Network analysis identifies wheat genes causally related to spike complexity, a key determinate of crop yield potential.
Abstract
The architecture of wheat (Triticum aestivum) inflorescence and its complexity is among the most important agronomic traits that influence yield. For example, wheat spikes vary considerably in the number of spikelets, which are specialized reproductive branches, and the number of florets, which are spikelet branches that produce seeds. The large and repetitive nature of the three homologous and highly similar subgenomes of wheat has impeded attempts at using genetic approaches to uncover beneficial alleles that can be utilized for yield improvement. Using a population-associative transcriptomic approach, we analyzed the transcriptomes of developing spikes in 90 wheat lines comprising 74 landrace and 16 elite varieties and correlated expression with variations in spike complexity traits. In combination with coexpression network analysis, we inferred the identities of genes related to spike complexity. Importantly, further experimental studies identified regulatory genes whose expression is associated with and influences spike complexity. The associative transcriptomic approach utilized in this study allows rapid identification of the genetic basis of important agronomic traits in crops with complex genomes.
Grains of cereal crops provide a major source of human diet and nutrition. Improving grain yield is a primary objective during crop domestication and a major goal of crop-breeding programs. Inflorescence (spike) architecture dictates the capacity for seed production in cereal crops, including wheat (Triticum aestivum), the world’s most widely grown cereal. In the archetypal bread wheat spike, the inflorescence meristem forms a limited number of lateral spikelet meristems (SMs) per rachis node, and a single terminal SM at the distal end. Each SM is indeterminate and typically produces two to four fertile florets that produce seeds (Fig. 1A; Supplemental Fig. S1; Bonnett, 1936; Fisher, 1973). Like maize (Zea mays) and rice (Oryza sativa), wheat yield per plant largely depends on the number of florets per spike and thus spike architecture. The numbers of SMs (and rachises) and florets per spikelet are major target traits for efforts aimed at improving wheat yield. In addition, the number of SMs per rachis may be increased to increase floret number, as in rare supernumerary spikelet variations.
Figure 1.
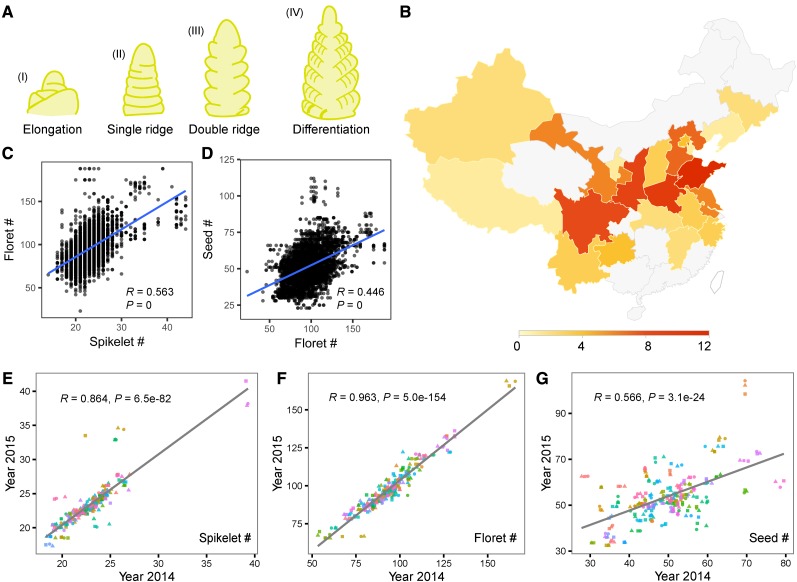
Spike complexity of 90 selected varieties from the Chinese mini core collection. A, Schematic diagram of main spike development in wheat. See Supplemental Figure S1 for scanning election micrographs. B, Geographical distribution of 90 selected winter wheat varieties in China. C, Scatter plot of spikelet number per spike against floret number per spike. D, Scatter plot of floret number per spike against seed number per spike. E to G, Scatter plot of average spikelet (B), floret (C), and seed (D) number per main spike from 2014 against the corresponding values from 2015. Each color denotes one variety, and the shape denotes the replicate. The gray line represents the regression trend calculated by the general linear model of each trait.
In rice, maize, and barley (Hordeum vulgare), multiple genes regulating spike development have been identified (Sreenivasulu and Schnurbusch, 2012; Tanaka et al., 2013). However, our understanding of wheat spike development remains rudimentary at the molecular level. In wheat, increased transcription levels of Q, an AP2-like gene, was markedly associated with spike compactness, suggesting that the Q gene may be implicated in spike development (Simons et al., 2006), although its role in spike complexity remains unclear. Feng et al. (2017) reported that a durum wheat ARGONAUTE1d gene mutant produced shorter spikes and fewer grains per spike than wild-type controls. In addition, recent studies have shown the mechanisms underlying the genetic regulation of rare supernumerary spikelet variations. For example, wheat Photoperiod1 (Ppd1) was identified as a regulator of paired spikelet formation (Boden et al., 2015). When Ppd1 is mutated, a secondary spikelet initiates immediately below a typical single spikelet in the same rachis node, thus forming a rare supernumerary spikelet variation. In another type of variation, one or more spikelets are replaced by long lateral branches, which form their own spikelets and florets. Mutations of the WFZP-A/BHt-A1 gene, encoding an AP2/ERF transcription factor, lead to such noncanonical spike branching (Derbyshire and Byrne, 2013; Dobrovolskaya et al., 2015; Poursarebani et al., 2015), which is similar to the branching produced by mutating its orthologs in maize, rice, and Brachypodium distachyon (Chuck et al., 2002; Komatsu et al., 2003). Although these recent breakthroughs shed light on the molecular mechanisms underlying rare supernumerary spikelet variations, little is known about genetic factors affecting the architecture of archetypal wheat spike, its complexity, and grain yield.
The allohexaploid common bread wheat genome is approximately 17 gigabases in size and consists of three sets of subgenomes (A, B, and D) derived from closely related species. An enormous amount of genomic sequencing has been performed to build a reference sequence for wheat (Brenchley et al., 2012; International Wheat Genome Sequencing Consortium, 2014), which enhanced our understanding of the wheat genome significantly. Nevertheless, the genetic complexity associated with wheat hampers map-based cloning and genome-wide association studies (GWAS). Although GWAS has been applied to wheat (for example, Guo et al., 2017; Maccaferri et al., 2015; Sun et al., 2017), it remains highly challenging to pinpoint causal genes within identified genetic loci by a GWAS approach.
With a long history of cultivation and artificial selection in diverse ecological zones, common wheat in China has a rich genetic diversity (He et al., 2001). A mini core collection of 231 Chinese wheat varieties, which is estimated to represent approximately 70% of the genetic diversity of the 23,090 varieties (Hao et al., 2008), widely used germplasm for wheat breeding in China. Based on geographical regions and allelic diversity previously reported by Hao et al. (2011), we selected 90 winter wheat varieties from 142 winter varieties within the mini core collection, including 74 landraces and 16 elite varieties (Fig. 1B; Supplemental Table S1), for transcriptome association analysis (Harper et al., 2012). By quantitatively correlating trait variation with variation in gene expression, we aimed to identify the gene regulatory networks underlying the development of spike architecture.
RESULTS
Variation in Spike Complexity among Mini Core Collection Varieties
We planted 90 wheat varieties at the same experimental site for 2 years with three replications. At least 60 seeds per variety per year were planted within each replication. We found that the 90 varieties have flowered at slightly different times (∼3 d). For each year, spikes were hand dissected from the main shoots of each variety at the same double ridge stage, when spikelet primordia occur between bract primordia at the middle part of the spike, after which we extracted mRNA for transcriptome sequencing. At the double-ridge stage, SMs have emerged to produce spikelets (Bonnett, 1936). In addition, florets are growing out from spikelets in the middle section, whereas new SMs are still being produced at the distal end at this stage (Fig. 1A; Supplemental Fig. S1). We also counted the number of spikelets, number of florets per spike, and number of seeds per spike 20 d after flowering (Supplemental Tables S2–S4). We observed positive correlations among the number of spikelets, florets per spike, and seeds per spike across the 90 selected wheat varieties (Fig. 1, C and D). This finding indicates that the number of spikelets, which is determined during early spike development, mainly controls the numbers of florets and grains per spike. Additionally, the number of spikelets and florets across the two planting years showed significantly high correlations (R = 0.864 and 0.963, P = 6.5e-82 and 5.0e-154, respectively), while a moderate correlation (R = 0.566, P value = 3.1e-24) was observed for the number of seeds, indicating that the latter trait is more influenced by the environment (Fig. 1, E–G).
Spike Transcriptome Variation among Varieties
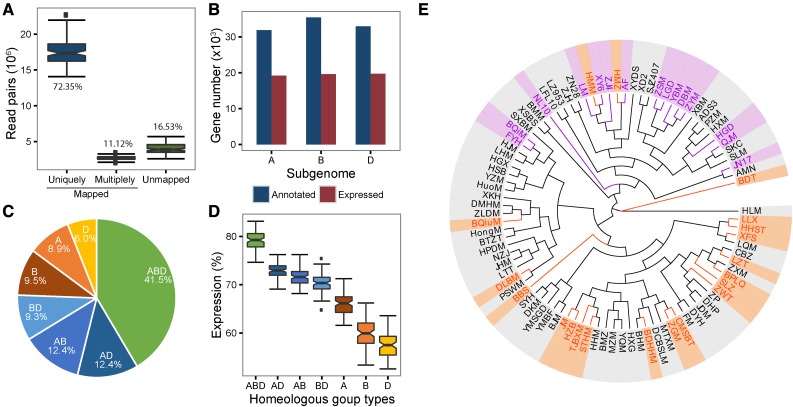
The RNA-seq reads for double-ridge stage spikes were mapped to the IWGSC genome survey sequence (International Wheat Genome Sequencing Consortium, 2014). On average, 17.5 million read pairs per variety sample were uniquely mapped to the genome (Fig. 2A; Supplemental Tables 5 and 6). Saturation analysis showed that the relative transcript abundance quantification error dropped below 5% when the resampling size was increased to 50%, indicating that the sequencing depth was adequate (Supplemental Fig. S2). Using a criterion of kilobases per million reads (RPKM) value ≥1 for gene expression, 58,494 of 100,344 annotated wheat genes were expressed in at least one variety, whereas 30,638 annotated wheat genes were expressed across all varieties (Fig. 2B; Supplemental Table S7). In addition, the number of transcripts derived from each subgenome was roughly equal (19,162 [32.8%] from subgenome A, 19,629 [33.7%] from subgenome B, and 19,703 [33.6%] from subgenome D; Supplemental Tables 8 and 9).
Figure 2.
Transcriptome analysis of 90 selected wheat varieties. A, Percentages of read pairs aligned to the reference genome sequence. B, The three subgenomes contribute roughly equal proportions of transcripts to the whole transcriptome. C, Proportion of genes classified to each type of HG. HGs were categorized based on the number of homeologous copies from the three subgenomes. D, Box plot of the expression ratios of different types of HGs. E, Phylogenetic tree of 90 selected wheat varieties. The branches labeled by orange belongs to variety set 1, which was grouped by expression pattern of GESs, and the purple ones belong to set 2. See Figure 3C for details.
Next, we identified subgenomic homeologs by considering both nucleotide homology (70% similarity) and chromosomal linearity, after which we categorized the 58,494 genes that were expressed in at least one variety into 24,230 homeologous groups (HGs) by following the method described by Pfeifer et al. (2014). Approximately 75% of the expressed genes were categorized into HGs containing at least two subgenomic counterparts (Fig. 2C; Supplemental Table S10), with 18,180 (41.5%) genes categorized into HGs containing three subgenomic homeologs (ABD type), 14,952 (31.1%) genes categorized into HGs containing two homeologs (AD, AB, or BD types), and 10,694 (24.4%) subgenome-unique genes (A, B, or D types). Among the three types of HGs, the ABD group has the highest proportion (approximately 80%) of expressed genes across the selected varieties, while the expressed proportions in the AB, BD, and AD groups range from 70% to 73% and the subgenome-unique genes have the lowest proportion of expressed genes (Fig. 2D). Based on the alignment of RNA-seq reads against the reference genomes, we identified 1,764,048 single nucleotide polymorphisms (SNPs) with minor allele frequency ≥3% and 236,849 indels in wheat transcripts (see “Materials and Methods”). A similarity was observed between structures of phylogenetic trees based on exonic SNPs and simple sequence repeat markers, respectively (Fig. 2E; Hao et al., 2011).
Transcriptome Association with Spike Complexity
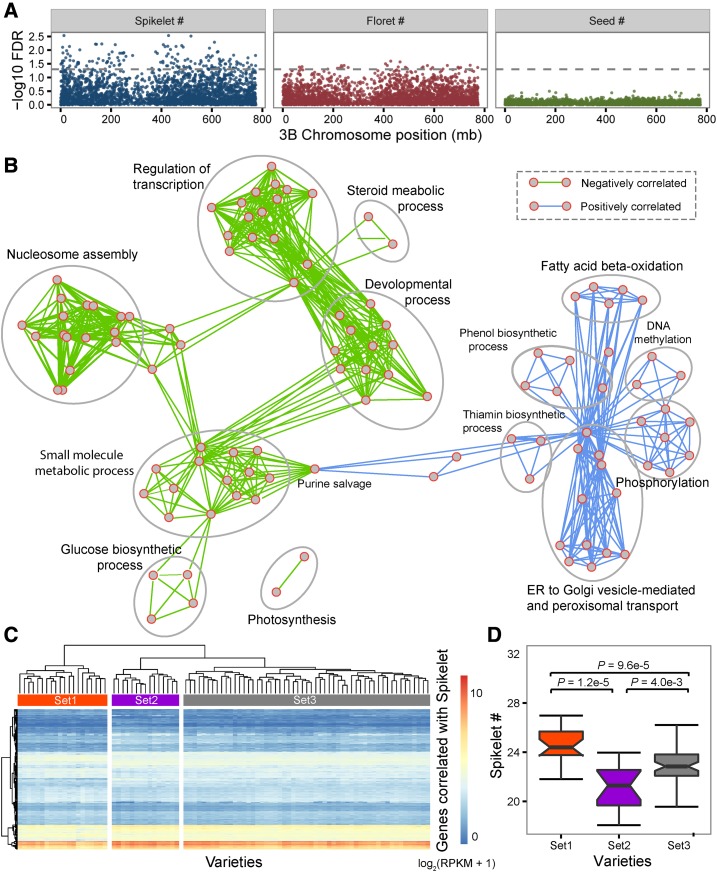
Further, we performed a transcriptome association analysis to correlate gene expression signatures (GESs) with trait variation. To summarize the observed spike trait values from three replicates of each variety over 2 years, we used the best linear unbiased estimation method, in which the variety setting was set as a fixed effect, whereas both year and the interaction between year and variety were set as random effects (Supplemental Tables 2–4). Next, we correlated gene expression levels with spike complexity traits across 90 varieties using Spearman's correlation coefficient. The expression levels of 1538 genes were significantly correlated (false discovery rate [FDR] ≤ 0.05) with the number of spikelets per spike, whereas the expression levels of 105 genes were significantly correlated (FDR ≤ 0.05) with the number of florets per spike. No gene showed expression correlation with the number of seeds per spike (Supplemental Table S11). In addition, correlation strength varied among the three studied traits. Spikelet number showed the strongest correlations with gene expression levels, followed by floret number and seed number per spike, as illustrated by the distribution of correlation significance for genes along each chromosome (Fig. 3A; Supplemental Fig. S3). As we sampled RNAs from spikes at the double-ridge stage, our data mainly reflect gene expression during spikelet formation, but not during seed development. Thus, the observation that the number of spikelets per spike showed the strongest correlation with expression levels was consistent with our experimental design. The finding that few GESs were correlated with the number of florets per spike or number of seeds per spike indicates that the transcriptomic context may have dramatically changed during the progression from spikelet initiation to floret initiation and seed formation.
Figure 3.
Transcriptome association analysis for spike complexity. A, Manhattan plot of the association significance between spike traits and gene expression abundance along chromosome 3B. The gray line shows the genome-wide significance level (FDR ≤ 0.05). B, Function enrichment network for GESs significantly associated with spikelet quantity (FDR ≤ 0.05). Enriched GO terms for negatively correlated genes are represented by nodes with green edges, and terms for positively correlated genes are represented by nodes with blue edges. C, The 1,538 GESs can be grouped into three distinct sets based on expression abundance. D, Box plot of the spikelet numbers of the three sets of wheat varieties.
To infer the functions of the 1,538 GESs found to be correlated with the number of spikelets per spike, we performed a Gene Ontology (GO) enrichment analysis to identify GO categories significantly enriched with GESs. The set of all expressed genes was used as the background set. Interestingly, GESs that were positively (754 GESs) and negatively (784 GESs) correlated with the number of spikelets per spike showed enrichment in distinct functional pathways. While the “fatty acid beta-oxidation,” “DNA methylation,” and “phenol and thiamin process” categories were enriched in positively correlated GESs, the “transcription and epigenetic regulation of development” and “energy storage processes” (including “photosynthesis and glucose biosynthesis” and “small molecule metabolic process”) categories were enriched in negatively correlated GESs (Fig. 3B). This pattern suggests a genome-wide transcriptional switch involving up- and down-regulation of multiple molecular pathways during the process of spikelet initiation.
Correlations of Phenotypic, Genotypic, and Transcriptomic Signatures
Based on the expression patterns of 1,538 GESs, we calculated the Euclidean distance matrix and hierarchically clustered 90 wheat varieties into three distinct sets by using the complete linkage method (Fig. 3C). Interestingly, the three sets of varieties showed different ranges of spikelet number per spike; wheat varieties in sets 1 and 2 have the highest and lowest spikelet numbers, respectively, while those in set 3 have a spikelet number average to that of sets 1 and 2 (Fig. 3D). This pattern strongly suggests that transcriptomic signatures are correlated with phenotypic quantity; this relationship can be used to discover trait-related genes.
To assess the consistency between expression variation and sequence variation, we combined transcriptomic signature-based variety sets with the transcript SNP-based phylogenetic tree (Fig. 2E). The grouping pattern based on sequence variation was partially correlated with that based on expression variation. Varieties in sets 1 and 2 were grouped separately, while the varieties in set 3 were spread between sets 1 and 2. This clear association between transcriptional patterns and spikelet number implies that a set of major regulators of spike complexity can be identified via transcriptome association analysis.
Correlation between GESs and Spikelet Number per Spike
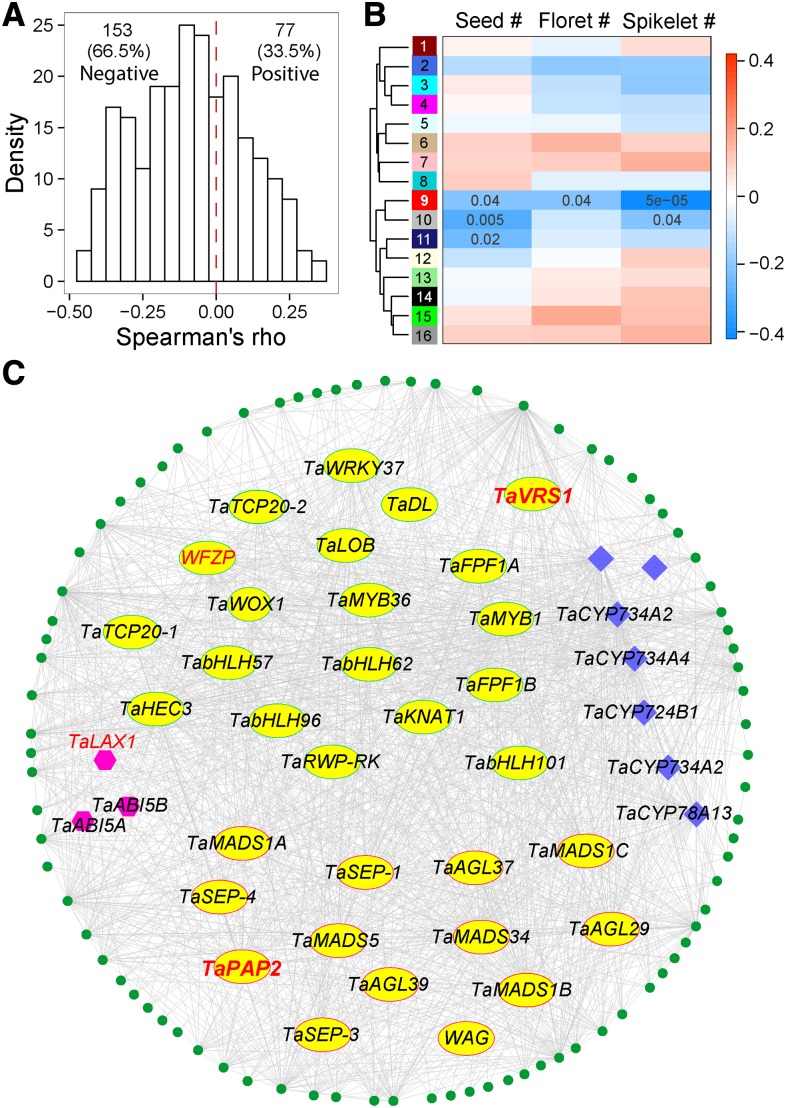
Among the 1538 GESs described above, we performed further analysis on a set of 230 wheat HGs orthologous to rice genes related to panicle and grain traits in spike development (Supplemental Table S12), according to the rice OGRO database (Yamamoto et al., 2012). Out of 230, 60 orthologous GESs showed statistically significant negative correlation with the number of spikelets, while 17 showed statistically significant positive correlation with the number of spikelets (P < 0.05; Fig. 4A; Supplemental Table S12). The greater number of negatively correlated GESs in our results suggests that a group of negative regulators may play important roles in switching off pathways that may positively influence spikelet number during spike development.
Figure 4.
Coexpression network analysis of GESs associated with spikelet number. A, Distribution of Spearman’s correlations for wheat genes homologous to rice genes previously reported as related to grain traits. B, Correlation heat map between subnetwork modules and spike traits. An eigen value of each module was computed by the WGCNA package to assess correlations between overall expression trends and spike traits. The color bar represents the scale of the Spearman’s correlation. C, Network of the red module (module 9) showing a significant negative correlation to all assessed spike traits.
To further screen for key regulatory genes, we performed gene coexpression network (GCN) analysis (Wisecaver et al., 2017). As homeologous genes among the three subgenomes tend to show similar expression patterns, this may introduce redundancy while calculating correlations. For this, we followed a previously published method in which the average expression values of homeologs are used to perform GCN analysis (Li et al., 2014; Pfeifer et al., 2014). First, we selected 7,945 genes with highly variable transcript abundance across the 90 varieties selected for this study (squared coefficient of variation [CV2] ≥ 0.25). A weighted GCN was constructed using the WGCNA package for R (Langfelder and Horvath, 2008), followed by decomposition of the network into 16 subnetwork modules. Each module contains a set of genes showing significant expression correlations with each other. An eigen value was calculated to represent the overall expression trend in each variety for each module. For each module, the correlations between the eigen value and the number of spikelets, florets, and seeds per spike were computed across 90 varieties (Fig. 4B). Notably, module 9 exhibited a significantly negative correlation with all three spike traits (P < 0.05), the strongest of which was with the spikelet number per spike (Spearman’s rho test, R = −0.4, P = 5.0e-5; Fig. 4C). Therefore, module 9 may harbor major regulators influencing the entire period of spike development, and thus strongly influencing wheat grain production.
Because transcription factors (TFs) are important direct modulators of gene expression, we focused on TF-encoding genes. Seven modules, which include 76.2% (6,052 of 7,945) of highly variable HGs, were enriched in TF-encoding genes (P ≤ 0.5, hypergeometric test). This observation suggests that TFs are involved in spike trait variations. Among the 16 modules, module 9 has the greatest degree of enrichment of TF-encoding genes (12.9% TFs compared to 4.4% TFs genome-wide, hypergeometric test, P = 6.4e-10; Supplemental Table S13). The intriguing features of module 9 led us to focus on this module for further analysis. Using a more stringent cutoff of adjacency ≥ 0.02, we refined module 9 to obtain a core subnetwork of 125 HGs (Fig. 4C; Supplemental Table S14). After manual curation, 32 of 125 (25.6%) core HGs were found to encode TFs, including 13 MADS-box TFs, which have been reported to be important regulators of floral development. In addition, seven genes belonging to the cytochrome P450 superfamily and three genes related to hormone signaling or transport were contained in the refined module.
Experimental Validation of Candidate Spike Regulators
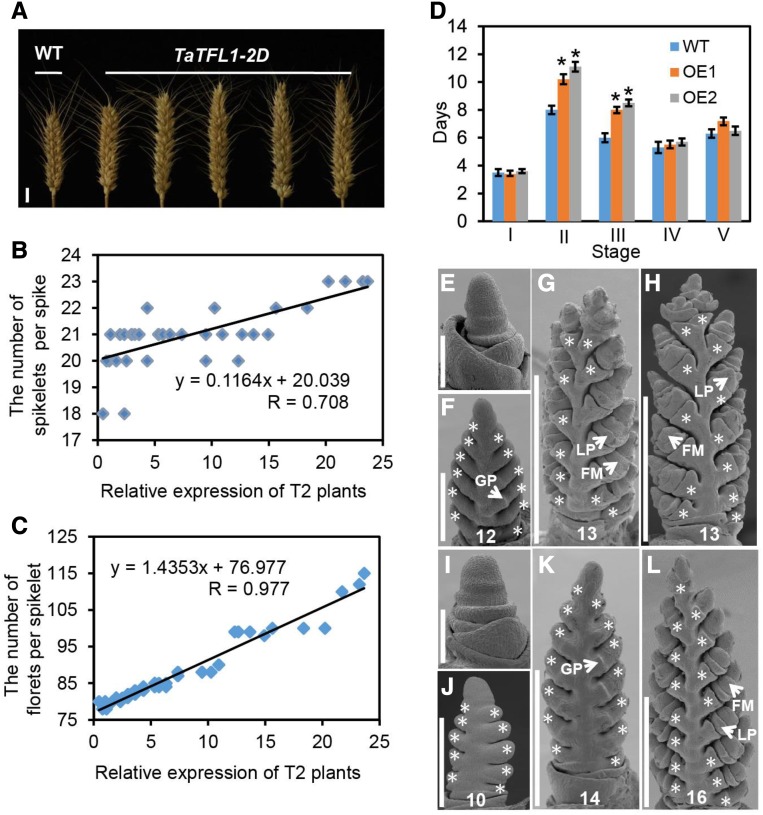
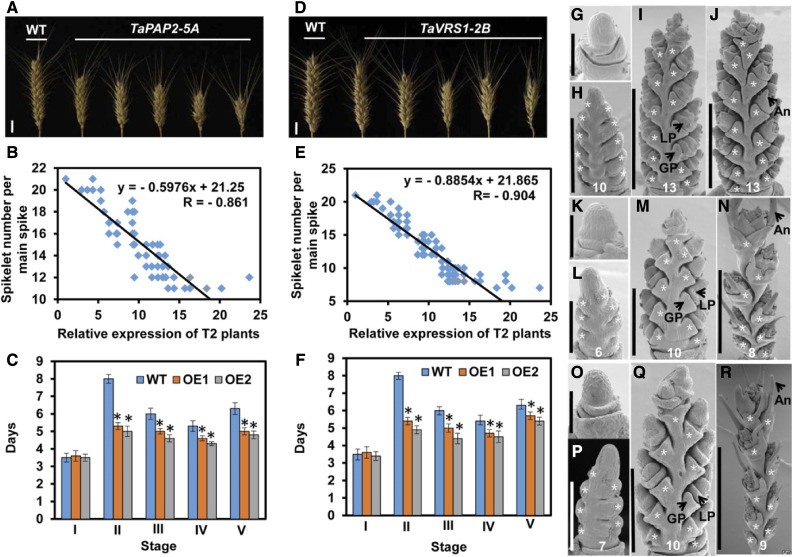
The results of the transcriptome analysis, especially the identified trait-correlated GESs, provide a rich resource of genes that could be important for efforts aimed at improving wheat grain production. As a proof of concept, we selected 10 genes for experimental verification (Supplemental Figure S4A). Among which, six genes have either expression significantly correlated with spikelet number (P ≤ 0.05), and/or present in module 9 (Supplemental Table S14). In detail, TaPAP2, WFZP, and TaLAX1 are found in both groups, TaRA3 and TaTFL1 are only correlated with spikelet number, and TaVRS1 is only found in module 9. All these six genes are annotated as development regulators by GO and/or MapMan. We further selected four additional genes (TaAPO1, TaLAX2, TaREV, and TaVT2) that are annotated as development regulators but have no expression correlation with spike complexity for experimental verification. As all candidates are HGs, we selected the homeolog showing the strongest correlation with spike traits for transgenic validation. We overexpressed the selected wheat genes in KN199, an elite hexaploid wheat variety with intermediate spike complexity. For each gene, we obtained more than 45 independent transformants, which were subjected to analysis of transgene expression levels and spike trait phenotypes. Overexpression of three out of the 10 tested genes significantly altered spike complexity. Overexpression of TaTFL1-2D increased spike complexity, while overexpression of TaPAP2-5A or TaVRS1-2B reduced spike complexity (Figs. 5A and 6, A and D). Notably, all these genes have expression correlation with spike complexity.
Figure 5.
Functional validation of TaTFL1-2D in the transgenic KN199 wheat line. A, Comparison of the spike complexity of KN199 and T2 transgenic TaTFL1-2D wheat plants. Scale bars, 1 cm. Positive correlation between spikelet number (B) and floret number (C) per main spike and TaTFL1 expression levels in T2 transgenic plants. D, Comparison of developmental duration between KN199 and T4 transgenic TaTFL1-2D lines (OE1 and OE2) at each stage. II, III, IV, and V indicate stage II, stage III, stage IV, and stage V, respectively. Data are the mean ± sd of 30 plants for each line. Student’s t test, *P < 0.05. Days, Days after a single ridge appearance; WT, wild type. E to H, Scanning electron micrographs of young spikes from KN199 plants at 3, 10, 15, and 21 d after a single ridge appearance. I to L, Scanning electron micrographs of young spikes from transgenic TaTFL1-2D plants at 3, 10, 15, and 21 d after a single ridge appearance. Scale bars, 200 μm (E and I), 300 μm (F and J), 500 μm (K), and 1 mm (G, H, and L). GP, glume primordium; LP, lemma primordium. The asterisks indicate spikelets. The number at the bottom represents the spikelet number per spike.
Figure 6.
Functional validation of TaPAP2-5A and TaVRS1-2B in the KN199 transgenic wheat line. Comparison of the spike complexity of KN199 plants with that of T2 transgenic TaPAP2-5A (A) and TaVRS1-2B (D) plants. Scale bars, 1 cm. Negative correlation between spikelet number per main spike and TaPAP2 (B) and TaVRS1-2B (E) expression levels in T2 transgenic plants. Comparison of developmental duration between KN199 plants and T4 transgenic TaTFL1-2D (C) and TaVRS1-2B (F) lines (OE1 and OE2) at each stage. I, II, III, IV, and V indicate stage I, stage II, stage III, stage IV, and stage V, respectively; WT, wild type. Data are the mean ± sd of 30 plants for each line. Days, Days after a single ridge appearance. G to J, Scanning electron micrographs of young spikes from KN199 plants at 2, 9, 15, and 22 d after a single ridge appearance. K to N, Scanning electron micrographs of young spikes from transgenic TaPAP2-5A plants at 2, 9, 15, and 22 d after a single ridge appearance. O to R, Scanning electron micrographs of young spikes from transgenic TaVRS1-2B plants at 2, 9, 15, and 22 d after a single ridge appearance. Scale bars, 200 μm (G, K, and O), 300 μm (H, L, and P), 500 μm (M and Q), 1 mm (I and J), and 3 mm (N and R). GP, glume primordium; LP, lemma primordium; An, awn. Student’s t test, *P < 0.05. The asterisks indicate spikelets. The number at the bottom represents the spikelet number per spike.
TaTFL1-2D, a member of positively correlated module 14, encodes a putative transcription cofactor sharing high protein sequence homology with Arabidopsis TFL1, a regulator of inflorescence meristem identity (Bradley et al., 1997). We found that the TaTFL1-2D transgene expression level was positively correlated with spikelet number, floret number, and seed number per spike (Fig. 5, B and C; Supplemental Fig. S4B). Our analysis of spike development in the transgenic lines indicated that the double-ridge stage and the following floret stage were significantly extended in TaTFL1-2D-overexpressing lines (Fig. 5D). During the double-ridge stage, SMs emerge to produce spikelets, while florets grow out from spikelets in the middle section (Fig. 1A). During the floret stage, most of the florets initiate glumes and lemmas to complete floret formation (Bonnett, 1936). The prolonged double ridge and floret stages in the TaTFL1-2D-overexpressing lines resulted in the formation of extra spikelets per spike and thus more florets per spikelet (Fig. 5, E–L), which considerably enhances spike complexity.
TaPAP2-5A and TaVRS1-2B, both contained in module 9, encode a MADS family transcription factor and a putative HD-ZIP family transcription factor, respectively. TaPAP2-5A shares high protein sequence homology with rice PAP2, an SM identity regulator (Kobayashi et al., 2010). TaVRS1-2B shares high protein sequence homology with barley Vrs1, a regulator of spikelet development (Komatsuda et al., 2007). Overexpressing either TaPAP2-5A or TaVRS1-2B reduced the spikelet number, floret number, and seed number per spike in a dosage-dependent manner (Fig. 6, A, B, D, and E; Supplemental Fig. S5, D, E, G, and H). In transgenic overexpressing lines, either TaPAP2-5A or TaVRS1-2B reduces the lengths of the double ridge stage, floret stage, stamen development stage, and anther development stage (Fig. 6, C and F). As a result, these transgenic plants develop spikes with fewer spikelets per spike and florets per spike in comparison with control lines transformed with empty vectors (Fig. 6, G–R). Whereas overexpressing TaPAP2-5A inhibits SM formation, consistent with the function of its rice ortholog, the effect of overexpressing TaVRS1-2B diverges from that of overexpressing its barley ortholog Vrs1. Barley Vrs1 suppresses lateral spikelet development to form rudimentary lateral spikelets (Komatsuda et al., 2007). In contrast, wheat TaVRS1-2B inhibits SM formation, as we did not detect rudimentary spikelets but found a reduced number of spikelets per spike in TaVRS1-2B-overexpressing lines. In addition, wheat TaVRS1-2B also inhibits floret meristem (FM) formation.
Taken together, the results of our transcriptome analysis reveal genes associated with and maybe causally related to spike complexity, providing a rich resource of genes that could be used to improve wheat grain yield. Appropriate expression of these genes can increase the number of productive spikelets, as well as the number of productive florets per spike, thus enhancing spikelet complexity and increasing the grain yield per plant.
DISCUSSION
Identifying genes conferring traits of agronomic importance is of enormous significance to crop improvement. Map-based cloning and GWAS in crops with relatively simple genomes, such as rice and maize, have been used to reveal agronomically important genes, but such study remains challenging in polyploid crops with complex genomes. In fact, our attempt to use genetic variations identified in the RNA-seq data to perform GWAS analysis did not yield any significant locus. To circumvent this difficulty, we used transcriptome association analysis and GCN analysis to correlate gene expression with trait variation. By using the recently released wheat genome sequence (International Wheat Genome Sequencing Consortium, 2014), we applied this analysis method to allohexaploid common wheat to study spike complexity.
Spike complexity determines the number of seeds per spike. Manipulation of spike complexity is a major strategy for improving yield potential. Whereas a large number of inflorescence regulators have been identified in other plant species, understanding of wheat spike development is relatively poor. Our transcriptome survey of a representative mini core collection identified a large number of potential regulators of wheat spike development. We identified a large number of candidate genes whose expression levels were positively or negatively associated with spike complexity. Although we quantified three traits related to spike complexity, namely the number of spikelets, florets per spike, and seeds per spike, we found the number of spikelets per spike had a by far larger number of positively and negatively correlated genes. This may reflect less environmental contribution but more genetic contribution to this trait. Thus, our coexpression analysis focused mostly on the number of spikelets per spike to obtain more reliable results.
Notably, the number of negative regulators of spike complexity was substantially greater than the number of positive regulators of spike complexity. We speculate that this finding may reflect a general inhibition of branching by the reproductive development program (Hagemann, 1990). As branching is the primitive condition for shoot growth, the onset of flowering significantly alters this development pattern from indeterminate to determinate, thus restricting branching ability and resulting in reduced complexity. Spike complexity depends on the remaining branching ability of the shoot apical meristem before its full termination into a flower. Most of the flowering genes are negative regulators of branching and show dominant expression after the floral transition; it is also conceivable that there are many negative regulators of spike complexity. Indeed, we identified many genes related to flower development, including those encoding MADS-box TFs in module 9, as putative negative regulators of spike complexity.
Importantly, our results demonstrate that overexpression of one of the three candidate genes, TaTFL1-2D, TaPAP2-5A, and TaVRS1-2B, affects inflorescence development in wheat and suggests that appropriate selection of alleles of favored expression or modifications in these genes could be used to increase wheat spike complexity and thus grain yield. Furthermore, our analysis identified wheat-specific functions of known spike regulators. For example, TaVRS1 overexpression negatively regulates spike branching by inhibiting SM initiation, whereas orthologous barley Vrs1 suppresses lateral spikelet outgrowth, but not SM initiation (Komatsuda et al., 2007). The neofunctionalization of TaVRS1 suggests fast evolution of the gene regulatory network underlying spike development in grasses. Detailed analysis of the spike development program in transgenic plants indicated that the duration of SM and FM formation stages are correlated with and are likely causal to spike complexity. This observation shows that the duration of remaining branching growth (before termination into flowers) is critical to the regulation of spike complexity. Our analysis also suggests that the processes of SM and FM formation utilize conserved regulatory modules, as all three genes that were experimentally validated in this study regulate both number of spikelets and number of florets, which are results of the two major branching events in wheat spike branching.
In addition to genetic factors, environmental adaption also has profound effects on yield. In this study, we used the same growth condition for all varieties to minimize environmental effects and to focus on genetic contributions to spike complexity. To this end, we selected only winter wheat varieties that can normally grow in Beijing. Future experiments in multiple sites would resolve the interaction between genetics and environment.
CONCLUSION
Early reproductive development in wheat is essential for grain number per spike, and hence the wheat yield potential. However, the allohexaploid wheat genome makes genetic dissection highly challenging. Recent breakthroughs in genome sequencing have enabled transcriptome analysis possible for this important food crop (Feng et al., 2017; Li et al., 2014). In this study, we analyzed the transcriptomes of young spikes in 90 winter wheat lines and correlated expression with variations in spike complexity traits (spikelet number per spike, floret number per spike, and seed number per spike). Together with weighted gene coexpression network analysis, we inferred candidate genes that may relate to spike complexity. Furthermore, experimental studies identified genes whose expression is not only associated with but also affects spike complexity. The associative transcriptomic method employed in this work may allow us to identify genetic basis of agronomically important traits in common wheat or other crops with complex genomes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
A total of 90 winter wheat (Triticum aestivum) varieties were grown at the Experimental Station of Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing for 2 consecutive years (planted in September in 2013 and 2014) with three replications and were arranged using Randomized Complete Block Design. For each replication, 60 wheat seeds per variety per year were sown in two 1.5-m-long rows with 25 cm row-row distance. For each variety in each replication, reproductive tissues at the early double ridge stage (see below and Fig. 1A) from 10 randomly selected spikes of the main shoot were collected for transcriptome analysis. At the early double-ridge stage, spikelet primordia occur between bract primordia at the middle part of a spike. For sample collection, leaves surrounding the young spike were removed by hand and the reproductive tissue (without stem) was cut with a sharp blade under a stereomicroscope to confirm developmental stage. In total, 30 spikes per variety per year were collected and the reproductive tissues were further sampled. The reproductive tissues were frozen in liquid nitrogen, after which total RNA was extracted using the RNA Miniprep Kit (Axygen). Equal amounts of total RNA from each year were pooled together for each variety. At least 3 µg of total RNA from each variety was used to construct a sequencing library using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs). Paired-end sequencing libraries with an insert size of ∼250 bp were sequenced on an Illumina HiSeq 2500 sequencer. The number of spikelets per spike, number of florets per spike, and the number of seeds per spike were also investigated for 10 randomly selected plants in each replication 20 d after flowering.
Evaluation of Wheat Spike Development Stages
The process of wheat spike development was considered as five separate stages (Bonnett, 1936; Fisher, 1973). In brief, the spike elongation stage (stage I) refers to the stage when the shoot apex growing point elongates, but its outline remains smooth. During the following single ridge stage (stage II), prematurely ceased foliar primordia appear as ridges surrounding the stem apex. The intervening tissue between two neighboring single ridges then enlarges to form an additional ridge (an SM) in conjunction with the subtending single ridge, marking the double-ridge stage (stage III). In the following differentiation stage (stage IV), glumes, followed by lemmas, initiate from SMs. Florets initiate as lateral swellings above the lemma initials. In the stamen and pistil stage (stage V), three stamens and one pistil emerge in the middle part of a spike, while upper spikelets develop floret primordia.
Read Alignment and Gene Expression Quantification
We downloaded the reference wheat genome sequence (IWGSC CSS + POPSEQ) and gene annotation from the Ensembl plant database (ftp://ftp.ensemblgenomes.org/pub/plants/release-28/). We also downloaded a recently released wheat genome assembly and the INSDC TGACv1 gene annotation. We mapped RNA-seq reads to both of the assemblies and gene annotation, as well as the prereleased IWGSC WGA v0.4 assembly that lacks gene annotation. We detected more expressed genes by using IWGSC CSS + POPSEQ (Supplemental Table S16), although INSDC TGACv1 and IWGSC WGA v0.4 gave slightly higher mapping rates of reads (Supplemental Table S17). We also found that aligning to IWGSC CSS + POPSEQ and INSDC TGAC v1 showed highly similar gene-expression profiles (Supplemental Fig. S6). Thus, we chose IWGSC CSS + POPSEQ as a reference genome and annotation, which are expected to give results similar to those of other assemblies.
For read alignment and expression quantification, we first removed low-quality reads, after that, we mapped the remaining reads to the reference genome using STAR version 2.4.2a (Dobin et al., 2013), allowing mismatches of 6 nucleotides at most on the paired-end reads. To eliminate false discovery of split junction reads, the intron length was set to 60 to 6000 nucleotides. Using HTSeq version 0.6.0, we counted uniquely mapped reads, normalized the read count by the trimmed mean of M values and transformed the results to reads per RPKM using edgeR version 3.12.1 (Robinson et al., 2010). Low abundance genes with an expression cutoff of RPKM ≥ 1 in at least one variety were removed from the set.
Phylogenetic Tree Construction
Construction of the phylogenetic tree was based on the SNPs identified from the RNA-seq data. First, we analyzed each variety using GATK version 3.3 (Van der Auwera et al., 2013; Supplemental Fig. S8). As the GATK workflow failed to report homozygous SNP sites before imputation, we recalled missing genotypes if they were covered by RNA-seq reads. After filtering out SNPs with minor allele frequency ≤ 3%, we imputed missing genotypes again using beagle version 4.1 (Browning and Browning, 2016). To obtain a set of representative SNPs for phylogenetic tree construction, we kept only SNPs anchored on chromosomes with LD ≤ 0.2. We randomly selected 20,000 SNPs for the construction of phylogenetic tree using maximum likelihood method implemented in DNAML (Felsenstein, 1989).
Identification of Homeologous Groups HGs
To avoid misidentification of HGs because of paralogous genes within the same subgenome, we considered both sequence homology and chromosomal linearity when one subgenomic homeolog had multiple best-hit counterparts in two other subgenomes. Specifically, we mapped the cDNA sequences of a wheat gene model with RPKM value ≥1 to the other two reference subgenomes using BLAT (Kent, 2002). As approximately 99% of annotated genes (exon + intron) have a size of less than 15,000 bp, we set the maximum intron size to 15,000 bp. Hits with identity less than 70% or that did not overlap with any annotated genes were filtered before identifying best-hit pairs. This procedure was performed repeatedly for each subgenome against the other two subgenomes. The HGs showing consistent one-to-one correspondence were retained and finally classified into seven categories, namely ABD, AB, AD, BD, A, B, and D type.
Gene Function Annotation Refinement and Enrichment Analysis
To gain a comprehensive gene annotation, we aligned wheat protein sequences to rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) using BLASTP with an e-value cutoff of 1e-5. We further curated a set of development regulators by referring the annotation of GO or MapMan. The GO annotation was downloaded from Ensembl Plant Biomart (https://plants.ensembl.org/biomart/martview), and MapMan pathway mappings were download form MapMan Store (http://mapman.gabipd.org/web/guest/mapmanstore; Usadel et al., 2009). To functionally categorize the genes positively or negatively correlated with spike complexity, we performed GO enrichment analysis using BiNGO version 3.0.3 (Maere et al., 2005) with hypergeometric test and considered terms with an FDR below 0.05 as significant and visualized the results as a network by EnrichmentMap version 2.2.1 (Merico et al., 2010).
Gene Coexpression Network Analysis
Scale-free coexpression network analysis was performed on log2 transformed RPKM values of expressed genes using the WGCNA package (v 1.51) in R (Langfelder and Horvath, 2008). An unsigned coexpression network was constructed for all pairwise Spearman correlations of gene expression. To weight highly correlated genes, we set the soft threshold power to 9, as determined by assessment of scale-free topology (Supplemental Fig. S7A). For network construction, we used a dynamic tree cutoff 0.20 to merge similar trees (Supplemental Fig. S7, B and C). To identify networks associated with spike-trait variables, we calculated the eige value of each module, after which Spearman’s rank correlation was calculated between the eigen value (overall expression trend of the genes in each module) and trait quantity.
Construction of Overexpressing Wheat Lines
Winter wheat variety Kenong 199 (KN199) was used to amplify gene sequences and generate transgenic wheat plants. To obtain transgenic wheat plants, the entire coding region of 10 genes were inserted separately into pUbi-pAHC25, a modified vector for wheat gene overexpression driven by the maize ubiquitin promoter (Wang et al., 2013). The resulting constructs were transformed into immature embryos of wheat variety KN199 by particle bombardment (Becker et al., 1994). At least 45 independent transformants were obtained and analyzed for transgene expression.
Spike Phenotype Analysis of Wheat Overexpression Lines
Seeds of transgenic lines (including lines transformed with an empty vector) were surface-sterilized in 2% NaClO for 15 min and rinsed overnight with flowing water, after which they were sown in soil and allowed to grow for 40 d in a 4°C environment. After 40 d, the seedlings were transferred to a greenhouse with long day condition (16-h-light/8-h-dark photoperiod, light intensity of 350 μmol photons m−2 s−1, ambient temperature of 22°C–25°C, and relative humidity of 60%–70%). Spike phenotypes were recorded for 30 to 45 randomly selected transgenic plants 20 d after flowering.
RT-qPCR
Total RNA was extracted from young leaves of transgenic overexpressing plants using the RNA Miniprep Kit (Axygen). First-strand cDNA was synthesized from 2 μg of DNase I-treated total RNA using the TransScript First-Strand cDNA Synthesis SuperMix Kit (TransGen) as recommended by the manufacturer and was stored at −20°C. RT-qPCR analysis was performed using the PrimeScript RT reagent Kit (TaKaRa) and a Bio-Rad CFX96 Real-time PCR detection system. Relative gene expression levels were determined using the method of Livak and Schmittgen (2001). As the nucleotide sequences of the homologous genes of TaTFL1, TaPAP2, and TaVRS1 were highly similar, gene-specific primers for TaTFL1-2D, TaPAP2-5A, and TaVRS1-2B were not designed. In this study, the relative expression levels of TaTFL1, TaPAP2, and TaVRS1 reflected the transcript abundance of the homologous genes of the three genes. We used β-TaTubulin mRNA as the internal control for RT-qPCR analysis. The primers used for RT-qPCR are listed in Supplemental Table S15.
Scanning Electron Microscopy
For scanning electron microscopy sem, young spikes from KN199 and T4 transgenic plants overexpressing TaTFL1-2D, TaPAP2-5A, and TaVRS1-2B at different stages were fixed overnight in 2.5% glutaraldehyde at 4°C. After dehydration in a series of ethanol solutions and substitution with 3-methylbutyl acetate, the samples were subjected to critical point drying, coated with platinum, and observed using a Hitachi S-3000N variable pressure scanning electron microscope.
Accession Numbers
The raw read data for this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRP091625.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Major early developmental stages of the wheat spike.
Supplemental Figure S2. Saturation analysis of transcriptomic depth.
Supplemental Figure S3. Transcriptome association analysis of spike complexity.
Supplemental Figure S4. Correlation of the expression levels of the three subgenomic copies of TaTFL1, TaPAP2, and TaVRS1 with spikelet quantity.
Supplemental Figure S5. Analyses of spike complexity for wild-type KN199 and transgenic wheat plants.
Supplemental Figure S6. Scatter plot of transcript abundance as determined by using two genome assemblies and annotations.
Supplemental Figure S7. Cutoffs used for coexpression network construction.
Supplemental Figure S8. Analytical workflow for this study and detailed pipeline used for variant calling.
Supplemental Table S1. Detailed information of 90 accessions used in the study.
Supplemental Table S2. The number of spikelets per main spike in 90 accessions.
Supplemental Table S3. The number of seeds per main spike in 90 accessions.
Supplemental Table S4. The number of florets per main spike in 90 accessions.
Supplemental Table S5. Summary of reads mapping.
Supplemental Table S6. Statistics of reads mapped to each subgenome.
Supplemental Table S7. Expression abundance of expressed genes in 90 accessions.
Supplemental Table S8. Statistics of the number of genes expressed in each variety.
Supplemental Table S9. Statistics of the number of expressed genes in each chromosome.
Supplemental Table S10. Statistics of the number of each type of HGs.
Supplemental Table S11. List of genes significantly correlated witch spikelet number.
Supplemental Table S12. Wheat genes homologous to rice spike development-related genes.
Supplemental Table S13. Transcription factor enrichment within each module classified by WGCNA.
Supplemental Table S14. Lists of genes in module 9 core subnetwork.
Supplemental Table S15. Primers used to generate transgenic wheat plants.
Supplemental Table S16. Statistics of reads mapped to different version of genome sequences.
Supplemental Table S17. Statistics of the number of expressed genes assessed using different version of genome sequences.
Acknowledgments
We thank Yanbao Tian (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for help with scanning electron microscopy.
Glossary
- SM
spikelet meristem
- GWAS
genome-wide association study
- HG
homeologous group
- SNP
single nucleotide polymorphism
- GES
gene expression signature
- GO
Gene Ontology
- GCN
gene coexpression network
- TF
transcription factor
- FM
floret meristem
Footnotes
This work was supported by the Chinese Academy of Sciences (grant XDA08020105), the Ministry of Agriculture of the People's Republic of China (Chinese Ministry of Agriculture) (2014ZX08010-002), the National Natural Science Foundation of China (grant 31430010), the National Program for Support of Top-Notch Young Professionals, and the State Key Laboratory of Plant Genomics.
Articles can be viewed without a subscription.
References
- Becker D, Brettschneider R, Lörz H (1994) Fertile transgenic wheat from microprojectile bombardment of scutellar tissue. Plant J 5: 299–307 [DOI] [PubMed] [Google Scholar]
- Boden SA, Cavanagh C, Cullis BR, Ramm K, Greenwood J, Jean Finnegan E, Trevaskis B, Swain SM (2015) Ppd1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat Plants 1: 14016. [DOI] [PubMed] [Google Scholar]
- Bonnett OT. (1936) The development of the wheat spike. J Agric Res 53: 445–451 [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, et al. (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR (2016) Genotype imputation with millions of reference samples. Am J Hum Genet 98: 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Derbyshire P, Byrne ME (2013) MORE SPIKELETS1 is required for spikelet fate in the inflorescence of Brachypodium. Plant Physiol 161: 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaya O, Pont C, Sibout R, Martinek P, Badaeva E, Murat F, Chosson A, Watanabe N, Prat E, Gautier N, et al. (2015) FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol 167: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Feng N, Song G, Guan J, Chen K, Jia M, Huang D, Wu J, Zhang L, Kong X, Geng S, et al. (2017) Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory genes. Plant Physiol 174: 1779–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JE. (1973) Developmental morphology of the inflorescence in hexaploid wheat cultivars with and without the cultivar norin 10 in their ancestry. Can J Plant Sci 53: 7–15 [Google Scholar]
- Guo Z, Chen D, Alqudah AM, Röder MS, Ganal MW, Schnurbusch T (2017) Genome-wide association analyses of 54 traits identified multiple loci for the determination of floret fertility in wheat. New Phytol 214: 257–270 [DOI] [PubMed] [Google Scholar]
- Hagemann W. (1990) Comparative morphology of acrogenous branch systems and phylogenetic considerations. II. Angiosperms. Acta Biotheor 38: 207–242 [Google Scholar]
- Hao C, Dong Y, Wang L, You G, Zhang H, Ge H, Jia J, Zhang X (2008) Genetic diversity and construction of core collection in Chinese wheat genetic resources. Chin Sci Bull 53: 1518–1526 [Google Scholar]
- Hao C, Wang L, Ge H, Dong Y, Zhang X (2011) Genetic diversity and linkage disequilibrium in Chinese bread wheat (Triticum aestivum L.) revealed by SSR markers. PLoS One 6: e17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, Hattori C, Werner P, Bancroft I (2012) Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol 30: 798–802 [DOI] [PubMed] [Google Scholar]
- He ZH, Rajaram S, Xin ZY, Huang GZ, editors (2001). A History of Wheat Breeding in China. CIMMYT, Mexico DF. [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Kent WJ. (2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130: 3841–3850 [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, et al. (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Liu D, Wu J, Zhao X, Hao M, Geng S, Yan J, Jiang X, Zhang L, Wu J, et al. (2014) mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: 1878–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Maccaferri M., Zhang J., Bulli P., Abate Z., Chao S., Cantu D., Bossolini E., Chen X., Pumphrey M., Dubcovsky J. (2015). A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 (Bethesda) 5: 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010) Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One 5: e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, Mayer KF, Olsen OA; International Wheat Genome Sequencing Consortium (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345: 1250091. [DOI] [PubMed] [Google Scholar]
- Poursarebani N, Seidensticker T, Koppolu R, Trautewig C, Gawroński P, Bini F, Govind G, Rutten T, Sakuma S, Tagiri A, et al. (2015) The genetic basis of composite spike form in barley and ‘Miracle-Wheat.’ Genetics 201: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Schnurbusch T (2012) A genetic playground for enhancing grain number in cereals. Trends Plant Sci 17: 91–101 [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Zhang F, Yan X, Zhang X, Dong Z, Cui D, Chen F (2017) Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol J 15: 953–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Pautler M, Jackson D, Hirano HY (2013) Grass meristems II: Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol 54: 313–324 [DOI] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M (2009) A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ 32: 1211–1229 [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013). From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, et al. (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot (Lond) 111: 1139–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Borowsky AT, Tzin V, Jander G, Kliebenstein DJ, Rokas A (2017) A global co-expression network approach for connecting genes to specialized metabolic pathways in plants. Plant Cell 29: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Yonemaru J, Yamamoto T, Yano M (2012) OGRO: The overview of functionally characterized genes in rice online database. Rice (N Y) 5: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]