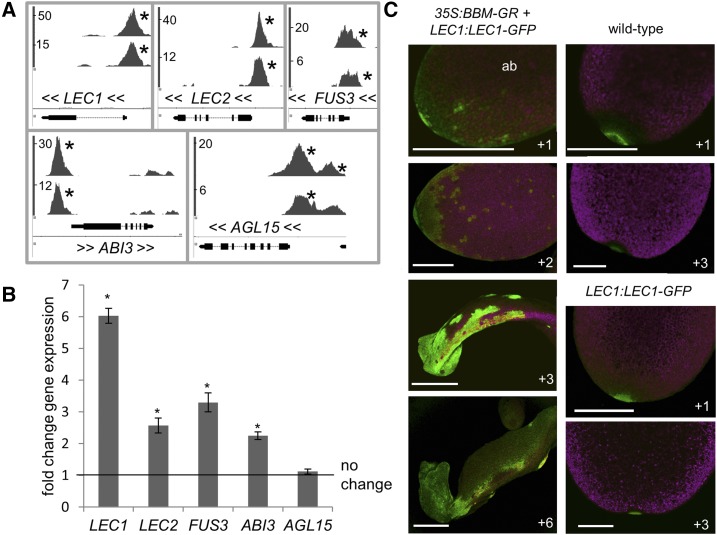
To understand BBM regulatory networks during embryogenesis, we identified genes that were bound by BBM in somatic embryo cultures using chromatin immunoprecipitation of BBM-YFP (BBM:BBM-YFP) and BBM-GFP (35S:BBM-GFP) fusion proteins followed by next-generation sequencing (ChIP-seq; Horstman et al., 2015). BiNGO analysis (Maere et al., 2005) of the top 1,000 potential target genes in these chromatin immunoprecipitation (ChIP) experiments was performed to identify overrepresented Gene Ontology categories (Supplemental Data Set S1; a selection of which is shown in Supplemental Table S1). Genes involved in auxin biosynthesis, transport, and signaling, as well root development, meristem initiation, and maintenance, were overrepresented in the ChIP-seq data sets, as expected from BBM’s function in the root and in line with studies of other AIL proteins (Horstman et al., 2014; Santuari et al., 2016). Genes involved in adaxial/abaxial polarity specification and shoot development also were overrepresented among BBM-bound genes (Supplemental Data Set S1; Supplemental Table S1). Notably, BBM bound to the promoter regions of genes with known functions in embryo identity and maturation, including the LAFL genes LEC1, LEC2, ABI3, and FUS3 (but not L1L), the MADS box transcription factor gene AGL15, which enhances somatic embryogenesis and functions in a positive feedback network with LEC2 (Harding et al., 2003; Braybrook et al., 2006; Zheng et al., 2009), and NF-YA9, which also induces somatic embryogenesis when overexpressed (Mu et al., 2013; Fig. 1A; Supplemental Data Set S1; Supplemental Table S1). Here, we focus our efforts on characterization of the interaction between BBM and members of the LAFL/AGL15 network. We confirmed BBM binding to the promoters of LEC1, LEC2, ABI3, and AGL15 by independent ChIP-qPCR experiments in somatic embryos (Supplemental Fig. S1A). We did not observe BBM binding to the FUS3 promoter, which is consistent with its lower and atypically shaped BBM ChIP-seq peak (Supplemental Fig. S1).
Figure 1.
BBM binds and transcriptionally regulates LAFL genes. A, ChIP-seq BBM-binding profiles for embryo-expressed genes: 35S:BBM-GFP (top profiles) and BBM:BBM-YFP (bottom profiles). x axis, Nucleotide positions (TAIR 10 annotation; black bars indicate exons, and lines indicate introns); y axis, ChIP-seq score (fold enrichment of the BBM-GFP/YFP ChIP to the control ChIP), as calculated by the CSAR package in Bioconductor; <<, direction of gene transcription; *, peaks with scores that are considered statistically significant (false discovery rate < 0.05). B and C, Transcriptional regulation of LAFL genes. Relative expression was determined by quantitative real-time reverse transcription-PCR (qPCR) in 35S:BBM-GR and Columbia-0 (Col-0) seeds 1 d after seed plating. Samples were treated for 3 h with dexamethasone (DEX) and cycloheximide (CHX; both at 10 µm). B, Error bars indicate se values of the three biological replicates. Statistically significant differences (asterisks) between DEX+CHX-treated 35S:BBM-GR and DEX+CHX-treated Col-0 were determined using Student’s t test (P < 0.01). C, LEC1:LEC1-GFP regulation by BBM. Samples were treated with 10 µm DEX 1 d after plating and imaged on subsequent days as indicated. The images show the adaxial sides of cotyledons, unless indicated (ab, abaxial side). The green signal in Col-0 and LEC1:LEC1-GFP cotyledon tips is autofluorescence. Bars = 250 µm.
ANT/AIL DNA-binding motifs were determined previously in vitro by SELEX and electrophoretic mobility shift assay (Nole-Wilson and Krizek, 2000; Yano et al., 2009; O’Malley et al., 2016; Santuari et al., 2016). MEME analysis (Bailey and Elkan, 1994) of in vivo BBM-bound regions identified an overrepresented sequence motif that resembles the ANT/AIL DNA-binding motif (Supplemental Fig. S1B; Santuari et al., 2016). The BBM-bound region in LEC1, LEC2, and FUS3 contains this BBM-binding motif, while the ABI3-bound region contains a slightly degenerate version thereof (Supplemental Fig. S1C). The BBM-binding motif was not found in the region bound by BBM in the AGL15 gene, which suggests that BBM binds AGL15 using a different motif or via an intermediate protein.
We determined whether BBM regulates LAFL/AGL15 gene expression during somatic embryo induction from imbibed seeds using a steroid (DEX)-regulated 35S::BBM-GR line in combination with qRT-PCR and reporter gene analysis. qRT-PCR analysis in the presence of DEX and the translational inhibitor CHX showed that LEC1, LEC2, FUS3, and ABI3 expression, but not AGL15 expression, was up-regulated after BBM-GR activation in imbibed seeds (Fig. 1B). LAFL/AGL15 genes were not differentially regulated upon BBM-GR activation in the same material that was used for ChIP (2,4-dichlorophenoxyacetic acid [2,4-D]-induced embryo cultures; data not shown). An explanation for this lack of transcriptional response could be the use of different promoters to drive BBM in the two experiments. BBM might only bind and activate LAFL/AGL15 genes during the induction/early stages of somatic embryogenesis (pBBM:BBM-YFP used for ChIP is expressed during early embryogenesis; Horstman et al., 2015) but not at later stages of somatic embryogenesis (p35S:BBM-GR used for transcription analysis [qRT-PCR] is expressed during late embryogenesis; Johnson et al., 2005).
Next, a LEC1:LEC1-GFP reporter (Li et al., 2014) was used to chart the dynamics of LEC1 expression during BBM-induced somatic embryogenesis (Fig. 1C). 35S:BBM-GR seedlings initially form somatic embryos on the cotyledon tip and later from the shoot apical meristem and cotyledon margins. LEC1-GFP was observed 1 d after BBM-GR activation, in small patches of cells on the abaxial side of the cotyledon (Fig. 1C, +1), and 1 d later at the cotyledon tip and in patches of cells on the adaxial cotyledon blade (Fig. 1C, +2). LEC1-GFP became stronger in the cotyledon tip and margin at the time when the tip began to swell (Fig. 1C, +3) and could be found later in the somatic embryos that formed at the cotyledon tip and at the margin (Fig. 1C, +6).
Our results demonstrate that BBM overexpression can ectopically activate LAFL gene expression during seed germination.
LAFL Proteins Modulate BBM-Induced Embryogenesis
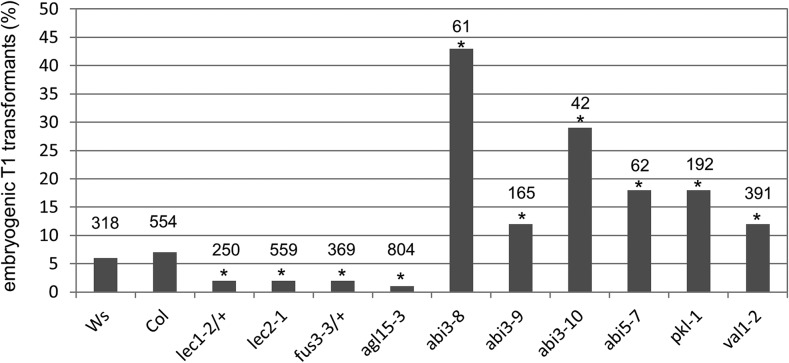
We investigated the genetic relationship between BBM and its LAFL/AGL15 gene targets. Since outcrossing BBM overexpression lines silences the BBM phenotype (Supplemental Fig. S2), we introduced the 35S:BBM-GR construct into the lec1-2, lec2-1, fus3-3, agl15-3, and abi3 (three alleles) mutant backgrounds by transformation (Fig. 2). The lec1-2 and fus3-3 seeds are desiccation intolerant (Meinke et al., 1994); therefore, heterozygous mutants (lec1-2/+ and fus3-3/+) were used for transformation.
Figure 2.
BBM-induced embryogenesis is modulated by LAFL genes. The percentage of primary embryogenic transformants obtained is shown after transformation of the 35S:BBM-GR construct to the wild-type Wassilewskija (Ws) or Col-0 background or the indicated mutant lines. Statistically significant differences (asterisks) in the number of embryogenic transgenic lines between the mutant and the corresponding wild-type line were determined using Pearson’s χ2 test (P < 0.05). The total number of transformants per line is indicated above each bar. Somatic embryo formation was not observed in any of the individual mutant backgrounds alone or in mutant + 35S:BBM-GR backgrounds in the absence of BBM-GR activation.
In wild-type Arabidopsis, 6% to 7% of the primary (T1) 35S:BBM-GR transformants were embryogenic when grown on DEX (Fig. 2). Transformation of the lec1-2/+, lec2-1, fus3-3/+, and agl15-3 mutants with the 35S:BBM-GR construct resulted in significantly reduced percentages of embryogenic seedlings compared with transformed wild-type seedlings (Fig. 2). We determined the genotype of the few embryogenic seedlings that were generated after transformation of 35S:BBM-GR to the lec1-2/+ and fus3-3/+ backgrounds. Only one of the embryogenic lec1-2/+ progeny contained the lec1-2 mutant allele, while none of the embryogenic fus3-3/+ progeny contained the fus3-3 mutant allele (Supplemental Table S2). To determine whether BBM-GR activation can induce somatic embryogenesis in a homozygous lec1-2 background, we rescued immature zygotic embryos from the single embryogenic lec1-2/+ 35S:BBM-GR line to bypass the lec1-2 desiccation-intolerant phenotype and cultured them on DEX-containing medium. Embryos were separated phenotypically into lec1-2 homozygous mutant and combined lec1-2 heterozygous/wild-type classes. Somatic embryos formed in lec1-2 heterozygous/wild-type seedlings but not in the homozygous lec1-2 seedlings (Supplemental Table S2).
These results suggest that LEC1, LEC2, and FUS3 are positive regulators of BBM-mediated somatic embryogenesis and that LEC1 and FUS3 are essential for this process. Surprisingly, we found that AGL15 also is a positive regulator of BBM-induced somatic embryogenesis, even though it is not transcriptionally regulated by BBM at the start of somatic embryo induction; AGL15 might be transcriptionally regulated by BBM at a later time point.
In contrast to the results obtained with the fus3, lec, and agl15 mutants, transformation of the 35S:BBM-GR construct to three different abi3 mutants enhanced the number of embryogenic seedlings (Fig. 2). abi3 is the only LAFL mutant that is insensitive to abscisic acid (ABA), and overexpression of ABI3 does not induce somatic embryogenesis (Parcy et al., 1994; Parcy and Giraudat, 1997). To separate the effects of ABA insensitivity and other embryo defects of abi3 mutants on the BBM phenotype, we tested another ABA-insensitive mutant, abi5-7, which does not show mutant embryo phenotypes other than ABA insensitivity (Nambara et al., 2002). As with the abi3 mutants, the abi5-7 mutant also enhanced the frequency of the BBM phenotype (Fig. 2), suggesting that BBM-mediated totipotency is suppressed by ABA signaling rather than by other functions of the ABI3/ABI5 proteins.
A number of regulatory proteins repress LAFL gene expression during the transition from seed to postembryonic growth. Seedlings with loss-of-function mutations in these proteins ectopically express LAFL genes and retain embryo identity (Jia et al., 2014). We determined whether loss-of-function mutants for two of these proteins, the CHD3 chromatin remodeler PKL and the B3 domain protein VAL1/HSI2, influence the penetrance of BBM-induced embryogenesis (Fig. 2). The pkl-1 and val1-2 (hsi2-5) mutants improved the efficiency of BBM-mediated somatic embryogenesis, as measured by the higher percentage of embryogenic primary transformants.
Together, the data show that members of the LAFL network, as well as their upstream negative regulators, are important direct and indirect components of the BBM signaling pathway.
AIL/PLT Proteins Promote Totipotency and Regulate LAFL Gene Expression
BBM is expressed in the embryo and the root meristem, where it regulates cell identity and growth along with other AIL proteins (Aida et al., 2004; Galinha et al., 2007). PLT1 and PLT2 induce spontaneous root organogenesis (Aida et al., 2004) and have roles in hormone-mediated regeneration (Kareem et al., 2015), while BBM and CHO1/EMK/AIL5/PLT5 (Tsuwamoto et al., 2010) are the only genes reported to induce somatic embryogenesis. We generated 35S:AIL overexpression lines for the six AIL genes that have not been reported to induce somatic embryogenesis when overexpressed, namely ANT, AIL1, PLT1, PLT2, PLT3/AIL6, and PLT7, and found that overexpression of all these genes except the phylogenetically distinct ANT and AIL1 (Kim et al., 2006) induced somatic embryogenesis in the primary transformants (Supplemental Fig. S3, A and B). A PLT2-GR fusion protein directly activated LEC1, LEC2, and FUS3 gene expression but not ABI3 and AGL15 expression (Supplemental Fig. S3C). Together, these data suggest extensive overlap in AIL protein function.
BBM- and PLT2-Mediated Somatic Embryogenesis Is Dose Dependent
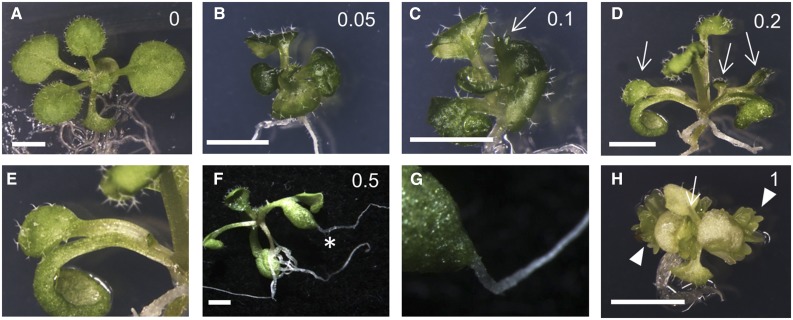
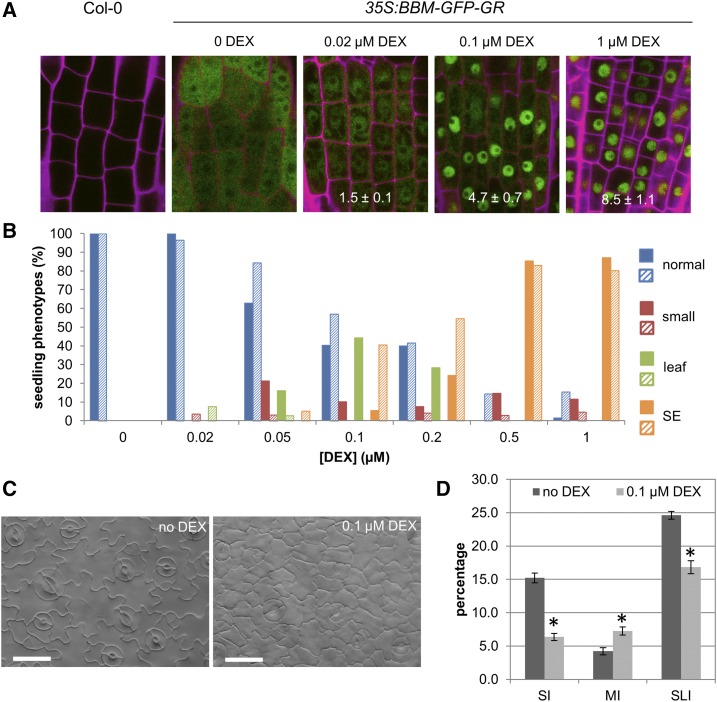
Ectopic AIL expression induces spontaneous adventitious growth, including somatic embryos, ectopic shoots and roots, and callus formation (Boutilier et al., 2002). These phenotypes are correlated with the amount of nucleus-localized BBM protein in DEX-treated 35S:BBM-(GFP)-GR lines (Figs. 3 and 4A). A relatively low BBM dose induced the formation of small seedlings with epinastic cotyledons and leaves (Figs. 3B and 4B) that showed reduced cell differentiation (Fig. 4, C and D). Intermediate DEX concentrations also induced ectopic trichome-bearing protrusions or ectopic leaves on their cotyledon petioles (Figs. 3, C–E, and 4B), and ectopic roots appeared after longer exposure to DEX (Fig. 3, F and G). A low frequency of seedlings with somatic embryos on their cotyledons (Fig. 3H) also was observed at intermediate DEX concentrations, and this phenotype became highly penetrant at the highest effective DEX concentration (Fig. 4B). PLT2 also directs the same dose-dependent overexpression phenotypes (Supplemental Fig. S4) as BBM overexpression, although ectopic root formation was evident earlier in the PLT2-GR lines than in the BBM-GR lines. These data suggest that AIL protein dose drives the developmental fate of regenerating tissues.
Figure 3.
BBM overexpression phenotypes are dose dependent. Phenotypes are shown for 35S:BBM-GR seedlings grown for 2 weeks (A–E and H) or 3 weeks (F and G) on the DEX concentration (µm) indicated in each image. A, A phenotypically normal seedling. B, A small seedling with epinastic leaves and cotyledons. C, A small, epinastic seedling with a trichome-bearing ectopic leaf (arrow) on the cotyledon petiole. D, A seedling with ectopic leaves on the petioles of both cotyledons (arrows). E, A magnified view of the ectopic leaf in D. F, A 35S:BBM-GR seedling with an ectopic root (asterisk). G, A magnified view of the ectopic root in F. H, A seedling with somatic embryos on the cotyledon margins (arrowheads). Bars = 2.5 mm.
Figure 4.
Quantification of BBM dose-dependent phenotypes. A, BBM-GFP-GR nuclear localization increases with increasing DEX concentration. The effect of DEX on BBM localization in roots of 35S:BBM-GFP-GR seedlings grown for 7 d in medium containing the indicated DEX concentration is shown. Non-DEX-treated (Col-0) roots are shown as a GFP-negative control. Subcellular BBM-GFP-GR localization was quantified for each DEX concentration by calculating the average nuclear-cytoplasmic GFP ratio (63–133 cells of five to eight roots per DEX concentration), which is indicated on the bottom of the images (±se). The average ratios are significantly different from each other (P < 0.01, Student’s t test). Green, GFP; magenta, propidium iodide. B, Frequency of phenotypes observed in 35S:BBM-GR seedlings from two independent transgenic lines (solid and hatched bars) grown for 2 weeks on medium containing different DEX concentrations (n = 100–350 seedlings). Leaf, Ectopic leaves; SE, somatic embryos. Seedlings that showed both ectopic shoots and somatic embryos were scored as SE. SE refers to both embryogenic tissue (smooth, swollen, bright green in color, and lacking trichomes) and cotyledons as well as histodifferentiated embryos. C, A relatively low BBM dose induces smaller and less-lobed leaf pavement cells compared with the control. The abaxial sides of cleared first leaves of 9-d-old 35:BBM-GR seedlings grown on medium without DEX (left) or with 0.1 μm DEX (right) are shown. Bars = 25 μm. D, Stomatal differentiation in DEX-treated 35:BBM-GR seedlings is reduced compared with untreated seedlings. In DEX-treated 35S::BBM-GR seedlings, fewer cells are committed to stomatal development, as reflected by the lower stomatal lineage index (SLI). Also, fewer mature stomata were found in leaves of DEX-treated seedlings (stomatal index [SI]), while the number of stomatal meristemoids was increased (meristemoid index [MI]). Error bars indicate se. Asterisks indicate statistically significant differences compared with the non-DEX-treated control (*, P < 0.05, Student’s t test).
BBM Promotes Context-Specific Embryogenesis
Previously, we identified direct BBM target genes in 4-d-old 35S:BBM-GR seedlings using microarray analysis (Passarinho et al., 2008). LAFL/AGL15 genes were not identified as BBM target genes in this study, and in general, there was little overlap between these seedling microarray-derived targets and the top 1,000 ChIP-seq-derived targets identified in somatic embryos (Supplemental Table S3). This discrepancy might be explained by the different tissues that were used in each study. Therefore, we examined the relationship between the developmental competence for BBM-mediated regeneration and LAFL transcription.
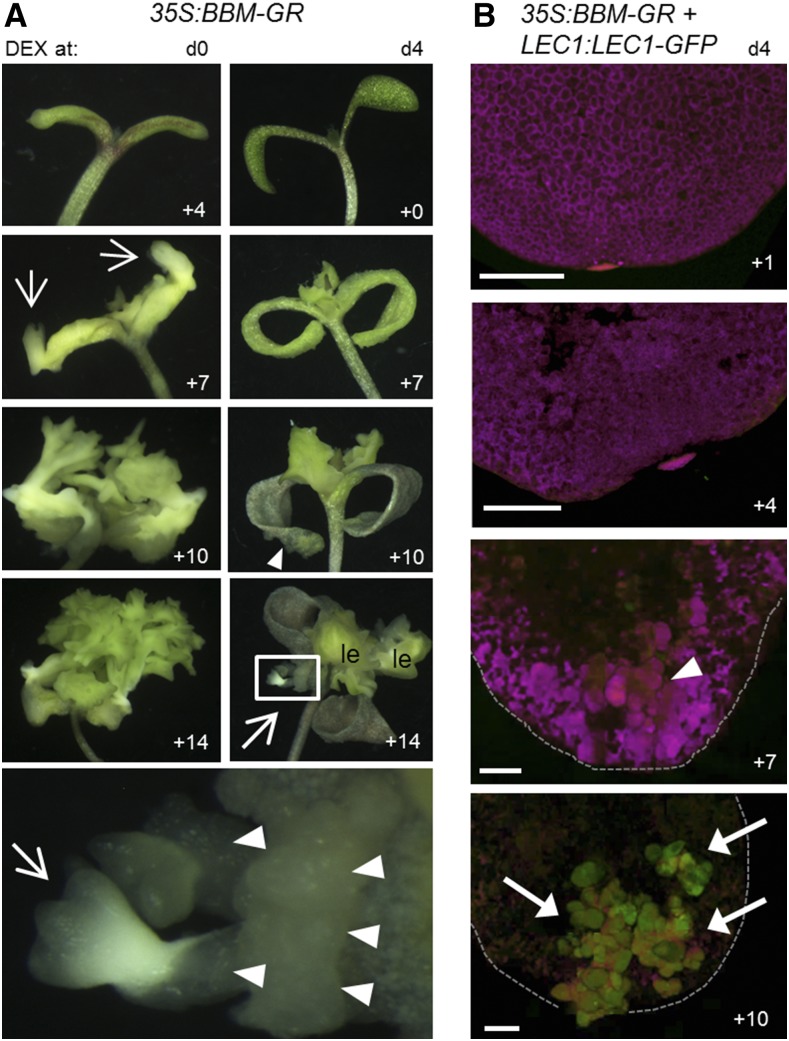
We activated BBM-GR at different time points before and after germination. When 35S:BBM-GR seeds were placed directly in DEX-containing medium before or during germination (days 0–2), 100% of the seedlings formed somatic embryos directly on their cotyledons after approximately 1 week (Fig. 5A; Supplemental Fig. S5A). By contrast, postgermination DEX treatment (days 3–4) induced callus formation on the adaxial side of the cotyledons of approximately 40% of the seedlings from which somatic embryos eventually developed (Fig. 5B; Supplemental Fig. S5A). We obtained similar results when we activated PLT2-GR before and after germination (Supplemental Fig. S5, B and C). Thus, AIL proteins induce somatic embryogenesis in two ways depending on the developmental stage of the explant: directly from cotyledons in a narrow window before germination, and indirectly via a callus phase after germination. In agreement with our previous microarray results, neither LEC1, LEC2, FUS3, nor ABI3 was expressed when BBM-GR (or PLT2-GR) was activated in postgermination seedlings, although AGL15 expression was up-regulated slightly under these conditions (Supplemental Fig. S5D). LEC1-GFP was only detected in this indirect pathway 10 d after DEX induction (Fig. 5B), when it was localized to globe-like embryos that emerged from the callus. The desiccation intolerance combined with the lack of or weak embryogenesis phenotypes of the lec1/fus3 35S:BBM-GR lines (see above) complicated further genetic analysis of the role of LEC1 and FUS3 in this indirect pathway.
Figure 5.
BBM-induced embryogenesis is context dependent. A, 35S:BBM-GR plants were cultured with 10 µm DEX starting at the dry seed stage (d0) or after germination (d4). The culture time after DEX application is indicated on the bottom right of each image. Arrowheads, Callus; arrows, somatic embryos/embryogenic tissue; le, callused leaf tissue. The bottom image is a magnification of the boxed region in the d4 +14 image. B, LEC1:LEC1-GFP regulation by BBM. Seedlings were treated with 10 µm DEX after germination (d4) and imaged on subsequent days as indicated. The images show the adaxial sides of cotyledons. Arrowhead, Callus on the distal end of the cotyledon blade; arrows, GFP-positive embryo clusters. The outline of the cotyledon margins is shown with dashed lines. Autofluorescence (magenta) was used to delineate the tissue. Bars = 250 μm.
Our results highlight the existence of a narrow developmental window of competence for direct embryogenesis and suggest that tissues outside this window require more extensive reprogramming (e.g. callus formation) before embryogenesis can be induced. LAFL genes are transcriptionally silenced after germination (Zhang et al., 2012; Zhou et al., 2013); therefore, these loci might only become transcriptionally accessible after redifferentiation of cotyledon cells to callus.
DISCUSSION
BBM-Mediated Embryogenesis Requires LAFL Genes
An increasing number of proteins are being identified that regulate cell totipotency in vivo and in vitro, including members of the LAFL network and AIL proteins (Horstman et al., 2014; Fehér, 2015). LAFL transcription is regulated at the chromatin level and by extensive transcriptional feedback loops between LAFL proteins. The interactions between LAFL proteins and other regulators of cell totipotency are less well known. Our data now place BBM/AIL proteins directly upstream of the LAFL/AGL15 genes. There is some evidence that LAFL proteins might act upstream of AILs: a Phaseolus vulgaris ABI3-like factor (PvALF) binds and activates Arabidopsis CHO1/EMK/AIL5/PLT5 (Sundaram et al., 2013), and Arabidopsis FUS was shown to bind BBM, PLT2, AIL6/PLT3, and PLT7, although transcriptional regulation was not investigated (Wang and Perry, 2013). In contrast, the lack of AIL deregulation after inducible overexpression of LEC1, ABI3, FUS3, or LEC2 (Braybrook and Harada, 2008; Yamamoto et al., 2010; Junker et al., 2012; Mönke et al., 2012) suggests that there is no direct feedback of LAFLs on AILs. BBM expression is reduced in lafl mutant seeds (Supplemental Fig. S6), but this genetic interaction could be indirect. Although BBM proteins appear to directly activate LAFL genes, the data are inconclusive regarding whether there are additional direct transcriptional feedback loops in the AIL-LAFL cell totipotency network.
LAFL/AGL15 proteins are required for BBM function, as BBM overexpression in lec1, lec2, fus3, and agl15 mutants either reduced or eliminated the capacity of seedlings to form somatic embryos. The reduced competence for somatic embryogenesis in these mutants could be explained in two ways: (1) the developmental defects in the mutants change the physiological state of the mature embryo/seedling in such a way that it is no longer responsive for BBM; or (2) BBM-induced embryogenesis relies on transcriptional activation of the LEC1, LEC2, FUS3, and AGL15 genes. Several lines of evidence support the latter scenario. First, we observed a reduced responsiveness to BBM in segregating lec1 and fus3 populations, which contained wild-type and heterozygous plants. However, the few embryogenic transformants in these populations were mainly wild types, suggesting that the lec1 and fus3 mutations affect BBM function in the heterozygote state. Heterozygous lec1 and fus3 mutants do not show obvious growth defects, suggesting that reduced LEC1 or FUS3 level (dose) in the heterozygous mutants, rather than a change in the physiological state of the tissue, reduces the response to BBM overexpression. Second, the abi3 mutant shows overlapping maturation defects with the other LAFL mutants (To et al., 2006; Roscoe et al., 2015), yet the abi3 mutations had the opposite effect on BBM-induced embryogenesis. Therefore, we hypothesize that the inability of BBM to induce LEC and FUS gene expression reduces the capacity for embryogenic growth in these mutants. This hypothesis is further strengthened by our observations that mutations in LAFL repressors enhance BBM-mediated embryogenesis, possibly by facilitating elevated LAFL gene expression.
Embryo Induction Is a Dose-Dependent Phenotype
It was suggested previously that the PLT2 protein regulates root meristem size and maintenance through a protein concentration gradient, with high, intermediate, and low AIL concentrations instructing stem cell fate, cell division, and differentiation, respectively (Galinha et al., 2007). AIL6/PLT3 overexpression also induces dose-dependent phenotypes in floral organs (Krizek and Eaddy, 2012). We showed that a high BBM/PLT2 dose induces embryogenesis, a lower dose induces organogenesis, and the lowest dose inhibits differentiation. Our overexpression data, therefore, support a general dose-dependent AIL output in plant tissues. This dose dependence complicates functional complementation studies of AILs, as even complementation with endogenous promoters (Galinha et al., 2007; Santuari et al., 2016) can lead to differences in expression levels among transformants and a range of developmental outcomes.
How dedifferentiation, organogenesis, and somatic embryogenesis are induced at successively higher AIL doses is not clear, but it likely reflects the endogenous roles of AIL proteins during embryo, organ, and meristem development. Mechanistically, AIL dose-dependent phenotypes could result from dose-dependent expression levels of the same set of target genes and/or from dose-specific activation of specific target genes. A transcription factor gradient can regulate the different sets of target genes through differences in binding site number and affinity (Rogers and Schier, 2011). Target genes with many or high-affinity binding sites are activated by low levels of the transcription factor, whereas genes with few or low-affinity binding sites are activated only at high transcription factor levels. Specificity also might be determined at the level of protein-protein interactions (Horstman et al., 2015). Defining the overlapping and unique target genes for each AIL transcription factor at different doses and the protein complexes in which they function will shed light on how the AIL dose directs specific developmental fates.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) lec2-1 (CS3868), lec1-2 (CS3867), fus3-3 (CS8014), agl15-3 (CS16479), and pkl-1 (CS3840) mutants were obtained from the Nottingham Arabidopsis Stock Centre. The val1-2 (hsi2-5; Sharma et al., 2013a), abi3-8, abi3-9, abi3-10, and abi5-7 (Nambara et al., 2002) mutants and the LEC1:LEC1-GFP marker (Li et al., 2014) were described previously. All mutants are in the Col-0 background, except lec1-2 and lec2-1, which are in the Wassilewskija background.
Seeds were sterilized with liquid bleach as described previously (Boutilier et al., 2002; Passarinho et al., 2008) and germinated on one-half-strength Murashige and Skoog medium containing 1% Suc and vitamins. DEX and CHX (both Sigma) were added to the medium as described in the text. Solid and liquid (rotary shaker, 60 rpm) cultures were kept at 21°C and 25°C, respectively (16-h-light/8-h-dark regime). lec1-2 35S:BBM-GR mutant embryos were rescued by excising them from sterilized siliques and allowing them to germinate on solid one-half-strength Murashige and Skoog medium containing 1% Suc and vitamins with DEX and kanamycin (for selection of the BBM transgene).
Vector Construction and Transformation
The 35S:BBM-GR construct was described previously (Passarinho et al., 2008). The ANT, PLT3/AIL6, PLT7, and PLT1 protein-coding regions were amplified from Arabidopsis Col-0 genomic DNA and the PLT2 protein-coding region from cDNA using the primers listed in Supplemental Table S4. 35S:AIL ectopic overexpression constructs were made using the Gateway binary vector pGD625 (Chalfun-Junior et al., 2005). The 35:BBM-GFP-GR construct was made using the Gateway-compatible destination vector pARC146 (Danisman et al., 2012). BBM-GFP used in the 35:BBM-GFP-GR construct was amplified from a BBM:BBM-GFP plasmid (Horstman et al., 2015). Constructs were introduced into wild-type or mutant lines by floral dip transformation (Clough and Bent, 1998).
Confocal Microscopy
Confocal imaging was performed as described previously (Soriano et al., 2014). Propidium iodide (10 µg mL−1) counterstaining (35S:BBM-GFP-GR roots) and autofluorescence (LEC1:LEC1-GFP cotyledons) were used to delineate the tissue. Both fluorophores were excited with a 532-nm laser and detected at 600 to 800 nm.
To quantify the subcellular BBM-GFP-GR localization in root, confocal images were made of the meristematic region of roots of 35S:BBM-GFP-GR seedlings grown for 7 d in medium containing different DEX concentrations. ImageJ was used to calculate the ratio of nuclear to cytoplasmic fluorescence by comparing the average fluorescence intensity in the nucleus with the average fluorescence intensity of an area of equal size in the cytoplasm.
Leaf Imaging and Quantification of Stomatal Development
The first leaf pairs of 9-d-old seedlings were placed overnight in 70% ethanol at 4°C, then transferred to 85% ethanol for 6 h, and subsequently to 3% bleach overnight or until imaging. Leaves were mounted in HCG solution (80 g of chloral hydrate, 10 mL of glycerol, and 30 mL of water) prior to imaging with a Nikon Optiphot microscope.
To calculate SI, eight images from the abaxial sides of cleared first leaves of 9-d-old 35:BBM-GR plants grown with or without DEX were analyzed (n = 125 and 350 cells per image). The SI, MI, and SLI were calculated as described previously (Peterson et al., 2013). SI = (number of stomata/(total number of stomata + nonstomatal epidermal cells)) × 100. For the SI, only mature stomata with a pore were counted. MI = (number of meristemoids/(total number of stomata + nonstomatal epidermal cells)) × 100. SLI = (number of stomata and stomata precursors/(total number of stomata + nonstomatal epidermal cells)) × 100.
ChIP-Seq
The previously published ChIP-seq data and data analysis (Horstman et al., 2015) can be downloaded from the Gene Expression Omnibus (GSE52400). The ChIP-seq experimental setup has been described (Horstman et al., 2015). Briefly, the experiments were performed using somatic embryos from either 2,4-D-induced BBM:BBM-YFP cultures (with BBM:NLS-GFP as a control) or a 35S:BBM-GFP overexpression line (with 35S:BBM as a control). Two independent ChIP-qPCR experiments on 2,4-D-induced BBM:BBM-YFP cultures were performed to validate the ChIP-seq results, using the same protocol as described previously (Horstman et al., 2015). A fold change was calculated by comparison with an unbound genomic control region (ARR6), and statistically significant differences between BBM-bound regions and a second unbound genomic control region (HSF1) were determined using Student’s t test (P < 0.05). The DNA primers are shown in Supplemental Table S4.
Expression Analysis
cDNA from Col-0 and 35S:BBM-GR seeds or seedlings (three biological replicates of each) were treated as described in the text and used for qPCR. qPCR was performed using the BioMark HD System (Fluidigm). The data were normalized against the SAND gene (Czechowski et al., 2005), and relative gene expression was calculated by comparison with similarly treated wild-type Col-0 or untreated 35S::BBM-GR (Livak and Schmittgen, 2001). The DNA primers are shown in Supplemental Table S4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. BBM binds to an ANT-like DNA-binding motif.
Supplemental Figure S2. Outcrossing BBM overexpression lines silences the BBM phenotype.
Supplemental Figure S3. Ectopic overexpression of AIL proteins induces somatic embryogenesis and activates LAFL expression.
Supplemental Figure S4. PLT2 ectopic overexpression induces dose-dependent phenotypes.
Supplemental Figure S5. AIL proteins induce context-dependent somatic embryogenesis.
Supplemental Figure S6. BBM expression is reduced in lafl mutant seeds.
Supplemental Table S1. BBM target genes.
Supplemental Table S2. Effects of the lec1 and fus3 mutant backgrounds on BBM-mediated somatic embryogenesis.
Supplemental Table S3. Overlap between BBM targets obtained using microarray and ChIP-seq analyses.
Supplemental Table S4. Oligonucleotide primers used in this study.
Supplemental Data Set S1. BBM target genes.
Acknowledgments
We thank Nirmala Sharma for the val1-2 (hsi2-5) mutant and Bas Dekkers for the abi3-8, abi3-9, abi3-10, and abi5-7 mutants.
Glossary
- ChIP-seq
chromatin immunoprecipitation followed by next-generation sequencing
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative real-time reverse transcription-PCR
- DEX
dexamethasone
- CHX
cycloheximide
- 2,4-D
2,4-dichlorophenoxyacetic acid
- ABA
abscisic acid
- Col-0
Columbia-0
- SLI
stomatal lineage index
- MI
meristemoid index
- SI
stomatal index
Footnotes
This work was funded by grants from the Technology Top Institute-Green Genetics and the Netherlands Organization for Scientific Research (NWO) program, NWO-Groen (project no. 870.15.110).
Articles can be viewed without a subscription.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In R Altman, D Brutlag, P Karp, R Lathrop, D. Searls, eds, Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp 28–36 [PubMed] [Google Scholar]
- Bandupriya HDD, Gibbings JG, Dunwell JM (2014) Overexpression of coconut AINTEGUMENTA-like gene, CnANT, promotes in vitro regeneration in transgenic Arabidopsis. Plant Cell Tiss Org Cult 116: 67–79 [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, et al. (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Harada JJ (2008) LECs go crazy in embryo development. Trends Plant Sci 13: 624–630 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC (2005) ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol Biol 57: 559–575 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Luo KM, Li ZG, Yang YW (2009) A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci 177: 43–48 [Google Scholar]
- Elhiti M, Stasolla C, Wang AM (2013) Molecular regulation of plant somatic embryogenesis. In Vitro Cell Dev Biol Plant 49: 631–642 [Google Scholar]
- El Ouakfaoui S, Schnell J, Abdeen A, Colville A, Labbé H, Han S, Baum B, Laberge S, Miki B (2010) Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol Biol 74: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér A. (2015) Somatic embryogenesis: stress-induced remodeling of plant cell fate. Biochim Biophys Acta 1849: 385–402 [DOI] [PubMed] [Google Scholar]
- Florez SL, Erwin RL, Maximova SN, Guiltinan MJ, Curtis WR (2015) Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol 15: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj MD. (2004) Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul 43: 27–47 [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 133: 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann I, de Lange B, Lambalk J, Angenent GC, Boutilier K (2011) Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep 30: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134: 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman A, Fukuoka H, Muino JM, Nitsch L, Guo C, Passarinho P, Sanchez-Perez G, Immink R, Angenent G, Boutilier K (2015) AIL and HDG proteins act antagonistically to control cell proliferation. Development 142: 454–464 [DOI] [PubMed] [Google Scholar]
- Horstman A, Willemsen V, Boutilier K, Heidstra R (2014) AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends Plant Sci 19: 146–157 [DOI] [PubMed] [Google Scholar]
- Jia H, McCarty DR, Suzuki M (2013) Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol 163: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Suzuki M, McCarty DR (2014) Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip Rev Dev Biol 3: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Degnan KA, Walker JR, Ingram GC (2005) AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J 44: 114–127 [DOI] [PubMed] [Google Scholar]
- Junker A, Mönke G, Rutten T, Keilwagen J, Seifert M, Thi TM, Renou JP, Balzergue S, Viehöver P, Hähnel U, et al. (2012) Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J 71: 427–442 [DOI] [PubMed] [Google Scholar]
- Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, et al. (2015) PLETHORA genes control regeneration by a two-step mechanism. Curr Biol 25: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Soltis PS, Wall K, Soltis DE (2006) Phylogeny and domain evolution in the APETALA2-like gene family. Mol Biol Evol 23: 107–120 [DOI] [PubMed] [Google Scholar]
- Krizek B. (2009) AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol 150: 1916–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. (1999) Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev Genet 25: 224–236 [DOI] [PubMed] [Google Scholar]
- Krizek BA. (2015) AINTEGUMENTA-LIKE genes have partly overlapping functions with AINTEGUMENTA but make distinct contributions to Arabidopsis thaliana flower development. J Exp Bot 66: 4537–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Eaddy M (2012) AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Mol Biol 78: 199–209 [DOI] [PubMed] [Google Scholar]
- Lelu-Walter MA, Thompson D, Harvengt L, Sanchez L, Toribio M, Pâques LE (2013) Somatic embryogenesis in forestry with a focus on Europe: state-of-the-art, benefits, challenges and future direction. Tree Genet Genomes 9: 883–899 [Google Scholar]
- Li H, Soriano M, Cordewener J, Muiño JM, Riksen T, Fukuoka H, Angenent GC, Boutilier K (2014) The histone deacetylase inhibitor trichostatin a promotes totipotency in the male gametophyte. Plant Cell 26: 195–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, et al. (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28: 1998–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Azhagiri A, Maliga P (2011) Transplastomics in Arabidopsis: progress toward developing an efficient method. Methods Mol Biol 774: 133–147 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo F, Gallard A, Pillot M, Jouannic S, Aberlenc-Bertossi F, Collin M, Verdeil JL, Tregear JW (2007) EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta 226: 1353–1362 [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Hong S, Liang Y, Zuo J (2013) Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant 6: 188–201 [DOI] [PubMed] [Google Scholar]
- Mudunkothge JS, Krizek BA (2012) Three Arabidopsis AIL/PLT genes act in combination to regulate shoot apical meristem function. Plant J 71: 108–121 [DOI] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P (2002) A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA (2000) DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res 28: 4076–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Tranby TL, Krizek BA (2005) AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol 57: 613–628 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. [DOI] [PubMed] [Google Scholar]
- Parcy F, Giraudat J (1997) Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J 11: 693–702 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Paek KY (2014) Bioreactor culture of shoots and somatic embryos of medicinal plants for production of bioactive compounds. In N Murthy, JJ Zhong, KY Paek, eds, Production of Biomass and Bioactive Compounds Using Bioreactor Technology. Springer, Dordrecht, The Netherlands, pp 337–368 [Google Scholar]
- Passarinho P, Ketelaar T, Xing M, van Arkel J, Maliepaard C, Hendriks MW, Joosen R, Lammers M, Herdies L, den Boer B, et al. (2008) BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol 68: 225–237 [DOI] [PubMed] [Google Scholar]
- Peterson KM, Shyu C, Burr CA, Horst RJ, Kanaoka MM, Omae M, Sato Y, Torii KU (2013) Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider SD Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KW, Schier AF (2011) Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol 27: 377–407 [DOI] [PubMed] [Google Scholar]
- Roscoe TT, Guilleminot J, Bessoule JJ, Berger F, Devic M (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol 56: 1215–1228 [DOI] [PubMed] [Google Scholar]
- Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L, Gorte M, Prasad K, Bao D, Timmermans-Hereijgers JL, et al. (2016) The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28: 2937–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Bender Y, Boyle K, Fobert PR (2013a) High-level expression of sugar inducible gene2 (HSI2) is a negative regulator of drought stress tolerance in Arabidopsis. BMC Plant Biol 13: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Shahzad A, Teixeira da Silva JA (2013b) Synseed technology: a complete synthesis. Biotechnol Adv 31: 186–207 [DOI] [PubMed] [Google Scholar]
- Soriano M, Li H, Jacquard C, Angenent GC, Krochko J, Offringa R, Boutilier K (2014) Plasticity in cell division patterns and auxin transport dependency during in vitro embryogenesis in Brassica napus. Plant Cell 26: 2568–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram S, Kertbundit S, Shakirov EV, Iyer LM, Jurícek M, Hall TC (2013) Gene networks and chromatin and transcriptional regulation of the phaseolin promoter in Arabidopsis. Plant Cell 25: 2601–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Wang HH, McCarty DR (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Yokoi S, Takahata Y (2010) Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol Biol 73: 481–492 [DOI] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol 51: 2031–2046 [DOI] [PubMed] [Google Scholar]
- Yang HF, Kou YP, Gao B, Soliman TMA, Xu KD, Ma N, Cao X, Zhao LJ (2014) Identification and functional analysis of BABY BOOM genes from Rosa canina. Biol Plant 58: 427–435 [Google Scholar]
- Yano R, Kanno Y, Jikumaru Y, Nakabayashi K, Kamiya Y, Nambara E (2009) CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol 151: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J (2012) The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol 159: 418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu CW, Yang S, Dong S, Ruan J, et al. (2013) HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25: 134–148 [DOI] [PMC free article] [PubMed] [Google Scholar]