A conifer glycosyltransferase is involved in the chemical resistance against spruce budworm, a major forest defoliator.
Abstract
Acetophenones are phenolic compounds involved in the resistance of white spruce (Picea glauca) against spruce budworm (Choristoneura fumiferiana), a major forest pest in North America. The acetophenones pungenol and piceol commonly accumulate in spruce foliage in the form of the corresponding glycosides, pungenin and picein. These glycosides appear to be inactive against the insect but can be cleaved by a spruce β-glucosidase, PgβGLU-1, which releases the active aglycons. The reverse glycosylation reaction was hypothesized to involve a family 1 UDP-sugar dependent glycosyltransferase (UGT) to facilitate acetophenone accumulation in the plant. Metabolite and transcriptome profiling over a developmental time course of white spruce bud burst and shoot growth revealed two UGTs, PgUGT5 and PgUGT5b, that glycosylate pungenol. Recombinant PgUGT5b enzyme produced mostly pungenin, while PgUGT5 produced mostly isopungenin. Both UGTs also were active in vitro on select flavonoids. However, the context of transcript and metabolite accumulation did not support a biological role in flavonoid metabolism but correlated with the formation of pungenin in growing shoots. Transcript levels of PgUGT5b were higher than those of PgUGT5 in needles across different genotypes of white spruce. These results support a role of PgUGT5b in the biosynthesis of the glycosylated acetophenone pungenin in white spruce.
The spruce budworm (SBW; Choristoneura fumiferiana) is one of the most destructive forest insect pests in North America. The foliage-feeding larvae of this lepidopteran insect affect several different conifer species, including white spruce (Picea glauca) as one of its major hosts. Outbreaks of SBW can change forest landscapes and ecosystems for decades as a consequence of canopy defoliation, which, in turn, can cause major economic losses to the affected regions (Chang et al., 2012; MacLean, 2016). SBW outbreaks are expected to increase in frequency and severity under conditions of climate change (Gray, 2013). Recently, a natural resistance phenotype of white spruce to SBW was identified in a population of trees in eastern Canada. Metabolite analysis showed that the presence of two phenolic compounds, the acetophenones pungenol (3,4-dihydroxyacetophenone) and piceol (4-hydroxyacetophenone), in white spruce foliage contributes to the resistance against SBW (Delvas et al., 2011; Parent et al., 2017). In the foliage of susceptible trees, the two acetophenones were detectable only in the form of the corresponding glycosides, pungenin and picein, which appear to be inactive against SBW. In contrast, the foliage of resistant trees also contained the biologically active aglycons pungenol and piceol.
The glycosylation of small molecules, which may modify their solubility, localization, and biological activity, is a common process in plant metabolism. For example, small molecules such as flavonoids, terpenoids, and benzoates are glycosylated by enzymes of the family 1 UDP glycosyltransferases (UGTs; Osmani et al., 2009). These UGTs comprise a diverse group of enzymes, which all contain a highly conserved 44-amino acid plant secondary product glycosyltransferase (PSPG) motif. The PSPG motif is important for interactions with the nucleotide-diphosphate-sugar (Gachon et al., 2005). Current knowledge of family 1 UGTs in plants is mostly based on work in angiosperms. In annotated dicot genomes, family 1 UGTs are present, with 85 to 241 members, while in monocot genomes, the size of this gene family may be even larger, with 147 to 730 members (Caputi et al., 2012). Members of the family 1 UGTs show high variability in their substrate specificity. Some of these enzymes glycosylate a broad range of acceptor molecules, while other UGTs act on only one or a few substrates (Osmani et al., 2009). Despite their large differences in acceptor preference and low sequence conservation, UGTs are conserved in their secondary and tertiary structures. The structure consists of two Rossmann fold domains joined by a linker. The N-terminal domain interacts with the acceptor, while the C-terminal domain contains the PSPG motif (Osmani et al., 2009). Substrate promiscuity can make it difficult to determine biologically relevant in vivo functions of UGTs (Song et al., 2015).
To our knowledge, only two different family 1 UGTs have been characterized from gymnosperms, the coniferyl alcohol UGTs from Norway spruce (Picea abies) and pine (Pinus strobus) and a Norway spruce flavonol UGT (Schmid and Grisebach, 1982; Heilemann and Strack, 1991; Steeves et al., 2001). Research on UGTs in gymnosperms has been limited due to the lack of genetic and genomic resources and the difficulty in recombinant expression of the encoded proteins. Recent advances in the development of genome and transcriptome resources for white spruce, a conifer, have enabled our ability to better explore this gene family in the background of a large gymnosperm genome (Birol et al., 2013; Warren et al., 2015).
In recent previous work on the acetophenones involved in white spruce resistance to SBW, we characterized the β-glycosidase PgβGLU-1, which releases the aglycons pungenol and piceol from their glycosides, pungenin and picein (Mageroy et al., 2015). Here, we describe the discovery and characterization of a counteracting acetophenone UGT that can produce the acetophenone glycoside pungenin.
RESULTS
UGT Discovery and Candidate Selection in Spruce Transcriptomes
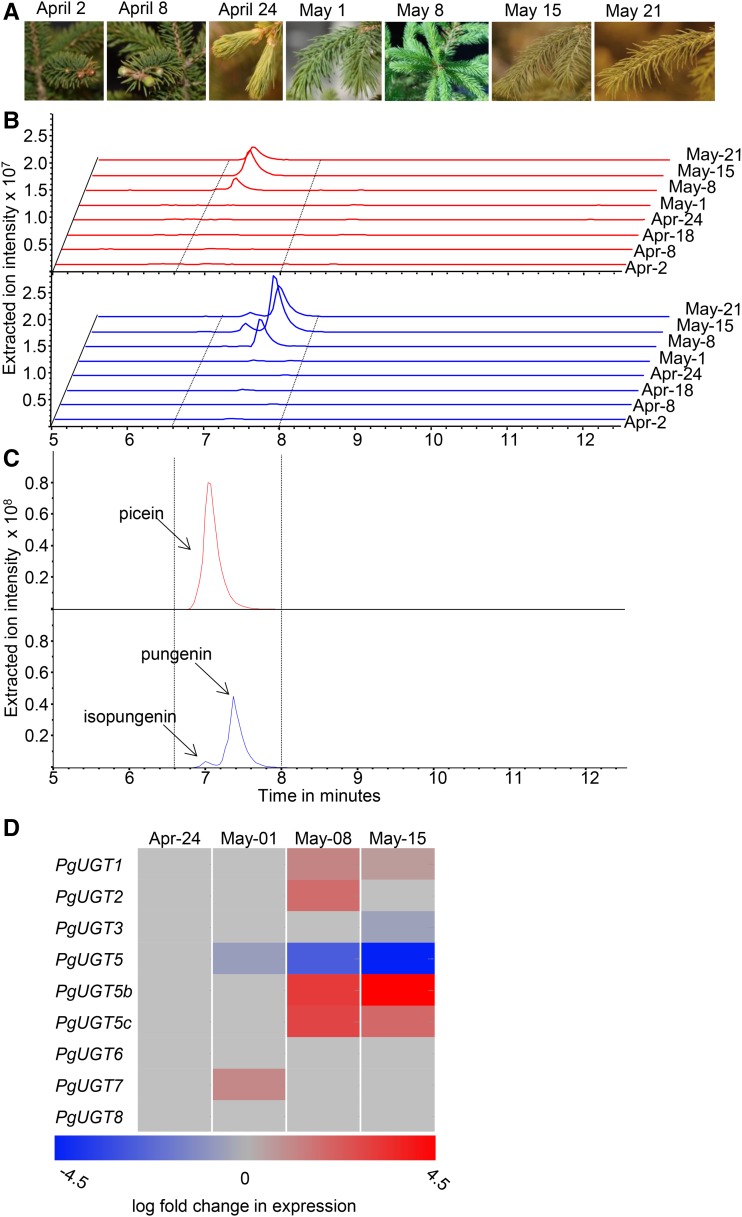
Previous work showed that the acetophenones pungenol and piceol, as well as their corresponding glycosides pungenin and picein, occur in foliage of white spruce genotypes resistant to SBW, while only the glycosides were detected in susceptible trees. The formation of these metabolites did not appear to be inducible by herbivore attack but may be under developmental control (Mageroy et al., 2015). We hypothesized that candidate UGTs with a role in acetophenone metabolism could be identified if their expression was correlated with the time course of acetophenone glycoside accumulation. To test this hypothesis, we analyzed metabolite and transcriptome profiles over a time course of shoot development during the early growing season. Samples were collected from a clonally propagated white spruce line (Pg653) at eight time points with weekly intervals and four biological replicates per time point (Fig. 1). The acetophenone glycosides pungenin and picein, as well as traces of isopungenin, became detectable at the May 8 time point, which was 4 to 5 weeks after the observed start of bud flush (Fig. 1).
Figure 1.
Profiles of acetophenone glycosides and PgUGT transcripts over the time course of early shoot development aided the selection of acetophenone UGT candidates. A, Samples were collected over the time course of shoot development beginning with the onset of bud burst until full shoot elongation. B, Liquid chromatography-mass spectrometry (LC-MS) extracted ion chromatograms (EIC) showed no detectable amounts of acetophenones prior to the May 8 time point, 4 to 5 weeks after the onset of bud flush. Picein is shown with the traces in red (EIC +137). Isopungenin and pungenin are shown with the traces in blue (EIC −313). C, The identities of acetophenones were confirmed by comparison with the LC-MS spectra of purified standards. D, The relative transcript abundance of PgUGTs over select time points of early shoot development was used to select candidate acetophenone UGTs. The heat map shows changes of transcript expression for each candidate PgUGT based on comparison of the transcript abundance at the three time points, May 1, May 8, and May 15, relative to the transcript abundance at the April 24 time point. Transcript abundance at the April 24 time point was set as the baseline for differential expression (DE) analysis. The time-course profile of PgUGT5b transcript accumulation was most similar to the time course of the accumulation profile of the acetophenone glycosides.
The transcriptomes of foliage collected at four time points were sequenced on the Illumina HiSeq platform with four biological replicates per time point. Time points were selected to cover 1 week (T1, April 24) and 2 weeks (T2, May 1) before the appearance of acetophenone glycosides and the first 2 weeks (T3, May 8; and T4, May 15) of acetophenone glycoside detection. Transcriptomes were analyzed for signatures of differential expression (DE) over the time course relative to transcript abundance at T1 (Supplemental Fig. S1). In total, 105, 1,465, and 3,803 transcripts were DE in T2 versus T1, T3 versus T1, and T4 versus T1, respectively, with an absolute log2 fold change ≥ 2 and adjusted P ≤ 0.01. To identify candidate UGTs, the data set of DE transcripts was screened for the PSPG motif (PROSITE no. PS00375), which identified 203 transcripts with greater than 98% amino acid identity in the conserved UGT motif. These candidate UGTs were screened further for sequences that had significant amino acid similarity (greater than 50%) to known angiosperm phenolic UGTs and hydroquinone UGTs, as such UGTs may contain active sites that can accommodate acetophenone binding. Transcripts with similarity to arbutin synthase were of particular interest, because this hydroquinone represents a simple benzene diol similar in structure to the spruce acetophenones.
To prioritize UGT candidates for functional characterization, we selected those with expression patterns similar to the pattern of increase of pungenin and picein across the time points T2 to T4 (Fig. 1). In total, seven candidate UGT transcripts were selected and were designated as PgUGT1, PgUGT2, PgUGT3, PgUGT5, PgUGT6, PgUGT7, and PgUGT8. For these UGTs, we assessed the expression in different organs and tissues using a BLASTN search against published white spruce transcriptomes for flushing buds, young buds, mature needles, xylem, and bark (Birol et al., 2013). All of the transcripts with matches of over 95% identity to PgUGT2, PgUGT3, PgUGT5, and PgUGT6 came from flushing bud, young bud, or mature needle transcriptomes (Supplemental Table S1). PgUGT1, PgUGT7, and PgUGT8 did not appear to have a preferential expression in young foliage (flushing and young buds) or mature needles. Next, we searched for sequences closely related to PgUGT1, PgUGT2, PgUGT3, PgUGT5, PgUGT6, PgUGT7, and PgUGT8 in organ- or tissue-specific transcriptomes of the related conifer Norway spruce (http://congenie.org), which included needles, stems, cones, wood, infected needles, galls, and girdled twigs (Nystedt et al., 2013). We considered transcripts with more than 98% nucleotide sequence identity to be the homologous Norway spruce genes corresponding to the PgUGTs. All homologs found in Norway spruce were most abundant in needle or shoot tissue, except for the PgUGT7 homolog, which was most abundant in galls (Supplemental Table S2). Overall, these results supported the preferential expression of most of our candidate gene set in foliage, which is the site of acetophenone accumulation.
UGT Full-Length cDNAs and Protein Expression in Escherichia coli
Full-length UGT cDNAs were amplified using RNA from Pg653 young shoots harvested 5 weeks after the beginning of bud flush. Upon sequencing of the PgUGT5 amplicons, we found three different genes that were amplified using the same primers, which were named PgUGT5, PgUGT5b, and PgUGT5c. Although they were amplified with the same primers, the three sequences shared less than 78% amino acid sequence identity (Supplemental Fig. S2). A total of nine candidate acetophenone UGT cDNAs, PgUGT1, PgUGT2, PgUGT3, PgUGT5, PgUGT5b, PgUGT5c, PgUGT6, PgUGT7, and PgUGT8, ranging from 1,395 to 1,515 nucleotides in length, were used for recombinant protein expression and enzyme characterization. The corresponding naming of these genes is according to the UDP Glucuronosyltransferase Nomenclature Committee (http://www.flinders.edu.au/medicine/sites/clinical-pharmacology/ugt-homepage.cfm; Supplemental Table S3). The cDNAs were cloned into a low-copy-number vector with a tetracycline-inducible promoter for E. coli expression. Protein expression was performed in C43 (DE3) cells containing the rare codon t-RNA plasmid pRARE. Proteins were purified using an N-terminal His tag and verified by SDS-PAGE and western blots showing protein expression in E. coli for all UGTs except for PgUGT8 (Supplemental Fig. S3).
Identification of UGTs That Are Active in the Glycosylation of the Acetophenone Pungenol
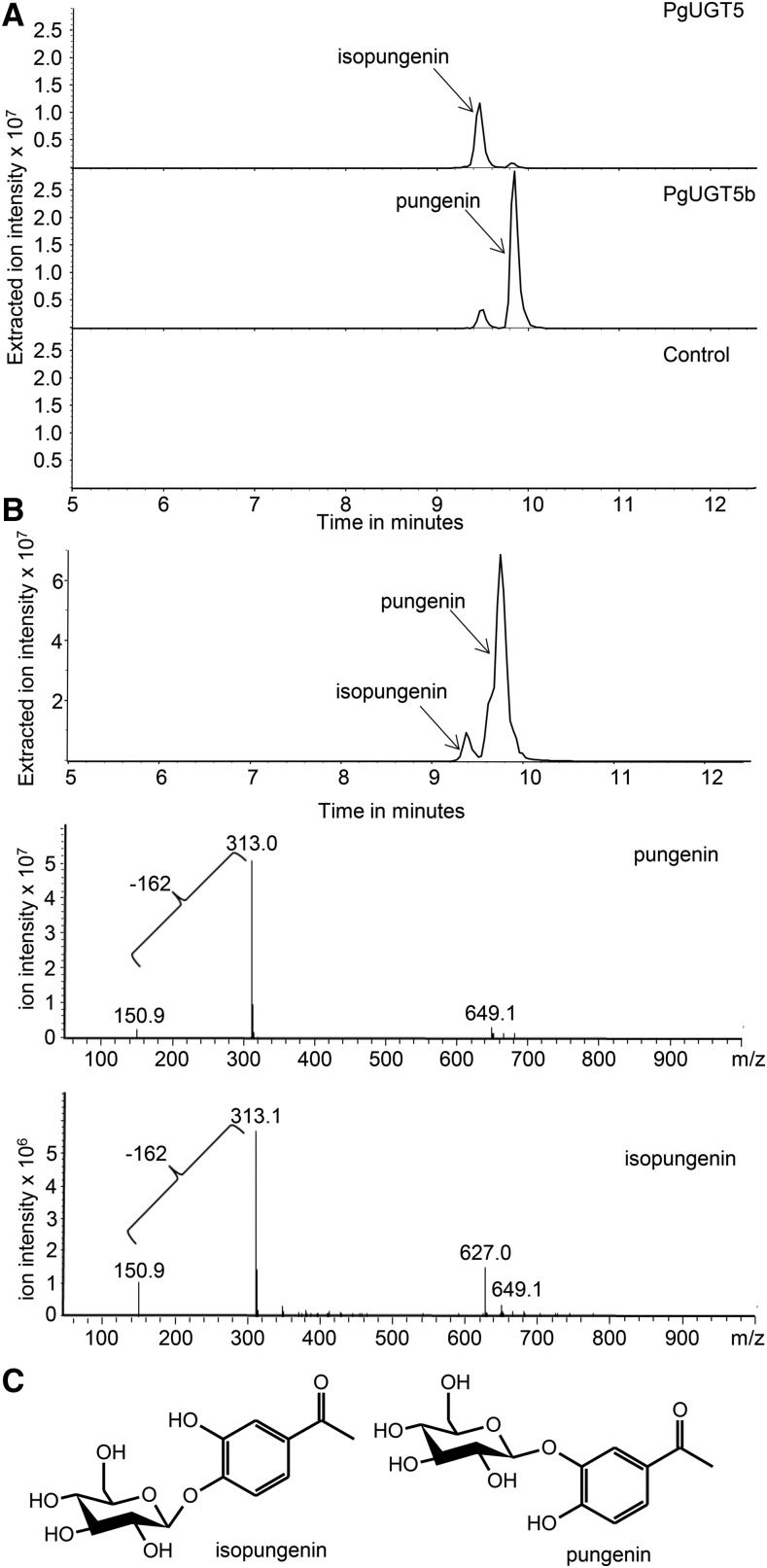
We performed enzyme assays with the expressed UGT proteins using UDP-Glc as the sugar donor and the acetophenone aglycons pungenol or piceol as acceptor substrates. These assays showed that PgUGT5 and PgUGT5b were active in the glycosylation of pungenol (Fig. 2), while the remaining UGTs were not active. None of the candidate UGTs showed activity with piceol. In assays with other phenylpropanoids, PgUGT1, PgUGT6, and PgUGT7 glycosylated the substrate quercetin (as did PgUGT5 and PgUGT5b), indicating that these three enzymes are active, but they apparently lack the ability to use pungenol or piceol as substrates (Supplemental Fig. S3). PgUGT5 and PgUGT5b showed different regioselectivity in the glycosylation of the hydroxyl groups of pungenol. PgUGT5 preferentially glycosylated the 4-hydroxyl group producing mostly isopungenin, while PgUGT5b preferentially glycosylated the 3-hydroxyl group producing mostly pungenin (Fig. 2). The pungenin and isopungenin products were verified using chemically synthesized authentic standards, which were confirmed by NMR.
Figure 2.
In vitro enzyme assays demonstrated the glucosyltransferase activity of PgUGT5 and PgUGT5b with pungenol as the substrate and preferential formation of isopungenin and pungenin, respectively. A, Extracted ion chromatograms from the LC-MS analyses of products of an in vitro enzyme assay performed with purified PgUGT5 and PgUGT5b and pungenol as the substrate. PgUGT5 predominantly glucosylated the 4-hydroxy group on the aromatic ring, producing isopungenin. PgUGT5b predominantly glucosylated the 3-hydroxy group, producing pungenin. B, Extracted ion chromatograms (−313) and mass spectra of authentic isopungenin and pungenin standards used to confirm the products of an in vitro enzyme assay performed with purified PgUGT5 and PgUGT5b. The bar showing the loss of −162 represents the loss of the Glc from pungenin or isopungenin. C, Structures of isopungenin and pungenin showing the difference in glucosylation position.
Substrate Specificity of PgUGT5 and PgUGT5b
Many UGTs are known to glycosylate multiple different substrates. To test the specificity of PgUGT5 and PgUGT5b for pungenol, the purified enzymes were tested for the preference of pungenol over other phenylpropanoid substrates using competition assays, where pungenol was used as a substrate in combination with (1) piceol, caffeic acid, and coumaric acid, (2) kaempferol, naringenin, and acetovanillone, or (3) resveratrol, quercetin, and 4-hydroxybenzoic acid. In these assays, PgUGT5 and PgUGT5b showed glycosylation activity only with kaempferol and quercetin in addition to pungenol (Supplemental Table S4). Both PgUGT5 and PgUGT5b produced a single quercetin glucoside, while three different products were formed with kaempferol by both enzymes in different ratios.
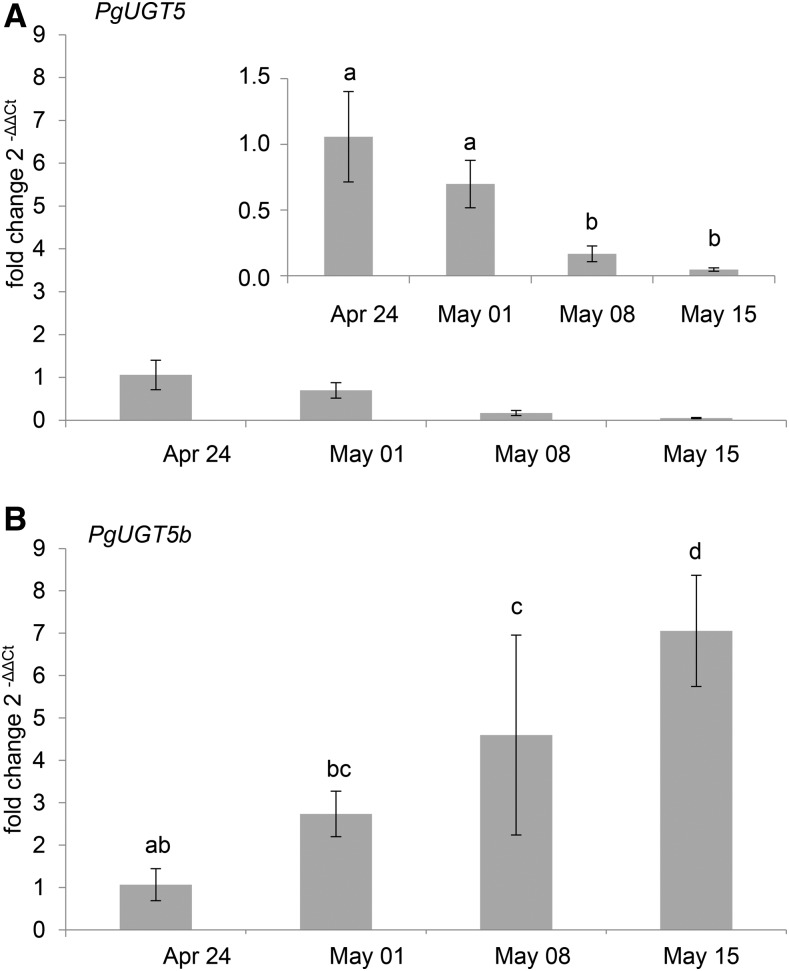
To assess the biological relevance of the observed activity of PgUGT5 and PgUGT5b with quercetin and kaempferol, we analyzed white spruce foliage samples over a time course of shoot development for the presence of quercetin and kaempferol glycosides and compared these with the time-course profiles of acetophenone glycosides and PgUGT5 and PgUGT5b transcripts. Acetophenone glycosides showed a developmental time course with an onset of accumulation during the early growing season (Fig. 1). Similarly, qRT-PCR analysis showed that PgUGT5b transcript abundance increased over the time course of shoot development (Fig. 3). In contrast, quercetin and kaempferol glycosides were already present early in shoot development prior to the increase of the PgUGT5b transcript levels (Supplemental Fig. S4). These results do not support a major role of PgUGT5b in the accumulation of quercetin and kaempferol glycosides in planta, since the accumulation of flavonoid glycosides preceded the increase of PgUGT5b transcript abundance. In contrast to PgUGT5b, transcript levels of PgUGT5 decreased over the time course of early shoot development (Fig. 3). Taken together, these results support a role of PgUGT5b in the glycosylation of pungenol in planta and do not support a biologically relevant role for PgUGT5b in the glycosylation of the alternative flavonoid substrates.
Figure 3.
qRT-PCR analysis of PgUGT5 and PgUGT5b transcript abundance showed DE over the time course of early shoot development. Total RNA extracted from tissue collected over the time course of early shoot development was used to determine the relative abundance of PgUGT5 and PgUGT5b transcripts. Transcript abundance detected for each time point was compared with the abundance at the April 24 time point. Results shown for each transcript and time point represent data from four biological replicates each with two technical replicates. Error bars represent sd. A, Transcript abundance of PgUGT5 decreased over the time course of early shoot development. The inset shows PgUGT5 expression on a smaller scale. B, Transcript abundance of PgUGT5b increased over the time course of shoot development. A two-way ANOVA followed by Tukey’s HSD test was used to determine significant differences (P < 0.01), indicated by the use of different letters.
In Planta Assessment of PgUGT5 and PgUGT5b
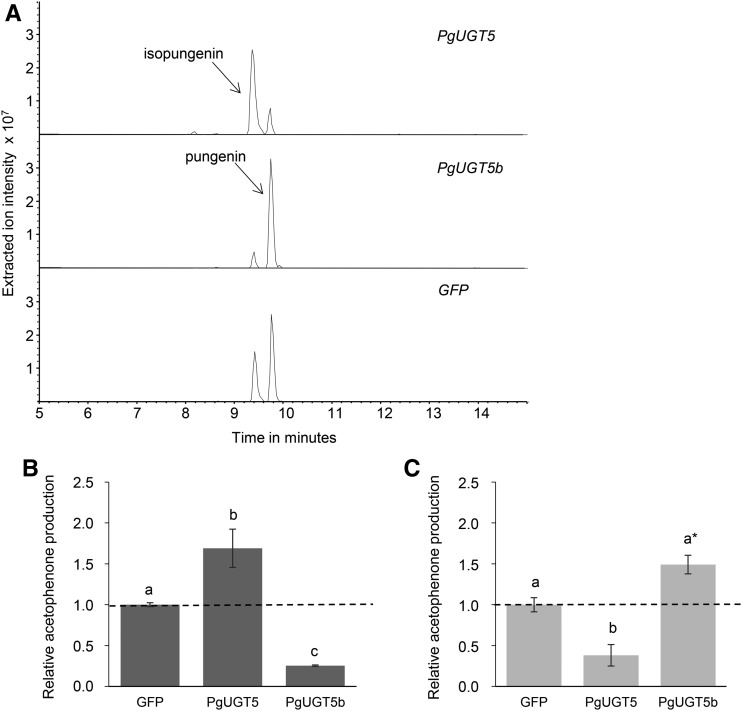
To further assess the functions of PgUGT5 and PgUGT5b in planta, we transformed 4-week-old Nicotiana benthamiana plants with plasmids for the transient expression of PgUGT5 and PgUGT5b. After 6 d, transformed leaves were infiltrated with pungenol and harvested after another 4 h for metabolite analysis. A baseline endogenous glycosylation of pungenol was observed in N. benthamiana plants that transiently expressed GFP as a control (Fig. 4). In agreement with results from the in vitro enzyme assays, transformation with PgUGT5 resulted in the preferential glycosylation of the 4-hydroxyl group of pungenol to produce isopungenin in planta, while transformation with PgUGT5b resulted in the preferential glycosylation of the 3-hydroxyl group to produce pungenin in planta (Fig. 4). When normalized to the endogenous glycosyltransferase baseline activity observed in GFP-transformed plants, transformation with PgUGT5 resulted in increased levels of isopungenin and a relative decrease in pungenin, while PgUGT5b had the opposite effect (Fig. 4).
Figure 4.
Pungenol glycosylation activity in planta demonstrated that PgUGT5 and PgUGT5b function as pungenol glycosyltransferases when expressed in N. benthamiana. Leaves of 4-week-old N. benthamiana plants were transiently transformed for PgUGT5, PgUGT5b, or GFP (control) expression. Six days following the transformation, leaves were infiltrated with purified pungenol. Four hours after pungenol infiltration, metabolites were extracted and analyzed for acetophenones by LC-MS. A, Extracted ion chromatograms (−313) of glucosylated pungenol showed that plants transiently transformed for PgUGT5 expression accumulated higher levels of isopungenin, while plants transiently transformed for PgUGT5b expression accumulated higher levels pungenin, relative to the background profile of these metabolites in plants transiently transformed for GFP expression. B and C, Amounts of isopungenin and pungenin produced in plants transiently transformed for PgUGT5 or PgUGT5b expression relative to their amounts produced in plants transiently transformed for GFP expression. The dashed lines represent the background levels of isopungenin and pungenin produced in plants transiently transformed for GFP expression due to endogenous N. benthamiana glycosylation activity. Error bars represent se of results obtained with four individual plants for each of the three different transformations (PgUGT5, PgUGT5b, or GFP). For each transiently transformed plant, three pungenol-infiltrated leaves were assayed. A two-way ANOVA followed by Tukey’s HSD test was used to determine significant differences (P < 0.05), indicated by the use of different letters. The asterisk in C denotes that P < 0.06.
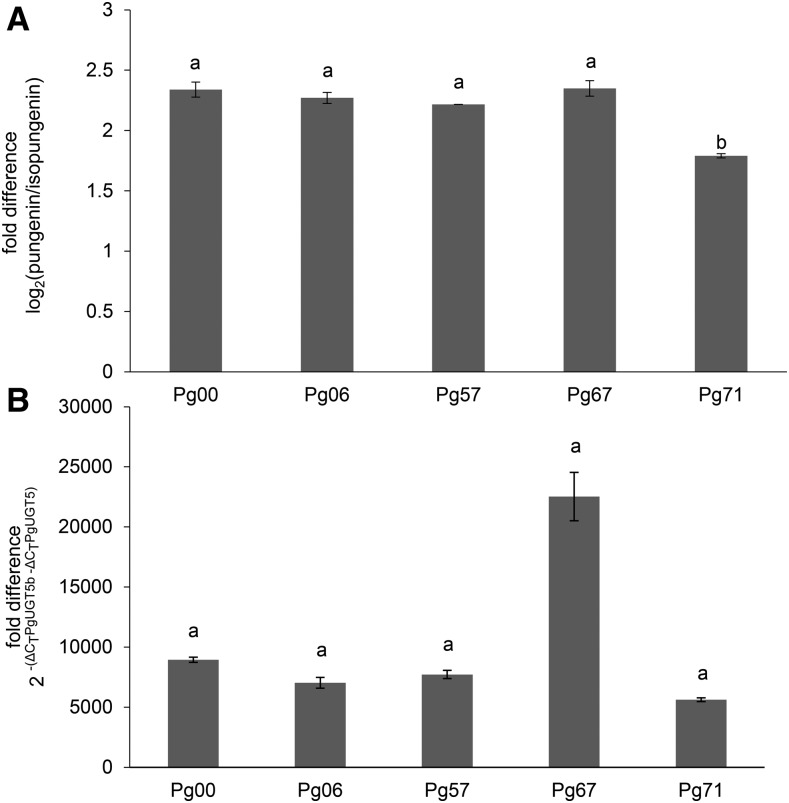
We also assessed the relative levels of pungenin and isopungenin in foliage of five different genotypes of white spruce and found pungenin consistently to be the more abundant of these two different pungenol glycosides (Fig. 5). Consistent with the higher ratio of pungenin compared with isopungenin, the PgUGT5b transcript also was more abundant than PgUGT5 (Fig. 5). The relatively higher abundance of pungenin and PgUGT5b in different white spruce genotypes agreed with the UGT products observed in the in vitro and in planta characterization of recombinant PgUGT5 and PgUGT5b.
Figure 5.
Across different genotypes of white spruce, a predominance of pungenin relative to isopungenin in foliage was consistent with the predominance of PgUGT5b relative to PgUGT5b transcript levels. A, Fold difference of pungenin to isopungenin detected in the foliage of each genotype. B, Fold difference in the abundance of PgUGT5b to PgUGT5 transcripts in the foliage of each genotype. Error bars represent se (n = 2). A two-way ANOVA followed by Tukey’s HSD test was used to determine significant differences (P < 0.05), indicated by the use of different letters. The apparent difference between Pg67 and the other lines in the relative abundance of PgUGT5b to PgUGT5 could not be confirmed as being statistically significant due to the limited number of replicates.
DISCUSSION
The reversible glycosylation of secondary metabolites is a common process in plant metabolism to inactivate and sequester reactive molecules, such as those that act as chemical defenses against pathogens or herbivores. UGTs involved in these processes have been characterized in several angiosperms, but very few UGTs involved in the glycosylation of small molecules have been described in gymnosperms. The recent development of genome and transcriptome assemblies for white spruce (Birol et al., 2013; Warren et al., 2015) made it possible to define large gene families, such as UGTs, in the extremely large space of an ∼20-Gb conifer genome. However, functional annotation of UGTs remains a major challenge.
Here, we were specifically focused on the discovery of UGT(s) that could glycosylate acetophenones that are involved in the resistance of white spruce against SBW. We first used a combination of metabolite profiling of acetophenone glycosides and analysis of UGT transcript expression profiles over the time course of shoot development as well as spatial patterns of UGT expression in different parts. This approach revealed a very large number of DE transcripts, which included over 200 (specifically, 203) candidate UGTs based on the presence of the PSPG motif. Importantly, this large number of DE UGTs was then reduced further to a number of high-priority candidates based on amino acid similarity (greater than 50%) to known angiosperm phenolic UGTs and hydroquinone UGTs. This critical step narrowed the number of candidates for functional characterization from over 200 different white spruce UGT transcripts that were detected by transcriptome analysis to nine PgUGTs. The identified candidates showed DE with increasing transcript abundance in shoots and foliage early in the growing season, preceding the accumulation of acetophenone glycosides, and preferential expression in foliage. Out of the nine candidates, only PgUGT5 and PgUGT5b were able to glycosylate the acetophenone pungenol in both in vitro and in planta assays. Other cloned PgUGTs were active with the flavonoid quercetin, but not with pungenol or piceol. Therefore, we concluded that the use of pungenol as a substrate is a specific feature of some PgUGTs, such as PgUGT5 and PgUGT5b, but is not a general function of all PgUGTs that can glycosylate phenylpropanoids. Additional PgUGT(s) that are active on piceol remain to be discovered.
Of the two pungenol PgUGTs, PgUGT5 produced mostly isopungenin, which is glycosylated at the 4-hydroxy position, while PgUGT5b produced mostly pungenol, which is glycosylated at the 3-hydroxy position. In white spruce foliage, pungenin is much more abundant than isopungenin, indicating that PgUGT5b is the more biologically prevalent pungenol UGT. Additionally, the transcript expression pattern of PgUGT5b matches the temporal pattern of pungenin appearance in developing shoots, increasing over development.
Many plant UGTs are promiscuous enzymes that accept multiple different substrates (Caputi et al., 2012). Therefore, it is not surprising that PgUGT5 and PgUGT5b also glycosylated substrates other than pungenol in vitro. The substrate promiscuity of plant UGTs also may explain our observation of pungenol glycosylation in N. benthamiana control plants injected with this compound, despite the fact that pungenol does not appear to occur naturally in detectable amounts in this plant. However, the in vitro activity of UGTs with multiple substrates may not fully reflect the in planta function of the enzyme, which also is affected directly by substrate availability (Song et al., 2015). This is likely the case with PgUGT5b. Although PgUGT5b had activity on kaempferol and quercetin, glycosylation of these flavonoids does not appear to be its primary function, as these flavonoid glycosides are present throughout shoot development, whereas PgUGT5b transcript expression and the accumulation of pungenin showed a developmental increase. We substantiated the roles of PgUGT5 and PgUGT5b in the formation of the pungenol glycosides isopungenin and pungenin using transient expression in N. benthamiana. This heterologous in planta assay system was used instead of a homologous system, because transgenic spruce would take several years to generate and observations in transgenic conifers can be ambiguous partly due to limitations in our understanding of gene expression and metabolic systems in these nonmodel species (Nagel et al., 2014).
Comparison of pungenin and isopungenin in five genotypes of white spruce showed that pungenin is the predominant isoform of glycosylated pungenol in the foliage. In the same five genotypes, PgUGT5b transcripts were much more abundant than PgUGT5 transcripts. The finding that pungenol is the predominant pungenol glucoside and PgUGT5b is the predominant pungenol glucosyltransferase in planta supports the identification of PgUGT5b as the most biologically relevant pungenol glucosyltransferase. During the time course of shoot development in the genotype Pg653, only pungenin was observed and not the pungenol aglycon. The ready glycosylation of pungenol may indicate that the formation of acetophenone glycosides is important for rapidly sequestering the potentially biologically more active acetophenone aglycon. The aglycon appears only in resistant white spruce genotypes that express the PgβGLU1 gene, the product of which acts as the glycosidase that cleaves acetophenone glycosides (Mageroy et al., 2015).
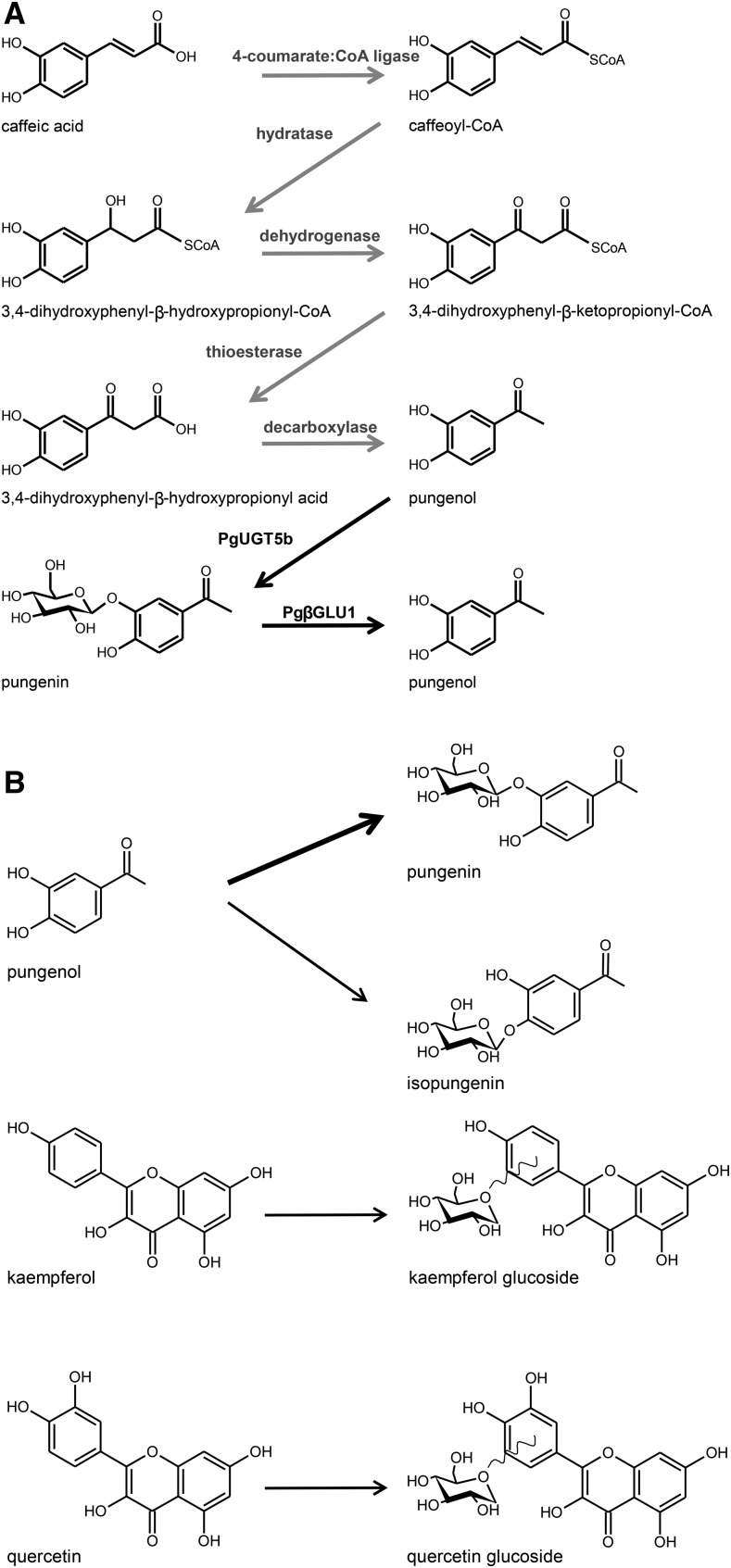
PgUGT5b represents the final step in the pungenin biosynthesis pathway in white spruce (Fig. 6). Its identification has filled a gap in our understanding of acetophenone metabolism in conifer defense and in plants in general (Negrel and Javelle, 2010). To the best of our knowledge, PgUGT5b is the first characterized UGT of plant defense metabolism that has been identified in a gymnosperm system.
Figure 6.
The proposed biosynthetic pathway of pungenol and the enzymatic activity of PgUGT5b. A, Proposed biosynthetic pathway of pungenol from caffeic acid based on feeding experiments with 14C-labeled precursors in spruce (Neish, 1959) and based on a predicted biosynthetic pathway for acetovanillone (Negrel and Javelle, 2010). The gray arrows represent proposed enzyme-catalyzed reactions that remain to be confirmed. The enzyme activity of PgβGLU1 was described previously by Mageroy et al. (2015). B, PgUGT5b utilizes pungenol, quercetin, and kaempferol as substrates. Pungenin appears to be the major product of PgUGT5b, as indicated by the thick black arrow. Isopungenin, quercetin glucosides, and kaempferol glucosides are additional products of PgUGT5b. The undulating line indicates that the exact position of glucosylation is not know for the quercetin glucosides or the kaempferol glucosides.
MATERIALS AND METHODS
Plant Materials
Clonally propagated white spruce (Picea glauca) genotype Pg653 was used for the shoot and foliage development time course (Klimaszewska et al., 2001). Four-year-old potted trees were moved into a greenhouse at the end of the winter dormancy period to induce bud flushing. Flushing shoots were collected once per week from undamaged trees with four replicates per time point over the time course of 7 weeks. In addition, foliage from five different white spruce genotypes, previously described by Mageroy et al. (2015), was collected from a plantation in Saint-Cyrille-de-Wendover, Quebec, Canada (45°930N, 72°520W).
Reagents
Pungenol (3,4-dihydroxyacetophenone) was purchased from TCI America. Kaempferol and quercetin dihydrate were purchased from TRC. 4-Hydroxybenzoic acid was purchased from Chromedex. All other reagents were purchased from Sigma-Aldrich.
Synthesis of Pungenin and Isopungenin Standards
Solvents and chemicals were of analytical grade and were purchased from Sigma‐Aldrich. Acetone was dried over molecular sieves. Analytical thin‐layer chromatography (TLC) was performed on Merck Silica Gel 60F254 (0.2 mm thickness on aluminum). TLC plates were visualized using UV light (254 or 365 nm), by immersion in 10% ammonium molybdate in 2 m H2SO4 or by an aqueous KMnO4 solution. 2,3,4,6-Tetra-O-acetyl-α-d-glucopyranosyl bromide (0.41 g, 1 mmol) was dissolved in 6 mL of CH2Cl2, then tetrabutylammonium bromide (0.64 g, 2 mmol) and 3,4-dihydroxyacetophenone (157 mg, 1.05 mmol) were added (Kröger and Thiem, 2007). After dissolution, 3 mL of a 1 m NaOH solution was added, and the biphasic mixture was stirred vigorously until the donor was consumed (4 h, 45 min). After phase separation, the aqueous phase was extracted two times with ethyl acetate. The combined organic phases were washed with brine, dried over MgSO4, and evaporated under reduced pressure. The crude product was purified by a silica gel column eluting with a gradient of CH2Cl2/ethyl acetate (9:1→2:1), yielding diglucosylated acetophenone (90 mg) and a mixture (88 mg) of 3,4-dihydroxyacetophenone, acetylated pungenin, and acetylated isopungenin. This complex mixture was acetylated under standard conditions. After dissolving in 2 mL of CH2Cl2, 100 µL of pyridine and 100 µL of acetic anhydride were added and the reaction mixture was stirred overnight. After evaporation under reduced pressure, the crude product was purified by another silica column eluting with a gradient of CH2Cl2/acetone (100:1→10:1), yielding 3,4-di-O-acetyl acetophenone (55 mg) and an inseparable mixture (49 mg) of per-O-acetylated pungenin and isopungenin. This inseparable mixture (49 mg) was dissolved in dry methanol (MeOH; 2 mL), and a 1 m sodium methoxide solution (20 µL) was added. The reaction mixture was stirred until analytical TLC monitoring indicated the consumption of starting materials. TLC was performed on Merck Silica Gel 60F254 (0.2 mm thickness on aluminum). TLC plates were visualized using UV light (254 or 365 nm) by immersion in 10% ammonium molybdate in 2 m H2SO4 or by an aqueous KMnO4 solution. Amberlyst 120 H+ was then added and filtered off after 5 min (pH ∼6–7), and the solvent was evaporated under reduced pressure. The crude product was purified by a silica gel column eluting with a gradient of CH2Cl2/MeOH (8:1→4:1), yielding a mixture (19 mg) of isopungenin and pungenin.
Chromatographic Separation of Synthetic Isopungenin and Pungenin
The HPLC system consisted of an Agilent Technologies quaternary solvent delivery pump (G1311A), an Agilent Technologies autosampler (G1313A), and a J&M Analytik photodiode array detector (detection at 200–700 nm). An LiChrospher 100 RP-18 liquid chromatography column (250 × 4.6 mm, 5-µm particle size; Merck) was used for the separation of isopungenin and pungenin with solvent A (water containing 0.1% formic acid) and solvent B (MeOH containing 0.1% formic acid). The following gradient was used: increase from 5% to 95% solvent B from 0 to 35 min; hold at 95% solvent B from 35 to 42 min; and decrease from 95% to 5% solvent B from 42 to 45 min. A prerun equilibration of 5 min was used. The flow rate was 1 mL min−1. The chromatography resulted in two peaks with retention times of 10.38 and 11.76 min. The same fractions from four HPLC runs were pooled, corresponding to each of the two product peaks, evaporated using a stream of nitrogen gas, and subjected to NMR analysis.
NMR Spectroscopy
1H-NMR, 1H-COSY, HSQC, and HMBC spectra were measured at 300 K on a Bruker Avance III HD 700 NMR spectrometer (Bruker Biospin) using a cryogenically cooled 1.7-mm TCI 1H13C probe. The operating frequency was 700.45 MHz for 1H and 176.13 MHz for 13C. MeOH-d4 was used as a solvent. Chemical shift values (δ) are referenced relative to the residual solvent signals of MeOH-d4 at δH 3.31 and δC 49.15. Coupling constants are in Hz. The residual HDO signal in the 1H-NMR spectra was suppressed using the NOESY solvent-presaturation (noesypr1d) pulse program.
3-(β-d-Glucopyranosyloxy)-4-Hydroxy Acetophenone (Pungenin)
Major constituent of HPLC peak retention time 11.76 min. UV (MeOH) λmax 200, 266, and 305 nm. 1H- NMR (700 MHz, MeOH-d4): δ 7.85 (1H, d, J = 2.2 Hz, H-2), 7.64 (1H, dd, J = 8.5, 2.2 Hz, H-6), 6.91 (1H, d, J = 8.5 Hz, H-5), 4.85 (1H, d, J = 7.5 Hz, H-1′), 3.92 (1H, dd, J = 12.1, 2.1 Hz, H-6′a), 3.73 (1H, dd, J = 12.1, 5.7 Hz, H-6′b), 3.53 (1H, overlap, H-2′), 3.49 (1H, overlap, H-3′), 3.47 (1H, overlap, H-5′), 3.42 (1H, dd, J = 9, 9 Hz, H-4′), 2.53 (3H, s, CH3). 13C NMR (175 MHz, MeOH-d4): δ 199.3 (C=O), 153.7 (C-4), 146.2 (C-3), 130.4 (C-1), 126.2 (C-6), 118.8 (C-2), 116.8 (C-5), 103.9 (C-1′), 78.3 (C-5′), 77.6 (C-3′), 74.7 (C-2′), 71.2 (C-4′), 62.3 (C-6′), 26.2 (CH3). The proportion of approximately 82% was determined from integral ratios of the 1H-NMR spectrum of the crude synthetic mixture. Incompletely assigned 1H-NMR data of pungenin in acetone-d6 (200 MHz) have been reported by Strunz et al. (1986).
4-(β-d-Glucopyranosyloxy)-3-Hydroxy Acetophenone (Isopungenin)
HPLC peak retention time 10.38 min. UV (MeOH) λmax 200, 224, 272, and 301 nm. 1H-NMR (700 MHz, MeOH-d4): δ 7.51 (1H, dd, J = 8.5, 2 Hz, H-6), 7.46 (1H, d, J = 2 Hz, H-2), 7.24 (1H, d, J = 8.5 Hz, H-5), 4.93 (1H, d, J = 7.5 Hz, H-1′), 3.91 (1H, dd, J = 12.1, 2.1 Hz, H-6′a), 3.72 (1H, dd, J = 12.1, 5.7 Hz, H-6′b), 3.54 (1H, overlap, H-2′), 3.49 (1H, overlap, H-3′), 3.47 (1H, overlap, H-5′), 3.42 (1H, dd, J = 9, 9 Hz, H-4′), 2.53 (3H, s, CH3). 13C NMR (175 MHz, MeOH-d4): δ 199.4 (C=O), 151.0 (C-4), 147.6 (C-3), 133.1 (C-1), 122.4 (C-6), 116.9 (C-5), 116.5 (C-2), 102.7 (C-1′), 78.3 (C-5′), 77.5 (C-3′), 74.6 (C-2′), 71.1 (C-4′), 62.2 (C-6′), 26.3 (CH3). The proportion of approximately 10% was determined from integral ratios of the 1H-NMR spectrum of the crude synthetic mixture. 1H- and 13C-NMR data in DMSO-d6 (300 and 75 MHz) have been reported by Lin and Lin (1997).
RNA Extraction and RNA Sequencing
Developing shoot samples collected over a time course from April 24, 2013, through May 15, 2013 (seven time points with four biological replicates per time point for a total of 28 samples), were ground in liquid nitrogen. Of each sample, 100 mg was used for RNA extraction with PureLink Plant RNA Reagent (ThermoFisher) according to the manufacturer’s instructions. A second aliquot of 100 mg of each ground tissue sample was used for metabolite analysis (see below). RNA quality was assessed with an Agilent Technologies 2100 Bioanalyzer using RNA nano chips. Based on the metabolite analysis, samples collected 2 weeks before and 2 weeks after (four time points with four biological replicates per time point for a total of 16 samples) were sent to the McGill University and Génome Québec Innovation Centre for RNA sequencing library preparation and HiSeq transcriptome sequencing. For each sample, approximately 300 ng per μL in a 10-μL total volume was provided for the preparation of libraries using the Illumina TruSeq RNA Library Prep Kit and HiSeq2000. Sixteen unstranded libraries were sequenced over four lanes resulting in 100-bp paired-end reads.
De Novo Transcriptome Assembly
Reads from the 16 RNA sequencing libraries were assessed for their quality using FastQC (Andrews, 2015). Bowtie2 (version 2.2.6; Langmead and Salzberg, 2012) was used to filter ribosomal sequences by mapping against Picea spp. rRNA downloaded from the National Center for Biotechnology Information (NCBI). Unmapped, nonribosomal reads were then trimmed for adapters using Trimmomatic (version 0.30; Bolger et al., 2014). BBMerge (version 34.92), part of the BBTools package, was used to merge overlapping paired-end reads into single-ended reads (Bushnell, 2015). Merged reads together with unmerged paired-end reads were assembled de novo with Velvet (version 1.2.10) and Oases (version 0.2.8) to produce 77,841 contigs with average length of 1,359 bp (Zerbino and Birney, 2008; Schulz et al., 2012). Contigs were clustered at the 99% sequence identity level using CD-HIT-EST (version 4.6.1; Fu et al., 2012) and condensed to 60,402 contigs. A total of 39,387 predicted peptides were generated from TransDecoder (version 2.0.1; Haas et al., 2013).
DE Analysis
DE analysis was performed with the voom/limma (Law et al., 2014) package in R using quantification results from Salmon (version 0.3.0), a rapid quantification tool and successor of Sailfish (Patro et al., 2014), with default parameters. Each of the three time points, May 1 (T2), May 8 (T3), and May 15 (T4), were compared with April 24 (T1) as a baseline for DE. A total of 105, 1,465, and 3,803 contigs were identified as differentially expressed in T2 versus T1, T3 versus T1, and T4 versus T1, respectively, with absolute log2 fold change ≥ 2 and adjusted P ≤ 0.01.
BLASTN Searches against Tissue-Specific Transcriptomes
The seven candidate UGT transcripts designated as PgUGT1, PgUGT2, PgUGT3, PgUGT5, PgUGT6, PgUGT7, and PgUGT8 were assessed for expression in different organs and tissues using BLASTN search against published white spruce transcriptomes for (1) flushing buds (SRX318119), (2) young buds (SRX318122), (3) mature needle (SRX318120), (4) xylem (SRX318121), and (5) bark (NCBI Sequence Read Archive: SRX318118; Birol et al., 2013) and against published Norway spruce (Picea abies) transcriptomes (http://congenie.org) for (1) needles, (2) stems, (3) cones, (4) wood, (5) infected needles, (6) galls, and (7) girdled twigs (Nystedt et al., 2013). The BLASTN parameters used were as follows: Match/Mismatch and Gap Costs = Match 2, Mismatch 3, Existence 5, Extension 2; Expectation value = 1.0E-20; Word size = 11. Transcripts with greater than 98% identity to a candidate UGT were considered homologs.
Metabolite Analysis
One hundred milligrams of ground tissue (see “RNA Extraction and RNA Sequencing” above) was placed in a vial with 1 mL of MeOH. The vial was capped and placed at 4°C with overnight shaking. The supernatant was removed and placed in a new vial. For LC-MS analysis, samples were diluted 1:10 by combining 100 μL of supernatant in 900 μL of MeOH. LC-MS analysis was performed using an LC-MSD-Trap-XCT_plus with an SB-C18, 15-cm column (Agilent Technologies). Solvent A was water with 0.2% (v/v) formic acid; solvent B was 100% (v/v) acetonitrile with 0.2% (v/v) formic acid. The following gradient was used: increase to 5% solvent B from 0 to 0.5 min; increase to 22% solvent B from 0.5 to 5 min; increase to 35% solvent B from 5 to 10 min; increase to 50% solvent B from 10 to 13 min; increase to 95% solvent B from 13 to 16 min; hold at 95% solvent B from 16 to 17 min; decrease to 5% solvent B from 17 to 17.1 min. The injection volume was 10 μL. The column flow rate was 0.8 mL min−1. Picein and pungenin were identified using the extracted ions 137(+) and 313(−), respectively.
UGT cDNA Cloning
cDNA was synthesized from RNA extracted from the tissue sample of the May 8 time point using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR, with double-stranded DNase according to the manufacturer’s instructions (ThermoFisher). Candidate UGT cDNAs were amplified using primers (Supplemental Table S5) designed according to UGT transcripts identified in the transcriptome assemblies with adaptors for cloning into either pASK-IBA37plus or pEAQ (IBA; Sainsbury et al., 2009). Resulting pASK-IBA37plus constructs were transformed into E. coli strain C43 pRARE, and pEAQ constructs were transformed into Agrobacterium tumefaciens strain AGL1 for transformation into Nicotiana benthamiana and transient expression. For candidate genes containing BsaI restriction sites, site-directed base changes were made using primers resulting in a synonymous mutation to remove the site.
Protein Expression and Purification
E. coli transformed with pASK-IBA37plus-UGT constructs was grown overnight in 5 mL of Luria broth with ampicillin at 37°C. Five-milliliter cultures were used to inoculate a 50-mL flask of Luria broth with ampicillin. Cultures were then grown at 16°C until they reached an OD600 of 0.5. Cultures were induced with 5 μL of anhydrous tetracycline (2 mg mL−1) and incubated overnight at 16°C. Cultures were centrifuged at 4,000g for 15 min. Supernatants were removed, and the pellets were resuspended in 1 mL of extraction buffer (amount per 10 mL: 50 mm Tris-HCl, pH 7.5, 10% glycerol, 10 mm MgCl2, 5 mm DTT, one-quarter-strength Pierce Protease Inhibitor Tablets, EDTA-free [ThermoFisher], 2 mg of lysozyme, and 1 μL of 25 units μL−1 benzonase [EMD Millipore]) and incubated on ice for 45 min. Cells were ruptured using sonication and then centrifuged at 4°C for 20 min at 13,500g. The supernatant was transferred into a new tube. DTT (1 mm) and 20 mm imidazole were added to the supernatant. The supernatant was incubated with nickel resin for 1 h at 4°C with rotation. Protein was purified on a gravity column and washed with 25 mL of wash buffer (50 mm Tris-HCl, pH 7.5, 10% glycerol, 1 mm DTT, and 20 mm imidazole). To elute protein off the column, 500 μL of elution buffer (50 mm Tris-HCl, pH 7.5, 10% glycerol, 1 mm DTT, and 200 mm imidazole) was added to a capped column, and the column was incubated for 10 min on ice before elution. Protein was then desalted with assay buffer (10 mm Tris-HCl, pH 7.5, 10% glycerol, and 1 mm DTT) using PD-25 desalting columns (GE). Protein was confirmed by SDS-PAGE using Coomassie Blue and western blots. Protein was quantified using Pierce Coomassie Plus (Bradford) reagent (ThermoFisher). Western blotting was performed using the His-Tag antibody HRP Conjugate Kit (EMD Millipore) and visualized with Clarity Western ECL Blotting Substrate (Bio-Rad).
UGT Enzyme Assays
To test for enzyme activity, 80 μL of purified protein was incubated with 2 mm UDP-Glc, 10 mm Tris-HCl, pH, 7.5, and 100 μm pungenol. The total reaction volume was 200 μL. Assays were incubated at 25°C for 3 h. Assays were stopped by the addition of 400 μL of ethyl acetate. Samples were vortexed well and centrifuged at 1,000g. The top layer was removed and dried under nitrogen gas. Samples were resuspended in 100 μL of MeOH for LC-MS analysis. Competition assays were carried out as in the test for enzyme activity assays except that the following acceptor substrates also were included: piceol, quercetin, naringenin, kaempferol, resveratrol, caffeic acid, coumaric acid, acetovanillone, and 4-hydroxy benzoic acid. LC-MS analysis was performed using an LC-MSD-Trap-XCT_plus with an SB-C18, 15-cm column (Agilent Technologies). Solvent A was water with 0.2% (v/v) formic acid; solvent B was 100% (v/v) acetonitrile with 0.2% (v/v) formic acid. The following gradient was used: increase to 5% solvent B from 0 to 0.25 min; increase to 20% solvent B from 0.25 to 4 min; increase to 25% solvent B from 4 to 12 min; increase to 40% solvent B from 12 to 14 min; increase to 90% solvent B from 14 to 21 min; hold at 90% solvent B from 21 to 24 min; decrease to 5% solvent B from 24 to 24.1 min. The column flow rate was 0.5 mL min−1. Picein and pungenin were identified using the extracted ions 137(+) and 313(−), respectively.
N. benthamiana Expression and in Planta Assays
A. tumefaciens transformed with pEAQ-UGT constructs was grown overnight at 28°C in 10 mL of Luria broth with kanamycin. Additionally, A. tumefaciens containing pEAQ-GFP and pBIN-p19 constructs also was grown under the same conditions as the positive control and to improve expression, respectively (Sainsbury et al., 2009). Cultures were centrifuged at 3,000g and resuspended in infiltration buffer (10 mm MES and 10 mm MgCl2) to an OD600 of 0.7. Each resuspended construct was mixed in a 1:1 ratio with resuspended pBIN-p19, resulting in the following solutions: pEAQ-UGT5 plus pBIN-p19, pEAQ-UGT5b plus pBIN-p19, and pEAQGFP plus pBIN-p19. Mixed solutions were allowed to sit at room temperature for 2 h. A. tumefaciens solutions were infiltrated into leaves of 4-week-old N. benthamiana plants using a 1-mL syringe. Three leaves of four individual plants per construct were infiltrated. After 6 d, leaves were infiltrated with a 500 μm solution of pungenol in infiltration buffer using the volume required to fill the void space of the leaf. After 4 h, leaves were harvested. The three infiltrated leaves from an individual plant were pooled and ground in liquid nitrogen. Ground tissue was extracted with 1 mL MeOH g−1 fresh weight. Metabolites were extracted overnight at 4°C with shaking. Samples were centrifuged at 4,000g. The supernatant was removed and placed into a new vial. Metabolites were analyzed with LC-MS as described for enzyme assays.
qRT-PCR
Metabolite analysis of various spruce genotypes was done as described previously for foliage from the time-course experiment. RNA also was extracted as described previously. qRT-PCR analysis of gene expression was performed using the qScript XLT One-Step RT-qPCR ToughMix (Quanta) with PrimeTime Mini qPCR (IDT) containing two primers and one probe per assay (Supplemental Table S5). Thioredoxin (GQ03413_J21) was chosen as the reference gene based on Raherison et al. (2015) and the shoot development time-course transcriptome analysis. Amplicons were confirmed by sequencing the PCR products.
Accession Numbers
Transcriptome libraries from the time course of shoot development are available in the NCBI/GenBank Sequence Read Archive (SRP069003). Nucleotide sequences for cloned PgUGTs are available in NCBI/GenBank (accession nos. KY963359, KY963360, KY963361, KY963363, KY963364, KY963365, KY963366, KY963367, and KY963368).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Work flow of transcriptome assembly and analysis.
Supplemental Figure S2. Sequence comparison of PgUGT5, PgUGT5b, and PgUGT5c.
Supplemental Figure S3. Expression and activity of PgUGT candidates.
Supplemental Figure S4. Flavonoid glycoside presence over shoot development.
Supplemental Figure S5. Schematic of isopungenin and pungenin chemical synthesis.
Supplemental Table S1. PgUGT homologs found in white spruce PG29 organ and tissue transcriptome libraries.
Supplemental Table S2. PgUGT homologs found in Norway spruce.
Supplemental Table S3. PgUGT official nomenclature.
Supplemental Table S4. Enzyme activity of PgUGT5 and PgUGT5b with different phenolic substrates.
Supplemental Table S5. Primer sequences for cloning and qRT-PCR.
Acknowledgments
We thank L. Madilao for technical assistance; Dr. S. Irmisch for helpful discussion; Dr. C. Goulet and Dr. F. Sainsbury for pEAQ-GG constructs and the A. tumefaciens strain; I. Giguére and Dr. G. Parent for white spruce genotype collections; and D. Baulcombe and Plant Bioscience Limited for providing the pBIN-p19 construct.
Glossary
- SBW
spruce budworm
- DE
differential expression
- TLC
thin‐layer chromatography
- NCBI
National Center for Biotechnology Information
- LC-MS
liquid chromatography-mass spectrometry
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Strategic Project Grant to J.B. and J.M. and Discovery Grant to J.B.) and funds received through Genome Canada, Genome British Columbia, Genome Quebec for the SMarTForests Project (to J.B. and J.M.) and the Spruce-Up (243FOR) Project (to J.B.). J.B. was supported, in part, by the UBC Distinguished University Scholar program. M.F. was supported by the Austrian Science Fond (FWF) with an Erwin Schrödinger Fellowship (J3293-B21).
Articles can be viewed without a subscription.
References
- Andrews S. (2015) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Yuen MM, Keeling CI, Brand D, Vandervalk BP, et al. (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29: 1492–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. (2015) BBMap short read aligner, and other bioinformatic tools. http://sourceforge.net/projects/bbmap/
- Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S (2012) A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J 69: 1030–1042 [DOI] [PubMed] [Google Scholar]
- Chang WY, Lantz VA, Hennigar CR, MacLean DA (2012) Economic impacts of forest pests: a case study of spruce budworm outbreaks and control in New Brunswick, Canada. Can J For Res 42: 490–505 [Google Scholar]
- Delvas N, Bauce É, Labbé C, Ollevier T, Bélanger R (2011) Phenolic compounds that confer resistance to spruce budworm. Entomol Exp Appl 141: 35–44 [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon CMM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10: 542–549 [DOI] [PubMed] [Google Scholar]
- Gray DR. (2013) The influence of forest composition and climate on outbreak characteristics of the spruce budworm in eastern Canada. Can J For Res 43: 1181–1195 [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann J, Strack D (1991) Flavonol glucosyl transferase from Norway spruce needles. Phytochemistry 30: 1773–1776 [Google Scholar]
- Klimaszewska K, Lachance D, Pelletier G, Lelu MAL, Seguin A (2001) Regeneration of transgenic Picea glauca, P. mariana, and P. abies after cocultivation of embryogenic tissue with Agrobacterium tumefaciens. In Vitro Cell Dev Biol Plant 37: 748–755 [Google Scholar]
- Kröger L, Thiem J (2007) Synthesis and evaluation of glycosyl donors with novel leaving groups for transglycosylations employing β-galactosidase from bovine testes. Carbohydr Res 342: 467–481 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Lin TC (1997) Two acetophenone glucosides, cynanonesides A and B, from Cynanchum taiwanianum and revision of the structure for cynandione A. J Nat Prod 60: 368–370 [Google Scholar]
- MacLean DA. (2016) Impacts of insect outbreaks on tree mortality, productivity, and stand development. Can Entomol 148: S138–S159 [Google Scholar]
- Mageroy MH, Parent G, Germanos G, Giguère I, Delvas N, Maaroufi H, Bauce É, Bohlmann J, Mackay JJ (2015) Expression of the β-glucosidase gene Pgβglu-1 underpins natural resistance of white spruce against spruce budworm. Plant J 81: 68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, Berasategui A, Paetz C, Gershenzon J, Schmidt A (2014) Overexpression of an isoprenyl diphosphate synthase in spruce leads to unexpected terpene diversion products that function in plant defense. Plant Physiol 164: 555–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrel J, Javelle F (2010) The biosynthesis of acetovanillone in tobacco cell-suspension cultures. Phytochemistry 71: 751–759 [DOI] [PubMed] [Google Scholar]
- Neish AC. (1959) Biosynthesis of pungenin from C14-labelled compounds by Colorado spruce. Can J Bot 37: 1085–1100 [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Møller BL (2009) Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70: 325–347 [DOI] [PubMed] [Google Scholar]
- Parent GJ, Giguère I, Germanos G, Lamara M, Bauce É, MacKay JJ (2017) Insect herbivory (Choristoneura fumiferana, Tortricidea) underlies tree population structure (Picea glauca, Pinaceae). Sci Rep 7: 42273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C (2014) Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 32: 462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raherison ESM, Giguère I, Caron S, Lamara M, MacKay JJ (2015) Modular organization of the white spruce (Picea glauca) transcriptome reveals functional organization and evolutionary signatures. New Phytol 207: 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7: 682–693 [DOI] [PubMed] [Google Scholar]
- Schmid G, Grisebach H (1982) Enzymic synthesis of lignin precursors: purification and properties of UDP glucose:coniferyl-alcohol glucosyltransferase from cambial sap of spruce (Picea abies L.). Eur J Biochem 123: 363–370 [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Gu L, Liu J, Zhao S, Hong X, Schulenburg K, Schwab W (2015) Functional characterization and substrate promiscuity of UGT71 glycosyltransferases from strawberry (Fragaria × ananassa). Plant Cell Physiol 56: 2478–2493 [DOI] [PubMed] [Google Scholar]
- Steeves V, Förster H, Pommer U, Savidge R (2001) Coniferyl alcohol metabolism in conifers. I. Glucosidic turnover of cinnamyl aldehydes by UDPG:coniferyl alcohol glucosyltransferase from pine cambium. Phytochemistry 57: 1085–1093 [DOI] [PubMed] [Google Scholar]
- Strunz GM, Giguère P, Thomas AW (1986) Synthesis of pungenin, a foliar constituent of some spruce species, and investigation of its efficacy as a feeding deterrent for spruce budworm [Choristoneura fumiferana (Clem.)]. J Chem Ecol 12: 251–260 [DOI] [PubMed] [Google Scholar]
- Warren RL, Keeling CI, Yuen MMS, Raymond A, Taylor GA, Vandervalk BP, Mohamadi H, Paulino D, Chiu R, Jackman SD, et al. (2015) Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J 89: 189–212 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]