Cell-fate determination in Arabidopsis roots utilizes lineage-dependent mechanisms at early stages and positional-dependent signaling at later stages.
Abstract
Tissue organization and pattern formation within a multicellular organism rely on coordinated cell division and cell-fate determination. In animals, cell fates are mainly determined by a cell lineage-dependent mechanism, whereas in plants, positional information is thought to be the primary determinant of cell fates. However, our understanding of cell-fate regulation in plants mostly relies on the histological and anatomical studies on Arabidopsis (Arabidopsis thaliana) roots, which contain a single layer of each cell type in nonvascular tissues. Here, we investigate the dynamic cell-fate acquisition in modified Arabidopsis roots with additional cell layers that are artificially generated by the misexpression of SHORT-ROOT (SHR). We found that cell-fate determination in Arabidopsis roots is a dimorphic cascade with lineage inheritance dominant in the early stage of pattern formation. The inherited cell identity can subsequently be removed or modified by positional information. The instruction of cell-fate conversion is not a fast readout during root development. The final identity of a cell type is determined by the synergistic contribution from multiple layers of regulation, including symplastic communication across tissues. Our findings underline the collaborative inputs during cell-fate instruction.
Organogenesis in plants requires a tight spatiotemporal regulation of cell division and cell-type specification (Bennett and Scheres, 2010; ten Hove et al., 2015; Radoeva and Weijers, 2014; Dong and Bergmann, 2010; Abrash and Bergmann, 2009; ten Hove and Heidstra, 2008). Our understanding of these two fundamental processes has been greatly advanced through using the model system, Arabidopsis (Arabidopsis thaliana) roots. The Arabidopsis root is composed of concentric rings of different cell files with stele, endodermis, cortex, and epidermis arranged from the inside to the outside (Benfey and Scheres, 2000; Dolan et al., 1993; Scheres et al., 1994). SHORT-ROOT (SHR) was identified as a key regulator of root radial patterning by directing the ground tissue formation (Helariutta et al., 2000; Nakajima et al., 2001). Both cortical and endodermal cell layers in ground tissue derive from the same cortex/endodermal initial (CEI) cells in the stem cell niche (Petricka et al., 2012; Van Norman et al., 2011; Petricka and Benfey, 2008). In the CEI and CEI daughter cells (CEID), SHR activates SCARECROW (SCR), and together both SHR and SCR turn on the expression of CYCLIN D6;1 (CYCD6;1), leading to the periclinal division in CEID and separating the endodermis and cortex (Sozzani et al., 2010; Cruz-Ramírez et al., 2012; Supplemental Fig. S1). Following the periclinal division, both SHR and SCR are thought be essential for determining the endodermal cell fate (Nakajima et al., 2001; Heidstra et al., 2004; Heo et al., 2011; Levesque et al., 2006; Cui et al., 2007; Long et al., 2015a, 2015b).
Recent studies have revealed a complex regulatory network involved in SHR-mediated division and cell-fate specification. However, most known regulators in the SHR pathway, such as SCR and BIRD family of zinc finger proteins, appeared to participate in the SHR-SCR feedback loop to restrict the functional scope of SHR to the CEID and its derivative endodermis (Cui et al., 2007; Long et al., 2015a, 2015b; Welch et al., 2007; Moreno-Risueno et al., 2015). An elegant model was also proposed to explain how SHR function is integrated with auxin signaling to provide spatial information for the periclinal division (Cruz-Ramírez et al., 2012). Interestingly, increased SHR movement is able to trigger extra cell division beyond the endodermis, suggesting SHR is likely functional in outer cell layers (Cui et al., 2007; Wu et al., 2014).
Here, we examined the ability of SHR to induce periclinal cell division and determine cell fate in a broader developmental context in Arabidopsis. Our results indicate that a conserved SHR-mediated regulatory network functions in most cell types outside of the stele. Using these artificially created supernumeral cell layers, we investigated how cell-fate commitment is spatiotemporally achieved in plants. Our results suggest that cell-fate acquisition in Arabidopsis roots is coordinately regulated by durable positional input and transient lineage inheritance. Furthermore, stele-derived symplastic signals participate in cell-fate acquisition of ground tissues. Our findings provide new insights into the long-standing question of cell-fate adoption and underline the crucial role of symplastic communication between tissues during the tissue formation in the root.
RESULTS
Mitotic Activity Is a Prerequisite of Conserved SHR Pathways across Different Tissues
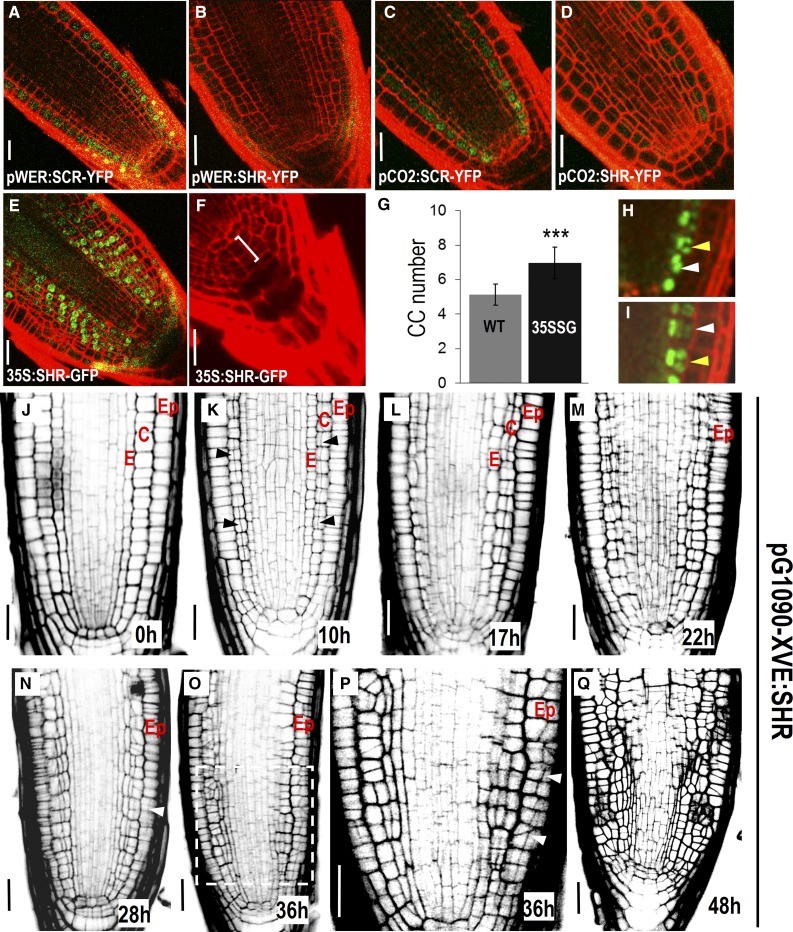
To examine the ability of SHR trigger periclinal cell division, we expressed SHR in different cell types using cell-specific promoters (pWER in epidermis and pCO2 in cortex). In both epidermis and cortex, SHR triggered periclinal cell divisions (Sena et al., 2004; Fig. 1, B and D). When constitutively expressed under the 35S promoter, SHR induced extensive periclinal divisions in the meristem outside of the stele (Sena et al., 2004; Fig. 1E). Interestingly, SHR was even able to activate periclinal division in columella stem cells (CSCs), leading to increased columella cell numbers (Fig. 1, F and G). In contrast, SHR direct target SCARECROW (SCR) was unable to influence the division pattern when misexpressed alone (Fig. 1, A and C), which is consistent with the previous studies showing that SCR forms a protein complex with SHR to promote periclinal cell division (Heidstra et al., 2004; Cui et al., 2007; Long et al., 2015b; Clark et al., 2016).
Figure 1.
Periclinal cell division triggered by SHR is independent of cell types. A to F, Misexpression of SHR (B and D–F) and SCR (A and C). Extra cell layers in collumella was marked by bracket in F. G, Quantification of cell layers in columella in wild-type (WT; n = 10) and 35S:SHR-GFP (35SSG) expressing roots (n = 12). Error bars indicate the sd from the mean. t tests indicate that there is a significant difference (P < 0.01) in columella cell layers between the wild type and 35SSG. H and I, pCO2:H2B-YFP in pG1090-XVE:SHR-expressing roots after 24 h of estradiol induction. Yellow arrows point to anticlinal cell division, and white arrows represent periclinal cell division. J to Q, Time course imaging of the root radial structure in pG1090-XVE:SHR-expressing roots with estradiol treatment for different time length (as labeled). Ep, Epidermis; C, cortex; E, endodermis. Black arrowheads in K and white arrowheads in N and P point to periclinal cell divisions. P is a zoomed view of boxed region in O. Scale bars = 20 μm.
To determine the competence of each individual cell type in response to SHR induction, we performed time course analysis of the division pattern in pG1090-XVE::SHR roots (Fig. 1, J–Q). pG1090-XVE is an inducible promoter that allows ectopic SHR expression upon estradiol treatment (Curtis and Grossniklaus, 2003). After 10 h in estradiol media, the cortex appeared to be the first cell layer that responded to SHR (Fig. 1K). Interestingly, the periclinal cell division started to occur in cells located far above the stem cell niche rather than in the initial cells. In agreement with this, we observed alternated occurrence of anticlinal division and periclinal division in one cell file, shown by the directional arrangement of telophase chromosomes of pCO2:H2B-YFP in pG1090-XVE:SHR roots (Fig. 1, H and I). Thus, SHR’s ability to induce periclinal cell division is not confined to the stem cell area. The epidermis, however, maintained normal cell division pattern until 28 h of treatment, when a few periclinal divisions became visible (Fig. 1N). After 36 h of treatment, the epidermis also exhibited extensive divisions (Fig. 1, O–Q). The asynchronous division suggested that different cell types possibly have distinct sensitivity to the SHR induction. However, we cannot rule out the possibility that pG1090-XVE:SHR had distinct expression levels or SHR had dissimilar stability in different cell types.
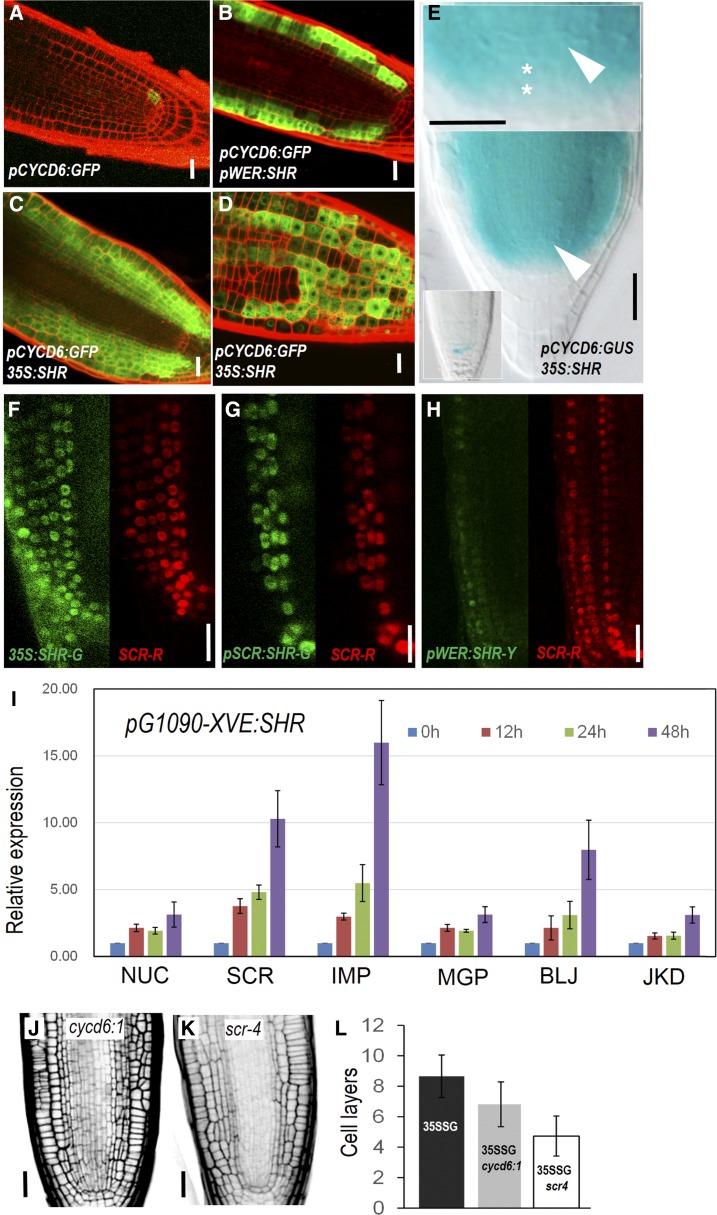
In CEID cells, SHR activates SCR, and together they up-regulate CYCD6;1, which coincides with the switch of division direction (Sozzani et al., 2010). To investigate the downstream mechanism in SHR-mediated periclinal divisions in cells outside of CEID, we observed the expression pattern of CYCD6;1 and SCR in SHR misexpression lines. In wild type, CYCD6;1 expression was confined in CEI and CEID (Sozzani et al., 2010; Fig. 2A). When SHR was misexpressed in the epidermis under the WEREWOLF (WER) promoter, we saw high levels of pCYCD6;1:GFP-GUS in the epidermis as well as its derivative cell layers (Fig. 2B). Consistent with this, 35S:SHR induced CYCD6 expression in almost all cell layers outside the stele (Fig. 2, C and D). In 35S:SHR/pCYCD6;1:GFP-GUS roots, we saw the clear activation of CYCD6 in CSC cells, which is in agreement with the division phenotypes in those cells (Fig. 2E).
Figure 2.
SHR-mediated regulatory network is conserved across different cell types. A to D, Expression of pCYCD6;1-GFP in different backgrounds. E, pCYCD6;1-GUS is activated in the CSC region in 35S:SHR-expressing roots. White arrowheads point to the QC, and asterisks represent the CSC cells. F to H, Expression of pSCR:SCR-mCherry (SCR-R) in different backgrounds. 35S:SHR-G, 35S:SHR-GFP; pSCR:SHR-G, pSCR:SHR-GFP; pWER:SHR-Y, pWER:SHR-YFP. I, qRT-PCR analysis of SHR targets in pG1090-XVE:SHR-expressing roots at different time points. NUC, NUTCRACKER; SCR, SCARECROW; IMP, IMPERIAL EAGLE; MGP, MAGPIE; BLJ, BLUEJAY; JKD, JACKDAW. Bars show mean ± sem of three biological replicates. J and K, Radial patterning of roots expressing 35S:SHR in cycd6;1 and scr-4 mutants. L, Quantitative comparison of cell layers outside of stele. 35SSG, p35S:SHR-GFP. Data represented are mean ± sd of 14 to ∼18 roots for each sample. Scale bars = 20 μm.
SCR, as a SHR direct target, is preferentially expressed in endodermis, CEI, CEID, and quiescent center (QC) in wild type (Di Laurenzio et al., 1996; Sabatini et al., 2003). Similar to CYCD6;1, SCR was also activated by SHR in all cells outside of stele. When SHR was constitutively expressed, pSCR:SCR-mCherry was seen in all cells with SHR-GFP (Fig. 2F). In multiple endodermal and epidermal layers induced by pSCR:SHR-GFP or pWER:SHR-GFP, SCR-mCherry exhibited almost the same pattern as SHR-GFP (Fig. 2, G and H). When SHR was inducibly activated, the expression level of most known SHR target genes including BIRD proteins was gradually enhanced, although with varied extent (Fig. 2I).
To investigate whether activation of SCR and CYCD6;1 is essential for the periclinal division, we examined the SHR-mediated division in scr-4 and cycd6;1 mutant backgrounds. In both mutants, the magnitude of ectopic divisions seemed to be affected (Fig. 2, J–L; Supplemental Fig. S2). The reduction of periclinal cell divisions was substantial in scr-4 but was only marginal in cycd6;1. In accordance with the phenotypes observed in cycd6;1 mutant, loss of function of CYCD6;1 could likely be compensated by unidentified redundant genes. Removal of SCR was unable to abolish, but did lessen the extent of, periclinal cell divisions upon SHR misexpression, suggesting that SCR enhances SHR ability to induce periclinal cell division.
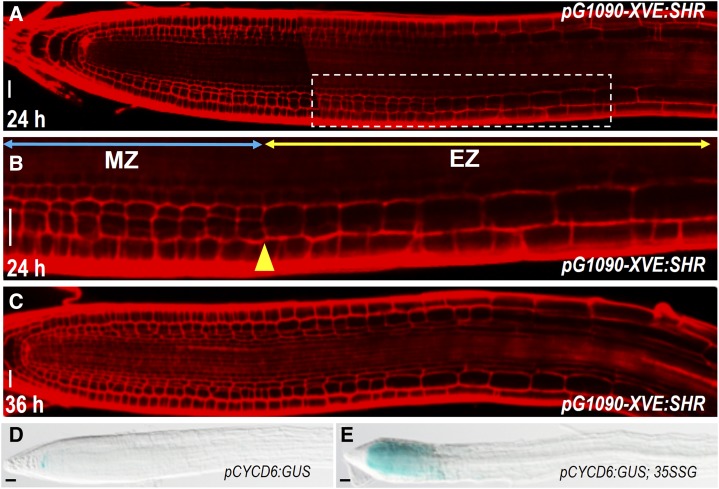
Although SHR activated periclinal cell divisions in most cells outside of the stele, the division appeared to be restricted to the meristem. In pG1090-XVE:SHR roots, the induction of periclinal cell division stopped precisely at the meristem-elongation zone junction (Fig. 3, A–C). This is different from PLETHORA2, which has been shown to trigger division in differentiated zone (Mähönen et al., 2014). In line with this, CYCD6;1 activation induced by SHR misexpression exhibited the similar developmental zone confinement (Fig. 3, D and E). Therefore, mitotic competence appeared to be a prerequisite for SHR function.
Figure 3.
Mitotic competence is a prerequisite for SHR induction. A to C, pG1090-XVE:SHR-expressing roots incubated in estradiol for 24 h (A and B) or 36 h (C). Note the periclinal cell division stopped at the junction of meristem zone (MZ) and elongation zone (EZ). This junction is marked by yellow arrowhead. Blue double-headed arrow indicates MZ, and yellow double-headed arrow represents EZ in B. D and E, Comparison of pCYCD6:GUS expression between wild-type root (D) and 35S:SHR-GFP root (35SSG; E). Scale bars = 20 μm.
These data reveal that SHR mediates periclinal cell division via a common pathway across different tissues. Activation of SHR can convert most mitotic active cells into periclinally dividing cells in a cell type-independent manner.
Cell-Fate Determination in Plants Depends on Both Lineage Inheritance and Position-Based Cell-Cell Communication
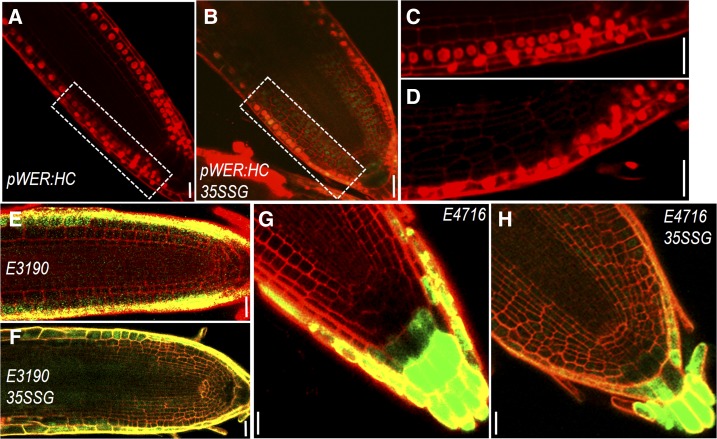
To better understand spatiotemporal dynamics of cell specification, we next observed fluorescent markers of different cell types and traced the dynamics of cell-fate transformation during extra cell layer formation induced by constitutively expressed SHR in Arabidopsis roots. We first examined the outermost cell layer by imaging a set of markers. The WER promoter is specifically active in the epidermis, the lateral root cap, and their initials (Fig. 4, A and C). With extensive periclinal cell divisions triggered by 35S:SHR-GFP, we saw no noticeable changes in WER:H2B-mCherry expression pattern (Fig. 4, B and D). Consistently, enhancer trap line E3190 (specifically expressed in epidermis and lateral root cap cells) was only activated in the outermost cell layers in 35S:SHR-GFP-expressing roots (Fig. 4, E and F). In the root cap, E4716 specifically marked the root cap peripheral layers, including border cells. There was no change in E4716 expression pattern when extra cell layers were created in columella region by SHR misexpression (Fig. 4, G and H). These results suggest that the epidermis maintains the cell fate despite of complex and altered tissue geometry.
Figure 4.
Epidermal cells maintain specialized fate despite complex and altered tissue geometry. A to D, pWER:H2B-mCherry (pWER:HC) expression in roots of wild type (A and C) and 35S:SHR-GFP (35SSG; B and D). C and D are zoomed view of boxed area in A and B, respectively. E and F, E3190 expression in roots of wild type (E) and 35SSG (F). G and H, E4716 expression in roots of wild type (G) and 35SSG (H). Scale bars = 20 μm.
Root stem cell niche (SCN) is located above the root cap. In the center of the SCN is the so-called the QC, which functions to maintain the stem cell status of the surrounding cells (Supplemental Fig. S1). When constitutively expressed, SHR induced ectopic cell divisions in SCN (Fig. 1F). To determine whether QC cells maintain their identity and function properly with aberrant anatomy in SCN, we observed QC-specific markers and starch staining (Lugol’s staining) that reflects columella differentiation status. Lugol’s staining showed that the starch accumulation pattern in the 35S:SHR-GFP-expressing roots is similar to that in wild type (Supplemental Fig. S3, A–F). There were extra cell layers seen in the CSC position of 35S:SHR-GFP roots. Interestingly, those extra cells showed no starch staining, suggesting that they likely resulted from CSC division (Supplemental Fig. S3, C and F). Both pWOX5:erGFP and QC25 in 35S:SHR-GFP roots exhibited wild-type-like expression patterns, indicating the QC properties were well maintained in these roots (Supplemental Fig. S3, G–J). In 35S:SHR root columella, markers including pPIN3:PIN3-GFP and enhancer trap line Q1630 also displayed the correct expression pattern (Supplemental Fig. S3, K–N). Taken together, these results support the dominant role of positional information in deciding cell fate in Arabidopsis roots. The relative position of a cell type can be precisely located within the root in spite of altered tissue structure.
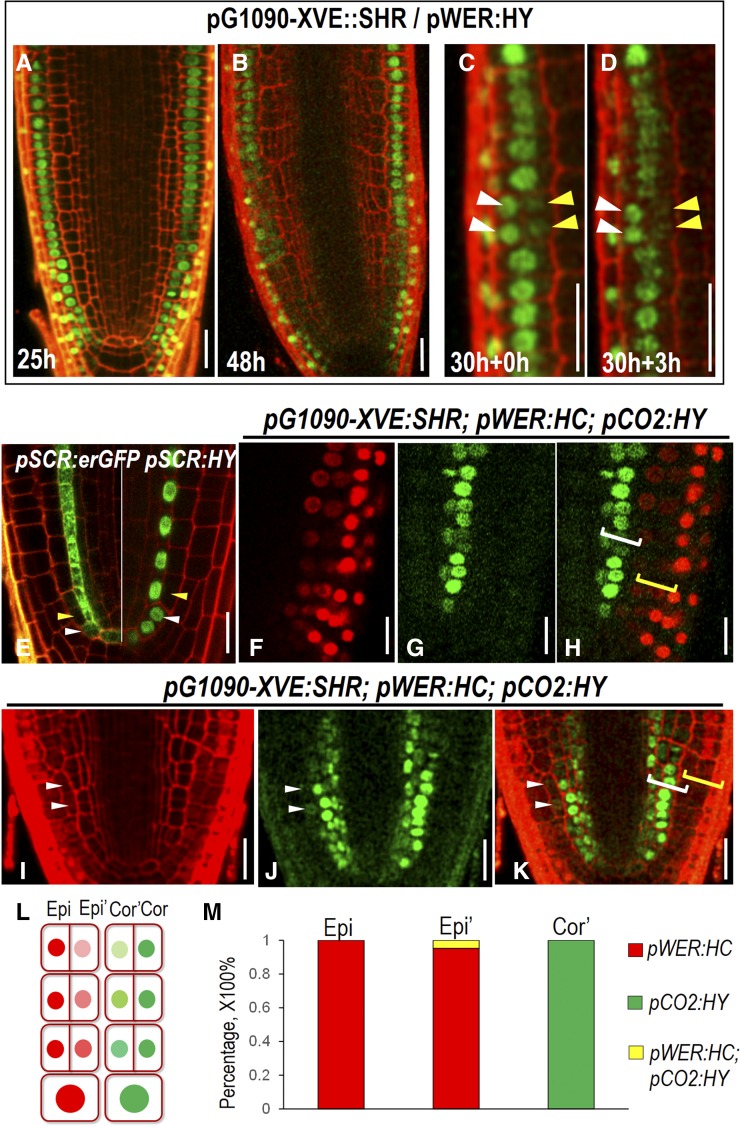
As the constitutive expression of SHR prevents the temporal resolution of the potential cell-fate transition, we made use of the inducible lines. Around 12 h after induction, pG1090-XVE::SHR started to promote the periclinal division in cortex, resulting in two juxtaposed cells, both of which exhibited the expression of cortex marker, pCO2:H2B-YFP (Supplemental Fig. S4A). With prolonged treatment by estradiol, the pCO2:H2B-YFP signal in outer layers tapered off (Supplemental Fig. S4B). And surprisingly, overall expression of pCO2:H2B-YFP in most cells became greatly reduced after 48 h SHR induction (Supplemental Fig. S4C). To rule out the variation among different roots, we performed a time course observation of pCO2:H2B-YFP in the same root. We pretreated pG1090-XVE::SHR expressing roots (n = 10) with estradiol for 20 h and then monitored the fluorescent intensity in different cell layers (Supplemental Fig. 4D). At this time point, the extra cell layer derived from cortex retained moderate levels of pCO2:H2B-YFP. After 3 h, we saw a clear drop of YFP signal in outer cortex layers (Supplemental Fig. S4E). This dynamic change of cell identity markers likely resulted from the action of positional information. The similar phenomenon was observed in the epidermis of pG1090-XVE::SHR roots. After 25 h estradiol induction, two juxtaposed cells expressing pWER:H2B-YFP became occasionally visible (Fig. 5A). After 48 h, many cells appeared to derive from epidermis still exhibited YFP signal (Fig. 5B). To see if cell-fate conversion occurred, we performed time course live imaging on a single root pretreated by estradiol for 30 h (Fig. 5, C and D). At 0 h of the imaging, H2B-YFP stayed at a high level in the inner cell layer that derived from periclinal cell division in epidermis. But pWER:H2B-YFP in the same cells became markedly reduced after 3 h, while the YFP signal in the juxtaposed layers maintained at the comparable level to that of 3 h ago (Fig. 5, C and D). One possibility is that the reduction of H2B-YFP in our observations was due to passive protein dilution by cell division rather than active cell identity selection. However, expression pattern of both pSCR:erGFP and pSCR:H2B-YFP roots seemed to argue against this possibility, in which the erGFP or YFP signal was clearly visible in cortex/endodermal initials but immediately lost in the first cortex (Fig. 5E). Thus, it is possible that switch on and off of a cell-specific marker is caused by an active fate conversion and is instructed by positional information.
Figure 5.
Cell-fate specification in roots with extra cell layers. A and B, Expression of pWER:H2B-YFP (pWER:HY) in pG1090-XVE::SHR roots at different estradiol induction time points. C and D, Time course observation of the same root expressing pG1090-XVE::SHR and pWER:HY at 30 hr estradiol induction (C) and 3 h after (D). White arrowheads point to the expression of pWER:HY in epidermis, and yellow arrowheads point to the expression of pWER:HY in the cell divided from epidermis. E, Expression of pSCR:erGFP (left) and pSCR:HY (right) in wild-type roots. White arrowheads point to CEI and CEID cells in which pSCR is still active. Yellow arrowheads point to the first cortex cell derived from the CEID in which pSCR activity is not seen. F to K, Expression of pWER:HC and pCO2:HY in pG1090-XVE:SHR-expressing roots after 48 h in estradiol. Note the separated expression zone of pWER:HC and pCO2:HY, marked by brackets in H and occasionally overlapped expression zone marked by white arrowheads in I to K. White brackets indicate the expression of pCO2:HY, and yellow brackets indicate the expression of pWER:HC. L, Scheme describing the separated expression zone shown in F to H. Epi, Epidermis; Epi’, extra cell layer derived from epidermis; Cor, cortex; Cor’, extra cell layer derived from cortex. M, Quantification of the percentage of cells expressing different markers in pG1090-XVE:SHR-expressing roots after 48 h in estradiol. Scale bars = 20 μm.
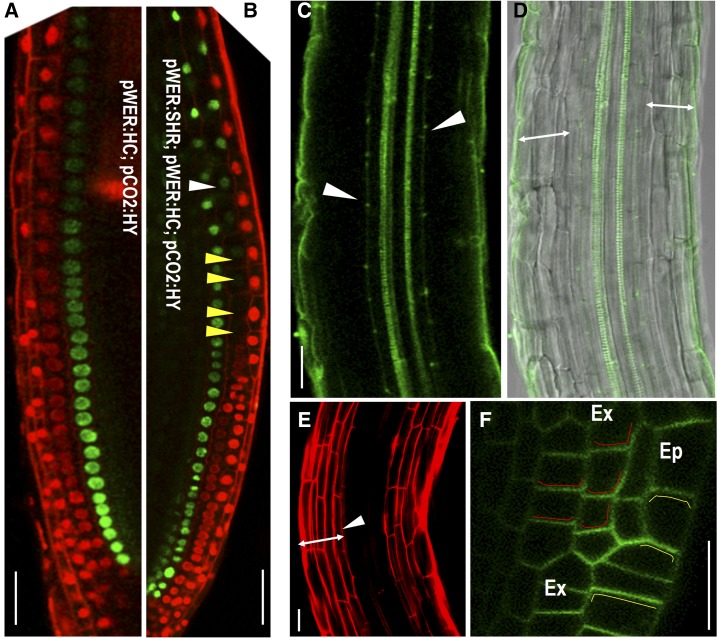
The expression of pWER:H2B-YFP faded away in extra cell layers originated from the epidermis while maintained at high level in the outermost cell layer. However, it is still difficult to know what cell types they became without additional markers. To address this question, we introduced both epidermal marker pWER:H2B-mCherry and cortex marker pCO2:H2B-YFP into pG1090-XVE::SHR lines. In normally patterned roots, pWER:H2B-mCherry and pCO2:H2B-YFP have distinct expression profiles without overlap. A 48 h incubation in estradiol triggered periclinal cell division in both cortex and epidermis of pG1090-XVE::SHR roots. However, we rarely detected overlapping expression of pWER:H2B-mCherry and pCO2:H2B-YFP (Fig. 5, F–M). The majority of the cells expressed the same reporters as the parental cells they originated from, indicating plant cells still have limited ability to maintain the cell-fate lineage (Fig. 5L). However, we often found that the CO2 promoter activity reduced when SHR was overexpressed (Supplemental Fig. S4C). To overcome this obstacle caused by overexpression, we specifically created a new cell layer only from epidermis by expressing SHR under pWER promoter. To examine the cell identity of this new cell layer, we simultaneously imaged pWER:H2B-mCherry and pCO2:H2B-YFP (Fig. 6A). As expected, the extra cell layers derived from epidermis that locate in the proximity of the stem cell niche expressed pWER:H2B-mCherry (Fig. 6B). But this expression tapered off, and as cells progress away from the position of the initial division, the cell fate was overridden by positional regulation and the extra cell file gradually adopted pCO2:H2B-YFP expression (Fig. 6B). Consistent with the loss of pWER:H2B-mCherry expression, we did not observe root hair initiation in extra cell layers divided from epidermis (Supplemental Fig. S4, F–H). As a weak expression of pCO2 can also be detected in newly formed endodermis in proximal meristem, we analyzed the functional features of the extra cell layers divided from epidermis by examining the presence of Casparian strip. Both the lignin autofluorescence staining and propidium iodide (PI) penetration assay showed that there was only a single layer of functional endodermis in pWER:SHR-expressing roots (Fig. 6, C–E). In addition, PIN2-GFP in extra cell layers of 35S:SHR roots showed basal side localization, which is the same polarity as in cortex (Fig. 6F).
Figure 6.
Lineage inheritance in the early stage of pattern formation is modified by positional information. A and B, Simultaneously imaging the expression of pWER:H2B-mCherry (pWER:HC) and pCO2:HY in roots of wild-type (A) and pWER:SHR-expressing line (B). Yellow arrowheads point to cells losing pWER:HC expression and white arrow head marks the starting cell that adopts the expression of pCO2:HY. C and D, Lignified Casparian strips visible as green autofluorescence of cell walls after clearing in pWER:SHR-expressing roots (marked by the white arrows). Double-headed arrows represent cell layers outside of endodermis. E, PI penetrated into all cell layers outside of endodermis (marked by the double-headed arrow). White arrowhead points to the position of endodermis where PI penetration was blocked. F, pPIN2:PIN2-GFP in the root expressing 35S:SHR. Ep, epidermis; Ex, extra cell layers. Red lines depict the basal side of PIN2-GFP in extra cell layers, and yellow lines depict the apical side of PIN2-GFP in epidermis. Scale bars = 20 μm.
Taken together, our results revealed that cell-fate acquisition in Arabidopsis roots is a combined process in which lineage inheritance plays a major role in the early stage as a cell is produced, but the lineage determinant can be gradually overridden by positional information. Interestingly, the fate conversion of cells outside of stem cell niche seemed to be a relatively slow readout of positional signaling in Arabidopsis roots.
SHR and Other Positional Cues Provide Independent Inputs in Cell-Fate Specification
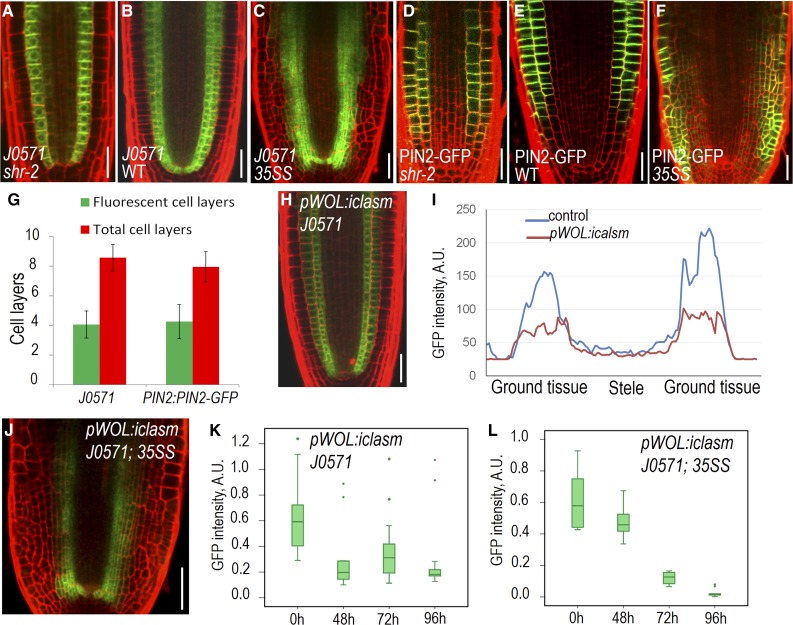
It was shown in previous studies and our observations that loss of SHR function was not accompanied by loss of ground tissue identity (Sozzani et al., 2010; Carlsbecker et al., 2010). In shr-2 mutants, the mutant cell layer (which is a single ground tissue layer in shr-2) still maintained the expression of J0571, a widely used ground tissue marker (Fig. 7, A and B). To determine whether SHR function is sufficient to confer ground tissue identity, we examined J0571 expression in root tissues ectopically expressing SHR. Unexpectedly, we only detected the J0571 expression in a limited number of extra cell layers created by 35S:SHR (Fig. 7, C and I). We further visualized another ground tissue-specific marker, E1839, and observed the expression was also restricted to part of the extra cell layers in mature zone (Supplemental Fig. S4, I and J). To understand the cell identity of these cells locating between J0571-expressing layers and the epidermis, we visualized pPIN2:PIN2-GFP, which is usually expressed in both epidermis and cortex cells (Fig. 7, D and E). In the supernumerary cell layers produced in 35S:SHR roots, PIN2-GFP was only visible in a few cell layers that are adjacent to the epidermis (Fig. 7, F and G). Interestingly, both J0571 and pPIN2:PIN2-GFP displayed a gradient pattern horizontally, with J0571 expression tapered off outward and PIN2-GFP declined inward. This suggests that there might be signals transmitted across the tissue to direct the cell fate of neighboring cells. To test this hypothesis, we utilized pWOL:icals3m system, which can block plasmodesmata (PD) within the stele upon estradiol induction (Vatén et al., 2011; Wu et al., 2016; Liu et al., 2017). As a result, the symplastic movement of signaling molecules out of the stele through PD was prevented in these roots. Once we blocked PD in stele, J0571 expression appeared much weaker (Fig. 7, H and I; Supplemental Fig. S5, A and B). Quantification of fluorescence intensity of J0571 indicated that the reduction of J0571 expression occurred in both endodermis and cortex. Compared to the wild type, J0571 fluorescence intensity dropped by 48% (n = 24, P = 5.81E-14; Student’s t test; Supplemental Fig. S5C) in endodermis and reduced by 72% (n = 25, P = 1.78E-20; Student’s t test; Supplemental Fig. S5D) in the cortex of pWOL:icals3m roots after 45 h estradiol induction. We further crossed pWOL:icals3m into 35S:SHR lines that also express J0571 marker. Compared to untreated lines, J0571-expressing cell layers in 35S:SHR roots narrowed down upon estradiol induction of pWOL:icals3m (Fig. 7J).
Figure 7.
SHR integrates stele-derived signals to foster ground tissue identity. (A–C) Expression of J0571 in the root tip of shr-2, wild-type (WT) and 35S:SHR (35SS). D to F, Expression of pPIN2:PIN2-GFP in the root tip of shr-2, wild type, and 35SS. G, Quantification of fluorescent cell layers in 35SS roots expressing J0571 or pPIN2:PIN2-GFP. Data represented are mean ± sd of 16 ∼ 20 roots for each sample. H, Expression of J0571 with the activation of pWOL:icals3m in stele by the treatment of estradiol for 48 h (I) Comparison of J0571 fluorescent intensity in wild-type control (shown in B) and pWOL:icals3m-expressing roots (shown in H) after 48 h estradiol treatment. J, Expression of J0571 in the root expressing 35SS and pWOL:icals3m with 48 h estradiol treatment. K and L, Time course quantification of J0571 with activation of pWOL:icals3m in the root of the wild type (K) and 35SS (L). n = 24 to 25 roots. Sample minimum, lower bar; lower quartile, box; median, middle cross line; upper quartile, box; sample maximum, upper bar; green dot, excluded outliers data. The difference between the 0 h and other time points is significant (P = 0.00052 for 48 h, 0.01374 for 72 h, and 0.00203 for 96 h in K; P = 0.00176 for 48 h, 1.0922E-11 for 72 h, and 1.97324E-13 for 96 h in L). Scale bars = 20 μm.
However, J0571 expression was not entirely abolished in the ground tissue with occluded stele, suggesting there might be gradient or dose-dependent effect of symplastic signals from the stele. To obtain the temporal information of J0571 expression change with disrupted symplastic signaling from the stele, we performed time course analysis of J0571 in both pWOL:icals3m and pWOL:icals3m/35S:SHR backgrounds. Although J0571 exhibited certain level of varied expression, the reduction trend appeared to be similar in both genetic contexts (Supplemental Figs. S6 and S7). The quantification indicated a significantly depleted J0571 fluorescence intensity after the symplastic communication between stele and outer cell layers was blocked (Fig. 7, K and L).
Together, our results suggest that stele-derived symplastic signaling contributes to the expression of J0571, the mostly used ground tissue marker. But the acquisition of the ground tissue cell fate presumably involves multiple layers of regulation. Although SHR is able to activate a group of downstream factors involved in specifying ground tissue, additional positional cues that are independent of SHR and mediated though PD need to participate in the regulation to confer the full ground tissue identity.
DISCUSSION
Cell-fate determination has long been a central question in development. In animals, lineage-based mechanisms for cell-fate determination play a major role in organogenesis. In plants, it is widely accepted that position-dependent cell-fate regulation is dominant. The evidence supporting this mostly derived from early surgical experiments and clonal analyses (Scheres, 2001; Kidner et al., 2000; Kim and Zambryski, 2005; Scheres et al., 2002). Since Arabidopsis was used as the model system, a large number of molecular and genetic tools, as well as nonintrusive techniques of cellular observation, have been developed. However, Arabidopsis has simplified structure in most organs. For example, a typical Arabidopsis root has only a single cortex layer, and most cell fates are specified in the root stem cell niche. This limits the dissection of cell-fate regulation under a more complex tissue context. In this paper, we examined the spatiotemporal regulation of cell fate in Arabidopsis roots with multiple cell layers formed in the epidermis and ground tissue. Our results provide an alternative perspective on cell-fate specification within a complex tissue beyond stem cell niche. This may reflect the situation in many other crop species, in which periclinal cell division in roots repeatedly occurs in meristem and the cell fate needs to be precisely determined.
SHR is a good example that asymmetric cell division and cell-fate determination are coordinated by a critical regulator. In the current prevailing model, SHR function in asymmetric cell division was only restricted in CEID and endodermis by the synergistic action of a group of interacting transcription factors (Cui et al., 2007; Long et al., 2015a, 2015b; Welch et al., 2007; Moreno-Risueno et al., 2015). A previous report also proposed that the competence to respond to SHR lies in the epidermal/LRC initials through induction of SCR. However, monocot SHR homologs seemed to move beyond the endodermis when expressed in the stele of Arabidopsis roots, inducing multiple layers of ground tissues (Wu et al., 2014). Thus, SHR could also function broadly in promoting periclinal cell division in various developmental contexts in addition to the CEI initials and stem cell region. This is confirmed in our misexpression of SHR analysis, in which SHR can broadly activate periclinal cell division in cells outside of its normal functional domain. When SHR was inducibly activated, the expression level of most known SHR target genes including BIRD family was gradually enhanced. Although the sensitivity to SHR induction appeared to vary in epidermis and cortex, SHR likely promotes periclinal cell division through a conserved pathway that includes most important components involved in SHR functions in CEID.
It is still unclear which components are indispensable for SHR-induced periclinal cell division. Removal of SCR was unable to abolish but did lessen the extent of periclinal division, suggesting that SCR participates in SHR regulation outside of endodermis. However, without SCR function, SHR failed to promote periclinal cell division in epidermis. This is consistent with the previous result showing blocked periclinal cell division induced by SHR in epidermis of scr-4 (Sena et al., 2004). SCR function seems to be essential for SHR ability to induce periclinal cell division only in epidermis. In scr-4 (the seedlings were less than 5 d old), SHR still induced periclinal cell division in the mutant cell layer. In addition to SCR, the mitotic competence appeared to be a prerequisite for the SHR function as the periclinal cell division triggered by SHR was restricted to the meristem.
Based on our results, part of a plant cell’s identity can be passed on from the parental cells to their progeny, which resembles the lineage-based mechanism. Our observations in the epidermis and ground tissues outside of the stem cell niche suggest that the cell-fate conversion is not a fast readout. In CEID, both endodermis and cortex appeared to adopt the respective fates immediately after periclinal division. Longer fate conversion time in epidermis and ground tissues suggests that position-based regulation could first remove the inherited identity of a cell to override the cell fate and promote the fate conversion. Thus, the actual cell-fate acquisition in Arabidopsis roots could be a dimorphic process in which lineage inheritance plays an important role in the early stage once a cell is produced, but the lineage determinant can be gradually overridden by the positional information. The hierarchical cellular states during cell-fate specification are not a unique case in plants. In fly and worm development, the expression of cell-specifying transcription factors is dictated by positional signaling in very early stages, but the stable lineage is achieved through the cell-autonomous regulation by polycomb group genes (Scheres, 2001). The order of regulatory cascade in animals appears to be opposite to our observation of cell-fate acquisition in plants. But in both cases, the existence of intermediate cells with mixed identity is possible. One interesting but unsolved question here in plants is how the positional information orchestrates cell-type specification by removing the inherited properties of the non-stem cell and conferring the new characteristics. In lineage-determined animal cells, cell identity conversion can be promoted via forced expression of lineage-specific transcription factors in various differentiated cell types (Chin, 2014). However, the efficiency of cell-fate reprogramming in a given population was low, and the completeness of fate conversion is still being questioned. Thus, the fundamental cellular principals and molecular mechanisms in both animals and plants are still awaiting elucidation.
The mobile transcription factor SHR has been proposed as one of the regulators of endodermal identity. But in this study, we found that SHR, while necessary, was not sufficient to confer endodermal cell fate. Expression of SHR in cells outside of endodermis failed to establish Casparian strip, a functional feature of endodermis. In addition to endodermis, we also have little knowledge of the specification of cortex identity. In this study, we found that stele-derived symplastic signaling contributes to the cell-fate determination in ground tissues. However, restricted expression of J0571 in 35S:SHR roots indicated that SHR alone was not sufficient to confer the ground tissue identity. Thus, it is possible that additional factors regulated by symplastic signaling from stele play roles in this process. Recently, two stele-derived peptides (CASPARIAN STRIP INTEGRITY FACTORS) were identified to move from stele outward to induce Casparian strip formation. Although CASPARIAN STRIP INTEGRITY FACTORS1/2 is unlikely the regulator of J0571 due to the blocked apoplastic path by Casparian strip, there might exist other uncharacterized factors from the stele that affect J0571 expression (Doblas et al., 2017; Nakayama et al., 2017). Interestingly, disruption of symplastic communication between stele and the outer cell layers did not entirely abolish J0571 expression. Since cells divided from epidermis can gradually adopt cortex fate, it is possible that signaling from epidermis also participates in this specification and jointly promotes ground tissue identity with signals from stele. In addition, signals that are independent of symplastic transport could also be involved in this process. But no matter how complex the tissue geometry is, the root appeared to precisely locate the epidermis and endodermis. Hence, the entire regulatory network that provides positional information for cell-fate determination in plants presumably involves multiple layers of regulation.
MATERIALS AND METHODS
Plant Materials and Growth Condition
The Arabidopsis (Arabidopsis thaliana) Columbia ecotype (Col-0) was used as the wild type throughout the experiments. The following marker lines—CO2:H2B-YFP, CYCD6;1:GFP-GUS, SCR:SCR-mCherry, WER:H2B-mCherry, E3190, E4716, QC25:GUS, WOX5:erGFP, PIN3:PIN3-GFP, Q1630, WER:H2B-YFP, SCR:H2B-YFP, J0571, PIN2:PIN2-GFP, E1839—were crossed into different transgenic plants or mutants. Homozygous lines were screened based on fluorescence, PCR genotyping, and the root phenotypes. After sterilization, the seeds were germinated after incubated for 2 d at 4°C in the dark. All plants were grown vertically on 0.5× Murashige and Skoog medium containing 0.05% (w/t) morpholinoethansulfonic acid monohydrate (pH 5.7), 1.0% (w/t) Suc, and 1.0% agar in a growth chamber at 23°C under a 16/8 h light/dark cycle. Plants were analyzed 6 to 7 d after plating unless otherwise stated.
Plasmid Construction and Plant Transformation
The 1,596-bp full-length cDNA of AtSHR was cloned into pDONR221 (Invitrogen) using BP recombination based on standard protocol (Invitrogen/Life Technologies). The destination vectors were modified from the previously reported pGreenBarT vector (Lee et al., 2006) according to traditional restriction digestion method described previously (Wu et al., 2014). All expression vectors were generated through LR Gateway reaction, and the resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101-pSouppMP. Arabidopsis (Col-0) was transformed following the floral-dip method. Transgenic plants were screened based on the resistance to glufosinate-ammonium (Basta) in soil. For all of the transgenes discussed, at least three independently transformed lines were analyzed, and one of them was chosen for further analysis.
Confocal Microscopy Imaging
Roots were mounted in 0.01 μg/mL propidium iodide (PI) in water. Roots tips were then examined using a 40× water-immersion lens on a Zeiss LSM 880 laser scanning confocal microscope with dual-channel setting of YFP and mCherry. Image quantification was performed using ImageJ 1.4.3 software. For fluorescence intensity analysis, we used the region selection function of ImageJ to create a region of interest where the fluorescence is typically seen. The average intensity of fluorescence was calculated by ROI manager and then used to calculate the ratio of relative fluorescence intensity. Representative images were collected from 10 to 25 roots with three biological replicates.
Staining and Chemical Treatments
For GUS staining, 5- to 7-d-old seedlings were incubated in the GUS (0.5 mg/mL) staining solution for 8 h at 37°C followed by the clearing in 70% ethanol. For starch staining, root tips were incubated in a 1:1 dilution of Lugol’s solution (Sigma-Aldrich) for 1 min, then briefly washed with water and mounted in the HCG solution (chloroacetaldehyde:water:glycerol = 8:3:1) for microscopy visualization. Samples were viewed using Nikon ECLIPSE Ni-U microscope connected to a Nikon DS-Ri 2 digital camera. Visualization of PI penetration and lignin autofluorescence was performed according to Alassimone et al. (2010).
For estrogen induction, 5- to 6-day-old seedlings were transferred to 0.5× Murashige and Skoog (Caisson) agar (Difco-BBL) plates containing 10 μm estradiol (Sigma-Aldrich) and the same medium containing the estradiol carrier (dimethyl sulfoxide) as controls.
Statistical Analysis
The Arabidopsis seedlings from three biological replicates were randomly chosen from each treatment. The data analysis was carried out by using spss17.0, and independent-sample t tests were used to determine the significance between the treatment and control group (P < 0.05). The box plots were exported by SPSS Data Editor.
qRT-PCR Analysis
cDNA was prepared from the total RNA extracted from the root tip (∼1 cm) of 6-d-old pG1090-XVE:SHR seedlings after 0, 12, 24, and 48 h estradiol incubation. qRT-PCR were performed on a Stratagen Mx3005P (Agilent Technologies) with the TransStart Top Green qPCR SuperMix (Transgen), according to the manufacturer’s instructions. In each run, three technical replicates were performed for each sample. The presented results are based on three biological replicates, and error bars represent sd of the mean of biological replicates. The primers of interested genes are as follows: MGP (5′-AAAGCAGAGGACGAGCAAAG-3′ and 5′-GGAATGAGCCTTCCAGTCAG-3′), NUC (5′-AGCTGCTGAAATTGGCGCTA-3′ and 5′-TGGTGGTTTGATCGGTGGAT-3′), JKD (5′-CAATGCATGCGCAAGGTCTAT-3′ and 5′-GAATTTGGAATTGGTGGTGGC-3′), LMP (5′-CAGTCACAAGCAACGACCCAT-3′ and 5′-CCATAATTCGTCCTCCACCAAA-3′), BLJ (5′-GTCCCCTAGCCTTTTCGACCTT-3′ and 5′-CGGTGCTCACAATTCCTCCAT-3′), SCR (5′-TTGAGAGCTGGAGGGAGAAA-3′ and 5′-CGTCCAAGCTGAAGCAGTGAGT-3′), and ACT2 (5′-CGCTGACCGTATGAGCAAAG-3′ and 5′-GAGATCCACATCTGCTGGAATG-3′) was used as the reference gene.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Cartoon summarizing the prevailing model for ground tissue patterning mediated by SHR pathway.
Supplemental Figure S2. The ability of SHR to trigger periclinal cell division relies on SCR.
Supplemental Figure S3. QC function and stem cells were well maintained in SHR ectopic expression lines.
Supplemental Figure S4. Examination of cell fate with misexpressed SHR.
Supplemental Figure S5. Blocked symplastic communication between stele and endodermis led to reduced J0571 fluorescent intensity in ground tissues.
Supplemental Figure S6. Time course observation of J0571 with the activation of pWOL:icalsm.
Supplemental Figure S7. Time course observation of J0571 in 35S:SHR roots with the activation of pWOL:icalsm.
Acknowledgments
We thank Ykä Helariutta, Ben Scheres, Ikram Blilou, Philip N. Benfey, Rosangela Sozzani, and Scott Poethig for materials, Zhenbiao Yang and anonymous reviewers for comments on the manuscript, and Lei Shi for technical assistance with the Zeiss 880 confocal microscope at the Cell Biology Core of Fujian Agriculture and Forestry University.
Glossary
- CEI
cortex/endodermal initial
- CEID
CEI daughter cell
- CSC
columella stem cell
- QC
quiescent center
- SCN
stem cell niche
- PD
plasmodesmata
Footnotes
Q.Y., P.L., N.L., H.W., and M.X. were supported by the 1000-Talents Program of China for young researchers (grant KXR15012A), by the National Science Foundation of China (31670184), and by a start-up fund from Fujian Agriculture and Forestry University awarded to S.W.
Articles can be viewed without a subscription.
References
- Abrash EB, Bergmann DC (2009) Asymmetric cell divisions: A view from plant development. Dev Cell 16: 783–796 [DOI] [PubMed] [Google Scholar]
- Alassimone J, Naseer S, Geldner N (2010) A developmental framework for endodermal differentiation and polarity. Proc Natl Acad Sci USA 107: 5214–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Scheres B (2000) Root development. Curr Biol 10: R813–R815 [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B (2010) Root development-two meristems for the price of one? Curr Top Dev Biol 91: 67–102 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MT. (2014) Reprogramming cell fate: A changing story. Front Cell Dev Biol 2: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Hinde E, Winter CM, Fisher AP, Crosti G, Blilou I, Gratton E, Benfey PN, Sozzani R (2016) Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 5: e14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, et al. (2012) A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N (2017) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dong J, Bergmann DC (2010) Stomatal patterning and development. Curr Top Dev Biol 91: 267–297 [DOI] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Heo JO, Chang KS, Kim IA, Lee MH, Lee SA, Song SK, Lee MM, Lim J (2011) Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci USA 108: 2166–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C, Sundaresan V, Roberts K, Dolan L (2000) Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211: 191–199 [DOI] [PubMed] [Google Scholar]
- Kim I, Zambryski PC (2005) Cell-to-cell communication via plasmodesmata during Arabidopsis embryogenesis. Curr Opin Plant Biol 8: 593–599 [DOI] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, Scheres B, Benfey PN (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu M, Liang N, Zheng Y, Yu Q, Wu S (2017) Symplastic communication spatially directs local auxin biosynthesis to maintain root stem cell niche in Arabidopsis. Proc Natl Acad Sci USA 114: 4005–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Goedhart J, Schneijderberg M, Terpstra I, Shimotohno A, Bouchet BP, Akhmanova A, Gadella TW Jr, Heidstra R, Scheres B, et al. (2015a) SCARECROW-LIKE23 and SCARECROW jointly specify endodermal cell fate but distinctly control SHORT-ROOT movement. Plant J 84: 773–784 [DOI] [PubMed] [Google Scholar]
- Long Y, Smet W, Cruz-Ramírez A, Castelijns B, de Jonge W, Mähönen AP, Bouchet BP, Perez GS, Akhmanova A, Scheres B, et al. (2015b) Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27: 1185–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B (2014) PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Sozzani R, Yardımcı GG, Petricka JJ, Vernoux T, Blilou I, Alonso J, Winter CM, Ohler U, Scheres B, et al. (2015) Transcriptional control of tissue formation throughout root development. Science 350: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y (2017) A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Benfey PN (2008) Root layers: Complex regulation of developmental patterning. Curr Opin Genet Dev 18: 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoeva T, Weijers D (2014) A roadmap to embryo identity in plants. Trends Plant Sci 19: 709–716 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Jung JW, Benfey PN (2004) A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Scheres B. (2001) Plant cell identity. The role of position and lineage. Plant Physiol 125: 112–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan L (2002) Root development. Arabidopsis Book 1: e0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P (1994) Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487 [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove CA, Heidstra R (2008) Who begets whom? Plant cell fate determination by asymmetric cell division. Curr Opin Plant Biol 11: 34–41 [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Lu KJ, Weijers D (2015) Building a plant: Cell fate specification in the early Arabidopsis embryo. Development 142: 420–430 [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Breakfield NW, Benfey PN (2011) Intercellular communication during plant development. Plant Cell 23: 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Lee C-M, Hayashi T, Price S, Divol F, Henry S, Pauluzzi G, Perin C, Gallagher KL (2014) A plausible mechanism, based upon Short-Root movement, for regulating the number of cortex cell layers in roots. Proc Natl Acad Sci USA 111: 16184–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, O'Lexy R, Xu M, Sang Y, Chen X, Yu Q, Gallagher KL (2016) Symplastic signaling instructs cell division, cell expansion, and cell polarity in the ground tissue of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 113: 11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]