Chromothripsis-like genome rearrangements cause somatic loss of fruit pigmentation and alter sexual reproductive development in grapevine.
Abstract
Grape (Vitis vinifera) color somatic variants that can be used to develop new grapevine cultivars occasionally appear associated with deletion events of uncertain origin. To understand the mutational mechanisms generating somatic structural variation in grapevine, we compared the Tempranillo Blanco (TB) white berry somatic variant with its black berry ancestor, Tempranillo Tinto. Whole-genome sequencing uncovered a catastrophic genome rearrangement in TB that caused the hemizygous deletion of 313 genes, including the loss of the functional copy for the MYB transcription factors required for anthocyanin pigmentation in the berry skin. Loss of heterozygosity and decreased copy number delimited interspersed monosomic and disomic regions in the right arm of linkage groups 2 and 5. At least 11 validated clustered breakpoints involving intrachromosomal and interchromosomal translocations between three linkage groups flanked the deleted fragments, which, according to segregation analyses, are phased in a single copy of each of the affected chromosomes. These hallmarks, along with the lack of homology between breakpoint joins and the randomness of the order and orientation of the rearranged fragments, are all consistent with a chromothripsis-like pattern generated after chromosome breakage and illegitimate rejoining. This unbalanced genome reshuffling has additional consequences in reproductive development. In TB, lack of sexual transmission of rearranged chromosomes associates with low gamete viability, which compromises fruit set and decreases fruit production. Our findings show that catastrophic genome rearrangements arise spontaneously and stabilize during plant somatic growth. These dramatic rearrangements generate new interesting phenotypes that can be selected for the improvement of vegetatively propagated plant species.
Somatic variation is a major source of diversity in multicellular organisms and is generated by unique mutation events affecting single dividing cells. Somatic mutations can spread by cell division to generate mutant sectors or cell lines derived from the original mutant cell. Somatic mutations range from single-nucleotide variation (SNV) and insertions-deletions (INDELs) to complex genome structural variation (SV) originating from chromosomal rearrangements (Li, 2016). Recently, the rapidly growing number of medical genomic studies has unveiled highly complex forms of SV that are collectively known as chromoanagenesis (Collins et al., 2017). Among them, the most commonly reported case is chromothripsis (for chromosome shattering), a cellular catastrophe initially described in human cancer that affects a few chromosomes (one to four) or chromosome arms and results in multiple clustered rearrangements (five or more breakpoints) and deletions after chromosome breakage and illegitimate rejoining (Stephens et al., 2011; Korbel and Campbell, 2013; Collins et al., 2017). Chromothripsis has been reported in mammals, but it has never been related to natural somatic variation in the plant kingdom.
Given their capacity for asexual propagation, plants may be better systems than animals in which to study somatic variation and its effects on somatic cells (Whitham and Slobodchikoff, 1981). Plant species lack a separated germ line, and somatic genetic variation represents an important source of phenotypic variation that may be adaptive and transmitted to the next sexual generation when present in the meristem cell layer giving rise to gametes (D’Amato, 1997). Somatic variation is especially relevant in plants such as long-living trees or vegetatively propagated species in which single genotypes are perpetuated for very long periods and can colonize vast extensions, which increase the likelihood of somatic mutation emergence and accumulation. This case is applicable to woody crops such as grapevine (Vitis vinifera), from which cultivar genotypes are asexually propagated by cuttings and perpetuated from the original sexual seedling that, in many cases, germinated several centuries ago (This et al., 2006). Somatic mutations generating new interesting phenotypes that are stabilized in grapevine plants as periclinal chimeras or that extend to all cell layers have been selected as new clones of wine grape cultivars or as new derivative cultivars (This et al., 2006; Pelsy et al., 2010; Torregrosa et al., 2011). Recently, genetic and genomic approaches identified SNV and new transposable element (TE) insertions responsible for emergent phenotypes in several grapevine somatic variants (Boss and Thomas, 2002; Fernandez et al., 2010, 2013; Battilana et al., 2011; Emanuelli et al., 2014).
Given their conspicuousness, berry color somatic variants have occasionally been selected and studied in grapevine (Kobayashi et al., 2004; Walker et al., 2006; Yakushiji et al., 2006; Furiya et al., 2009). Grape color results from the accumulation of anthocyanins generally in the berry skin (Boss et al., 1996), which is genetically controlled by a major locus on linkage group (LG) 2 (Doligez et al., 2006). This locus colocalizes with a cluster of tandemly repeated VviMybA genes (Wong et al., 2016). Functional VviMybA1 and VviMybA2 genes are essential for grape pigmentation, whereas the presence of a Gret1 retrotransposon insertion in the promoter of VviMybA1 along with a small INDEL causing a frame shift in VviMybA2 are responsible for a null allele of the grape color locus that is frequently in homozygosis in white berry cultivars (Kobayashi et al., 2004; Lijavetzky et al., 2006; This et al., 2007; Walker et al., 2007). Black berry cultivars heterozygous for the null allele occasionally display grape color variants with either red/gray or white berries depending on whether only the L2 or both L1 and L2 meristem cell layers, respectively, carry mutations at the color locus (Walker et al., 2006; Furiya et al., 2009; Vezzulli et al., 2012; Migliaro et al., 2014; Pelsy et al., 2015). Initial characterization of red/gray and white berry somatic variants of Cabernet Sauvignon and Pinot Noir using Southern blots showed that the absence of anthocyanins in the berry was related to deletion of the functional allele of the color locus (Walker et al., 2006; Yakushiji et al., 2006). Recently, the use of loss of heterozygosity (LOH) analyses along LG 2 suggested that the size of these deletions can be quite variable in different berry color somatic variants (Vezzulli et al., 2012; Migliaro et al., 2014; Pelsy et al., 2015). Unfortunately, no information is available yet on the breakpoints delimiting these deletions, which is required to understand their mutational origin.
Tempranillo Blanco (TB) is a white berry somatic variant that originally appeared as a bud sport mutant of Tempranillo Tinto (TT) (Martinez et al., 2006). Like many black berry grapevine cultivars, TT is heterozygous for the color locus functional allele, and TB has been proposed to appear by deletion of this allele (Ibáñez et al., 2012; Migliaro et al., 2014). To understand the origin of the TB variant, we carried out SV analysis after whole-genome sequencing (WGS) of TT and TB lines. SV breakpoints were confirmed by DNA sequencing of specific amplicons, and self-cross progeny were used for haplotype phasing and transmission analysis. Globally, the results show the relevance of complex SV as a driver of clonal variation in plants.
RESULTS
Hemizygous Deletions around the Color Locus Associate with Anthocyanin Biosynthesis Blockage in TB
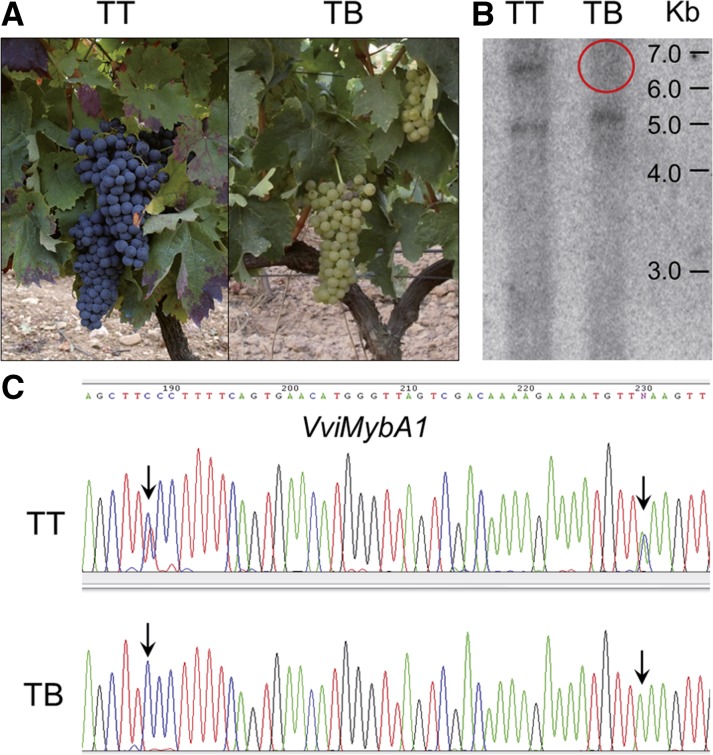
TB is a grapevine cultivar generated by vegetative propagation of the white-berried mutant phenotype that appeared as a spontaneous bud sport in a TT plant (Martinez et al., 2006; Fig. 1A). In TB, the presence of a hemizygous deletion eliminating the functional allele of the VviMybA1 gene on the color locus at LG 2 was confirmed using DNA-blot hybridization of DNA extracted from both TT and TB plants (Fig. 1B). The sequence of a specific amplicon corresponding to part of the VviMybA1 gene showed LOH in TB, further confirming the presence of a hemizygous deletion affecting at least this gene in TB (Fig. 1C). Given the high heterozygosity of grapevine cultivars (Laucou et al., 2011), the extent of the deletion in TB was first assessed using primer pairs (Supplemental Table S1) intended for LOH analysis of amplified fragments along the putatively affected region in LG 2. This analysis showed that putative hemizygous deletions extended outside of the MybA gene cluster in LG 2 (Supplemental Table S2). Unfortunately, the efficiency of this method to identify the loss of one allele is limited to originally heterozygous genomic regions, and, coincident with other studies (Migliaro et al., 2014), many tested amplicons only provided monomorphic sequences in the TT original cultivar, with no polymorphic site detected after position chr2:15,861,181 (Supplemental Table S2) in the PN40024 12X.0 reference genome assembly (https://urgi.versailles.inra.fr/Species/Vitis/Data-Sequences/Genome-sequences).
Figure 1.
Initial characterization of a TB white grape variant line. A, TT and TB plant phenotypes. Mature plants show the absence of anthocyanin pigments in TB bunches. B, Southern blot for a VviMybA1 probe showing the loss of one copy in TB. Only the smallest positive fragment, corresponding to the nonfunctional allele with a predicted ApaI target site within the Gret1 retroelement, is kept in TB. C, Loss of VviMybA1 heterozygosity in TB. Only one VviMybA1 haplotype is kept in TB according to the LOH at original heterozygous single-nucleotide polymorphisms (SNPs) in TT.
To globally predict the extension of hemizygous regions in TB irrespective of the heterozygosity level in TT, the GrapeGen GeneChip cDNA-based microarray (Lijavetzky et al., 2012) was initially used for comparative genomic hybridization (CGH). Significant copy number variation (CNV) was detected for probe sets corresponding to 31 annotated genes, and all but one involved decreased copy number in TB compared with TT (Supplemental Table S3), 14 of them mapping on LG 2 around the color locus and extending to regions that are homozygous in TT according to the amplicon analysis (Supplemental Fig. S1). Thus, both copy number decrease and LOH supported hemizygosity around the LG 2 color locus in TB. Unexpectedly, the signal of probe sets corresponding to 14 genes mapping on LG 5 decreased significantly in TB too (Supplemental Table S3). Plotting of signal log ratios along reference genome chromosomes suggested the existence of three hemizygous regions of approximately 1 Mb each in LG 5 of TB (Supplemental Fig. S1). Thus, CGH microarrays suggested complex SV in the TB genome involving multiple deletions in LGs 2 and 5. However, the use of a cDNA-based microarray did not provide enough resolution to identify the limits of this sequence variation, which required a WGS comparison.
WGS Detects Interspersed Monosomic and Disomic Regions in LGs 2 and 5 of TB
To delimit SV occurrence in TB, WGS was performed on both Tempranillo lines using 90-nucleotide paired-end reads. A total of ∼209 × 106 clean reads per cultivar were obtained (Supplemental Table S4). A similar alignment rate was obtained in both Tempranillo lines, with more than 90% coverage of the reference genome in an average effective depth of 30× (Supplemental Table S5).
Heterozygosity Loss
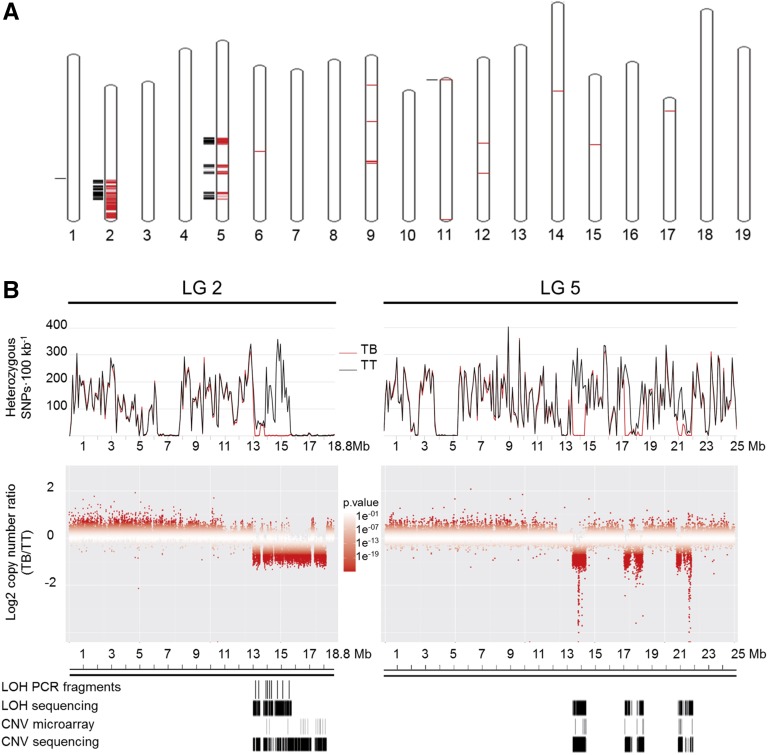
According to the same logic followed previously for selected amplicons, LOH analysis from WGS data was used to widely detect the loss of one allele at original heterozygous sites. Coincident with microarray-based CGH results, large LOH regions were detected in TB LGs 2 and 5, collectively spanning 4.3 Mb (Fig. 2; Supplemental Table S6). Additionally, two small windows were detected in LGs 1 and 11, spanning 760 and 1,027 bp (Fig. 2A; Supplemental Table S6). Monomorphic segments were detected for both TT and TB in LG 2 (6.3–7.7 and 15.8–18.8 Mb) and LG 5 (3.8–5.2 and 12.6–12.9 Mb; Fig. 2B), precluding their LOH analysis in TB.
Figure 2.
Genome-wide scanning of LOH and copy number decrease in TB. A, Genome-wide representation of regions with significant LOH or copy number decreases in TB. Significant intervals were mapped to chromosomes of the PN40024 12X.0 reference genome using the V. vinifera whole-genome tool of EnsemblPlants (http://plants.ensembl.org/Vitis_vinifera/Location/Genome). Black outer lines show chromosome segments with significant LOH in TB. Red inner lines show chromosome segments with significantly decreased copy numbers in TB. B, Close-up of heterozygosity and CNV comparison between TT and TB resequencing data mapping on LGs 2 and 5. Top, Heterozygosity levels in TT (black traces) and TB (red traces). Middle, Copy number ratios between TB and TT. Windows are represented in increasing red color tones corresponding with CNV significance P values. Bottom, Comparison of LOH and CNV segments in TB detected by WGS and other approaches.
CNV
To identify CNV in TB at the whole-genome scale irrespective of the heterozygosity level, we compared the number of reads aligned to the 12X.0 reference genome between the two Tempranillo lines. Large areas with significant copy number decreases in TB were detected in LGs 2 and 5 (Fig. 2A; Supplemental Table S7). In LG 2, significantly lower copy number in TB extended to at least three alternating segments located between 13 and 18.2 Mb, close to the 3′ end of the chromosome (Fig. 2B). The extension of deletions to TT homozygous regions observed in CGH microarrays was confirmed by the CNV analysis of WGS data. The lower copy number in LG 5 of TB comprised at least three alternating segments located between 13.4 and 21.9 Mb, also interspersed with segments showing unaltered copy number (Fig. 2B). In LGs 2 and 5, the overlap between LOH and decreased copy number strongly indicated that both events result from the deletion of fragments leaving monosomic chromosome regions in TB. Similarly, the small LG 11 region with LOH in TB also showed a copy number decrease (Fig. 2A). A few other small regions with reductions of copy number in TB were detected in other LGs (Fig. 2A). Although microarray and LOH analyses did not confirm them as hemizygous deletions, specific positions showing LOH were detected within decreased copy number fragments that were interspersed in LG 9 between positions 14,733,335 and 14,920,269 (Supplemental Table S7). Altogether, these results support the presence of large hemizygous regions in LGs 2 and 5 of TB. The discontinuity of hemizygosity in these regions indicates the existence of multiple chromosome breaks in the origin of the TB genome. Following the same significance values (see “Materials and Methods”), no parallel significantly increased copy number regions were detected in LGs 2 and 5 of TB, which indicates that duplications are not involved in this SV.
SV Junction Sites Reveal Multiple Interchromosomal and Intrachromosomal Translocations in the Genome of TB
WGS reads were explored to delimit the breakpoints of fragments deleted in TB and to elucidate the nature of the SV. Discordant pair-end mapping (PEM) is useful to detect SV breakpoints from short-read sequencing data (Korbel et al., 2007; Marroni et al., 2014). Thus, TB-specific discordant PEM and soft-clipped reads with respect to the grapevine reference genome revealed six SV junctions in the TB genome not present in TT. Collectively, they comprised six breakpoints in LG 2, five in LG 5, and one in LG 9 (Table I).
Table I. List of SV junctions predicted in the genome of TB by discordant read mapping analysis.
| SV Junction | SV Class | Breakpoint Positions Mapped in the Reference Genome | |
|---|---|---|---|
| Inv | Inversion | chr2:13,025,297 | chr2:17,391,598 |
| T1 | Translocation | chr2:13,537,963 | chr9:14,760,326 |
| T2 | Translocation | chr2:13,764,277 | chr5:14,363,763 |
| T3 | Translocation | chr2:17,143,691 | chr5:21,919,555 |
| T4 | Translocation | chr2:18,226,646 | chr5:20,858,083 |
| T5 | Translocation (intrachromosomal) | chr5:13,404,539 | chr5:18,450,318 |
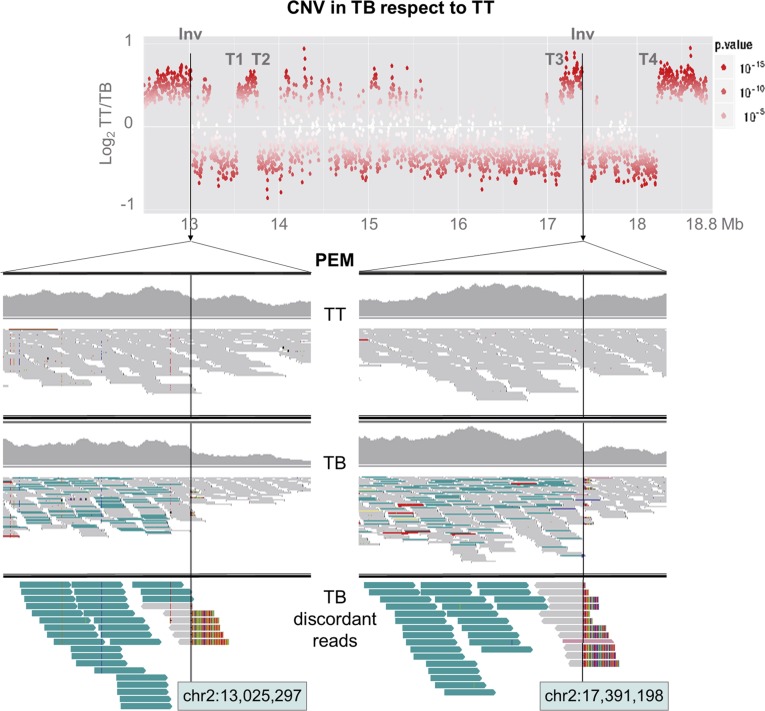
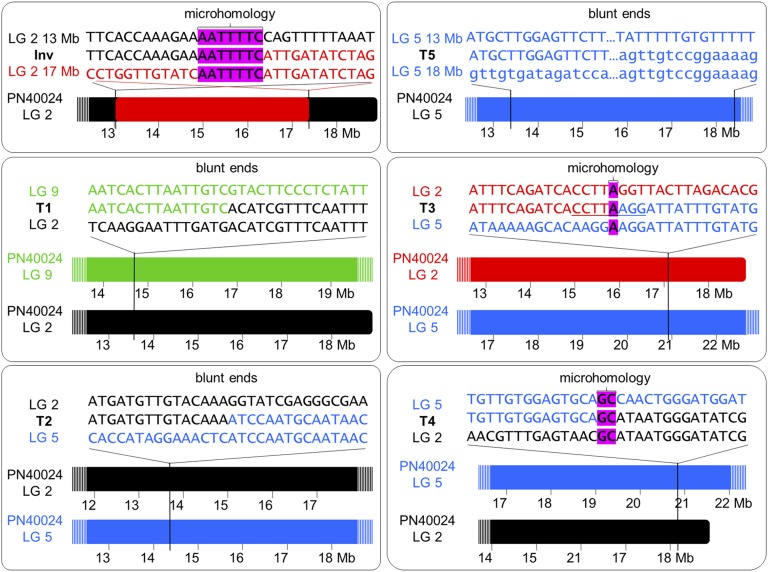
An inversion junction in LG 2 (Inv) was predicted by the identification in TB but not in TT of read pairs mapping on LG 2 with mates separated one from another by more than 4.36 Mb (Fig. 3). Mates at both positions displayed the same alignment orientation, which is compatible with an inversion (Korbel et al., 2007; Rausch et al., 2012). TB-specific soft-clipped reads were detected at both flanks of the junction (Fig. 3). BLASTN identified that the unaligned extreme of these soft-clipped reads matches to the 5′ extreme of the mate inversion flank, unveiling the putative inversion junction breakpoint at nucleotide level resolution (between positions chr2:13,025,297 and chr2:17,391,598; Table I). Similarly, discordant reads identified four SV junctions with flanks in different chromosomes of the reference genome (T1–T4; Table I). These rearrangements were interpreted as interchromosomal translocations and involved LGs 2 and 5 in all cases except T1, involving LGs 2 and 9. For the T5 event, discordant alignment identified an SV junction joining LG 5 fragments originally separated by ∼5 Mb, which is compatible with an intrachromosomal translocation (Supplemental Fig. S2). Interestingly, one flank of all predicted SV breakpoints showed decreased copy number (Fig. 3; Supplemental Fig. S2), suggesting that these recombinations involved hemizygous deletion bridges. In this manner, T2 and T3 breakpoints delimit a deleted fragment 3.38 Mb in LG 2 (chr2:13,764,277–chr2:17,143,691), which contained the allele of the grape color locus that is lost in TB (Fig. 4A). Furthermore, the existence of additional SVs in TB is reasonable, considering that discordant read analysis could not detect the breakpoints for at least one obvious copy number discontinuity in LG 5 and a likely one in LG 9 (Fig. 4A), whereas missing copy number-neutral breakpoints also are possible.
Figure 3.
Inversion breakpoints in TB LG 2. The graph at top shows a sharp decay in copy number at the 3′ flank of the two Inv breakpoints. The other graphs show reads aligned at both Inv breakpoint sites in TT and TB (images obtained from Integrative Genomics Viewer). All aligned reads are displayed in the IGV first (TT) and second (TB) upper windows, while only discordant reads from TB are displayed in the lower window. Reads are represented as arrows showing the orientation of paired-end mates. From the PEM analysis, read pairs with abnormally increased insert size are colored in turquoise and were detected only in TB. These read pairs are sense oriented, as expected in an inversion junction. Soft-clipped reads spanning the breakpoint show an unaligned extreme in colors.
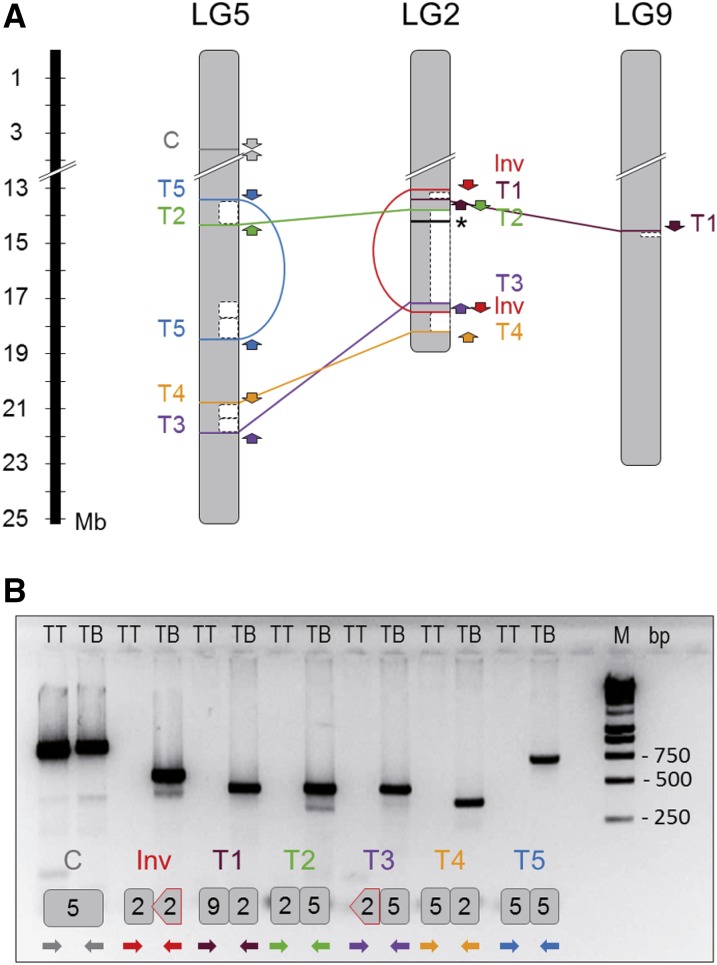
Figure 4.
Mapping and validation of SV junctions detected in the genome of TB. A, Scheme depicting SV breakpoints detected in the TB genome. Breakpoints detected by discordant read analysis from WGS data are mapped on the affected chromosomes according to the PN40024 12X.0 reference genome assembly. One inversion (Inv), four interchromosomal translocation (T1–T4), and one intrachromosomal translocation (T5) junctions were predicted. Hemizygous chromosome regions detected by LOH and/or CNV from WGS data are indicated with white boxes. PCR primers used for validation are represented by arrows according to their mapping positions and orientation with respect to the reference genome (Inv, red; T1, brown; T2, green; T3, orange; T4, purple; T5, blue; the arrow head indicates the 3′ end of the primer). The asterisk indicates the position of the cluster of MybA genes at the grape color locus on LG 2. B, Agarose gel electrophoresis image showing the PCR amplification product obtained using primer pairs designed to amplify Inv and T1 to T5 SV junctions. Primer pairs designed for the TB junctions as well as the positive control (C) were tested for TB and TT. In the gel image, below the two electrophoresis wells corresponding to the same primer pair combination, a scheme illustrating the expected amplicon in TB is shown. Chromosome fragments involved in the rearrangement junction included in the amplicon are represented in the scheme. Inverted fragments are represented as arrow-ended blocks. The alignment of PCR primers colored as in A is depicted by arrows below the amplicon. M, DNA size marker; TB, TB gDNA was used as a template; TT, TT gDNA was used as a template.
Experimental Validation of Genome SVs in TB
To validate predicted SV junctions, small contigs (450–900 bp) were reconstructed in silico from the sequences of the discordant reads that predicted each junction. Then, we ran PCR using primer pairs (Supplemental Table S1) with mates mapping at each flank of the predicted SV junction breakpoint. No amplicon was expected in TT, given the incompatible orientation between primer pairs in Inv, their excessive alignment distance in Inv and T5, or their alignment in different chromosomes in T1 to T4 (Fig. 4A). As expected, amplification was obtained from TB but not from TT genomic DNA (gDNA) for all primer pairs except for the positive control (in a region without SV) that was amplified in both genotypes (Fig. 4B). The actual sequences of the amplicons obtained in TB were checked by capillary electrophoresis Sanger sequencing (Supplemental Table S8). BLAST alignment of these sequences against the 12X.0 reference genome confirmed the fragments and breakpoints/junctions predicted by WGS in the absence of point polymorphisms compared with TT. Thus, these results validate all SV junctions identified in TB from the WGS comparison. Furthermore, their presence and stability were confirmed by obtaining the same amplicons in five additional TB lines that were tested, including the most direct descendant of the original bud sport that is conserved, a first-round propagation plant (Supplemental Fig. S3; Supplemental Table S9). The same amplicons were obtained using either gDNA isolated from berry flesh (L2) or skin (L1 contaminated with L2) tissues of TB, suggesting that the same genome rearrangement was present in both meristem cell layers of TB bud sport. Despite analyzing five additional TT accessions likely belonging to clonal lineages close to the original TT ancestor, no amplicon corresponding to SV junctions identified in TB was obtained in any tested TT line (Supplemental Fig. S3). These results suggest that all described genome rearrangements appeared together associated with the emergence of berry color variation and have been stable over successive mitotic growth and propagation cycles.
Rearrangements in TB Are Characterized by Nonhomologous Breakpoint Joins
To understand the forces driving the rearrangements originating the variant genome of TB, we searched for repetitive sequences and motifs in SV junctions. In four of them (T1–T3 and T5), one junction flank matched, with high similarity score, to fragments of Gypsy-27, VHARB-N2, VLINE1, and VIHAT3 grapevine-specific TEs, respectively (Table II). These TE-like sequences were found only in one breakpoint donor site for each of these SV junctions (Supplemental Fig. S4), precluding the existence of nonallelic homologous recombination in the origin of the rearrangements. However, the involvement of these TE sequences in TB SV through an uncertain mechanism cannot be completely ruled out, given that they appear to be enriched within the sequenced SV junctions (Table II). On the other hand, no match with any repeat element deposited in RepBase was obtained for the Inv junction. Nevertheless, a 7-bp microhomology was observed between both breakpoints of this event (Fig. 5; Supplemental Table S8). Furthermore, 1-bp microhomology in the context of an 8-bp palindrome (CCTTAAGG) was present at the T3 junction site, while a 2-bp microhomology was found in T4. Blunt ends with no homology were detected for T1, T2, and T5 junctions (Fig. 5).
Table II. Features of breakpoint junctions sequenced in derivative chromosomes of TB.
| Sequenced Breakpoint Junction | Sequenced Length | Microhomology in Junction | Breakpoint Flank | Repetitive Elements | Repetitive Element in Total Sequenced Junctions | Repetitive Element in the Grapevine Reference Genome |
|---|---|---|---|---|---|---|
| T1 | 730 bp | Blunt ends | T1_LG2 | Gypsy-27_VV-I | 8.9% | 1.19% |
| T1_LG9 | – | |||||
| T2 | 621 bp | Blunt ends | T2_LG2 | VHARB-N2_VV | 7.1% | 0.10% |
| T2_LG5 | – | |||||
| T3 | 529 bp | 1 bp in an 8-bp palindrome | T3_LG2 | – | ||
| T3_LG5 | VLINE1_VV | 5.2% | 1.91% | |||
| T4 | 437 bp | 2 bp | T4_LG2 | – | ||
| T4_LG5 | – | |||||
| T5 | 807 bp | Blunt ends | T5_Proximal | VIHAT3 | 10.9% | 0.12% |
| T5_Distal | – | |||||
| Inv | 647 bp | 7 bp | Inv_Proximal | – | ||
| Inv_Distal | – |
Figure 5.
Nature of breakpoint junction sequences detected in TB. For each validated SV junction site, close-ups of breakpoint and junction site sequences are shown at top (LG 2 with regular orientation in black, LG 2 with inverted orientation in red, LG 5 in blue, and LG 9 in green). Breakpoints in the context of the PN40024 12X.0 reference genome assembly chromosomes are represented at bottom.
Linkage of Rearrangements in a Single Copy of the Affected Chromosomes and in Silico Reconstruction of the TB Derivative Genome
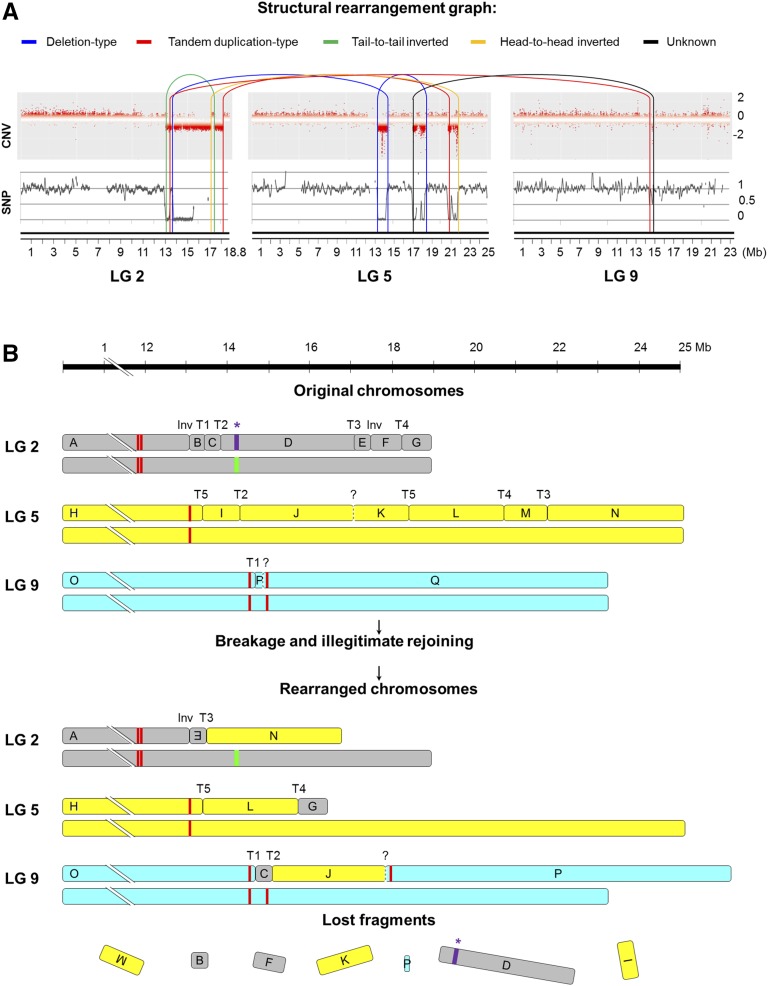
Trying to reconstruct the haplotype phasing of rearrangements in the TB genome, we carried out high-throughput genotyping of a TT self-cross progeny (S1) comprising 78 siblings. S1 individuals were genotyped using GrapeReSeq Illumina 18K SNP Infinium chips (Houel et al., 2015). Subsequently, the haplotype of each TT homologous chromosome in LGs 2 and 5 was reconstructed according to the data of 286 SNP markers that were heterozygous in TT (Supplemental Table S10). The genotype of TB at these SNP positions was inferred from our WGS data, and comparison with the two TT haplotypes showed that all hemizygous deletions detected in TB were linked in a single homologous copy of LGs 2 and 5 (Supplemental Table S10). In view of that and considering detected CNV, LOH, and breakpoint junctions (Fig. 6A), we could in silico reconstruct the TB derivative chromosomes in a single homologous copy of the affected LGs (Fig. 6B). In this reconstruction, we hypothesized an additional translocation junction that was not identified in the PEM analysis to connect hemizygous deletion breakpoints detected by the CNV analysis at LGs 5 and 9 (Figs. 2B and 4A). In fact, although not passing all the significance thresholds in the SV analysis, read pairs supporting this translocation join, and delimiting a deletion of an ∼16-kb fragment in LG 9 of TB according to CNV, were detected from sequencing read alignments. Altogether, 13 breakpoints and 16 chromosome fragments were considered to reconstruct the three derivative chromosomes of TB (Fig. 6B). According to this model, no other undetected breakpoint would be required to explain this catastrophic rearrangement.
Figure 6.
DNA structural rearrangement graph and predicted rearranged chromosomes in TB. A, Structural rearrangement graph depicting CNV, LOH, and rearrangement joins and types in the affected LGs. CNV is shown as log2 (copy number ratio TB/TT) calculated for each window of the affected chromosomes by CNV-seq. SNP heterozygosity level is depicted as log2 (heterozygous SNPs per 100-kb ratio TB/TT). Connections between rearranged fragments are depicted by curved lines colored as indicated at the top of the image according to join types observed in the paired-end alignment analysis. B, Diagram depicting the original and the predicted rearranged orders of chromosome fragments affected by the chromothripsis-like restructuring that originated the variant genome of TB. Deleted fragments in TB were predicted by LOH and CNV from WGS data, and their extremes were confirmed by PCR of breakpoint junctions in most cases. Fragment order in TB derivative chromosomes was predicted considering the six deleted fragments and the six breakpoint junction sequences detected in the discordant read analysis that were validated by PCR (Inv, inversion; T, translocation). Question marks represent an additional breakpoint junction that was hypothesized from WGS read pairs that collectively did not pass all the SV analysis significance thresholds. According to this model, no other undetected breakpoint would be required to explain this catastrophic rearrangement. Rearrangements are represented in a single copy of the affected chromosomes according to the linkage of deleted fragments in the same chromosome haplotype, which was inferred from a TT self-cross progeny segregation analysis and WGS data of TB. In purple, the asterisk indicates the segment comprising the cluster of MybA genes belonging to the grape color locus functional allele, which is present in the heterozygous state in TT and is lost after genome rearrangements in TB. The white allele is represented by the green line. Red lines denote centromeric repeats according to predictions on the grapevine reference genome (Di Gaspero and Foria, 2015; Genoscope Web site).
The rearrangement pattern detected in TB resembles the features described for chromothripsis events (Stephens et al., 2011; Korbel and Campbell, 2013; Collins et al., 2017). Accordingly, computational and statistical analyses proposed for the inference of chromothripsis events were carried out, confirming that all the criteria are fulfilled in this grapevine somatic variant. (1) TB is characterized by interspersed regions with monosomic and disomic copy number states flanked by clustered breakpoints in the right arm of LGs 2 and 5, in the absence of significant copy number gain along the affected chromosome regions (Figs. 2 and 4). A goodness-of-fit test confirmed (95% confidence in the Kolmogorov-Smirnov test) that PCR-validated breakpoints are not distributed exponentially along LGs 2 and 5, which is in agreement with clustering rather than random distribution of breakpoints along the rearranged chromosomes as described for chromothripsis. (2) Rearrangements are linked in a single homologous copy of LGs 2 and 5 (Supplemental Table S10), which, along with SV data, allowed us to reconstruct and walk the tentative single copy of derivative chromosomes following a head-to-tail sequence of fragments (Fig. 6), as expected in a chromothripsis scenario (Korbel and Campbell, 2013). (3) Simulation analyses confirmed the randomness of the predicted fragment order in TB derivative chromosomes. A rearrangement order value of 81 was estimated for TB considering fragment order coordinates as described by Korbel and Campbell (2013). This value is within the normal distribution or randomness interval (68.9 ± 15.7) estimated by three independent rounds of 108 Monte Carlo simulations. (4) The proportion of join types in validated SV junctions is a probable result (P = 0.703) of a multinomial distribution with equal probability for each of the four possible join types (Fig. 6A), suggesting that fragments were randomly joined as expected in a single catastrophic event (Stephens et al., 2011; Korbel and Campbell, 2013). (5) The DNA sequence of the breakpoint joins validated in TB shows a lack of homology (Fig. 5), which is a hallmark of breakage-repair mechanisms like those involved in chromothripsis (Kloosterman et al., 2012; Malhotra et al., 2013). Altogether, the rearrangement pattern of the TB derivative genome is compatible with the characteristics that have been described for chromothripsis events.
Genetic Alterations Caused by the Genome Reshuffling
The genome of TB contains predicted hemizygous areas that include 165 and 148 annotated genes in LGs 2 and 5, respectively, according to the grapevine 12X V1 gene annotations (http://genomes.cribi.unipd.it/). In addition to gene dose decrease, and considering the characteristically high heterozygosity of the grapevine genome (Velasco et al., 2007), such a hemizygous state might have phenotypic consequences due to the loss of functional alleles in contrasting loci, as is the case for the color locus (Fig. 1). In this regard, TB bears 48 and 100 predicted hemizygous genes located in LGs 2 and 5, respectively, that show heterozygous polymorphisms in TT. In LG 2, alternative alleles with respect to the PN40024 reference genome were found only for three hemizygous genes. This low rate of variation indicates that the copy present in TB regions involving hemizygous deletions in LG 2 displays a genotype very close to that existing in the white-berried PN40024 quasihomozygous line used for the reference genome, which is homozygous for the MybA locus null allele. This result further confirms the loss of the color locus functional allele in TB, precluding anthocyanin pigmentation in the berries of this variant. In contrast, 73 hemizygous genes in LG 5 displayed variant alleles in TB with respect to the reference (Supplemental Table S11). Variant alleles with possible deleterious changes remaining hemizygous in TB were specifically searched. Although no major effect variants relative to the reference genome were detected, nonsynonymous changes were predicted for 21 genes (Supplemental Table S11). Moreover, three annotated genes (VIT_02s0087g00180, VIT_05s0102g00090, and VIT_09s0070g00940) were truncated by Inv, T3, and T1 breakpoints, respectively. The single presence of these alleles might result in specific TB phenotypes.
Reduced Sexual Transmission of Rearranged Chromosomes Associated with a Decreased Fruit Yield
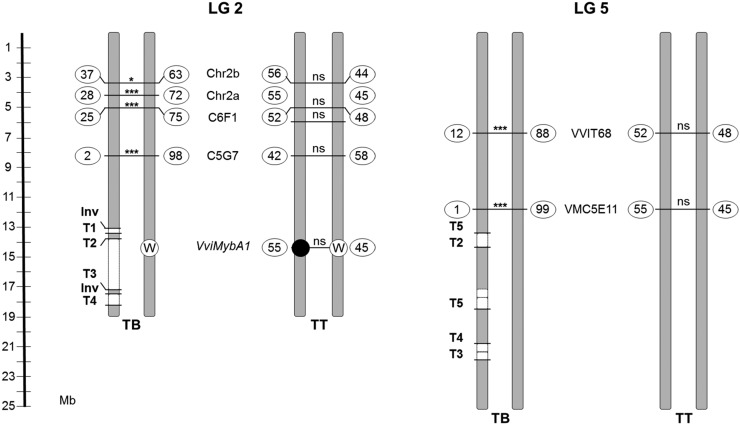
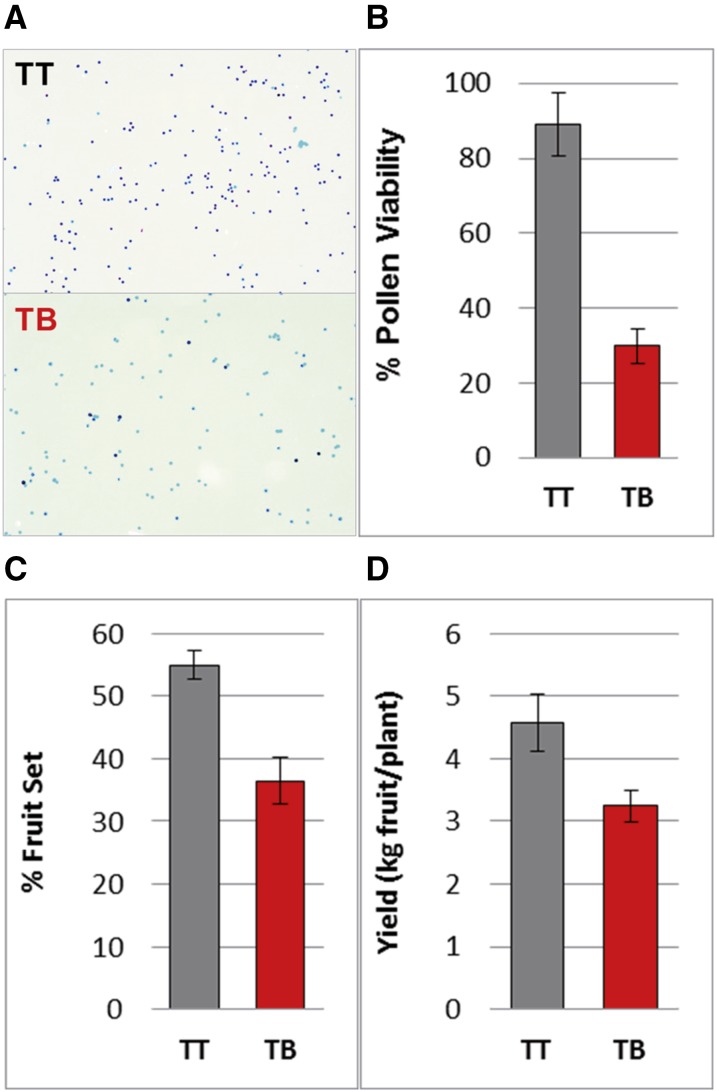
Complex and unbalanced chromosomal rearrangements found in the genome of TB might lead to defects during sexual reproduction (Pellestor, 2014). This hypothesis was tested specifically by studying the sexual transmission of LGs 2 and 5 in selfed progeny of TT and TB, consisting of 38 and 46 individuals, respectively. For LG 2, four heterozygous microsatellite markers were selected outside of the rearranged region. The allelic distribution observed in the progeny was different for TT and TB (frequencies inside the circles in Fig. 7). In TT, the two alleles segregated with a similar frequency for every marker studied, as expected for Mendelian inheritance (nonsignificant χ2). Nevertheless, in TB, distorted transmission rates were identified, as every marker analyzed showed one allele with a significantly lower frequency than expected for Mendelian inheritance. The frequency of the disfavored allele decreased as the marker was closer to the chromosome rearranged region. From the genotype frequencies in the TB progeny, the two putative haplotypes in TB were inferred, showing that all the disfavored alleles were linked in the same haplotype. The VviMybA1 gene also was genotyped by the independent amplification of the functional and nonfunctional alleles (Fig. 7). The study of genotypic frequencies for the closest microsatellite and VviMybA1 in the TT progeny allowed establishing that the disfavored haplotype in the TB progeny was that carrying the functional allele for VviMybA1 in TT and, thus, the haplotype linked to SV and deletions in TB. For LG 5, a similar lack of transmission of one haplotype was observed when two heterozygous microsatellite markers were analyzed in the two S1 progeny. Thus, segregation analysis further supports that SV affected only one copy of each chromosome. Rearranged chromosome segments are not transmitted to the next generation or are transmitted at a very low frequency. This lack of transmission was associated with a reduction of about 59% of male gamete viability in TB with respect to TT (Fig. 8), which suggests that derivative chromosomes lack genes that are essential for the viability of the haploid phase. This sterility rate would be consistent with the loss of essential genes for microgametophyte viability in both LGs 2 and 5. The effect of complex chromosome rearrangements on gamete viability might result in additional agronomical alterations present in TB, considering that reduced pollen viability correlated with 44% and 29% drops in fruit set rate and fruit yield, respectively (Fig. 8).
Figure 7.
Segregation of molecular markers at LGs 2 (left) and 5 (right) in TB and TT selfed progeny. Markers are mapped to the PN40024 reference genome chromosomes. Absolute frequencies (%) for each allele are shown inside circles, and the significance value of χ2 tests for the difference between the two allele frequencies is indicated for every marker (*, P < 0.05; ***, P < 0.0001; ns, not significant). White and black circles represent white and black berry color alleles in the VviMybA1 locus, respectively. Validated inversion (Inv) and translocation (T1–T5) breakpoints are depicted as well. Deleted chromosome segments detected by CNV analysis from WGS data are depicted as white boxes and linked in the same chromosome copy as determined by haplotype phasing using a TT self-cross progeny.
Figure 8.
Pollen viability, fruit set, and yield in TB and TT. A, TT and TB pollen after Alexander’s staining. Dark pollen is viable, while empty (pale) pollen is sterile. B to D, Percentage of pollen viability (B), fruit set rate (C), and fruit yield (D) in both Tempranillo lines. The data depicted correspond to means ± sd. All data were recorded in the experimental vineyard of Finca La Grajera during the 2015 season. Similar results were obtained in two other seasons.
DISCUSSION
Complex Unbalanced Genome Rearrangements Caused the Loss of the Berry Color Locus Functional Allele
Deletions resulting in grapevine white berry variants have been described previously (Walker et al., 2007; Vezzulli et al., 2012; Migliaro et al., 2014; Pelsy et al., 2015). However, the mutational mechanisms generating these deletions remain uncertain. So far, only gene replacement has been proposed as the mechanism causing LOH and originating new white berry alleles in Pinot somatic variants by mitotic recombination between chromosome 2 homologs linked to deletion of the color locus functional allele (Pelsy et al., 2015). Nonetheless, we would consider that, since CNV analysis of the variant lines was not performed in that study, the presence of hemizygous deletions instead of gene replacement cannot be excluded in the origin of the described Pinot alleles. In the TB white berry variant, Southern blotting and WGS comparisons with its black berry ancestor, TT, confirmed the presence of hemizygous deletions leaving at the MybA gene cluster corresponding to the berry color locus a single copy corresponding to the null allele (Fig. 1B; Supplemental Table S11). The parallel copy number decrease observed indicates that LOH in these TB chromosome regions results directly from their hemizygous state (Fig. 2), as described in other Pinot and Cabernet Sauvignon grape color variants (Walker et al., 2006; Yakushiji et al., 2006).
Genome-wide deep sequencing analyses uncovered a much more complex SV in the origin of the TB berry color variant than was identified previously in any other grapevine somatic variant. Hemizygous segments detected in LGs 2 and 5 of TB are flanked by breakpoints corresponding to several translocations and inversions (Table I; Fig. 6A). These events collectively resulted in the hemizygous deletion of more than 8 Mb (Fig. 2), ∼1.7% of the grapevine haploid genome (Jaillon et al., 2007). The same hemizygous deletions were detected repeatedly using different approaches (Figs. 1, 2, and 4; Supplemental Figs. S1 and S3; Supplemental Table S2), and the corresponding breakpoints were confirmed experimentally irrespective of the clonal lineage proximity of TB and TT accessions to the original TB bud sport (Supplemental Fig. S3). Thus, all identified SV events are characteristic of the TB bud sport and stable in subsequent multiplication cycles and, therefore, do not seem to correspond to the accumulation of secondary mutations. Remarkably, the longest deleted fragment in TB was that containing the color locus functional allele (∼3.4 Mb between T2 and T3 breakpoints in chromosome 2; Table I).
For WGS, we used whole leaves to obtain DNA assuming that both L1 and L2 meristem layers were mutated in TB, as reported before for this and other grapevine white berry variants (Walker et al., 2006; Vezzulli et al., 2012; Migliaro et al., 2014; Pelsy et al., 2015). The existence of independent chimeric mutations in L1 and L2 involving the loss of the color locus functional allele was observed in a white berry variant of Pinot (Pelsy et al., 2015). However, in TB, the frequency of WGS discordant reads detected in SV junctions appears close to 50% (Fig. 3), while no read for a second allele was identified in SNP positions showing LOH (Supplemental Table S11). These results indicate that the cell lineage in which the complex genome rearrangement appeared had colonized L1 and L2 cell layers in the meristem that generated the TB bud sport.
Genome Reshuffling in TB Shows Hallmarks of Chromosome Breakage and Illegitimate Fusion
The existence of SV has been reported among the few grapevine cultivars sequenced so far at the whole-genome level when compared with the PN40024 reference sequence (Jaillon et al., 2007; Velasco et al., 2007; Da Silva et al., 2013; Di Genova et al., 2014; Cardone et al., 2016; Xu et al., 2016). Moreover, apart from TE insertions, major SVs were not considered in the previous comparison of whole-genome sequences performed between four clones of Pinot Noir (Carrier et al., 2012). Our study provides the opportunity to identify possible mutational causes of somatic SV in grapevine by directly comparing the derivative genome of TB with the original genome in TT (Fig. 6B). In this manner, as described in “Results,” an analysis of the rearrangement pattern detected in TB indicates that it fulfills all the criteria proposed for the inference of a chromothripsis event (Stephens et al., 2011; Korbel and Campbell, 2013; Collins et al., 2017). Although the cellular origin of chromothripsis is still controversial (Sorzano et al., 2013), both breakage-fusion-bridge (BFB) cycles and the degradation of lagged chromosomes during mitosis inside of micronuclei are mechanisms that have been proposed as possible triggers (Li et al., 2014; Zhang et al., 2015). Proof for the micronuclei origin of chromothripsis arose from the observation that missegregated anaphase chromosomes are enclosed in extranuclear bodies (micronuclei), where they can undergo fragmentation and illegitimate repair before their restitution into the nucleus in one to three consecutive cell division cycles (Crasta et al., 2012; Zhang et al., 2015; Ly et al., 2017). Similar complex rearrangements associated with micronuclei formation and dependent on the DNA-repair machinery were observed during the process of genome elimination taking place after the hybridization of transgenic Arabidopsis (Arabidopsis thaliana) plants expressing mutant forms of H3 histones (Tan et al., 2015), which indicates that cellular mechanisms involved in chromothripsis are present in plants. In TB, the lack of homology or the microhomology of 1 to 7 bp that characterize SV junctions (Fig. 5) is compatible with nonhomologous end joining at blunt ends and microhomology-mediated end joining repair mechanisms of double-strand breaks generating these junctions (Jones and Jallepalli, 2012; Sinha et al., 2016).
The fact that the left arm of chromosomes 2 and 5 is protected from rearrangement in TB might contradict the idea of a simultaneous breakage of the two chromosomes and subsequent random repair inside of a micronucleus. Rather, the type of chromothripsis pattern found in TB has been argued to be generated by BFB cycles (Sorzano et al., 2013), a mechanism initially proposed in plants (McClintock, 1941). BFB cycles can generate chromothripsis-like patterns after chromosome fusions, producing dicentric chromosomes that are broken at random positions between the two centromeres by the action of the mitotic spindle and, then, rejoined and broken again in successive cell divisions that produce the deletion of fragments segregated in different daughter cells (Sorzano et al., 2013). In TB, the putative pseudodicentric nature of the grapevine chromosome 9 (Di Gaspero and Foria, 2015) might have precipitated the causal genome instability, as the two breakpoints detected in LG 9 fall within the small region between the two predicted centromeres (Fig. 6B). Irrespective of the cellular origin of this chromothripsis-like conformation, the causal genome instability appears to be stabilized in the TB derivative cultivar (Supplemental Fig. S3). Therefore, the case of TB provides evidence that these complex unbalanced somatic events lead to genetic and phenotype diversity that can be exploited as a source of novel traits in vegetatively propagated plants.
Consequences of Complex Unbalanced Chromosome Rearrangements as a Source of Somatic Variation in Plants
SV is a common driver of genome evolution in sexually reproduced plants (Cao et al., 2011; Chia et al., 2012; Qi et al., 2013) and a source of genetic diversity and new traits in vegetatively propagated plant crops (Terol et al., 2015). Resembling the TB derivative cultivar, chromoanagenesis events can rapidly produce dramatic genome reshuffle, resulting in multiple molecular and phenotype alterations (Leibowitz et al., 2015). At the gene level, the partial monosomy of large chromosome segments yields more than 300 hemizygous genes in TB. In highly heterozygous species such as grapevine, monosomy may unmask the expression of deleterious alleles originally present in a heterozygous state. In TB, this is the case for the berry color locus at which rearrangements caused deletions that left only the null allele (Fig. 1B; Migliaro et al., 2014). Other nonsynonymous SNVs originally heterozygous in TT are present in the hemizygous state in TB (Supplemental Table S11), which might account for additional phenotype variation in this cultivar.
At the reproductive level, the sexual fitness of TB seems to be compromised by the genome reshuffling. Segregation distortion in a TB S1 progeny indicates a lack of transmission of rearranged chromosome arms (Fig. 7). Similar lack of transmission linked to meiotic recombination might have been mistaken as a proof of somatic gene replacement in grape color variants of Pinot (Pelsy et al., 2015). However, considering the proportion of the hemizygous genome and the reduction of pollen viability observed in TB (Figs. 2 and 8), it is more likely to assume that highly rearranged chromosomes like those present in TB are deleterious for the resulting unbalanced haploid gametes (Pellestor, 2014). Different levels of sterility also were observed in Arabidopsis lines with shattered chromosomes (Tan et al., 2015). Although sexual reproduction is not necessary for the maintenance of vegetatively propagated crops, gametophyte viability is required for proper fruit set (Iyer, 1966); thus, complex SV may have agronomic consequences in decreasing fruit yield, as observed in TB (Fig. 8).
CONCLUSION
A whole-genome scale approach was helpful to show that, in the TB white grape variant, the deletion of the grape color locus functional allele is linked to a much more complex genome rearrangement than was observed previously in any other grapevine somatic variant. To our knowledge, this is the first study delimiting the deletion of this allele at a base resolution level. In TB, this deletion occurred associated with an unbalanced chromoanagenesis event entailing an illegitimate rejoining of fragments after multiple chromosome breakage. Taken together, our results show that complex chromosome rearrangements with chromothriptic features naturally emerge and stabilize during plant vegetative growth. Although possibly reducing sexual fitness and compromising fruit and seed production, these complex somatic events rapidly generate new phenotypes that can be selected and vegetatively propagated. Therefore, complex unbalanced genome rearrangements that emerged during somatic growth might be relevant for the genetic improvement and evolution of clonally propagated plant species such as woody crops.
MATERIALS AND METHODS
Plant Material
Molecular biology and viticultural experiments, respectively, were carried out using materials collected from the two ancestral TB multiplication grape (Vitis vinifera) vineyards (TB-ICVV2 and TB-ICVV3 accessions; Supplemental Table S9). These vineyards are located in Finca Valdegón (in Agoncillo, La Rioja, Spain) and Finca La Grajera (in Logroño, La Rioja, Spain), respectively. As a representative accession, TT clone RJ51 (the most cultivated TT clone in the Rioja DOC region) from both sites was used as respective control. Both plots belong to the Grapevine Germplasm Collection of the Instituto de Ciencias de la Vid y del Vino (ICVV; ESP-217) and are maintained under the same agronomical conditions. All plants are grafted in Richter-110 rootstock, trellised in a double cordon Royat system, and cultivated in a similar way. The genotypes of nine microsatellite loci located in different chromosomes were obtained as described elsewhere (Ibáñez et al., 2009), confirming that TB matches the genotype of TT (Supplemental Table S12). Another five TB and five TT accessions from diverse origins were used to assess the presence of SV breakpoints (Supplemental Table S9).
Selfed progeny of TT-RJ51 and TB-ICVV2 accessions were generated in 2008, consisting of 78 and 46 individuals, respectively. These progeny were maintained ungrafted in pots at Finca Valdegón.
DNA Extraction
In all experiments, total gDNA was isolated from young leaves using the DNeasy Plant Mini Kit (Qiagen) according to the protocol described by the manufacturer. Additionally, mature fruits of TT-RJ51 and TB-ICVV1 were harvested in 2016, and gDNA was obtained separately from berry skin and flesh. Independent DNA samples were used for each technique: microsatellite genotyping, Southern blot, GrapeGen GeneChip hybridization, Illumina sequencing, and PCR amplification.
Southern Blot
DNA-blot hybridization analysis was performed as described by Sambrook et al. (1989). Briefly, gDNA from TT and TB was digested with ApaI, transferred to a nylon membrane, and hybridized to a VviMybA1 probe as described previously (Lijavetzky et al., 2006). The VviMybA1 probe was generated by PCR amplification with primers Ps (5′-TCACGGGGTTTAGAAAGTGG-3′) and Pas (5′-ATCAATTGGGGAATTGGTGA-3′) using TT gDNA as a template. The hybridized membrane was scanned with a STORM PhosphorImager (Molecular Dynamics).
LOH Analysis in Specific Amplicons
To analyze LOH in TB, primer pairs for 30 amplicons were designed along LG 2 (Supplemental Table S1). PCR amplifications were carried out using Taq DNA Polymerase (Qiagen) as recommended by the manufacturer. PCR products were purified with ExoSAP-IT (USB Products Affymetrix) according to the manufacturer’s instructions and then used for Sanger sequencing at the Genomic Unit of Parque Científico de Madrid with the same primers used for amplification.
CNV Analysis by GeneChip Hybridization
Two biological replicates of TT and TB were hybridized to the GrapeGen GeneChip (Lijavetzky et al., 2012). For each replicate, gDNA samples (10 µg each) were fragmented to an average size of 0.5 kb with a sonicator Labsonic U set at 50% intensity and repeating duty cycle of 0.5, four pulses of 10 s each. Labeling of DNA was performed as recommended in the GeneChip Whole Transcript Double-Stranded Target Assay (Affymetrix). Briefly, fragmented DNA was amplified, purified, and biotin terminal labeled with the dsDNA Terminal Labeling Kit (Affymetrix) following recommendations. Finally, 7.5 µg of labeled DNA was used for the hybridization of microarrays according to the Affymetrix GeneChip Expression Analysis Technical Manual. CEL files were RMA normalized, and differential probe set hybridization between TB and TT was assessed using the RankProd package from Bioconductor (Breitling et al., 2004). A cutoff value of 1.8-fold change absolute value and P < 0.01 were established to detect genes with genomic CNV. To this end, the annotation of the GrapeGen GeneChip according to 12X V1 gene predictions from Lijavetzky et al. (2012) was used.
Genome Resequencing and Computational Comparative Analyses
Sequencing and Preprocessing
Two gDNA libraries were built from a sample of young leaves from one individual of TT-RJ51 and another from TB-ICVV2 (second-round multiplication plant). The gDNA of each individual was fragmented randomly. After electrophoresis, DNA fragments of ∼470 bp were gel purified. Adapter ligation and DNA cluster preparation were then performed and subjected to sequencing. Each Tempranillo line was sequenced in a different lane at Beijing Genomics Institute facilities using the Illumina HiSeq 2000 sequencing system. On each sample, a total of ∼209 × 106 paired-end reads of 90-nucleotide length were obtained (∼18.8 Gb of total sequence per sample; Supplemental Table S4).
Sequence data were processed by removing the adapter sequence from reads and subsequently taking out the reads with low quality (Phred quality ≤ 5 in 50% or more of the positions of the read) to obtain the clean data described in Supplemental Table S4. The final Q20 of each lane was above 95%. Two files containing the clean reads in FastQ format, corresponding to TT and TB samples, were submitted to an in-house pipeline composed of several bash shell scripts (available upon request to the authors). These scripts used shell commands, Perl scripts, and open-source programs to process the reads, produce the alignments, select particular subsets of alignments for the PEM study, and carry out the CNV study and the variant calling. The pipeline also included a prediction of effects for the retrieved variants and a scanning for LOH regions in TB. Each specific procedure is described below.
Alignment
Each FastQ file was aligned to the grapevine PN40024 12X.0 reference genome assembly using BWA version 0.5.9-r16 with the option sampe (for paired ends) and default parameters (Li and Durbin, 2009). Approximately 76% of the reads aligned with the reference genome (Supplemental Table S5). Redundant reads comprising the same span were then removed using the rmdup option of SAMtools (Li et al., 2009). To improve the alignments by minimizing the number of artifactual mismatching bases due to the close presence of INDELs with respect to the reference genome, local realignment was then done using GATK version 1.0.5777 (tools RealignerTargetCreator and IndelRealigner; McKenna et al., 2010). To increase the ability of SV breakpoint detection, another script was used to split each of the 90-nucleotide clean reads into two fragments of 45 nucleotides. The new sets of FastQ files containing the 45-nucleotide split reads also were aligned as described above and used for SV junction detection.
Variant Calling and LOH Genome Scanning
Files containing read alignments of TT and TB in BAM format were used to search for Tempranillo line-specific polymorphisms (SNPs and INDELs). Aligned reads were preprocessed as described above, and moreover, reads with mapping quality < 40 after local realignment in IndelRealigner were discarded too. Variant calling on each line was carried out using the BCFtools utility from SAMtools for the whole genome and the HaplotypeCaller tool from the GATK package for LGs 2 and 5. Initially, for both tools, VCF files independently comparing each Tempranillo line with the grapevine 12X.0 reference genome were generated. In order to remove false positives, the following filters and cutoff thresholds were used: polymorphisms were considered when 15 or more reads covered the position and the variant sequence was observed in 35% or more for heterozygous sites and 90% or more with two or fewer reads of the nonvariant allele for homozygous sites. Finally, the resulting TT and TB sets of heterozygous polymorphisms identified in SAMtools were binned in 100-kb bins, and these bins were reported and plotted for each LG. To delimit segments of LOH along the TB genome, intervals of three or more consecutive heterozygous sites in TT (according to the filters described above) that fulfilled the criteria of homozygous sites in TB were considered. To estimate the genotype in TB irrespective of the variation relative to the reference genome, the sequence at these positions was obtained from the compilation in the pileup file of the alignments. In these cases, a Perl script was constructed to detect homozygous sites in TB according to eight or more reads coverage and 90% or greater frequency of the most frequent allele and two or more for alleles other than the most frequent one.
GATK HaplotypeCaller also was used for variant calling in LGs 2 and 5, and the results were cross-checked with those obtained from SAMtools + BCFtools. Although the results were highly equivalent, HaplotypeCaller was used to study the effect of polymorphisms involving LOH in hemizygous regions of TB by assuming that this application could perform better on INDEL detection (Hwang et al., 2015). The same filters described for the output of SAMtools were used to detect TT polymorphic sites with LOH in TB (heterozygous in TT and homozygous in TB). In this case, to estimate the genotype of TB for nonvariant LOH sites, the variant calling was repeated after merging the reads from the two lines. The genotype in the nonvariant line was inferred by deducting the reads of the variant line from the total of both. Finally, the effect of detected polymorphisms was predicted using SnpEff version 2.0.3 (Cingolani et al., 2012) according to the grapevine 12X V1 gene predictions from CRIBI (http://genomes.cribi.unipd.it/) and the functional annotation and classification of genes described by Grimplet et al. (2012).
Genome Resequencing CNV
CNV-seq (Xie and Tammi, 2009) was applied using TB and TT as test and reference, respectively, on the corresponding files containing the initial position of each aligned read relative to the reference genome (called hits files by the authors of CNV-seq). Significance values were set in CNV-seq to log2 (copy number TB/TT) ≤ −0.5 and P ≤ 0.00001 for each chromosomal window and four or more consecutive sliding windows for annotating a CNV zone. The CNV zones detected by CNV-seq with P ≤ 0.05 and log2 (copy number TB/TT) ≤ −0.5 or ≥ 0.37 were considered as significantly decreased or increased, respectively, in copy number in TB relative to TT.
SV Breakpoint Search
PEM and soft-clipped reads indicative of the presence of SV junctions were searched specifically in the two SAM files containing the TB and TT reads, respectively, aligned to the PN40024 12X.0 reference genome. The two alignment sets of whole 90-nucleotide reads and reads split into two 45-nucleotide fragments were processed equally in parallel. First, read pairs with mapping quality = 0 in BWA were removed, as this indicates mapping to multiple locations. From the remaining set of aligned reads and using bash shell commands (grep and awk) and the SAMtools merge utility, putative chromosome rearrangement breakpoint sites were searched by extracting two subsets of paired-end reads with discordant mate alignment: (1) read pairs with increased insert size indicative of intrachromosomal translocations or large deletions: absolute value of insert size (TLEN field in the SAM file) > 4 times the median TLEN value of all aligned pairs in the sample; and (2) read pairs with mates in different chromosomes indicative of interchromosome translocations (RNEXT field different from = or RNAME value in the SAM file). To determine the orientation of the rearranged chromosome fragments comprising putative breakpoints, paired-end reads with unexpected mate orientation alignment were searched from the previous two subsets. Two different discordant orientations were considered: (1) both mates with unexpected orientation (FLAG field of the SAM file with values = 81, 161, 97, or 145); and (2) only one mate with unexpected orientation (FLAG with values 65, 129, 113, or 177). An additional subset comprising soft-clipped reads (only partially mapping to the reference in one read extreme) was searched for in genomic regions with discordant mate alignment 1 or 2. A read containing CIGAR alignment operation S was the criterion used to select soft-clipped reads.
To select breakpoints distinguishing TB and TT genomes, BEDtools (Quinlan and Hall, 2010) of TB-specific discordant reads were extracted to delimit SV candidate breakpoints, which were inspected visually using Integrative Genomics Viewer (version 2.2) software (Thorvaldsdóttir et al., 2013).
The nonclipped part of TB-specific soft-clipped reads mapping on candidate breakpoint areas was aligned to the PN40024 12X.0 genome assembly using BLASTN suite (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch).
Analysis of Repetitive Sequences at Breakpoints
For each SV junction detected in TB, a consensus breakpoint junction sequence was built from the sequence of discordant reads and submitted to RepeatMasker version 4.0.3 (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). V. vinifera was the DNA source, and ABBlast/WUBlast was the search engine to query for matches with repeat elements deposited in RepBase database update 2013/04/22 (http://www.girinst.org/repbase), which includes all described repeat elements specific of V. vinifera. TE-like sequences were searched by BLAST of RepBase sequences against (1) the 12X.0 reference genome in regions where TB SV breakpoints mapped and (2) TB consensus sequences at SV junctions. The structure and domains of detected TE sequences were studied with National Center for Biotechnology Information Conserved Domain Database tools (http://www.ncbi.nlm.nih.gov/cdd/).
Validation of SV Junctions
PCR primers were designed at each flank of genomically detected SV junctions (Supplemental Table S1). The consensus breakpoint junction sequence reconstructed from discordant reads was used as a template for primer pair design. Primer specificity was checked by BLAST against the PN40024 12X grapevine reference genome assembly in the Genoscope Web site (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/). These primer pairs were tested for PCR amplification using as a template gDNA from different TB and TT accessions (Supplemental Fig. S3; Supplemental Table S9). PCRs were carried out using MyTaq DNA Polymerase (Bioline, Meridian Life Science) following the manufacturer’s protocol. The VviDXS (VIT_05s0020g02130) gene, mapping on a presumably disomic region of LG 5 in both TT and TB, was included in the experiment as a positive control using primers described elsewhere (Emanuelli et al., 2010). An aliquot of the amplification product was subjected to electrophoresis through 1% agarose gels to check for the presence and size of PCR products. PCR products were sequenced as described above.
SNP Chip Genotyping and Haplotype Phasing
Total gDNA from 78 TT S1 progeny individuals was genotyped using the GrapeReSeq Illumina 18K SNP Infinium as described elsewhere (Houel et al., 2015). SNP genotype data were processed as described in the GrapeReSeq project (https://urgi.versailles.inra.fr/Species/Vitis/GrapeReSeq_Illumina_20K). To reconstruct the original phasing of the two haplotypes in TT, segregation for each pair of adjacent SNP loci represented in the chip was studied, assuming that the most frequent genotype combination in the progeny corresponded to the parental haplotypes. The genotype of TB at SNP positions represented in the chip mapping on LGs 2 and 5 was obtained from our WGS data set. The haplotype remaining in TB at positions involving LOH in this variant was compared with the two haplotypes reconstructed in TT to infer the homologous copy affected by each deletion.
Sexual Transmission Analysis
A series of markers were genotyped in a set of TT and TB self-cross S1 progeny to study the sexual transmission of each homologous chromosome at LGs 2 and 5. Four heterozygous microsatellite markers in both Tempranillo lines mapping on LG 2 were selected: Chr2b, Chr2a, C6F1, and C5G7. Genotyping of VviMybA1 alleles in TT, TB, and TT self-cross S1 progeny was carried out as described previously (Lijavetzky et al., 2006). Primer pairs a-d3 and b-d were used to amplify null and functional alleles, respectively. Allele segregation was determined for each locus and progeny, and their deviation from expected values (0.5) was evaluated through χ2 tests. This statistical test was performed using Microsoft Excel software. Genotypic segregation for each pair of adjacent loci was studied to determine microsatellite haplotypes, assuming that the most frequent double homozygous genotypes in the progeny correspond to the parental haplotypes. Joint genotypic segregation of C5G7 and VviMybA1 was used to establish the haplotypes at LG 2. For LG 5, two microsatellite markers were selected, VVIT68 and VMC5E11, and the procedure was similar to that described for LG 2.
Fruit Production and Pollen Viability
Experiments were carried out at Finca La Grajera during 2015. In this vineyard plot, three rows (∼50 plants each) of TB plants are interspersed by three rows of TT plants. Each row was used as an independent biological replicate, and four plants per replicate were analyzed. Pollen was collected simultaneously from TT and TB inflorescences (one per plant) at 50% bloom. Pollen viability was analyzed using Alexander’s modified staining as described previously (Royo et al., 2016). For each Tempranillo line, more than 1,000 pollen grains per biological replicate were analyzed. Fruit set percentage was calculated as the rate between the number of ripened fruits per cluster at maturity and the number of flowers at flowering time in the same cluster (one cluster per plant was analyzed). The number of flowers per inflorescence was estimated by bagging the inflorescence before the onset of flowering and counting flower caps inside the bag after fruit set. Fruit yield was estimated as the total mass of grape bunches per plant.
Accession Numbers
GrapeGen GeneChip and WGS data are deposited in the National Center for Biotechnology Information under Gene Expression Omnibus GSE80801 and Sequence Read Archive SRP065756 (BioProject PRJNA301084) accession numbers, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Microarray-based CNV in a TB white berry somatic variant.
Supplemental Figure S2. Intrachromosome translocation in TB LG 5.
Supplemental Figure S3. Validation of TB SV junctions in additional Tempranillo accessions.
Supplemental Figure S4. Transposon and repetitive sequences detected at TB breakpoints sites.
Supplemental Table S1. Oligonucleotides used as primers for PCR amplification.
Supplemental Table S2. Heterozygosity analysis in PCR-amplified regions around the color locus in LG 2.
Supplemental Table S3. GrapeGen GeneChip probe sets showing differential CGH between TB and TT.
Supplemental Table S4. Summary of resequencing data production (clean data).
Supplemental Table S5. Summary of resequencing read alignments.
Supplemental Table S6. Genome regions showing LOH in TB.
Supplemental Table S7. Genome windows showing copy number decrease in TB.
Supplemental Table S8. Sanger capillary electrophoresis-obtained sequences of PCR amplicons confirming breakpoint junctions predicted by genome resequencing in TB.
Supplemental Table S9. Origins of Tempranillo accessions.
Supplemental Table S10. Haplotype phasing of fragments deleted in LGs 2 and 5 of TB.
Supplemental Table S11. List of predicted hemizygous variant sites in the TB genome and functional effect.
Supplemental Table S12. Microsatellite genotyping identification.
Acknowledgments
We thank Drs. Karel H.M. van Wely and Daniel Trujillano for thoughtful comments and feedback and Guillermo Juárez and Silvia Hernáiz for technical assistance; we also thank the genomics service of the CNB-Consejo Superior de Investigaciones Científicas for running the CGH hybridizations.
Glossary
- SNV
single-nucleotide variation
- INDEL
insertion-deletion
- SV
structural variation
- TE
transposable element
- LG
linkage group
- LOH
loss of heterozygosity
- TB
Tempranillo Blanco
- TT
Tempranillo Tinto
- WGS
whole-genome sequencing
- CGH
comparative genomic hybridization
- CNV
copy number variation
- PEM
pair-end mapping
- Inv
inversion junction in LG 2
- gDNA
genomic DNA
- SNP
single-nucleotide polymorphism
- BFB
breakage-fusion-bridge
Footnotes
This work was partially supported by the project GrapeReSeq from the Spanish Ministry of Education and Sciences (EUI2008-03752), by the European Union Seventh Framework Programme (KBBE Contract 311775 Innovine), and by the Spanish Ministry of Economy (MINECO) (BIO2014-59324-R). This work also benefited from the networking activities within the funded European Cooperation in Science and Technology (COST) Action FA1106 QualityFruit.
References
- Battilana J, Emanuelli F, Gambino G, Gribaudo I, Gasperi F, Boss PK, Grando MS (2011) Functional effect of grapevine 1-deoxy-D-xylulose 5-phosphate synthase substitution K284N on Muscat flavour formation. J Exp Bot 62: 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32: 565–569 [DOI] [PubMed] [Google Scholar]
- Boss PK, Thomas MR (2002) Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416: 847–850 [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Cardone MF, D’Addabbo P, Alkan C, Bergamini C, Catacchio CR, Anaclerio F, Chiatante G, Marra A, Giannuzzi G, Perniola R, et al. (2016) Inter-varietal structural variation in grapevine genomes. Plant J 88: 648–661 [DOI] [PubMed] [Google Scholar]
- Carrier G, Le Cunff L, Dereeper A, Legrand D, Sabot F, Bouchez O, Audeguin L, Boursiquot JM, This P (2012) Transposable elements are a major cause of somatic polymorphism in Vitis vinifera L. PLoS ONE 7: e32973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia JM, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, Elshire RJ, Gaut B, Geller L, Glaubitz JC, et al. (2012) Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet 44: 803–807 [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, Mason T, Pregno G, Dorrani N, et al. (2017) Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amato F. (1997) Role of somatic mutations in the evolution of higher plants. Caryologia 50: 1–15 [Google Scholar]
- Da Silva C, Zamperin G, Ferrarini A, Minio A, Dal Molin A, Venturini L, Buson G, Tononi P, Avanzato C, Zago E, et al. (2013) The high polyphenol content of grapevine cultivar tannat berries is conferred primarily by genes that are not shared with the reference genome. Plant Cell 25: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gaspero G, Foria S (2015) Molecular grapevine breeding techniques. In Reynolds AG, ed, Grapevine Breeding Programs for the Wine Industry. Woodhead Publishing, Oxford, pp 23–37 [Google Scholar]
- Di Genova A, Almeida AM, Muñoz-Espinoza C, Vizoso P, Travisany D, Moraga C, Pinto M, Hinrichsen P, Orellana A, Maass A (2014) Whole genome comparison between table and wine grapes reveals a comprehensive catalog of structural variants. BMC Plant Biol 14: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doligez A, Adam-Blondon AF, Cipriani G, Di Gaspero G, Laucou V, Merdinoglu D, Meredith CP, Riaz S, Roux C, This P (2006) An integrated SSR map of grapevine based on five mapping populations. Theor Appl Genet 113: 369–382 [DOI] [PubMed] [Google Scholar]
- Emanuelli F, Battilana J, Costantini L, Le Cunff L, Boursiquot JM, This P, Grando MS (2010) A candidate gene association study on Muscat flavor in grapevine (Vitis vinifera L.). BMC Plant Biol 10: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli F, Sordo M, Lorenzi S, Battilana J, Grando MS (2014) Development of user-friendly functional molecular markers for VvDXS gene conferring Muscat flavor in grapevine. Mol Breed 33: 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Chaïb J, Martinez-Zapater JM, Thomas MR, Torregrosa L (2013) Mis-expression of a PISTILLATA-like MADS box gene prevents fruit development in grapevine. Plant J 73: 918–928 [DOI] [PubMed] [Google Scholar]
- Fernandez L, Torregrosa L, Segura V, Bouquet A, Martinez-Zapater JM (2010) Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. Plant J 61: 545–557 [DOI] [PubMed] [Google Scholar]
- Furiya T, Suzuki S, Sueta T, Takayanagi T (2009) Molecular characterization of a bud sport of Pinot Gris bearing white berries. Am J Enol Vitic 60: 66–73 [Google Scholar]
- Grimplet J, Van Hemert J, Carbonell-Bejerano P, Díaz-Riquelme J, Dickerson J, Fennell A, Pezzotti M, Martínez-Zapater JM (2012) Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res Notes 5: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houel C, Chatbanyong R, Doligez A, Rienth M, Foria S, Luchaire N, Roux C, Adivèze A, Lopez G, Farnos M, et al. (2015) Identification of stable QTLs for vegetative and reproductive traits in the microvine (Vitis vinifera L.) using the 18 K Infinium chip. BMC Plant Biol 15: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Kim E, Lee I, Marcotte EM (2015) Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep 5: 17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez J, Muñoz-Organero G, Zinelabidine LH, de Andrés MT, Cabello F, Martínez-Zapater JM (2012) Genetic origin of the grapevine cultivar Tempranillo. Am J Enol Vitic 63: 549–553 [Google Scholar]
- Ibáñez J, Vélez MD, de Andrés MT, Borrego J (2009) Molecular markers for establishing distinctness in vegetatively propagated crops: a case study in grapevine. Theor Appl Genet 119: 1213–1222 [DOI] [PubMed] [Google Scholar]
- Iyer GS. (1966) Induction of pollen sterility in grapes (Vitis vinifera). Vitis 5: 433–445 [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jones MJ, Jallepalli PV (2012) Chromothripsis: chromosomes in crisis. Dev Cell 23: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, et al. (2012) Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep 1: 648–655 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ (2013) Criteria for inference of chromothripsis in cancer genomes. Cell 152: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. (2007) Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laucou V, Lacombe T, Dechesne F, Siret R, Bruno JP, Dessup M, Dessup T, Ortigosa P, Parra P, Roux C, et al. (2011) High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor Appl Genet 122: 1233–1245 [DOI] [PubMed] [Google Scholar]
- Leibowitz ML, Zhang CZ, Pellman D (2015) Chromothripsis: a new mechanism for rapid karyotype evolution. Annu Rev Genet 49: 183–211 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XQ. (2016) Natural attributes and agricultural implications of somatic genome variation. Curr Issues Mol Biol 20: 29–46 [PubMed] [Google Scholar]
- Li Y, Schwab C, Ryan S, Papaemmanuil E, Robinson HM, Jacobs P, Moorman AV, Dyer S, Borrow J, Griffiths M, et al. (2014) Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature 508: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonell-Bejerano P, Grimplet J, Bravo G, Flores P, Fenoll J, Hellín P, Oliveros JC, Martínez-Zapater JM (2012) Berry flesh and skin ripening features in Vitis vinifera as assessed by transcriptional profiling. PLoS ONE 7: e39547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky D, Ruiz-García L, Cabezas JA, De Andrés MT, Bravo G, Ibáñez A, Carreño J, Cabello F, Ibáñez J, Martínez-Zapater JM (2006) Molecular genetics of berry colour variation in table grape. Mol Genet Genomics 276: 427–435 [DOI] [PubMed] [Google Scholar]
- Ly P, Teitz LS, Kim DH, Shoshani O, Skaletsky H, Fachinetti D, Page DC, Cleveland DW (2017) Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat Cell Biol 19: 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Lindberg M, Faust GG, Leibowitz ML, Clark RA, Layer RM, Quinlan AR, Hall IM (2013) Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res 23: 762–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F, Pinosio S, Morgante M (2014) Structural variation and genome complexity: is dispensable really dispensable? Curr Opin Plant Biol 18: 31–36 [DOI] [PubMed] [Google Scholar]
- Martinez J, Vicente T, Martinez T, Chavarri JB, Garcia-Escudero E (2006) Una nueva variedad blanca para la D.O.Ca. Rioja: el Tempranillo Blanco [A new white variety for the DOC Rioja: ‘Tempranillo Blanco’]. In 29th OIV World Congress of Vine and Wine: 4th General Assembly of the OIV. Organisation Internationale de la Vigne et du Vin, Logrono, Spain [Google Scholar]
- McClintock B. (1941) The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaro D, Crespan M, Muñoz-Organero G, Velasco R, Moser C, Vezzulli S (2014) Structural dynamics at the berry colour locus in Vitis vinifera L. somatic variants. Aust J Grape Wine Res 20: 485–495 [Google Scholar]
- Pelsy F, Dumas V, Bévilacqua L, Hocquigny S, Merdinoglu D (2015) Chromosome replacement and deletion lead to clonal polymorphism of berry color in grapevine. PLoS Genet 11: e1005081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelsy F, Hocquigny S, Moncada X, Barbeau G, Forget D, Hinrichsen P, Merdinoglu D (2010) An extensive study of the genetic diversity within seven French wine grape variety collections. Theor Appl Genet 120: 1219–1231 [DOI] [PubMed] [Google Scholar]
- Pellestor F. (2014) Chromothripsis: how does such a catastrophic event impact human reproduction? Hum Reprod 29: 388–393 [DOI] [PubMed] [Google Scholar]
- Qi J, Liu X, Shen D, Miao H, Xie B, Li X, Zeng P, Wang S, Shang Y, Gu X, et al. (2013) A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet 45: 1510–1515 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo C, Carbonell-Bejerano P, Torres-Pérez R, Nebish A, Martínez Ó, Rey M, Aroutiounian R, Ibáñez J, Martínez-Zapater JM (2016) Developmental, transcriptome, and genetic alterations associated with parthenocarpy in the grapevine seedless somatic variant Corinto bianco. J Exp Bot 67: 259–273 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sinha S, Villarreal D, Shim EY, Lee SE (2016) Risky business: microhomology-mediated end joining. Mutat Res 788: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano CO, Pascual-Montano A, Sánchez de Diego A, Martínez-A C, van Wely KH (2013) Chromothripsis: breakage-fusion-bridge over and over again. Cell Cycle 12: 2016–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. (2011) Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EH, Henry IM, Ravi M, Bradnam KR, Mandakova T, Marimuthu MP, Korf I, Lysak MA, Comai L, Chan SW (2015) Catastrophic chromosomal restructuring during genome elimination in plants. eLife 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Ibañez V, Carbonell J, Alonso R, Estornell LH, Licciardello C, Gut IG, Dopazo J, Talon M (2015) Involvement of a citrus meiotic recombination TTC-repeat motif in the formation of gross deletions generated by ionizing radiation and MULE activation. BMC Genomics 16: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This P, Lacombe T, Cadle-Davidson M, Owens CL (2007) Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor Appl Genet 114: 723–730 [DOI] [PubMed] [Google Scholar]
- This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22: 511–519 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregrosa L, Fernandez L, Bouquet A, Boursiquot JM, Pelsy F, Martínez-Zapater JM (2011) Origins and consequences of somatic variation in grapevine. In Adam-Blondon AF, Martinez-Zapater JM, Kole C, eds, Genetics, Genomics, and Breeding of Grapes. Science Publishers, Jersey, British Isles, pp 68–92 [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, et al. (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli S, Leonardelli L, Malossini U, Stefanini M, Velasco R, Moser C (2012) Pinot blanc and Pinot gris arose as independent somatic mutations of Pinot noir. J Exp Bot 63: 6359–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DA, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Robinson SP (2006) Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Mol Biol 62: 623–635 [DOI] [PubMed] [Google Scholar]
- Whitham TG, Slobodchikoff CN (1981) Evolution by individuals, plant-herbivore interactions, and mosaics of genetic variability: the adaptive significance of somatic mutations in plants. Oecologia 49: 287–292 [DOI] [PubMed] [Google Scholar]
- Wong DC, Schlechter R, Vannozzi A, Höll J, Hmmam I, Bogs J, Tornielli GB, Castellarin SD, Matus JT (2016) A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res 23: 451–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Tammi MT (2009) CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gao Z, Tao J, Jiang W, Zhang S, Wang Q, Qu S (2016) Genome-wide detection of SNP and SV variations to reveal early ripening-related genes in grape. PLoS ONE 11: e0147749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushiji H, Kobayashi S, Goto-Yamamoto N, Tae Jeong S, Sueta T, Mitani N, Azuma A (2006) A skin color mutation of grapevine, from black-skinned Pinot Noir to white-skinned Pinot Blanc, is caused by deletion of the functional VvmybA1 allele. Biosci Biotechnol Biochem 70: 1506–1508 [DOI] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D (2015) Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]