AUXIN BINDING PROTEIN1 is required for auxin-dependent protoplast swelling in Arabidopsis.
Abstract
Members of the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) family are known auxin receptors. To analyze the possible receptor function of AUXIN BINDING PROTEIN1 (ABP1), an auxin receptor currently under debate, we performed different approaches. We performed a pharmacological approach using α-(2,4-dimethylphenylethyl-2-oxo)-indole-3-acetic acid (auxinole), α-(phenylethyl-2-oxo)-indole-3-acetic acid (PEO-IAA), and 5-fluoroindole-3-acetic acid (5-F-IAA) to discriminate between ABP1- and TIR1/AFB-mediated processes in Arabidopsis (Arabidopsis thaliana). We used a peptide of the carboxyl-terminal region of AtABP1 as a tool. We performed mutant analysis with the null alleles of ABP1, abp1-c1 and abp1-TD1, and the TILLING mutant abp1-5. We employed Coimbra, an accession that exhibits an amino acid exchange in the auxin-binding domain of ABP1. We measured either volume changes of single hypocotyl protoplasts or hypocotyl growth, both at high temporal resolution. 5-F-IAA selectively activated the TIR1/AFB pathway but did not induce protoplast swelling; instead, it showed auxin activity in the hypocotyl growth test. In contrast, PEO-IAA induced an auxin-like swelling response but no hypocotyl growth. The carboxyl-terminal peptide of AtABP1 induced an auxin-like swelling response. In the ABP1-related mutants and Coimbra, no auxin-induced protoplast swelling occurred. ABP1 seems to be involved in mediating rapid auxin-induced protoplast swelling, but it is not involved in the control of rapid auxin-induced growth.
“Working with AUXIN BINDING PROTEIN1 (ABP1) is like walking in a minefield.” This notion by an anonymous referee in 2001 has never been more valid than today. Even though ABP1 was the first auxin receptor candidate ever identified (Löbler and Klämbt, 1985a, 1985b; Shimomura et al, 1986; Hesse et al., 1989; Palme et al., 1990), its function has always been a matter of controversy (Jones, 1990; Hertel, 1995; Venis, 1995). Rapid electrophysiological auxin responses as well as auxin-induced protoplast swelling have been shown to be mediated by ABP1 (Barbier-Brygoo et al., 1991; Rück et al., 1993; Iino et al., 2001; Steffens et al., 2001). It has been convincingly shown that the members of the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) receptor family (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005) trigger auxin-induced gene expression and hypocotyl growth (Fendrych et al., 2016) through enhanced expression of SMALL AUXIN UP RNA genes (Spartz et al., 2017). Some other responses appear to be too rapid to depend on transcription (Scherer, 2011). ABP1 was frequently proposed as a receptor candidate for a nontranscriptional pathway for rapid auxin-induced effects, but convincing molecular and biochemical evidence is still elusive. One observation suggesting an important role for ABP1 was the embryo-lethal phenotypes of homozygous abp1-1 and abp1-1s knockout lines (Chen et al., 2001; Tzafrir et al., 2004). In these mutants, embryo development ceased at globular embryo stage (Chen et al., 2001). While this finding pointed to a pivotal role of ABP1 in elongation growth and plant development, it made it very difficult to analyze the function of the protein. Complex strategies to circumvent embryo lethality, like induced knockdown of ABP1 expression or induced expression of anti-ABP1 antibodies, have been developed (Chen et al., 2001; Braun et al., 2008). Recent reports, however, convincingly demonstrated that the embryo-lethal phenotypes of the former abp1 knockouts was simply due to off-target effects on the neighboring BELAYA SMERT gene (Dai et al., 2015; Michalko et al., 2015). Not surprisingly, the ABP1 knockout alleles abp1-c1 and abp1-TD1 generated by CRISPR/CAS9 technology or by T-DNA insertion did not display any conspicuous phenotype (Gao et al., 2015). These observations also explained the relatively mild phenotype of the abp1-5 TILLING mutant, which, unfortunately, also appears to be prone to a large number of off-target mutations (Enders et al., 2015). Nevertheless, the abp1-5 mutant exhibits an amino acid exchange from His to Tyr at position 94 in the auxin-binding domain, potentially resulting in an inhibition of indole-3-acetic acid (IAA) binding to ABP1.
A strategy to elucidate the function of ABP1 independent of mutant analysis is the usage of chemical auxin analogs (Pufky et al., 2003). The halogenated IAA derivative 5-fluoroindole-3-acetic acid (5-F-IAA) as well as α-(phenylethyl-2-oxo)-IAA (PEO-IAA) and α-(2,4-dimethylphenylethyl-2-oxo)-IAA (auxinole) have been suggested to act differentially on the ABP1 and TIR1/AFB signaling pathways (Robert et al., 2010). IAA and 5-F-IAA, but not PEO-IAA, induced the expression of auxin-dependent genes via the TIR1/AFB pathway (Robert et al., 2010). In contrast, PEO-IAA, but not 5-F-IAA, was suggested to be an agonist of ABP1-dependent pathways. As a complement to genetic analysis, the use of these small molecules that specifically modulate aspects of auxin signal transduction represents an attractive tool for auxin biology. In order to analyze the role of ABP1 and/or TIR1/AFB signaling in hypocotyl growth of Arabidopsis (Arabidopsis thaliana), we used the analogs mentioned above.
Structure/function approaches are prominent tools to analyze auxin action in Arabidopsis seedlings. A gene expression profiling study that used six different natural or synthetic auxins to identify the complete auxin-induced transcriptome of etiolated Arabidopsis seedlings at early time points, such as 20 min after treatment, is one example (Pufky et al., 2003). Protoplast swelling has been shown previously to depend on ABP1, as indicated by the action of various ABP1-specific immunological tools in the response (Steffens et al., 2001; Yamagami et al., 2004; Dahlke et al., 2009). Here, we characterize the activity of auxinole, PEO-IAA, and 5-F-IAA in the protoplast-swelling assay (Steffens and Lüthen, 2000) and, in comparison with the classical auxin response, fast hypocotyl elongation of Arabidopsis. As ABP1 action has been implicated in the early phase of auxin action, we measured hypocotyl growth at a high temporal resolution. In addition, we performed a computational docking study with ABP1 and TIR1 from Arabidopsis to understand the binding properties of the auxin analogs used. The phenotypical analysis was complemented by the use of available ABP1 mutants and the analysis of an Arabidopsis accession carrying an amino acid exchange in the auxin-binding domain of ABP1.
RESULTS
PEO-IAA Induces an IAA-Like Protoplast Expansion, Whereas the TIR1/AFB Agonist 5-F-IAA Does Not Induce This Cellular Response
Earlier investigations had demonstrated that the natural auxin IAA induced swelling of protoplasts derived from the growing region of etiolated Arabidopsis hypocotyls (Steffens et al., 2001). Immunological tools had suggested a role for ABP1 in the protoplast-swelling response (Steffens et al., 2001; Yamagami et al., 2004). In this study, we chose a pharmacological approach in an attempt to discriminate between ABP1 and TIR1/AFB signaling pathways. To that end, we performed single-cell analysis with the three auxin derivatives 5-F-IAA, PEO-IAA, and auxinole.
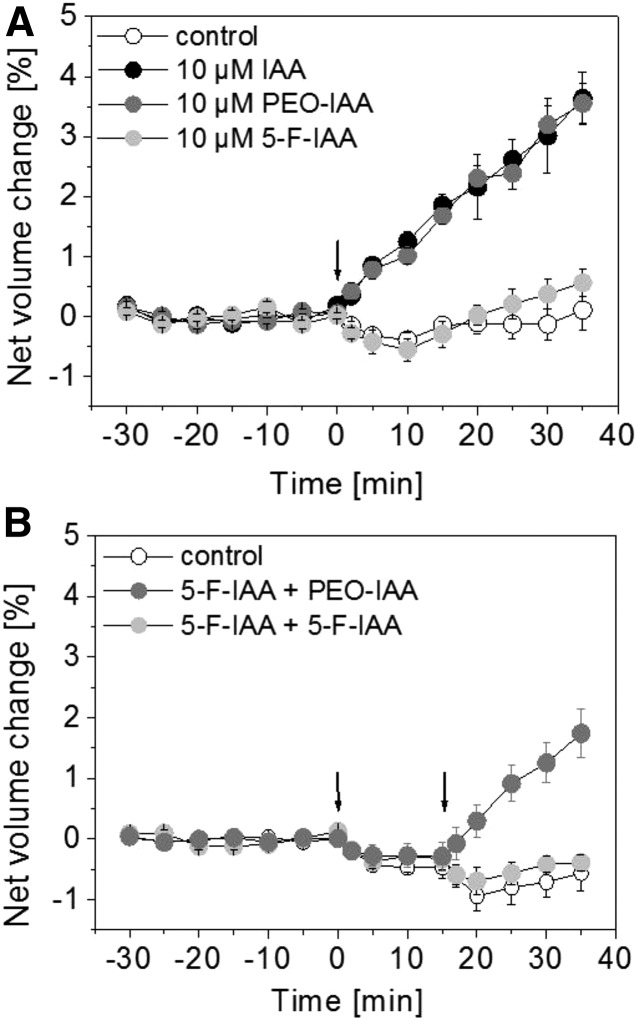
As shown earlier, protoplasts respond with a rapid swelling after IAA treatment (Fig. 1A), whereas the net volume of control protoplasts remains nearly constant around 0% over the course of the experiment (Fig. 1A). To analyze if the synthetic auxin analog PEO-IAA has an effect on protoplast expansion, PEO-IAA was applied at a concentration of 10 µm (Fig. 1A). The net volume of protoplasts increased in an IAA-like manner just after application (Fig. 1A). The fungal toxin fusicoccin (FC) was used as a control (Table I; Steffens and Lüthen, 2000). The reactions induced by IAA and FC are comparable to the change in net volume induced by PEO-IAA (Table I). Treatment with auxinole, another auxin analog with a structure similar to PEO-IAA (Hayashi et al., 2012), also resulted in an increase in net volume at a concentration of 10 µm (Table I). The halogenated auxin derivative 5-F-IAA at a concentration of 10 µm induced no obvious change in net volume (Fig. 1A). This lack of activity is not due to an unspecific inhibition of protoplast swelling by 5-F-IAA, as PEO-IAA induced swelling in protoplasts in the presence of 5-F-IAA (Fig. 1B).
Figure 1.
IAA and PEO-IAA induce protoplast expansion, and 5-F-IAA does not. A, Protoplasts were treated with 10 µm IAA or the same concentration of PEO-IAA or 5-F-IAA or buffer as a control at 0 min (indicated by the arrow). IAA and PEO-IAA induced a similar increase in net volume. Results are averages ± se from a minimum of n = 5 protoplasts from two independent experiments. B, Competition experiments with 5-F-IAA plus PEO-IAA and 5-F-IAA plus 5-F-IAA treatment. First, 10 µm 5-F-IAA was applied at 0 min (first arrow). The second application is indicated by the second arrow at 15 min. PEO-IAA (10 µm) still induced protoplast swelling when 5-F-IAA was applied first. As a control, buffer was applied at the second time point. Values are means ± se from a minimum of n = 7 protoplasts from two independent experiments.
Table I. Comparison of the change in net volume (%) of protoplasts after 30 min of treatment with IAA, PEO-IAA, auxinole, and 5-F-IAA at a concentration of 10 µm, FC at a concentration of 1 µm, and untreated controls.
Values are averages ± se from a minimum of n = 3 protoplasts. Different letters indicate statistically distinct values (ANOVA, P < 0.05).
| Treatment | Change in Net Volume after 30 min |
|---|---|
| Control | −0.1 ± 0.3 a |
| IAA | +3.0 ± 0.6 b |
| PEO-IAA | +3.2 ± 0.3 b |
| Auxinole | +2.1 ± 0.7 b,c |
| 5-F-IAA | +0.4 ± 0.3 c |
| FC | +3.0 ± 0.3 b |
The two synthetic auxin analogs PEO-IAA and auxinole induced a rapid and auxin-like swelling response indicating that protoplast swelling may be mediated by ABP1. This response on the cellular level may not depend on auxin-induced gene expression via the TIR1/AFB signaling pathway; in fact, both substances were identified in a screen for inhibitors of TIR1/AFB signaling.
Auxin-Induced Protoplast Swelling Is Not Altered in a Quadruple Mutant Line Lacking TIR1, AFB1, AFB2, and AFB3
To test the role of TIR1 and three other members of the TIR1/AFB family of auxin receptors, we performed the protoplast expansion assay with the quadruple mutant tir1-1 afb1-1 afb2-1 afb3-1 (Dharmasiri et al., 2005a, 2005b). In these plants, auxin-induced gene expression is strongly suppressed. Protoplasts derived from hypocotyls of the quadruple mutant respond with a rapid swelling after treatment with 10 µm IAA (Supplemental Fig. S1), whereas control protoplasts did not respond with an expansion over the course of the experiment (Supplemental Fig. S1). This result indicates that TIR1, AFB1, AFB2, and AFB3 are not involved in IAA-induced protoplast swelling.
Hypocotyl Elongation Growth and Protoplast Swelling Differ in Agonist Specificity
It had been shown previously that IAA-induced growth of etiolated Arabidopsis hypocotyls and protoplast swelling are early auxin responses (Steffens et al., 2001). To better understand how the auxin response is mediated at the organ level, we performed hypocotyl growth measurements of etiolated Arabidopsis seedlings with 5-F-IAA and PEO-IAA.
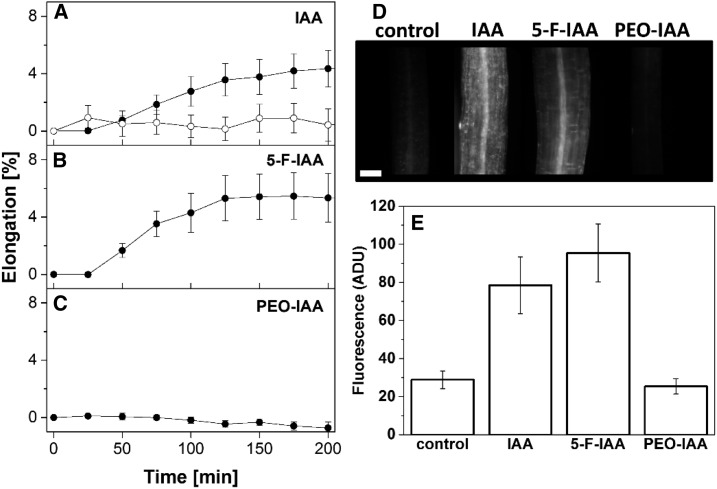
For the growth analysis, we chose 10 µm IAA for comparison (Fig. 2A). This concentration rapidly enhanced the growth of etiolated, auxin-depleted hypocotyl segments (Fig. 2A). 5-F-IAA, which had been suggested to selectively activate the TIR1/AFB signaling pathway, induced an IAA-like growth response at the same concentration (Fig. 2B). PEO-IAA, despite its IAA-like effect on protoplast swelling, did not trigger any significant hypocotyl elongation (Fig. 2C) when compared with an untreated control (Fig. 2A).
Figure 2.
IAA and 5-F-IAA induce hypocotyl elongation, and PEO-IAA does not. A to C, Time courses of growth of etiolated hypocotyl segments. Segments were untreated controls (white circles) or treated with 10 µm of the indicated substances (black circles) at 0 min. While IAA (A) and 5-F-IAA (B) trigger rapid growth responses, PEO-IAA (C) did not induce segment elongation. Values are means ± se from n = 5 to 8 segments per experiment. D, Fluorescence micrographs of DR5rev:GFP plants treated for 24 h with 10 µm of the substances indicated. Typical images of a hypocotyl segment are shown. Bar = 100 µm. E, Photometry of the fluorescence G channel signal of calibrated images ± se. While IAA (n = 20) and 5-F-IAA (n = 7) trigger GFP expression, PEO-IAA (n = 7) is inactive. Values from untreated controls also are shown (n = 18). ADU, Analog-to-digital units.
The halogenated auxin derivative 5-F-IAA induced an IAA-like growth response of etiolated hypocotyls, indicating that this reaction on the organ level may require an activation of the TIR1/AFB signaling pathway. In fact, the activity on the DR5rev:GFP reporter gene system in hypocotyls showed the same specificity pattern as the growth response in hypocotyls (Fig. 2, D and E). IAA and 5-F-IAA induced strong expression, while PEO-IAA was inactive. These results demonstrate that the specificity patterns of protoplast swelling on one side and hypocotyl elongation and gene expression on the other are different. While PEO-IAA induced swelling, it triggered neither growth nor expression of the GFP reporter. 5-F-IAA is a very active auxin in the growth response and in gene expression, but it does not induce any protoplast swelling.
5-F-IAA and PEO-IAA Are Potential Ligands of ABP1 and TIR1
Our physiological results on single cells indicate that the inhibitor of TIR1 signaling PEO-IAA may bind another protein, ABP1 being the prime candidate, to transduce the signal into the cell. To that end, we performed a computational docking study. We analyzed the mode and the probability of binding of PEO-IAA, auxinole, IAA, and 5-F-IAA to ABP1 and to TIR1 from Arabidopsis. For comparison, we also analyzed the binding of 1-naphthaleneacetic acid (1-NAA). It has been discussed earlier that there are different binding affinities of 5-F-IAA and PEO-IAA to the auxin-binding proteins of Arabidopsis (Hayashi et al., 2012).
The crystal structures of ZmABP1 (Woo et al., 2002) and AtTIR1 (Tan et al., 2007) are known, and the binding of 1-NAA and IAA was modeled. AtABP1 was used for the calculation of the docking scores. For AtABP1, a homology model exists that used ZmABP1 (1lrh; Woo et al., 2002) as a template. Their sequence identity is 63%. We used the crystal structure of AtTIR1 (2p1o; Tan et al., 2007) that also exists in the RCSB Protein Data Bank (Berman et al., 2000) in a 1-NAA-bound form. Modeling was performed without Aux/IAA proteins that form a complex with TIR1 upon auxin binding (Calderón Villalobos et al., 2012). The possible binding mode of the natural auxin IAA in AtABP1 (Supplemental Fig. S2A), as predicted by docking with Molecular Operating Environment (MOE), confirmed that the carboxyl group is coordinated by a zinc ion, which is complexed by three His residues (Supplemental Fig. S2B). The aforementioned and additional docking studies performed here indicated that there should not be any large differences in the affinity of natural and synthetic auxins tested to bind AtABP1 and AtTIR1 (Table II). Docking scores after second rescoring with the GBVI/WSA ΔG scoring function in MOE suggest that IAA (Table II) and 1-NAA have the same probability to bind to both auxin receptor proteins in Arabidopsis. General binding of the large auxin analog auxinole (Supplemental Fig. S2) is not altered in AtABP1 or AtTIR1 in comparison with binding of IAA (Table II). PEO-IAA binding to AtABP1 is reduced slightly in comparison with IAA binding, but it should still be possible (Table II).
Table II. Docking scores after second rescoring with the GBVI/WSA ΔG scoring function in MOE of the docked ligands into ABP1 and TIR1.
The absolute docking scores for the substance can only be used for comparison of the binding affinities within each single auxin-binding protein.
| Treatment |
Docking Score |
|
|---|---|---|
| ABP1 | TIR1 | |
| IAA | −12.252 | −6.763 |
| PEO-IAA | −10.527 | −7.261 |
| Auxinole | −13.353 | −7.495 |
| 5-F-IAA | −12.254 | −6.658 |
| 1-NAA | −12.629 | −6.828 |
The isosurface of the auxin-binding domains and the protein structures in general were supposed to be identical in the docking study performed here independently of which substance was modeled. Therefore, these docking studies suggest that the auxin analogs 5-F-IAA, auxinole, and PEO-IAA are able to bind to both ABP1 and TIR1 from Arabidopsis. Conclusions on protein structural changes possibly important for signal transduction cannot be drawn from this study.
C-Terminal Peptides of ABP1 Mimic IAA- and PEO-IAA-Induced Protoplast Swelling
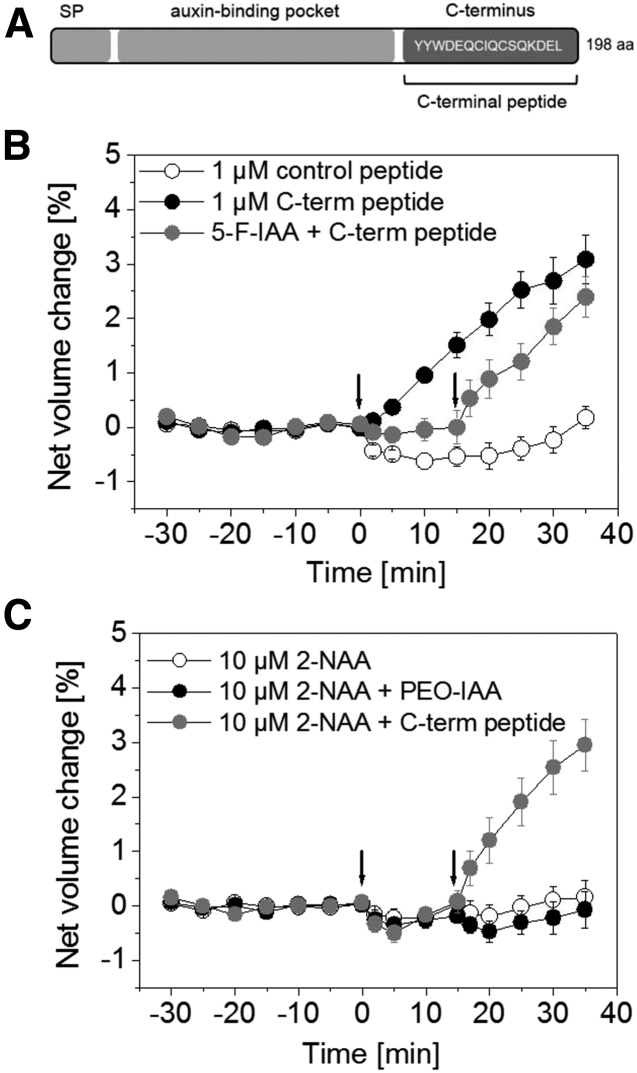
By investigating the action of ABP1, we tested the hypothesis that PEO-IAA acts on protoplast swelling via ABP1. Tools such as C-terminal peptides of ZmABP1 or AtABP1 had been used in earlier studies to analyze the role of ABP1 in various fast auxin-related responses (Rück et al., 1993; Thiel et al., 1993; Steffens et al., 2001). It had been suggested that ABP1 attaches to an interacting membrane protein via its C terminus; therefore, C-terminal peptides have been proposed to exert similar effects to an IAA-activated ABP1 complex. In this study, we used a synthetic peptide consisting of the last 16 amino acids of the C-terminal region of AtABP1 in our protoplast-swelling assay (Fig. 3A).
Figure 3.
The C-terminal peptide of ABP1 induces an IAA-like swelling response. A, Model of ABP1 protein (not in scale) with the predicted signal peptide (SP), auxin-binding pocket, and the C terminus with the ER retention signal KDEL. The sequence of the C-terminal peptide used in this study is highlighted. It consists of the last 16 amino acids (aa) of ABP1. B, Protoplasts were treated with 1 µm of the C-terminal peptide (black circles) or with 1 µm of a control peptide (white circles) at 0 min (first arrow). A competition experiment using 10 µm 5-F-IAA plus 1 µm of the C-terminal peptide also is shown (gray circles). 5-F-IAA was applied at 0 min (first arrow) and the peptide at 15 min indicated by the second arrow. The C-terminal peptide induced a rapid increase in net volume also in the presence of 5-F-IAA. Results are means ± se from a minimum of n = 7 protoplasts from at least two experiments. C, Double treatments were performed to analyze if blocking of the auxin-binding domain of ABP1 by 2-NAA had an effect on the response of PEO-IAA or the C-terminal peptide. All protoplasts were treated with 10 µm 2-NAA at 0 min (first arrow). Protoplast swelling did not occur in 2-NAA-treated protoplasts (first arrow). PEO-IAA at a concentration of 10 µm and the C-terminal peptide at a concentration of 1 µm were applied at 15 min (second arrow). The response of PEO-IAA was inhibited by preincubation with 2-NAA, while swelling still occurred in the presence of the C-terminal peptide. Results are means ± se from a minimum of n = 9 protoplasts from at least two independent experiments.
The auxin agonist activity of the C-terminal peptide is shown in Figure 3, B and C. After the application of 1 µm peptide, the net volume of the protoplasts increased (Fig. 3B). This observation confirmed earlier protoplast experiments in Arabidopsis, where a C-terminal peptide lacking the last three amino acids, DEL, induced a rapid protoplast expansion (Steffens et al., 2001). These results indicate that the signaling pathway leading to protoplast swelling involves ABP1. A control peptide with a sequence not related to ABP1 did not induce protoplast expansion (Fig. 3B).
In addition, we performed a double treatment with 10 µm 5-F-IAA and 1 µm of the C-terminal peptide to address the question of whether 5-F-IAA unspecifically impairs the ability of the protoplasts to swell. Again, protoplasts treated with 5-F-IAA remained nearly constant in net volume, but they responded immediately to a subsequent application of C-terminal peptide with a rapid protoplast expansion (Fig. 3B). The net volume of protoplasts treated with the C-terminal peptide increased rapidly. This demonstrates that protoplasts are still able to swell in the presence of 5-F-IAA.
The inactive auxin analog 2-naphthaleneacetic acid (2-NAA) binds to ABP1 (Edgerton et al., 1994), but the net volume of protoplasts treated with 10 µm 2-NAA remained control like (Fig. 3C; Steffens and Lüthen, 2000). We performed double treatments with protoplasts that were preincubated for 15 min with 2-NAA before a second application with PEO-IAA or the C-terminal peptide followed (Fig. 3C). 2-NAA inhibited the increase in net volume of PEO-IAA-treated protoplasts completely, while protoplast swelling induced by the C-terminal peptide did not decrease. These results indicate that the binding of PEO-IAA to the auxin-binding domain of ABP1 is a prerequisite for protoplast swelling and that 2-NAA blocks this binding.
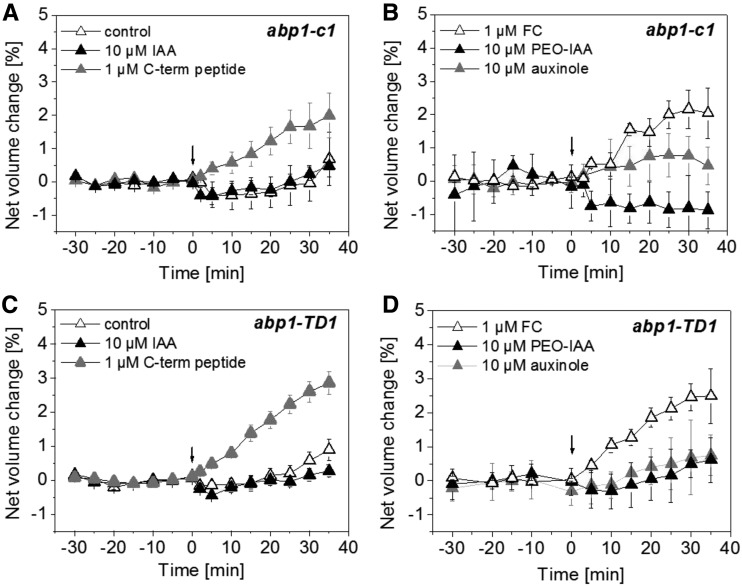
Auxin-Induced Protoplast Swelling Is Abolished in abp1 Mutants, and ABP1 Peptides Can Complement the Mutant Phenotype
The results presented so far and earlier immunological studies strongly suggested that ABP1 was functionally involved in auxin-induced protoplast swelling. To test this hypothesis, we performed the protoplast-swelling assay with the CRISPR/CAS9 line abp1-c1 and the T-DNA insertion line abp1-TD1 (Gao et al., 2015). Both lines were shown previously to not contain any detectable ABP1 transcript and ABP1 protein, as both are null alleles (Gao et al., 2015). We applied 10 µm IAA (Fig. 4, A and C) or 10 µm PEO-IAA and 10 µm auxinole (Fig. 4, B and D) to protoplasts derived from the hypocotyl of both abp1 mutants. IAA-induced protoplast swelling was abolished completely in both abp1 null mutants (Fig. 4, A and C), indicating that ABP1 is a prerequisite in this cellular response. Also, PEO-IAA and auxinole both did not trigger protoplast expansion. We applied the C-terminal peptide (Fig. 4, A and C) and FC (Fig. 4, B and D) as controls. The C-terminal peptide induced a fast protoplast-swelling response in both abp1 null mutants in the absence of any auxin (Fig. 4, A and C), indicating that the downstream signaling pathway is not affected in these abp1 plants. FC triggered protoplast expansion, indicating that the abp1 null mutant protoplasts are still able to change their volume in response to effectors acting independently of auxin signaling (Fig. 4D).
Figure 4.
IAA-induced protoplast swelling is dependent on the presence of ABP1. A and C, Protoplasts of abp1-c1 (A) and abp1-TD1 (C) were not treated (white triangles) or treated with 10 µm IAA (black triangles) or 1 µm C-terminal peptide (gray triangles) at 0 min (indicated by the arrows). IAA did not induce protoplast swelling. The C-terminal peptide was used as a control and induced protoplast expansion. B and D, Protoplasts of abp1-c1 (B) and abp1-TD1 (D) were treated with 1 µm FC (white triangles), with 10 µm PEO-IAA (black triangles), or with 10 µm auxinole (gray triangles) at 0 min (indicated by the arrows). PEO-IAA and auxinole did not induce protoplast swelling. FC was used as a control and induced protoplast expansion. Results are means ± se of a minimum of n = 6 protoplasts from at least two independent experiments.
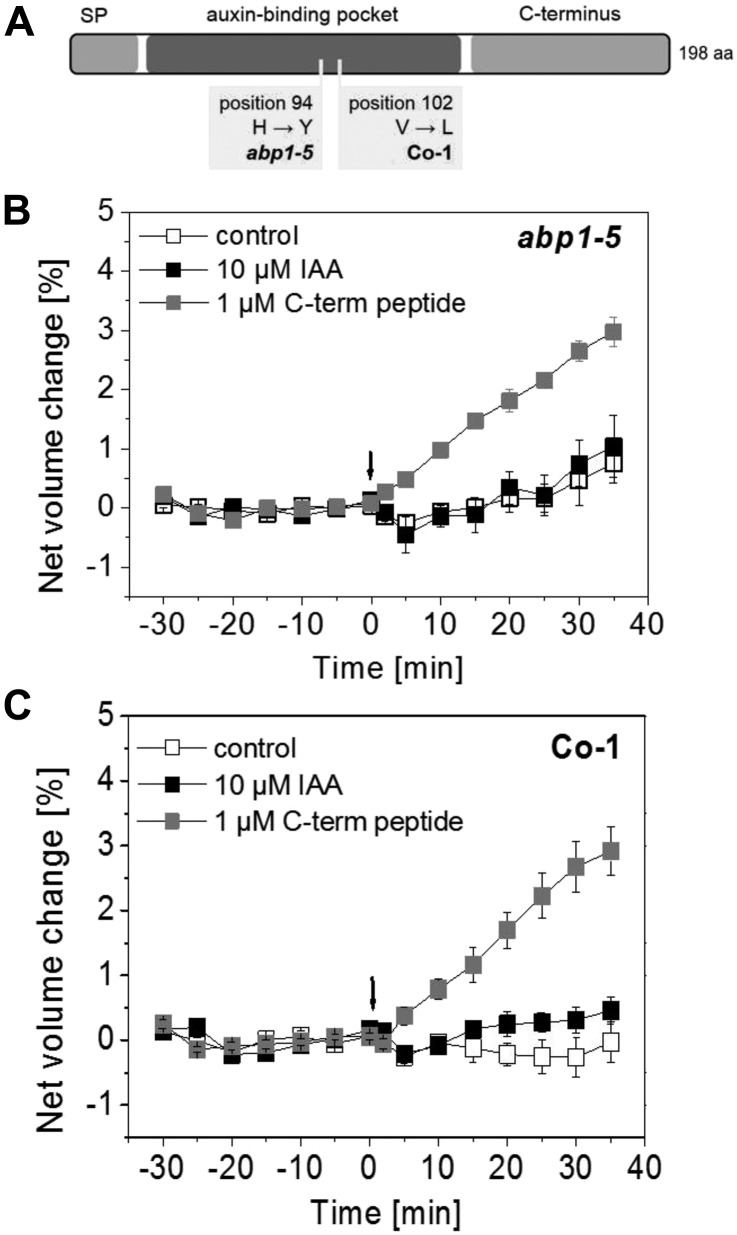
Alterations in the Auxin-Binding Domain of ABP1 Prevent Auxin-Induced Protoplast Swelling in abp1-5 and Coimbra
As an additional test of the view that ABP1 is needed for protoplast swelling, we performed protoplast-swelling analysis with abp1-5, a weak abp1 allele. This TILLING mutant is defective in the auxin-binding domain of ABP1 because of a His-to-Tyr point mutation at position 94 (Xu et al., 2010; Fig. 5A). This mutation leads to the loss of the coordinative Zn2+ ion assumed to result in a lack of auxin binding (Lars-Oliver Essen, personal communication). Figure 5B shows that protoplasts derived from hypocotyls of abp1-5 did not swell in response to 10 µm IAA. Protoplasts from abp1-5 still responded to the C-terminal peptide with a swelling response, indicating that the auxin signaling pathway in general is not affected in this mutant (Fig. 5B).
Figure 5.
IAA does not induce swelling of abp1-5 and Co-1 hypocotyl protoplasts. A, Model of ABP1 protein (not in scale) indicating the amino acid (aa) exchanges in abp1-5 and Co-1. Both alterations occur in the auxin-binding pocket. SP, Signal peptide. B and C, Protoplasts of abp1-5 (B) and Co-1 (C) treated with 10 µm IAA at 0 min (indicated by the arrows) did not respond with rapid swelling. C-terminal peptide at a concentration of 1 µm was applied as a control. It induced a fast swelling response in B and C. Results are means ± se of a minimum of n = 7 protoplasts from at least two independent experiments.
In addition, we performed a comparison of ABP1 sequences in available Arabidopsis accessions (http://signal.salk.edu/atg1001/3.0/gebrowser.php) with the aim to identify accessions with exchanges in amino acid sequences in the auxin-binding domain of ABP1. The ABP1 protein from Columbia-0 (Col-0; NP_192207.1, AT4G02980.1) was used as a consensus sequence. It exhibits a 33-amino acid N-terminal signal peptide followed by a region with at least three important amino acids (Supplemental Fig. S3). One is glycosylated Asn-46, and the others are Gly-59 and Gln-81, which are involved in auxin binding. The highly conserved box a starts at position 89 with Thr and Pro. The two His residues in positions 92 and 94 and Glu-97 coordinate the Zn2+ ion that is important for auxin binding (Woo et al., 2002). The region that comprises the amino acids in positions 106 to 148 is not highly conserved. There is glycosylated Asn-130 and another His-141 that coordinates the Zn2+ ion. Box b is highly conserved, but the function of this domain that comprises amino acids 149 to 161 is unknown. One Ile separates box b from box c in Arabidopsis. Amino acids of box c were described to have a function in auxin binding, especially the Pro-182 at the end of this domain. Box a, box b, and box c comprise the auxin-binding pocket (Fig. 5A). The C-terminal region is separated by two Tyr residues and starts with Trp-185, which functions in auxin binding. The C terminus of ABP1 ends with an endoplasmic reticulum (ER) retention signal that explains the predominant location of this protein in the ER.
We identified eight ecotypes exhibiting single-nucleotide polymorphisms (SNPs) in ABP1. Out of these, four natural accessions contain an SNP in the putative auxin-binding region. One of those is Coimbra (Co-1). In this accession, Val-102 is changed to Leu-102 (Fig. 5A). The function of this amino acid in box a is unknown. The ABP1 protein of Co-1 belongs to another protein variant group than ABP1 from Col-0 (Supplemental Table S1). Additionally, we performed comparisons of TIR1 and AFB1 to AFB5 to understand if there are amino acid exchanges in this auxin coreceptor family in Co-1 (Supplemental Table S1). TIR1, AFB1, AFB3, and AFB5 proteins from Co-1 belong to the same protein variant group as the proteins from Col-0, whereas AFB2 and AFB4 belong to another protein variant group (Supplemental Table S1).
To analyze if auxin binding to Val-102 is a prerequisite for fast protoplast swelling, we performed the analysis in the natural accession Co-1 (Fig. 5C). Protoplasts derived from Co-1 hypocotyls did not respond with a swelling reaction to 10 µm of the natural auxin IAA. Figure 5C shows that the protoplasts are still capable of swelling in response to the C-terminal peptide at a concentration of 1 µm. Taken together, these results indicate that auxin binding to the auxin-binding domain of ABP1 is important for protoplast swelling.
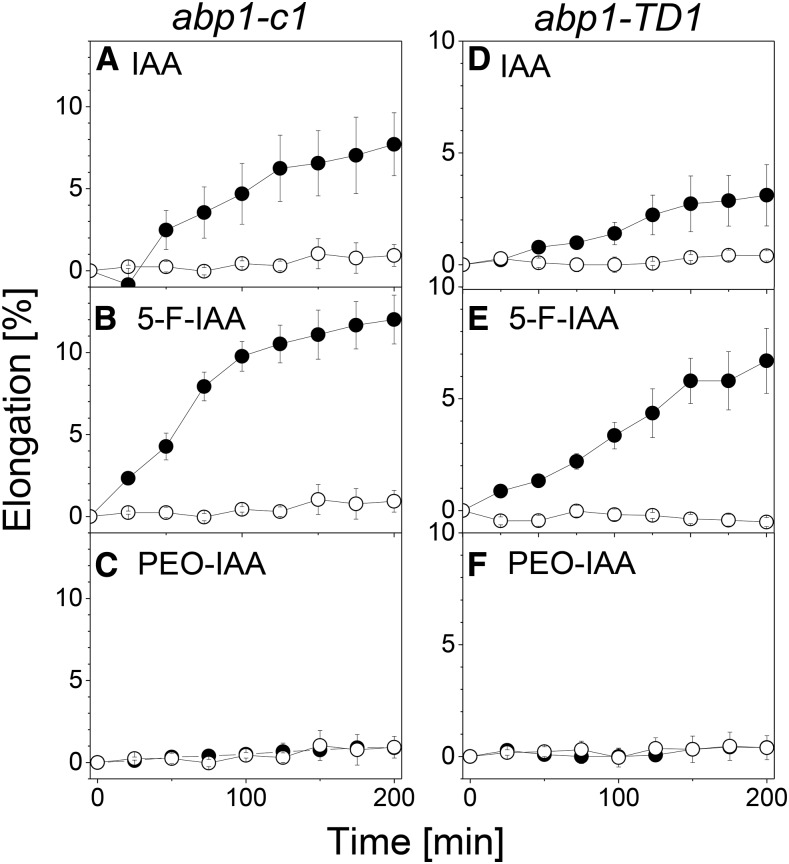
abp1-c1 and abp1-TD1 Respond to Active Auxins with Rapid Hypocotyl Elongation Growth
To test the hypothesis that ABP1 just triggers the early phases of auxin-induced growth, we treated hypocotyls of abp1-c1 and abp1-TD1 with IAA and the various auxin analogs. As shown in Figure 6, rapid elongation was triggered in the same way as in the wild type: 10 µm IAA (Fig. 6, A and D) and 10 µm 5-F-IAA (Fig. 6, B and E) rapidly induced elongation growth, while no such activity could be observed for 10 µm PEO-IAA (Fig. 6, C and F). Within the limits of the temporal resolution of our measurements, there were no differences in lag phase of the responses to IAA and 5-F-IAA in comparison with the wild type (Fig. 2). The variations in the amplitude visible in comparison with Figure 2 were within the range of variations often seen between different genotypes of Arabidopsis, but both abp1 mutants were clearly able to respond to IAA and the agonist of the TIR1/AFB pathway 5-F-IAA. We conclude that the pattern of activity of these test substances is identical in abp1 and wild-type hypocotyls and that rapid auxin-induced hypocotyl growth occurs independently of the presence of ABP1.
Figure 6.
The rapid growth response in hypocotyls to auxin does not depend on ABP1. Rapid growth responses to IAA and auxin analogs are shown in excised auxin-depleted hypocotyls of abp1-c1 (A–C) and abp1-TD1 (D–F). More explanations are given in the legend to Figure 2. In both abp1 mutants, IAA and 5-F-IAA induce growth, while PEO-IAA is not active. Values are means ± se from n = 8 segments per experiment.
Sequence Polymorphisms and Sequence Divergence of ABP1 and Auxin Coreceptors
To examine whether signatures of natural selection are present in ABP1 or the TIR1/AFB genes, we analyzed sequence polymorphism levels within Arabidopsis and sequence divergence levels using Arabidopsis lyrata as the outgroup (Table III). Since, within one population, alleles might not have been fixed, dN/dS ratios could possibly be misleading without considering species-specific genome-wide patterns. Accounting for this, we used a background data set for Arabidopsis as the empirical distribution (Janitza et al., 2012). The high ABP1 dN/dS ratio of 6.529 (empirical-like dN, low dS) would argue for positive selection acting on ABP1 within Arabidopsis; however, considering the empirical distribution (21,329 annotated gene models), this dN/dS ratio is not significant. In contrast to ABP1, the TIR1/AFB genes (low dN, empirical-like dS) showed a significant difference favoring negative selection acting on this gene family within Arabidopsis (Supplemental Fig. S4). The kA/kS ratios for both ABP1 and the TIR1/AFB genes showed no significant difference from 19,775 A. thaliana-A. lyrata ortholog pairs (Supplemental Fig. S5).
Table III. Comparison of within- and between-species nucleotide diversity of auxin-binding genes.
Mean nucleotide diversity (dN/dS) and nucleotide divergence (kA/kS) of auxin (co)receptor genes are shown. The McDonald and Kreitman test (MK Test) results are shown before/after masking all codons with a minor allele frequency lower than 5%. Asterisks indicate significant differences from empirical distribution or significant distinction from neutrality based on the McDonald and Kreitman test (*, significant after Benjamini and Hochberg correction at P < 0.05). For the calculation of empirical ratios, all values not available and infinite values were excluded (dN/dS, 818 from 21,329 gene models; kA/kS, 0 from 19,775 ortholog pairs), as indicated by #.
| Genes | dN | dS | dN/dS | kA | kS | kA/kS | MK Test |
|---|---|---|---|---|---|---|---|
| ABP1 | 11.1E-4 | 1.7E-4 | 6.529 | 0.015 | 0.128 | 0.121 | 0.03*/0.69 |
| TIR1 | 0.5E-4 | 17.2E-4 | 0.029 | 0.015 | 0.117 | 0.128 | 0.87/1.00 |
| AFB1 | 0.7E-4 | 33.7E-4 | 0.021 | 0.018 | 0.123 | 0.150 | 0.06/0.69 |
| AFB2 | 4.4E-4 | 68.0E-4 | 0.065 | 0.005 | 0.189 | 0.029 | 0.03*/0.69 |
| AFB3 | 2.7E-4 | 20.6E-4 | 0.131 | 0.013 | 0.164 | 0.078 | 0.30/0.69 |
| AFB4 | 9.9E-4 | 90.7E-4 | 0.109 | 0.027 | 0.132 | 0.204 | 0.48/0.53 |
| AFB5 | 1.5E-4 | 76.5E-4 | 0.020 | 0.007 | 0.133 | 0.049 | 1.00/0.69 |
| TIR1/AFBs | 3.1E-4* | 52.5E-4 | 0.058* | 0.014 | 0.157 | 0.101 | – |
| Empirical | 18.5E-4 | 90.3E-4 | 0.412# | 0.044 | 0.206 | 0.232# | – |
To further evaluate the diverse patterns for dN/dS and kA/kS ratios that are based on complete coding sequences, we applied a sliding window approach that can highlight regions under different selection constraints. Here, ABP1 under three window sizes (30, 90, and 150 bp) shows a higher interspecific peak as compared with the intraspecific peak, which was not observed for the TIR1/AFB genes (Supplemental Fig. S6). In addition to mutation rates, dN/dS and kA/kS ratios higher than 1 would indicate positive selection and values lower than 1 would indicate negative selection, so the McDonald and Kreitman test was applied. This test was performed to reject the null hypothesis that the observed mutations are neutral. For ABP1 and AFB2, a statistical differentiation from a neutral evolution model using A. lyrata as the outgroup, with neutrality index values higher than 1 favoring purifying or balancing selection, was observed. However, after masking all codons with a minor allele frequency lower than 5%, this significance vanished (Table III). To further illustrate the differences between Arabidopsis subpopulations, haplotype networks were visualized for ABP1 and the TIR1/AFB genes (Supplemental Figs. S7–S10), showing that ABP1, TIR1, AFB1, and AFB3 have a rather centralized haplotype network whereas AFB2, AFB4, and AFB5 are more scattered. For AFB4, even the Col-0 and Co-1 protein variant is not the major haplotype form (Supplemental Fig. S10).
DISCUSSION
Although auxin is the phytohormone known for the longest time, the question of how auxin is perceived and how the auxin signal is transduced inside the cell is still a matter of debate. Especially, it is not clear if ABP1 plays a role as an auxin receptor aside from the TIR1/AFB auxin receptors. It has been established that the latter proteins perceive responses for gene induction, and it was proposed that ABP1 might trigger rapid, growth-related responses at the plasma membrane or elsewhere in the cell (Scherer, 2011). Thus, the question was raised whether ABP1 is a genuine auxin receptor. But what features make a protein a receptor? According to the elementary definition, “Cells have proteins called receptors that bind to signaling molecules and initiate a physiological response” (http://www.nature.com/scitable/topicpage/cell-signaling-14047077), ABP1 shares at least some of these features, thereby supporting the interpretation that this protein may be a promising auxin receptor candidate. Clearly, ABP1 binds auxin, as demonstrated by physiological and structural studies (Hesse et al., 1989; Woo et al., 2002). Furthermore, ABP1 binds auxin at concentrations that were described to be physiological, with an optimal pH at around 5 to 6. Several groups attempted to provide experimental evidence to support the hypothesis that ABP1 is located at the apoplastic side of the plasma membrane (Jones and Herman, 1993; Diekmann et al., 1995), but convincing evidence points to a predominant location of ABP1 in the lumen of the ER (Campos et al., 1993; Jones and Herman, 1993; Tian et al., 1995; Henderson et al., 1997; Xu et al., 2014; Adamowski and Friml, 2015). Since the auxin-binding conditions in the ER are not perfect, it cannot be excluded that a small amount of ABP1, possibly below detection by current protein biochemical methods, may escape the ER and emerge at the plasma membrane, thereby exhibiting features as a putative extracellular receptor candidate responsible for fast nontranscriptional auxin responses. However, convincing biochemical evidence to support this interesting hypothesis is still lacking.
Auxin-Induced Protoplast Swelling Depends on ABP1
Recently, a number of proposed phenotypes of abp1 mutants were either shown to be mimicked by off-target mutations (Enders et al., 2015; Michalko et al., 2016) or could not be confirmed in the CRISPR/CAS9 abp1-c1 line and the T-DNA insertion line abp1-TD1 (Gao et al., 2015). The general impression was that these newly available abp1 mutants lacked any phenotype, putting into question any role of ABP1. Here, we show that TIR1, AFB1, AFB2, and AFB3 are not required for auxin-induced rapid protoplast expansion, whereas protoplasts isolated from a large set of abp1 mutants do not display the typical swelling response to auxin. To our knowledge, this is the first phenotype reported for the new abp1 null alleles. It supports the earlier view that protoplast swelling depends on ABP1, which was based on the activity of anti-ABP1 antibodies and C-terminal peptides of ABP1 on this auxin effect (Steffens et al., 2001; Yamagami et al., 2004; Dahlke et al., 2009). These immunological studies provide indirect evidence that ABP1 could be extracellularly located; however, endocytic uptake of antibodies and peptides might alternatively cause intracellular responses. In addition, ABP1 was discussed to mediate auxin-induced responses that occur within seconds or the first 1 min, such as a rapid change in plasma membrane potential and potassium fluxes (Rück et al., 1993; Thiel et al., 1993). Both reactions are the basis of protoplast expansion.
We convincingly demonstrate that two abp1 knockout lines generated by two different strategies fail to respond to the native auxin IAA and to PEO-IAA and auxinole in the protoplast-swelling assay. We also demonstrate that the abp1-5 mutant does not respond to auxin in this assay. This TILLING line was recently shown to contain a large number of off-target mutations (Enders et al., 2015), including one in phytochrome B, as well as a number of other mutations that might obscure phenotypes and lead to misinterpretations. Also, phenotypes reported earlier in conditional knockdown lines could not be confirmed in the newly available abp1-c1 and abp1-TD1 lines (Gao et al., 2015; Michalko et al., 2016).
In an additional approach, we compared ABP1 sequences of different Arabidopsis ecotypes to identify accessions with naturally occurring variations in the ABP1 protein. We show that Co-1, an ecotype containing a point mutation in the auxin-binding domain of ABP1, also is impaired in protoplast swelling. To complement our studies, we performed analysis on nucleotide diversity, neutrality tests, and haplotype distribution tests. As compared with the sister species A. lyrata, ABP1 and the TIR1/AFB genes show no evidence of directional selection nor a clear signature of purifying or balancing selection after masking minor alleles. TIR1/AFB genes are conserved within Arabidopsis, as indicated by significant low interallelic dN/dS ratios. However, for ABP1 and TIR1/AFB genes, some isolated regions show spikes in interallelic dN/dS and interspecies kA/kS ratios that might imply differentiated functional alleles or directional selection (Bakker et al., 2006). No obvious region shows spikes for both parameters for ABP1, and the decreased synonymous nucleotide diversity might be linked to codon usage efficiency, but this needs to be evaluated further. ABP1 consists of one major centralized haplotype, whereas the Co-1 accessions might represent a slightly deleterious protein variant.
However, the fact that the set of mutations investigated in this study created consistent phenotypes in protoplast swelling makes it very unlikely that off-target mutations play a role in the findings presented here, and it shows that the fast protoplast expansion indeed is linked to ABP1.
PEO-IAA and 5-F-IAA Target Different Auxin Pathways
We also demonstrate that protoplast swelling differs from rapid growth in the specificity patterns of synthetic auxin analogs. We show that PEO-IAA is a strong auxin in the protoplast response while it is inactive in terms of promoting hypocotyl growth and does not trigger gene expression, as visualized by DR5rev:GFP activity. The latter finding is not surprising, as PEO-IAA and auxinole were identified originally as inhibitors of TIR1 signaling (Hayashi et al., 2012). On the other hand, 5-F-IAA has a strong growth-promoting activity while not exerting any protoplast volume changes. Similar to IAA, 5-F-IAA also increases lateral root formation, reduces primary root length, and induces DR5-driven gene expression, indicating that 5-F-IAA activates TIR1/AFB signaling. Our bioinformatic modeling analyses may give a hint that IAA, PEO-IAA, auxinole, and 5-F-IAA might bind strongly to ABP1. However, binding probability alone is not a sufficient parameter to predict the possible action of a substance. This is shown by the fact that IAA, 5-F-IAA, PEO-IAA, and auxinole share a similar binding activity for TIR1 but are known to exert very different effects on TIR1/AFB-dependent gene expression. While PEO-IAA and auxinole do not induce gene expression or even inhibit the TIR1/AFB pathway, the other substances listed are active or very active auxins. Physiological activity strongly depends on the way the substance inserts and is oriented into its binding pocket. Further docking studies that take the isosurface of the auxin-binding pocket of ABP1 into account will help to elucidate if the binding of these substance may result in conformational changes.
However, several phenotypes identified in the alcohol-induced abp1 knockdown lines have not been confirmed in the now available abp1-c1 and abp1-TD1 plants (Gao et al., 2015) and may depend on off-target effects (Michalko et al., 2016). Here, we show that the same specificity pattern (IAA = PEO-IAA > > 5-F-IAA) appears in protoplast swelling. We also demonstrate this swelling response to be affected in a number of abp1 mutants not related to the alcohol-induced abp1 knockdown lines. Hence, some of the published observations made for these plants may really be due to the knockdown of ABP1. Therefore, these individual effects should be thoroughly reinvestigated in the new abp1 knockout lines.
Rapid Growth Responses to Auxin Are Not Related to ABP1 Signaling
Auxin-induced growth is a rapid response occurring after a lag phase of 10 to 20 min. It has been argued that this very short delay indicates the involvement of a nontranscriptional pathway, possibly mediated by ABP1 (Scherer, 2011). Gao et al. (2015) already showed that auxin-induced root growth inhibition is not altered in the abp1-c1 mutant. They measured the effect after 2 d of auxin administration, which is not representative for a rapid effect. Interestingly, 5-d-old etiolated seedlings of the abp1-c1 and abp1-TD1 mutants used in this study for the protoplast-swelling assay exhibit significantly reduced fresh weight (Supplemental Fig. S11), which may suggest that ABP1, although it does not affect the rapid growth response, might be related to long-term developmental processes. The lack of an auxin-induced swelling response in the ABP1 mutants and the lower fresh weight also could point to a role of ABP1 in osmoregulation and the regulation of water relations in general.
In this study, we show that the classical positive rapid growth response of etioliated hypocotyls to the natural auxin IAA is not altered in the abp1 mutants, even if measured at a high temporal resolution. Also, we show that the specificity pattern of ABP1-related effects observed in protoplast swelling is not observed in the growth response; in contrast, 5-F-IAA is a powerful inducer of elongation growth in hypocotyls, while PEO-IAA is not active (IAA = 5-F-IAA > > PEO-IAA). Summing up, we conclude that ABP1 is not involved in auxin-induced hypocotyl elongation or in the early growth phases. Instead, TIR1 and other AFBs may be the candidates for the growth-relevant auxin receptor, as their ligand specificity is well reflected in the data reported here. This interpretation has very recently been strengthened by studies of early 1-NAA- and IAA-induced hypocotyl growth (Fendrych et al., 2016; Spartz et al., 2017), but the possibility of yet another growth-relevant receptor cannot be entirely excluded.
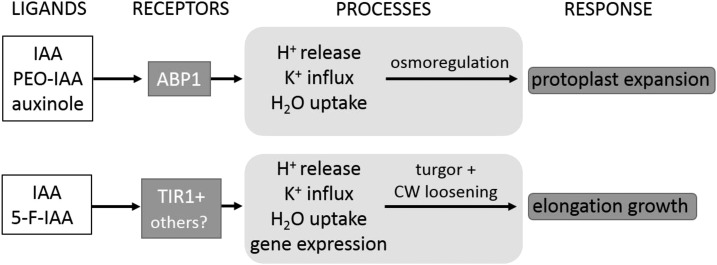
Protoplast Swelling and Growth Are Governed by Different Processes of Volume Control
This investigation shows striking differences in the auxin specificity of two types of auxin-dependent processes that both result in rapid increases of cell volume: protoplast expansion and elongation growth. This appears astonishing at first glance but not so surprising at the second, as both responses differ in their general biophysical properties. Although both types of volume increase require water uptake, the driving forces of both responses may be very different from a biophysical view. Elongation growth in intact organs is turgor driven and involves cell wall loosening (Lockhart, 1965; Cosgrove, 1993). The observed volume changes in protoplasts rather reflect an auxin-dependent type of osmoregulation (Iino et al., 2001), while the mechanical properties of the cell wall may be the main target of auxin on the organ level. Our data indicate that both aspects of volume regulation are mediated by different pathways (Fig. 7). The proton pump itself has been suggested to be activated directly by ABP1 (Rück et al., 1993), and potassium channels (Thiel et al., 1993) and aquaporins (Hachez et al., 2013) may be additional targets of the ABP1 pathway, while the TIR1/AFB pathway may address targets like expansins (McQueen-Mason and Cosgrove, 1995), xyloglucan endotransferases, or other cell wall-loosening enzymes. Many of these targets are dependent on an acidic cell wall pH, being regulated in part by the activity of the proton pump, which, by itself, also is a target of auxin action. There may be considerable cross talk between both pathways, as potassium channels (Philippar et al., 1999) and the H+-ATPase (Hager et al., 1991; Rober-Kleber et al., 2003) have been shown to be regulated both transcriptionally and directly (Rück et al., 1993; Thiel et al., 1993) by auxin.
Figure 7.
ABP1 mediates fast protoplast swelling, and TIR1 or other receptor candidates are important for hypocotyl growth. The model highlights that osmotically driven protoplast expansion is dependent on H+ pump activity, K+ influx, and water uptake. This fast ABP1-mediated response is triggered by IAA, PEO-IAA, and auxinole. IAA- and 5-F-IAA-induced hypocotyl growth are dependent on turgor pressure and cell wall (CW) loosening, processes regulated by TIR1 and putative other auxin receptor proteins.
CONCLUSION
Our study strengthens the view that ABP1 mediates rapid, nontranscriptional auxin responses related to osmoregulation (Fig. 7). The involvement of ABP1 in the growth response remains far from proven, especially as the biophysical causes for auxin- and FC-induced growth responses on the organ level, and the volume changes observed in protoplasts, are thought to be quite different. This matter may be clarified by a critical comparison of the responses of auxin signal transduction mutants at the single-cell level and at the organ level, in order to explore the similarities between auxin-induced effects and effects altered by the immunological tools of ABP1.
Receptors are defined by initiating a physiological response. As auxin initiates rapid, ABP1-dependent protoplast swelling, ABP1 may indeed be called a receptor for auxin.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Experiments were carried out using Arabidopsis (Arabidopsis thaliana) ecotypes Col-0 and Co-1 (Co-1.SALK/1-198). Other lines used in this study are in the Col-0 background (abp1-5, abp1-c1, abp1-TD1, and DR5rev:GFP [Nottingham Arabidopsis Stock Centre identifier N9361]) or in the Col-0/Wassilewskija background (tir1-1 afb1-1 afb2-1 afb3-1). For protoplast experiments, Arabidopsis seeds were surface sterilized for 15 min in 2% (w/v) sodium hypochlorite, washed five times with autoclaved water, and laid out under sterile conditions. Seedlings were grown in the dark at 22°C for 5 d on plates containing 0.5× Murashige and Skoog medium (Murashige and Skoog, 1962) and 1% (w/v) Suc solidified with 0.7% (w/v) phytagel. For growth experiments, seedlings were grown in the dark for 3 d on moist paper. If not stated otherwise, the experiments were performed with Col-0.
Protoplast-Swelling Assay
Protoplasts were isolated from hypocotyls of 5-d-old etiolated seedlings as described earlier (Steffens et al., 2001; Stührwohldt et al., 2011). Vital protoplasts with visible cytoplasmic streaming were used for the experiments. Briefly, photographs of single protoplasts were taken over the indicated times using a camera on a light microscope. After the experiment, cross-sectional areas of each protoplast were measured using the image-processing program ImageJ (Schneider et al., 2012). Assuming a spherical shape of the protoplasts, we were able to calculate the volume of each protoplast. Untreated control protoplasts slowly decreased in volume with linear time function (Steffens and Lüthen, 2000). This endogenous shrinkage was corrected, and changes in net volume were calculated for each protoplast (Steffens and Lüthen, 2000). Detailed description is given in Supplemental Materials and Methods S1 and S2. FC, IAA, PEO-IAA, 5-F-IAA, and auxinole were dissolved in buffer consisting of 10 mm KCl, 10 mm MES, 1 mm CaCl2, and 0.002‰ DMF. A C-terminal peptide was derived from the last 16 amino acids of AtABP1 with the sequence YYWDEQCIQCSQKDEL (Genosphere Biotechnologies). A control peptide with a sequence not related to ABP1 consists of the same amino acids in the following order: ECDKLYQCQIEWYDSQ.
Hypocotyl Growth Measurements
Hypocotyl growth measurements were carried out as described elsewhere (Schenck et al., 2010; Fraas et al., 2014). Briefly, hypocotyl segments of 3-d-old etiolated seedlings were cut below the hook and depleted of endogenous auxin by immersing them in auxin-free medium consisting of 10 mm KCl and 1 mm CaCl2 for 1.5 h. Segments were transferred onto a 24-well plate filled with 0.5 mL of a solution with or without IAA or synthetic auxin analogs as indicated. Image acquisition was performed by photographing segments using a binocular microscope and measuring them manually using ImageJ software as described elsewhere (Schenck et al., 2010).
Docking Studies of AtABP1 and AtTIR1
Rigid docking was performed with MOE (2014.09; Chemical Computing Group). For the docking, the crystal structure (1lrh) of Zea mays ABP1 (ZmABP1), the model of Arabidopsis ABP1 (AtABP1), and the crystal structure of Arabidopsis TIR1 (2p1o) were used. Hydrogens were added to the respective protein with the Protonate 3D routine in MOE. The homology model of AtABP1 was minimized according to the Amber12:EHT force field in the presence of 1-NAA and the zinc ion in the binding pocket. The ligand 2D chemical structures were drawn in Marvin Sketch (Marvin 6.1; ChemAxon), and a conformational search was performed with MOE in order to generate 3D conformations. The number of conformations was limited to a maximum of 100 per ligand, and duplicate conformations (root-mean-square deviation < 0.25 Å) were removed. A pharmacophore query was built based on the properties of 1-NAA bound to ZmABP1 (1lrh) and consisted of two hydrogen bond acceptors, to mimic the carbonyl oxygens complexing the zinc ion, and an aromatic moiety. The docking protocol employed the pharmacophore placement method and the London ΔG scoring function. Duplicate poses were removed automatically based on their hydrogen bond and hydrophobic patterns; moreover, poses with positive binding free energy as predicted by the GBVI/WSA ΔG scoring function also were removed. The top-ranked poses were used for visual inspection. The images of the docking poses were generated with PyMOL (PyMOL Molecular Graphics System, version 1.2r3pre; Schrödinger).
Identification of Accessions Defective in the Auxin-Binding Domain of ABP1 from Arabidopsis
The Web site http://signal.salk.edu/atg1001/3.0/gebrowser.php (Arabidopsis 1001 genome project; Schmitz et al., 2013) was used for the identification of an SNP in the ABP1 protein of various Arabidopsis accessions. The ABP1 protein from Col-0 (NP_192207.1, AT4G02980.1) was used as a consensus sequence. The accession Co-1 (Co-1.SALK/1-198) was identified to harbor a nonsynonymous SNP, which was validated by Sanger sequencing from genomic DNA, and was used for the above-mentioned protoplast experiments.
Nucleotide Diversity, Neutrality Tests, and Haplotype Distributions
To calculate nucleotide diversity (dN, dS) and nucleotide divergence (kA, kS) for ABP1 (AT4G02980.1), TIR1 (AT3G62980.1), AFB1 (AT4G03190.1), AFB2 (AT3G26810.1), AFB3 (AT1G12820.1), AFB4 (AT4G24390.2), and AFB5 (AT5G49980.1), coding sequence alignments were generated using the Arabidopsis Col-0 accession (TAIR10) annotation as a reference. The alignments that were used for nucleotide diversity and divergence calculations consist of the Col-0 sequence information, the Co-1 accession (Arabidopsis 1001 genome project; Schmitz et al., 2013), 80 diverse Arabidopsis accessions (filtered_variant.txt.gz for each accession mapped on the corresponding annotation; Cao et al., 2011), and the corresponding best BLAST+ (Camacho et al., 2009) Arabidopsis lyrata (version 1.0; Hu et al., 2011) hit obtained from Phytozome version 11 (Goodstein et al., 2012) as an outgroup. These coding sequence alignments were translated and grouped into protein variation groups (Supplemental Table S1) and were further used to calculate nucleotide diversity and nucleotide divergence for the complete coding region and with a sliding window approach (sizes, 30, 90, and 150 bp; step, 9 bp) with the software suite DnaSP version 5.10.01 (Librado and Rozas, 2009). For background comparison against all TAIR10-annotated gene models, see Janitza et al. (2012). The results from the McDonald and Kreitman test conducted with DnaSP version 5.10.01 were further corrected for multiple testing (Benjamini and Hochberg, 1995). For haplotype network visualization (Supplemental Figs. S7–S10), the above-mentioned alignments were realigned in a codon-aware manner adding best BLAST+-derived Brassica rapa (FPsc version 1.3), Eutrema salsuginea (version 1.0; Yang et al., 2013), and Capsella rubella (version 1.0; Slotte et al., 2013) coding sequences obtained from Phytozome version 11. Accession locations were obtained from https://easygwas.ethz.ch (Grimm et al., 2012), and TCS haplotype networks (Clement et al., 2002) were generated and plotted with popart (http://popart.otago.ac.nz) for ABP1 and the pairs TIR1-AFB1, AFB2-AFB3, and AFB4-AFB5.
DR5:GFP Imaging
Etiolated seedlings of DR5rev:GFP were grown as described above, incubated in 10 mm KCl and 1 mm CaCl2, and treated with the auxins as indicated. After 24 h, photographs were taken using an Olympus BHS fluorescence microscope equipped with a 10× SPLAN Apo objective. A Canon 450D camera was used to take the photographs (20-s exposure time). Calibrated RAW images were debayered using Fitswork (www.fitswork.de), and luminance values in the G-channel of the resulting RGB image were measured and corrected for background signal using the same software.
Statistics
Statistical analyses were performed with Minitab (Minitab). Comparison of means was analyzed for statistical significance with an ANOVA and Tukey’s test. Constant variance and normal distribution of data were verified before statistical analysis, and the P value was set to P < 0.001 if one of both conditions was not met.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ABP1 (AT4G02980.1), TIR1 (AT3G62980.1), AFB1 (AT4G03190.1), AFB2 (AT3G26810.1), AFB3 (AT1G12820.1), AFB4 (AT4G24390.2), and AFB5 (AT5G49980.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. IAA induces protoplast expansion in tir1-1 afb1-1 afb-2-1 afb3-1.
Supplemental Figure S2. Docking study of ABP1 and TIR1 from Arabidopsis.
Supplemental Figure S3. Sequence alignment of ABP1 proteins from the model species Zea mays, Oryza sativa, and Arabidopsis.
Supplemental Figure S4. Nucleotide diversity among 80 natural accessions of Arabidopsis.
Supplemental Figure S5. Nucleotide divergence of ABP1 and TIR1/AFBs between Arabidopsis and A. lyrata.
Supplemental Figure S6. Nucleotide diversity and divergence sliding window plots.
Supplemental Figure S7. ABP1 TCS haplotype network.
Supplemental Figure S8. TIR1-AFB1 TCS haplotype network.
Supplemental Figure S9. AFB2-AFB3 TCS haplotype network.
Supplemental Figure S10. AFB4-AFB5 TCS haplotype network.
Supplemental Figure S11. Images and fresh weights of 5-d-old wild-type (Col-0), abp1-c1, and abp1-TD1 seedlings.
Supplemental Table S1. Protein variant groups of ABP1, TIR1, and AFB1 to AFB5, sample identifiers, latitude, longitude, and region for various Arabidopsis accessions.
Supplemental Materials and Methods S1. Detailed explanation of the protoplast procedure.
Supplemental Materials and Methods S2. Examples of time series of single protoplasts.
Acknowledgments
We thank Ken-ichiro Hayashi (Okayama University of Science) for providing PEO-IAA and auxinole, Kjeld Engvild (Roskilde) for providing 5-F-IAA, Yunde Zhao (University of California) for providing seeds of abp1-c1 and abp1-TD1, and Mark Estelle (University of California) for providing seeds of the quadruple mutant tir1-1 afb1-1 afb2-1 afb3-1. We thank Christian Michalski, Agnes Damm, and Meike Eckel (Marburg University) for excellent technical assistance and Lars-Oliver Essen (Marburg University) for fruitful discussion on ABP1 docking studies. We are grateful to the Chemical Computing Group for making the program MOE available to us.
Glossary
- FC
fusicoccin
- 1-NAA
1-naphthaleneacetic acid
- MOE
Molecular Operating Environment
- 2-NAA
2-naphthaleneacetic acid
- ER
endoplasmic reticulum
- Col-0
Columbia-0
- Co-1
Coimbra
- SNP
single-nucleotide polymorphism
Footnotes
This work was supported by BMBF Microsystems, the Excellence Initiative of the German Federal and State Governments (EXC 294), SFB746, and Deutsches Zentrum für Luft und Raumfahrt. S.F.’s contribution is part of a Ph.D. thesis supported by the Appuhn-Stiftung (Hamburg, Germany).
References
- Adamowski M, Friml J (2015) PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EG, Toomajian C, Kreitman M, Bergelson J (2006) A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18: 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier-Brygoo H, Ephritikhine G, Klämbt D, Maurel C, Palme K, Schell J, Guern J (1991) Perception of the auxin signal at the plasma membrane of tobacco mesophyll protoplasts. Plant J 1: 83–93 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser 57: 289–300 [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, Fleming AJ (2008) Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20: 2746–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos N, Schell J, Palme K (1993) Aminoterminal signal sequences of maize auxin-binding proteins are involved in import in the ER. Plant Cell Physiol 34: 211–220 [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM (2001) ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev 15: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Snell Q, Walker P, Posada D, Crandall K (2002) TCS: estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium. IEEE, http://dx.doi.org/10.1109/IPDPS.2002.1016585 [Google Scholar]
- Cosgrove DJ. (1993) Water uptake by growing cells: an assessment of the controlling roles of wall relaxation, solute uptake, and hydraulic conductance. Int J Plant Sci 154: 10–21 [DOI] [PubMed] [Google Scholar]
- Dahlke RI, Lüthen H, Steffens B (2009) The auxin-binding pocket of auxin-binding protein 1 comprises the highly conserved boxes a and c. Planta 230: 917–924 [DOI] [PubMed] [Google Scholar]
- Dai X, Zhang Y, Zhang D, Chen J, Gao X, Estelle M, Zhao Y (2015) Embryonic lethality of Arabidopsis abp1-1 is caused by deletion of the adjacent BSM gene. Nat Plants 1: 15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Diekmann W, Venis MA, Robinson DG (1995) Auxins induce clustering of the auxin-binding protein at the surface of maize coleoptile protoplasts. Proc Natl Acad Sci USA 92: 3425–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton MD, Tropsha A, Jones AM (1994) Modelling the auxin-binding site of auxin binding protein 1 of maize. Phytochemistry 35: 1111–1123 [Google Scholar]
- Enders TA, Oh S, Yang Z, Montgomery BL, Strader LC (2015) Genome sequencing of Arabidopsis abp1-5 reveals second-site mutations that may affect phenotypes. Plant Cell 27: 1820–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J (2016) TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraas S, Niehoff V, Lüthen H (2014) A high-throughput imaging auxanometer for roots and hypocotyls of Arabidopsis using a 2D skeletonizing algorithm. Physiol Plant 151: 112–118 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y (2015) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA 112: 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Greshake B, Kleeberger S, Lippert C, Stegle O, Schölkopf B, Weigel D, Borgwardt K (2012) easyGWAS: an integrated interspecies platform for performing genome-wide association studies. arXiv 1212.4788
- Hachez C, Besserer A, Chevalier AS, Chaumont F (2013) Insights into plant plasma membrane aquaporin trafficking. Trends Plant Sci 18: 344–352 [DOI] [PubMed] [Google Scholar]
- Hager A, Debus G, Edel HG, Stransky H, Serrano R (1991) Auxin induces exocytosis and the rapid synthesis of a high-turnover pool of plasma-membrane H+-ATPase. Planta 185: 527–537 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H (2012) Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem Biol 7: 590–598 [DOI] [PubMed] [Google Scholar]
- Henderson J, Bauly JM, Ashford DA, Oliver SC, Hawes CR, Lazarus CM, Venis MA, Napier RM (1997) Retention of maize auxin-binding protein in the endoplasmic reticulum: quantifying escape and the role of auxin. Planta 202: 313–323 [DOI] [PubMed] [Google Scholar]
- Hertel R. (1995) Auxin-binding protein 1 is a red herring. J Exp Bot 46: 461–462 [Google Scholar]
- Hesse T, Feldwisch J, Balshüsemann D, Bauw G, Puype M, Vandekerckhove J, Löbler M, Klämbt D, Schell J, Palme K (1989) Molecular cloning and structural analysis of a gene from Zea mays (L.) coding for a putative receptor for the plant hormone auxin. EMBO J 8: 2453–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino M, Long C, Wang X (2001) Auxin- and abscisic acid-dependent osmoregulation in protoplasts of Phaseolus vulgaris pulvini. Plant Cell Physiol 42: 1219–1227 [DOI] [PubMed] [Google Scholar]
- Janitza P, Ullrich KK, Quint M (2012) Toward a comprehensive phylogenetic reconstruction of the evolutionary history of mitogen-activated protein kinases in the plant kingdom. Front Plant Sci 3: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM. (1990) Do we have the auxin receptor yet? Plant Physiol 80: 154–158 [Google Scholar]
- Jones AM, Herman EM (1993) KDEL-containing auxin-binding protein is secreted to the plasma membrane and cell wall. Plant Physiol 101: 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Löbler M, Klämbt D (1985a) Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). I. Purification by immunological methods and characterization. J Biol Chem 260: 9848–9853 [PubMed] [Google Scholar]
- Löbler M, Klämbt D (1985b) Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). II. Localization of a putative auxin receptor. J Biol Chem 260: 9854–9859 [PubMed] [Google Scholar]
- Lockhart JA. (1965) An analysis of irreversible plant cell elongation. J Theor Biol 8: 264–275 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ (1995) Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol 107: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalko J, Dravecká M, Bollenbach T, Friml J (2015) Embryo-lethal phenotypes in early abp1 mutants are due to disruption of the neighboring BSM gene. F1000 Res 4: 1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalko J, Glanc M, Perrot-Rechenmann C, Friml J (2016) Strong morphological defects in conditional Arabidopsis abp1 knock-down mutants generated in absence of functional ABP1 protein. F1000 Res 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Palme K, Feldwisch J, Hesse T, Bauw G, Vandekerckhove J, Schell J (1990) Isolation of auxin-binding protein 1 and molecular analysis of the axr gene from Zea mays (L.). In Roberts J, Kirk C, Venis M, eds, Hormone Perception and Signal Transduction in Animals and Plants. Company of Biologists, Cambridge, UK, pp 299–313 [Google Scholar]
- Philippar K, Fuchs I, Luthen H, Hoth S, Bauer CS, Haga K, Thiel G, Ljung K, Sandberg G, Bottger M, et al. (1999) Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc Natl Acad Sci USA 96: 12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufky J, Qiu Y, Rao MV, Hurban P, Jones AM (2003) The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct Integr Genomics 3: 135–143 [DOI] [PubMed] [Google Scholar]
- Rober-Kleber N, Albrechtová JTP, Fleig S, Huck N, Michalke W, Wagner E, Speth V, Neuhaus G, Fischer-Iglesias C (2003) Plasma membrane H+-ATPase is involved in auxin-mediated cell elongation during wheat embryo development. Plant Physiol 131: 1302–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al. (2010) ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rück A, Palme K, Venis MA, Napier RM, Felle RH (1993) Patch-clamp analysis establishes a role for an auxin-binding protein in the auxin stimulation of plasma-membrane current in Zea mays protoplasts. Plant J 4: 41–46 [Google Scholar]
- Schenck D, Christian M, Jones A, Lüthen H (2010) Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol 152: 1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer GFE. (2011) AUXIN-BINDING-PROTEIN1, the second auxin receptor: what is the significance of a two-receptor concept in plant signal transduction? J Exp Bot 62: 3339–3357 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, McCosh RB, Chen H, Schork NJ, et al. (2013) Patterns of population epigenomic diversity. Nature 495: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura S, Sotobayashi T, Futai M, Fukui T (1986) Purification and properties of an auxin-binding protein from maize shoot membranes. J Biochem 99: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, Steige K, Platts AE, Escobar JS, Newman LK, et al. (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45: 831–835 [DOI] [PubMed] [Google Scholar]
- Spartz AK, Lor VS, Ren H, Olszewski NE, Miller ND, Wu G, Spalding EP, Gray WM (2017) Constitutive expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in tomato confers auxin-independent hypocotyl elongation. Plant Physiol 173: 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Feckler C, Palme K, Christian M, Böttger M, Lüthen H (2001) The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J 27: 591–599 [DOI] [PubMed] [Google Scholar]
- Steffens B, Lüthen H (2000) New methods to analyse auxin-induced growth. II. The swelling reaction of protoplasts: a model system for the analysis of auxin signal transduction? Plant Growth Regul 32: 115–122 [Google Scholar]
- Stührwohldt N, Dahlke RI, Steffens B, Johnson A, Sauter M (2011) Phytosulfokine-α controls hypocotyl length and cell expansion in Arabidopsis thaliana through phytosulfokine receptor 1. PLoS ONE 6: e21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Thiel G, Blatt MR, Fricker MD, White IR, Millner P (1993) Modulation of K+ channels in Vicia stomatal guard cells by peptide homologs to the auxin-binding protein C terminus. Proc Natl Acad Sci USA 90: 11493–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Klämbt D, Jones AM (1995) Auxin-binding protein 1 does not bind auxin within the endoplasmic reticulum despite this being the predominant subcellular location for this hormone receptor. J Biol Chem 270: 26962–26969 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venis MA. (1995) Auxin-binding protein 1 is a red herring: oh no it isn’t. J Exp Bot 46: 463–465 [Google Scholar]
- Woo EJ, Marshall J, Bauly J, Chen JG, Venis M, Napier RM, Pickersgill RW (2002) Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J 21: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H, et al. (2014) Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami M, Haga K, Napier RM, Iino M (2004) Two distinct signaling pathways participate in auxin-induced swelling of pea epidermal protoplasts. Plant Physiol 134: 735–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Jarvis DE, Chen H, Beilstein MA, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, et al. (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]