Abstract
Purpose
Motor function and recovery after stroke likely rely directly on the residual anatomical connections in the brain and its resting-state functional connectivity. Both structural and functional properties of cortical networks after stroke are revealed using multimodal magnetic resonance imaging (MRI). Specifically, functional connectivity MRI (fcMRI) can extract functional networks of the brain at rest, while structural connectivity can be estimated from white matter fiber orientations measured with high angular-resolution diffusion imaging (HARDI). A model that marries these two techniques may be the key to understanding functional recovery after stroke. In this study, a novel set of voxel-level measures of structurofunctional correlations (SFC) was developed and tested in a group of chronic stroke subjects.
Methods
A fully automated method is presented for modeling the structure-function relationship of brain connectivity in individuals with stroke. Brains from ten chronic stroke subjects and nine age-matched controls were imaged with a structural T1-weighted scan, resting-state fMRI, and HARDI. Each subject's T1-weighted image was nonlinearly registered to a T1-weighted 152-brain MNI template using a local histogram-matching technique that alleviates distortions caused by brain lesions. Fractional anisotropy maps and mean BOLD images of each subject were separately registered to the individual's T1-weighted image using affine transformations. White matter fiber orientations within each voxel were estimated with the q-ball model, which approximates the orientation distribution function (ODF) from the diffusion-weighted measurements. Deterministic q-ball tractography was performed in order to obtain a set of fiber trajectories. The new structurofunctional correlation method assigns each voxel a new BOLD time course based on a summation of its structural connections with a common fiber length interval. Then, the voxel's original time-course was correlated with this fiber-distance BOLD signal to derive a novel structurofunctional correlation coefficient. These steps were repeated for eight fiber distance intervals, and the maximum of these correlations was used to define an intrinsic structurofunctional correlation (iSFC) index. A network-based SFC map (nSFC) was also developed in order to enhance resting-state functional networks derived from conventional functional connectivity analyses. iSFC and nSFC maps were individually compared between stroke subjects and controls using a voxel-based two-tailed Student's t-test (alpha = 0.01). A linear regression was also performed between the SFC metrics and the Box and Blocks Score, a clinical measure of arm motor function.
Results
Significant decreases (p < 0.01) in iSFC were found in stroke subjects within functional hubs of the brain, including the precuneus, prefrontal cortex, posterior parietal cortex, and cingulate gyrus. Many of these differences were significantly correlated with the Box and Blocks Score. The nSFC maps of prefrontal networks in control subjects revealed localized increases within the cerebellum, and these enhancements were diminished in stroke subjects. This finding was further supported by a reduction in functional connectivity between the prefrontal cortex and cerebellum. Default-mode network nSFC maps were higher in the contralesional hemisphere of lower-functioning stroke subjects.
Conclusion
The results demonstrate that changes after a stroke in both intrinsic and network-based structurofunctional correlations at rest are correlated with motor function, underscoring the importance of residual structural connectivity in cortical networks.
Keywords: Stroke, Structural connectivity, Functional connectivity, Structurofunctional correlation
1. Introduction
Resting-state functional magnetic resonance imaging and diffusion MRI together provide unprecedented insight into the structure-function relationships associated with changes in brain connectivity after stroke. This information might be useful for prognosis after stroke and could help to personalize rehabilitation strategies for individual patients. Consequently, the purpose of this study was 1) to develop a novel connectivity model that marries structural and functional connectivity analyses, and 2) to demonstrate that this new technique provides unique information about subject-specific changes in brain connectivity that may be used to develop new imaging biomarkers for functional recovery.
Diffusion magnetic resonance imaging has provided potentially useful tools for detecting changes in white matter structure and structural connectivity following stroke. One such tool, high angular-resolution diffusion imaging (HARDI) (Tuch et al., 2002) measures the diffusion coefficient within each voxel for many directions. From these directional data, an orientation distribution function (ODF) of white matter fibers within a voxel can be estimated with techniques such as q-ball imaging (Tuch, 2004). Diffusion MRI tractography uses these directional diffusivity measurements to model white matter fiber pathways. Deterministic tractography approaches propagate modeled fibers along the principle direction of diffusion (Zhang et al., 2009), while probabilistic approaches add random perturbations to estimate a distribution of structural connections to a voxel (Behrens et al., 2007, Behrens et al., 2003). Measures of structural connectivity of the brain can then be calculated based on these tractography models, and applied to the brain of stroke subjects. Structural connectivity analyses automatically locate changes in fiber pathways after stroke (Yeh et al., 2013), and retained connectivity is associated with higher motor function (Buch et al., 2012, Crofts et al., 2011, Koch et al., 2016).
Resting-state functional magnetic resonance imaging (rs-fMRI) can be used to measure low-frequency oscillations in cortical signals that are associated with functional connectivity of the brain (Biswal et al., 1995, Fox, 2010). In this approach, measurements are made while the subject lies in a scanner with their eyes closed, making it attractive for clinical use. The rs-fMRI data are often analyzed using an independent components analysis (ICA) to automatically extract sets, or networks, of voxels that follow a common pattern of signal change (Beckmann and Smith, 2004, Beckmann and Filippini, 2009, Du and Fan, 2013, Park et al., 2014, Rytty et al., 2013). Each independent component includes a spatial volume of voxel contributions and a common signal time-course. When applied to stroke, rs-fMRI connectivity analysis has provided evidence of cortical reorganization associated with motor recovery (Chen and Schlaug, 2016, van Meer et al., 2010, Wang et al., 2010), with the most common finding of reduced interhemispheric functional connectivity between sensorimotor cortices (Carter et al., 2010, Rehme and Grefkes, 2013, Tang et al., 2016, Urbin et al., 2014). These studies are promising; rs-fMRI might have clinical use in predicting outcomes and developing therapeutic interventions based on monitoring of cortical reorganization (Carter et al., 2012, Ovadia-Caro et al., 2014).
Multimodal MRI techniques that combine diffusion and functional MRI approaches provide the opportunity to relate brain tract structure to functional networks (Honey et al., 2010). In healthy individuals, there is a strong correspondence between the default-mode network, a distinct brain network that demonstrates strong functional connectivity at rest, and the density of anatomical connections between its nodes (Hagmann et al., 2008). This same concept that anatomical connections, measured using diffusion imaging, underlie functional connectivity extends throughout the brain (Honey et al., 2009). An important factor in this relationship between anatomical connections and functional connectivity is distance between nodes of interest. Specifically, an inverse relationship has been observed between node distance and functional connectivity (Sporns, 2011).
The objective of the current study was to combine diffusion MRI and rs-MRI information into a single analysis of structurofunctional connectivity. A number of previous studies assessing both structural and functional connectivity have compared structural and functional connectivity matrices obtained separately from diffusion MRI and fMRI respectively (Damoiseaux and Greicius, 2009). More recently, a combined structural-functional connectome (Horn et al., 2014) has been described using correlations between the structural and functional connections of all voxels of the brain. An important observation from these studies is that functional connectivity is observed between regions that are not connected by major white matter tracts. Fjell et al. (2017) explored this concept in detail, calculating functional connectivity of brain regions with and without direct structural connections, noting that changes in functional connectivity are not strongly correlated with changes in direct structural connections with aging (Fjell et al., 2017). Thus, accounting for indirect structural connections could be an important factor in determining brain function, especially after an injury (Kalinosky et al., 2013). In general, combining structural and functional connectivity information might be particularly useful in describing changes in functional connectivity following localized brain lesions of structural networks, such as those associated with stroke (Rehme and Grefkes, 2013). Here we describe a new automated analysis that expresses functional connectivity in the context of structural connectivity while avoiding constraints on either individual analysis. Our approach first calculates the functional connectivity for all structurally connected voxels of the brain. We then identify average levels of functional connectivity to a voxel for regions that are structurally connected at similar distances. A novel structurofunctional correlation (SFC) that identifies maximum functional connectivity to a voxel across these distances can then be obtained. This technique produced a voxel-level intrinsic metric (iSFC) and an enhanced resting state network (RSN) map (nSFC). We tested this approach in ten stroke subjects and nine age-matched controls to validate the structurofunctional connectivity measures through correlation with clinical assessment of arm motor function.
2. Methods
2.1. Data collection
2.1.1. Subject recruitment and functional testing
Ten individuals with chronic post-stroke hemiparesis (4 female, age 66.7 ± 7.94 years, at least 1.1 years since stroke), and nine age-matched control subjects (5 female, age 64.2 ± 7.73 years) participated in this study. The experimental protocol was approved by the Institutional Review Boards of Marquette University and the Medical College of Wisconsin, and written consent was obtained from each subject. Inclusion criteria included a history of stroke that occurred no < 6 months prior to recruitment. Stroke subject information is provided in Table 1. Subjects with no ability to perform supination, pronation, ulnar deviation, or radial deviation of the wrist were excluded. Note that exact dates of the stroke, treatment history, or any other medical records were not collected. Control subjects were comprised of individuals without history of stroke or other neurological impairments that were age-matched (within 3 years) and gendered-matched to the stroke subjects.
Table 1.
Stroke subject information.
| Subject ID | S04 | S05 | S07 | S08 | S10 | S12 | S14 | S15 | S16 | S18 |
|---|---|---|---|---|---|---|---|---|---|---|
| Sex | M | M | F | M | M | F | M | F | F | M |
| Age | 65 | 57 | 83 | 66 | 69 | 65 | 64 | 80 | 62 | 66 |
| Lesion location | Cort | Subc | BS | Subc | Cort | Cort | Cbl | Subc | Subc | Subc |
| Lesion side | R | L | L | L | L | R | L | L | L | L |
| Arm affected | L | R | R | R | R | L | L | R | R | R |
| Dominant post | R | R | L | R | R | R | R | R | R | R |
| Dominant pre | L | R | R | R | R | Amb | R | R | R | R |
| Wolf motor | 71 | 72 | 38 | 72 | 75 | 54 | 58 | 74 | 75 | 74 |
| Fugl-meyer | 124 | 106 | 82 | 122 | 124 | 97 | 120 | 126 | 124 | 122 |
| Box & Blocks (ND) | 24.5 | 62.5 | 8 | 41 | 75.5 | 14.5 | 13 | 57 | 50 | 64 |
Cort = Cortical, Subc = Subcortical, BS = Brainstem, Cbl = Cerebellum, Amb = Ambidextrous, R = Right, L = Left.
A certified physical therapist evaluated each stroke subject with three clinical tests of motor function, including the upper extremity (UE) portion of the Fugl-Meyer Assessment (Fugl-Meyer et al., 1975), the Box and Blocks Test of Manual Dexterity (Mathiowetz et al., 1985) and the Wolf Motor Function Test (Wolf et al., 2001) for upper extremity motor ability. In the Box and Blocks Test, a top-open plywood box is divided into two square compartments by a partition 15.2 cm in height. While sitting in front of the box, the subject uses one hand to move as many wooden blocks as possible from one square compartment to the other in 60 s. Every stroke subject repeated the test twice with each hand, and the # of blocks was used as the score. In right-handed healthy adults of ages 20–75, Box and Blocks performance is 76.8 ± 11.5 blocks in 60 s (Mathiowetz et al., 1985). The Fugl-Meyer Assessment quantifies impairment in motor function, sensation, and balance in stroke patients with hemiplegia, and the upper extremity score ranges from 0 to 126. Lower scores signify greater level of impairment. The Wolf Motor Function Test (WMFT) uses time-based tasks to evaluate upper extremity movement. Some of these tasks include lifting a can or pencil, moving hand or forearms onto the table, stacking checkers, and folding a towel (Wolf et al., 2008). Wolf motor scores range from 0 to 76, with a higher score indicating greater function.
2.2. MRI scans
Every subject was screened for MRI safety according to the Medical College of Wisconsin Institutional Review Board before entering the magnetic environment. An axial T1-weighted anatomical image was acquired using a fast spoiled gradient recalled (SPGR) pulse sequence, with TE: 3.2 ms, TR: 8.16 ms, flip angle: 12°, prep time: 450, bandwidth: 22.73, FOV: 240 mm, 156 1 mm slices, and matrix size: 256 × 240. Next, an axial q-ball high angular resolution diffusion imaging (HARDI) sequence was acquired with a single-shot echo planar imaging sequence, including 5 b = 0 images, 150 diffusion-weighted directions, SENSE parallel imaging, TE: 72.3 ms, TR: 5700 ms, FOV: 250 mm, matrix size: 128 × 128 resampled to a 256 × 256 grid (2 mm pixel width/height), and 57 2.5 mm slices. The 2.5 mm slice thickness was needed for cerebellum coverage in subjects with larger heads. For functional MRI, an axial view gradient-echo echo-planar sequence was acquired with TE: 25 ms, TR: 2000 ms, flip angle: 77°, FOV: 240 mm × 240 mm, 41 slices with 3.5 mm thickness. Subjects were instructed to stay awake with the eyes closed for the 6-min resting-state fMRI scan, during which 180 frames were acquired in 2 s intervals. Supplementary Fig. S1 shows an axial slice from the T1-weighted image of each stroke subject that includes the lesion location.
2.3. MRI data processing
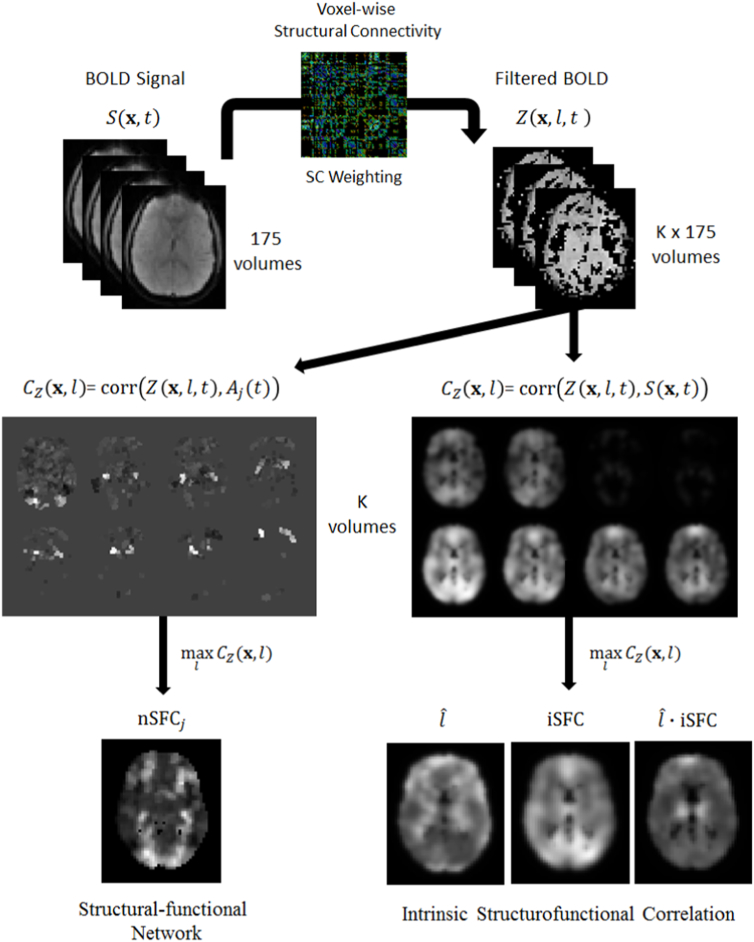
A summary of the MRI processing pipeline is shown in Fig. 1 and Table S1. Briefly, the MELODIC tool was used to decompose the fMRI BOLD signal, S(x,t) into 83 independent components, each with a spatial map, Mj(x,t) and time-course Aj(t). Diffusion MRI data were fit to a q-ball fiber orientation model, and tractography was performed in native subject space to reconstruct fiber pathways. Each subject's anatomical T1-weighted image was spatially normalized to MNI space, and affine registration was performed in native subject spaces to align anatomical images with diffusion and mean BOLD images. Fiber trajectories were transformed into MNI space, and a fiber-distance averaged BOLD signal, Z(x,t), was calculated for each voxel. Intrinsic structurofunctional correlation (iSFC) was calculated as the maximum temporal correlation of Z(x,t) with voxel BOLD signals. The iSFC can have values between 0 and 1. High iSFCmax indicates that for some fiber length, , a strong correlation in BOLD fluctuations exists between a voxel and the ensemble of its structurally connected voxels at that distance. On the other hand, a low iSFCmax near zero occurs only if a voxel is uncorrelated with its anatomical connections at all fiber lengths. In this case, the voxel's direct structural connections do not account for the variability in its BOLD signal. Enhanced resting-state network maps (nSFCj) were derived by maximizing correlations of Z(x,t) with independent component time-courses Aj(t). Again, values ranged from 0 to 1, with higher values indicating an enhanced connectivity to the standard network. A more detailed description of each step in the data processing follows.
Fig. 1.
Pipeline overview for calculating struturofunctional correlation (SFC) metrics. The cleaned BOLD signal S(x, t) from the MELODIC analysis was processed with fiber-based averaging to compute signal Z(x, t, l). Next, Z(x, t, l) was correlated with each voxel's original BOLD signal and the time-course of each resting-state network, giving two CZ correlations. The CZ correlations were maximized by fiber-length to derive the intrinsic (iSFC) and network-based (nSFC) metrics.
2.4. Image registration and lesion-side normalization
Fully automated techniques were used for intersubject and intermodality image registration. MRI data were also corrected to place lesions on the left side of the brain. Lesion-side normalization was performed prior to registration by flipping each row of data in the x-dimension in stroke subjects with right lesions. Anatomical T1-weighted images were registered to the 152-MNI template from fMRI of the Brain Software Library (FSL). First, the FLIRT tool from FSL (Jenkinson and Smith, 2001) was used to perform linear affine registration to translate and rotate the brain. The anatomical images were then deformed to MNI space using an ITK implementation of the Maxwell's demons algorithm (Thirion, 1998).
Each subject's anatomical image was registered to native diffusion MRI and fMRI spaces. Fractional anisotropy (FA) was calculated from the diffusion coefficients (Basser et al., 1994), and the FA map was flipped and registered to the subject's flipped T1-weighted image using a 12-parameter affine registration. The fMRI data were similarly flipped, and the mean BOLD image was registered to the anatomical image. Registration transforms were concatenated in order to map each subject's BOLD and diffusion images to MNI coordinates. Following tractography described in subsequent sections, fiber trajectories were similarly warped into MNI space. In subjects with right-side lesions, fibers were first flipped over the mid-sagittal plane.
2.5. Resting-state network calculation
Resting-state networks (RSNs) were determined from raw BOLD time-course data using the Multivariate Exploratory Linear Decomposition into Independent Components (MELODIC) Version 3.14 available with the FSL (www.fmrib.ox.ac.uk/fsl). All 19 subjects were time concatenated for a single group ICA. The data were high-pass filtered with a cutoff of 100 s (Marchini and Ripley, 2000). Before time-concatenation, five TRs were discarded from each subject, leaving 175 volumes over 350 s. The functional image volumes were motion corrected using the MCFLIRT implementation (Jenkinson et al., 2002). Slice-time correction was applied using linear interpolation. Skull-stripping was automatically performed with the brain extraction tool (BET) (Smith, 2002), and the data were spatially smoothed with a 4 mm full-width half-max Gaussian kernel. The brain mask was used to exclude non-brain voxels from the remaining analyses. All subjects were spatially normalized to an anatomical MNI standard template using a 12-parameter affine registration implemented in FLIRT (Jenkinson and Smith, 2001). The voxel BOLD times series were demeaned, variance normalized, and whitened.
The number of independent components was estimated using a Bayesian approach described in (Minka, 2000). Using Probabilistic Independent Component Analysis (Beckmann and Smith, 2004), the whitened time data were projected onto a 83-dimension subspace. A fixed-point method (Hyvarinen, 1999) decomposed the data into 83 independent components that accounted for variability in temporal, spatial, and subject domains. The spatial components were normalized by the variance of the residuals (Beckmann and Smith, 2004).
The ICA method decomposes the BOLD data S(x, t) as a function of voxel x and time t into spatiotemporal vectors with independent spatial maps and time-courses. The jth network has a spatial map, Mj(x), and time-course, Aj(t). As described by Joel et al. (Joel et al., 2011), these independent components are related to the original BOLD signal as
| (1) |
Once the group independent components were calculated, a dual regression (Beckmann and Filippini, 2009) was used to estimate subject-specific spatial maps and time courses. Components with vertical stripes in the axial view were associated with motion and excluded from further analysis. 45 of the 83 components were excluded due to high amounts of frequency content above 0.1 Hz or were determined to be related to anatomical artifacts. The MELODIC tool was used to regress these components out of each subject's original fMRI data.
2.6. Structural connectivity analysis
The high-angular resolution diffusion-weighted images were processed with the Diffusion Toolkit to estimate the fiber orientation and reconstruct fiber tracts (Wang et al., 2007). At each voxel, q-ball reconstruction was performed with 181 reconstruction points and 150 measurement points. Since access to the fiber trajectories was required for our model, the deterministic tractography module was used from the Diffusion Toolkit. Diffusion MRI tractography was performed in the subject's native space. Stopping criteria included an angle threshold of 35°. At each voxel, 30 seeds were randomly distributed and fiber trajectories were reconstructed. This path was lesion-side corrected and warped into MNI space using a concatenation of the subject's intermodality registration and anatomical intersubject registration. These fibers were stored for later data analyses.
2.7. Structurofunctional correlation
In this study, we were interested in the level of functional connectivity to the ensemble of structural connections at different fiber distances from a voxel. Every fiber endpoint voxel was treated as a potential network node. All processing that combines structural and functional connectivity was performed in MNI space. Thus, both BOLD data and fiber trajectories were both spatially normalized using the anatomical image registration. Note that voxels were not excluded based on apriori knowledge of lesion location. By including only fiber endpoints, gray matter voxels with direct damage or lost structural connections due to a lesion are inherently excluded from the calculation of metrics defined below.
Rather than analyzing the functional connectivity of a voxel to every structurally connected location, signal averaging was first performed across connections of a similar fiber path distance. Note that this model makes an assumption that functionally connected nodes in a network can be both distinguished and grouped by fiber distance in a meaningful way. Also note that since averaging was performed with fiber length fixed, metrics within this framework may be less affected by dependence of functional connectivity on fiber distance as reported in (Sporns, 2011).
To calculate structurofunctional correlation, first consider V(x , l) as a map that provides a set of N voxel locations, {y1, y2, … , yN}, that are structurally connected to voxel x by fibers with length l. Although N will vary with x, we write it as a constant in subsequent equations for simplicity. Let S(x, t) be the BOLD signal at time t and physical location x, with x ∈ ℝ3. Spatial averaging of S across all voxels included in Vgives a new weighted time signal, Z(x, l, t). This structurally connected mean BOLD signal is defined as
| (2) |
The fiber-length BOLD correlation, CZ, of a voxel x and an arbitrary function f(t) at length l is
| (3) |
Maximizing CZ with respect to fiber length gives the structurofunctional correlation (SFC), which represents the greatest fixed-length correlation of f(t) with the structurally connected voxels to x. Here, we denote as the fiber length that maximizes CZ, given as
| (4) |
Furthermore, the SFC is
| (5) |
2.8. Intrinsic and network-based SFC metrics
The first metric introduced is intrinsic structural-functional connectivity (iSFC), which is defined as the maximum temporal correlation between a voxel's BOLD signal with the fiber-length averaged BOLD signal of its structural connections. In the case that f(t) in Eq. (5) is the original fMRI signal at x, then SFC provides the functional connectivity of voxel x to its own structural connections. The intrinsic structurofunctional correlation (iSFC) is defined as
| (6) |
As a correlative measure, iSFC can have values between − 1 and 1. High iSFC indicates that for some fiber length, a strong correlation in BOLD fluctuations exists between a voxel and the ensemble of its structurally connected voxels at that distance. On the other hand, an iSFC with absolute value near zero occurs only if a voxel is not correlated with its anatomical connections at any fiber length. In this case, the voxel's direct structural connections do not account for the variability in its BOLD signal. There are multiple potential causes for this scenario. First, resting-state activity may be influenced by indirect connections via multiple neuron pathways. Additionally, a voxel may have a similar number inhibitory and excitatory functional connections at a fixed fiber length.
Function f(t) can also be selected as the time-course signal Aj(t) of the jth network from the independent component analysis. Effectively, SFC is then the maximum BOLD correlation of x’s structural connections with the network time-course. We define the network-based structurofunctional correlation (nSFC) as
| (7) |
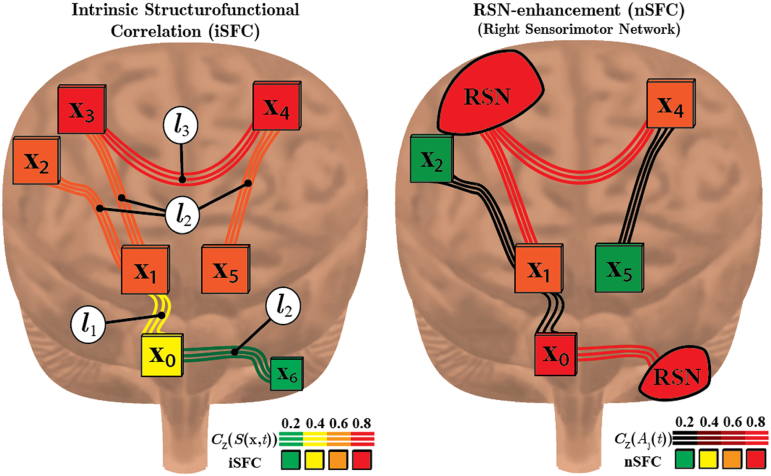
In example Fig. 2a, the iSFC is shown for multiple voxels within the brain. Voxel x1 is connected to cortical voxels x2 and x3 by fibers with length l2. The fiber-length BOLD correlation CZ(x1, l2) of function f(t) = S(x1, t) is its correlation with mean BOLD signal (S(x2, t) + S(x3, t))/2, shown to be 0.6. Likewise, at fiber length l1, x1 is structurally connected to brainstem voxel x0. Then, CZ(x1, l1) is the correlation of S(x1, t) with S(x0, t), given as 0.4. The iSFC(x1) is the maximum of these two correlations, being 0.6. Similarly, since x3 and x4 are exclusively connected by fibers with length l3, then CZ(x3, l3) and CZ(x4, l3) are both equal to the functional connectivity between x3 and x4, being near 0.8 as indicated by the red fibers and fill color.
Fig. 2.
iSFC and nSFC Example. Diagram depicting the intrinsic (a) and network-based (b) structural-functional correlation (SFC). The SFC provides a maximum temporal correlation, CZ, of a function f(t) to the structural connections of each voxel xi. a) Letting f(t) be the original BOLD signal of xi, SFC becomes a voxel-based intrinsic connectivity measure (iSFC). b) On the other hand, SFC represents an enhanced resting-state network if f(t) is the time-course of an RSN. Shown here is the left sensorimotor RSN with red filled regions correlated with Aj(t) in the right sensorimotor cortex and left cerebellum. Voxels x1 and x4 have high nSFC since they are structurally connected to regions with BOLD signals correlated with Aj(t).
Fig. 2b demonstrates the network-based SFC (nSFC) for the right sensorimotor network. Voxels within the red-filled regions in the right sensorimotor cortex and left cerebellum have BOLD signals that are highly correlated with the sensorimotor network time-course A(t). These regions are nodes within the sensorimotor network or its internetwork functional connections. Since voxels x0, x1, and x4 have direct structural connections to the RSN nodes, they will have increased nSFCj, even though some may not be originally a member of the sensorimotor network. Thus, nSFC enhances voxels that have direct structural connections to the nodes of a resting-state functional network.
2.9. Implementation of iSFC
The signal Z(x, l, t) was calculated independently using Eq. (2) for each time frame, t, and fiber length interval, lk. The full range of fiber lengths was chosen such that the longest projection fibers in the control group would fall within the highest bin. Shown in Fig. S2, the distribution of geodesic fiber length was calculated in every subject from reconstructed fiber pathways from diffusion MRI tractography. The maximum fiber lengths originating from the cortex across all control subjects was near 22 cm. Thus, six intervals were centered between 2, 4, 7, 10, 13, 16, 19, and 22 cm. A 4-D dataset with the same dimensions as the original fMRI data was created to store the filtered data for each fiber length interval. The BOLD signal S(x, t) was estimated for each gray matter fiber endpoint. Specifically, the signal at the endpoint of each tract was defined as the mean of the BOLD signals across the three endmost points of the fiber. Values at each of these three fiber points were extracted from the original fMRI signal by trilinear interpolation within MNI space. At each voxel, x, the fiber-length BOLD signal Z(x, l, t) was calculated by averaging S(x, t) of all structurally connected voxels. Z(x, l, t) was calculated for fibers within the every specified length interval l.
According to Eqs. (3), each voxel's intrinsic fiber-based correlation CZ(x, l, S(x, t)) was calculated for every fiber length interval l. Next, using Eq. (4), the fiber length, , maximizing CZ was determined. Following Eqs. (5), (6), the iSFC at each voxel was calculated as the .
2.10. Calculation of structurofunctional RSN maps
Resting-state networks that were analyzed in this study included the posterior default-mode network (DMN), the prefrontal network (anterior DMN), the primary visual network, the ipsilesional and contralesional sensorimotor networks, the bilateral cerebellar network, and the basal ganglia network. These networks were identified from the independent components from the MELODIC analysis.
In order to validate the level of network map enhancement, the nSFC was compared to the correlation coefficient between BOLD signal S(x, t) and each independent component time course Aj(t). Note that the independent component spatial maps, Mj(x), in Eq. (6) are not correlation coefficients. The correlation of the network's timecourse with the original BOLD signal provided a network map with the greatest weight given to the jth network. We denote this baseline correlation as RSNj. Based on in Eq. (7) we then use a similar notation to define the enhanced correlation as nSFCj. The difference between the Fisher-transformed nSFCj and RSNj was then used as a measure of network enhancement. The original and enhanced correlation maps were visualized for comparison using 3D isosurfaces at multiple isovalues.
2.11. Statistical analysis of SFC metrics
Voxel-based based analysis was used to measure differences in five metrics between stroke subjects and age-matched controls. Motion parameters were used as covariates when calculating all metrics. First, intrinsic measures iSFC, , and their product were calculated in all subjects. Next, the conventional RSN map, RSNj, and the SFC-enhanced map, nSFCj, were calculated for seven networks of interest. These included the primary visual, the default-mode, left and right sensorimotor, prefrontal, cerebellar, and basal ganglia networks. The Fischer transform was used to normalize nSFCj and RSNj for statistical testing. Stroke and control groups were compared by merging the groups together and computing Box and Blocks contrast maps for each metric. Box and Blocks scores were generated for control subjects by using population statistics based on age in healthy subjects (Mathiowetz and Weber, 1985). An F-test with ∝ = 0.001 was used to calculate the significance of the regression slope of each contrast. In order to correct the p-values for multiple comparisons, the 3dclustsim tool in AFNI was used to determine a minimum cutoff cluster volume of 700 mm3, corresponding to a corrected p-value of 0.05. From these correlation-based clusters, the mean metric value was calculated for each subject. Posthoc Student's t-tests (∝ = 0.05) were performed on cluster means between stroke and control groups. A linear regression between cluster mean and Box and Blocks was performed with stroke patients only. Cluster-based post-hoc tests were further corrected for multiple cluster comparisons using false discovery rate (q < 0.05).
3. Results
3.1. Control iSFC maps and group differences
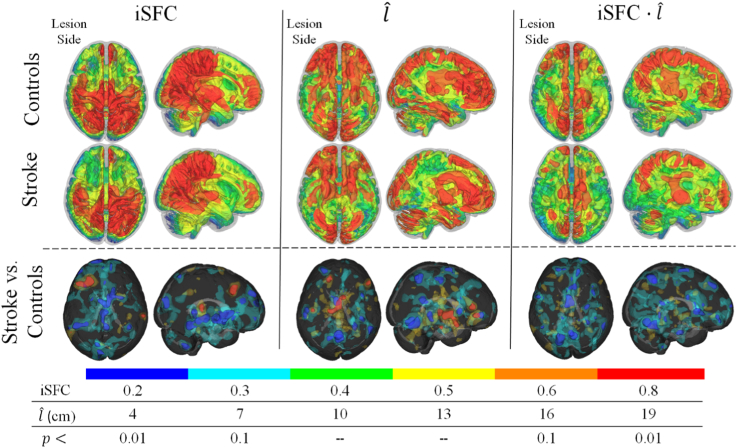
Intrinsic structurofunctional connectivity was characterized in each brain using iSFC, , and the product of these two metrics, iSFC, from Eqs. (4), (5), (6). The greatest iSFC was observed in the posterior parietal and sensorimotor regions of the brain. Shown by isosurfaces of the iSFC metric in Fig. 3 (top left), the voxels with greatest iSFC were located within the precuneus, anterior cingulate gyrus, and sensorimotor cortex. In turn, voxels having the greatest fiber length that maximizes functional connectivity () were within the prefrontal, posterior parietal cortex, and cerebellum regions (top center images in Fig. 3). The product of iSFC and , which gives greater structurofunctional weighting to long connections, demonstrated greatest values in the prefrontal and parietal portions of the cingulate gyrus. These three structurofunctional metrics were next compared between stroke and control groups.
Fig. 3.
Intrinsic structurofunctional correlation (iSFC) in stroke and controls. On the left, group differences in the structurofunctional correlation metric (iSFC) indicative of a voxel's maximum functional connectivity to structurally connected neighbors at a fixed fiber distance are shown. Isosurfaces of distance associated with the maximum correlation ( are shown in the center column. On the right, the iSFC product displays the group averages and differences for the product of the first two metrics. The bottom row shows clusters of significantly different voxels (P < 0.01) with a minimum cluster size of 700mm3. Blue clusters indicate significant decreases in stroke, compared to controls. Red clusters indicate significant increases. Lesion side is the left in these images. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
There were multiple brain areas in stroke subjects with significantly lower intrinsic structurofunctional connectivity (P < 0.01) compared to controls. Specifically, there were three main regions of the brain that demonstrated significant decreases in iSFC (P < 0.01; t values corrected for multiple comparisons based on cluster size) in stroke compared to control brains. These regions included the right lateral cerebellum, the midbrain, and thalamus. Conversely, there were increases in iSFC within the left dorsolateral prefrontal cortex (Fig. 3, left columns). Stroke subjects also had significant differences in the fiber distance, , associated with maximum intrinsic functional connectivity to a voxel. Stroke subjects had lower within the superior parietal lobule and precuneus, while was greater in stroke subjects within the thalamus, contralesional inferior temporal gyrus, and ipsilesional inferior prefrontal cortex. (Fig. 3, middle columns). The product of the maximum functional connectivity and the fiber distance associated with that connectivity was significantly lower (P < 0.01) within the superior parietal lobule, the supplementary motor area, the medial prefrontal cortex, the posterior portion of the inferior temporal gyrus, and the contralesional inferior frontal gyrus (Fig. 3, right column).
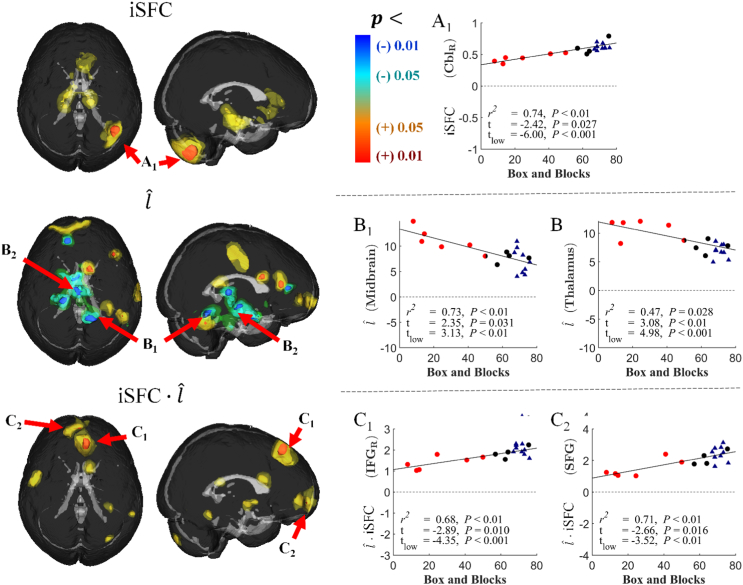
Voxel-wise linear regression was performed between each metric and Box and Blocks score to locate regions associated with upper extremity function (Fig. 4 and Table 2). An F-test was used to calculate a P-value at each voxel, and clusters of significant voxels (P < 0.01) are reported. The mean metric value of each cluster was compared between stroke and controls with a Student's t-test, and linearly regressed with Box and Blocks score. These clusters for iSFC, , and their product are listed in Table 2. For each stroke subject, a Box and Blocks z-score was calculated based on age-specific normative data in healthy individuals (Mathiowetz et al., 1985). Six subjects with z < − 3 were classified as lower functioning. These subjects are shown in Fig. 4 with red-colored data points, whereas the other stroke subjects are encoded in black. One main spatial cluster in the right cerebellum had a mean iSFC that was correlated with the Box and Blocks Score (Fig. 4a). The left portion of Fig. 4a shows the location of the cluster, color coded with the P-value for significance of the correlation. The cerebellum iSFC was correlated with Box and Blocks score (P < 0.01), and the cluster mean was significantly different between the stroke group and controls (P = 0.027). The low functioning (red markers in scatter plots) did have a significantly different iSFC within this cluster (P < 0.001). Spatial contrasts in iSFC with Wolf Motor and Fugl-Meyer Assessment scores were similar to the Box and Blocks contrasts, as shown in supplementary Fig. S3. However, p-values did not survive multiple comparisons corrections. We believe this was largely due to a ceiling effect on the Fugl-Meyer and Wolf Motor scores, with over half of the stroke subjects achieving near perfect scores.
Fig. 4.
Scatter plots of clinical correlations with iSFC. Spatial maps and scatterplots of the correlation between structurofunctional metrics and the Box and Blocks score. Spatial maps indicate the location of significantly correlated clusters, color coded by P-value after correction for multiple comparisons. The scatterplots show the individual subject data and regression line for the correlation analyses. t-values indicate the comparison between the stroke subjects (circles) and controls (triangles), where tlow indicates the t value for comparison of the stroke subjects with non-normal Box and Block score (red triangles) and controls. A. The significant cluster (P < 0.01, corrected) with a positive structurofunctional (iSFC) correlation with Box and Blocks was located in the right cerebellum. B. Significant negative correlations between and Box and Blocks score were observed in the thalamus and right cerebellum. C. Correlations between iSFC and Box and Blocks score were observed in the superior frontal gyrus (SFG).
Table 2.
Localized stroke-related differences and behavioral contrasts with the intrinsic structurofunctional connectivity (iSFC) metrics. Voxel-wise contrasts with Box and Blocks scores were calculated for iSFC, the maximum-correlation fiber-length, and fiber-length specific functional connectivity (SFC). Pearson correlation coefficients were thresholded (p < 0.01, uncorrected) to identify significant clusters, which were corrected for multiple comparisons by imposing a minimum size of 700mm3 (p < 0.05, corrected). The MNI coordinate of the maximum behavioral correlation is reported for each cluster. In order to test for group differences within each correlation cluster, the mean metric value was calculated within each cluster for every subject. Post-hoc t-tests and Box and Blocks correlations were performed on the means and were corrected for multiple comparisons using false-discovery rate (q < 0.05).
| Metric | Cluster |
Stroke vs controls |
Box & Blocks |
|||||
|---|---|---|---|---|---|---|---|---|
| ROI | x | y | z | nVox | t (all) | t (low) | R | |
| iSFC | Cbl_R | 42 | − 61 | − 40 | 30† | − 2.419†† | − 6.005†† | 0.86†† |
| iSFC | SFG_L | − 10 | 75 | 12 | 34 | − 2.819†† | − 4.284†† | 0.60†† |
| Midbrain | − 6 | − 13 | − 12 | 37 | 2.349†† | 3.127†† | − 0.85†† | |
| Thal | − 22 | − 21 | − 4 | 25 | 3.085†† | 4.985†† | − 0.69†† | |
| MTG_L | − 66 | − 29 | − 8 | 52 | 1.533†† | 2.438†† | − 0.75†† | |
| SFG_R | 2 | 39 | 44 | 44 | − 2.660†† | − 3.524†† | 0.84†† | |
| IFG_R | 34 | 15 | 4 | 14 | − 2.894†† | − 4.350†† | 0.82†† | |
| Amyg_L | − 22 | − 225 | − 20 | 106 | − 2.882†† | − 3.648†† | 0.69†† | |
| CZ(l = 1) | SFG_L | 538 | − 169 | 40 | 111 | − 3.885†† | − 5.102†† | 0.62†† |
| CZ(l = 3) | PFC_L | 346 | − 145 | 12 | 78 | − 2.932†† | − 4.575†† | 0.66†† |
| CZ(l = 1) | PFC_L | − 6 | − 145 | 16 | 54 | − 2.538†† | − 4.581†† | 0.82†† |
| CZ(l = 2) | MFG_L | 146 | − 193 | 56 | 51 | 4.254†† | 4.947†† | − 0.68†† |
| CZ(l = 1) | Cbl_R | 46 | − 269 | − 44 | 41 | − 3.123†† | − 5.813†† | 0.71†† |
| CZ(l = 2) | AG_L | 130 | − 277 | 40 | 30 | 1.818†† | 4.324†† | − 0.84†† |
| CZ(l = 2) | Midbrain | 170 | − 237 | − 12 | 30 | − 1.959†† | − 3.287†† | 0.75†† |
p < 0.05.
p < 0.01.
Maximum correlation fiber-length , shown in Fig. 4, there were clusters with both positive and negative significant correlations with Box and Blocks score. Positively correlated clusters were located within the inferior temporal gyrus, cingulate gyrus, and superior frontal gyrus. Clusters with a negative correlation between mean and Box and Blocks score were in the left middle temporal gyrus, thalamus, and midbrain. Across all stroke subjects, the mean was significantly different in clusters that were correlated with Box and Blocks score (P < 0.01). The lower functioning subjects were significantly different from controls within these same clusters. Two clusters with significant positive correlations between Box and Blocks and the mean product of the iSFC and were located within the medial prefrontal cortex (P < 0.01). Furthermore, the cluster means were also significantly lower in stroke subjects (P < 0.01).
3.2. Stroke-related differences in fiber-length averaged BOLD correlations
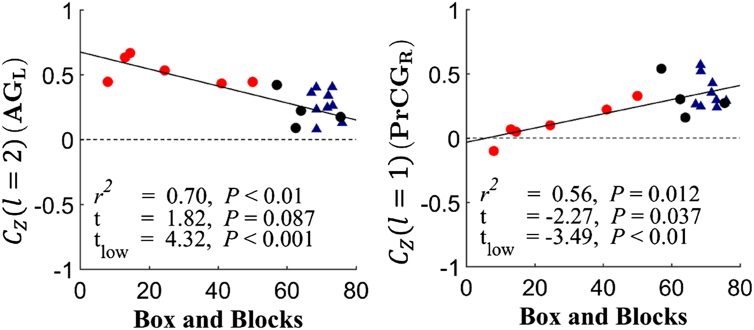
Voxel-wise differences in CZ between stroke subjects and age-matched controls reveal that intrinsic structurofunctional correlations can be delineated at specific fiber lengths. As shown in Fig. 5 for fiber distance between 40 and 70 mm (l = 1), the precentral gyrus had an increased functional connectivity (P < 0.001) with its structural neighbors for stroke subjects. Likewise, stroke subjects had lower functional connectivity (P < 0.001), correlated with Box and Blocks (R2 = 0.58), between the contralesional lateral orbitofrontal cortex and its neighbors at a 70–100 mm (l = 2) fiber length.
Fig. 5.
Scatter plots showing fiber-length BOLD correlation group differences. Correlations between CZ at l = 2 and Box and Blocks score for stroke subjects (circles) and controls (triangles) are shown for two significant regions. a) Stroke subjects have increased functional connectivity of the left (ipsilesional) angular gyrus (AG) with surrounding structural connections within fiber distance interval l = 1. b) The right (contralesional) precentral gyrus (PrCG) has decreased functional connectivity at fiber distance interval l = 2.
3.3. Behavioral correlations of enhanced resting-state network spatial maps
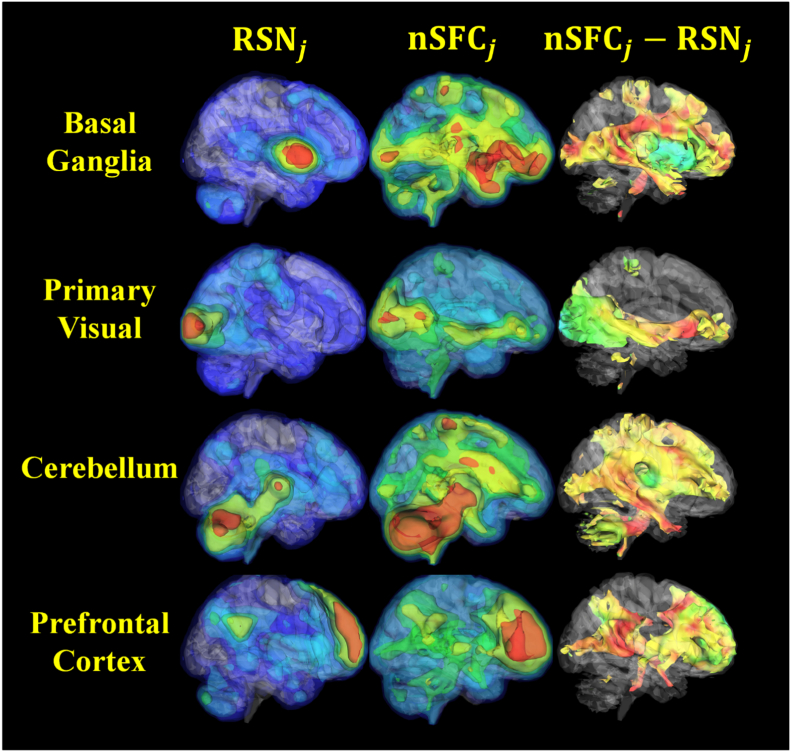
The network-based structurofunctional correlation (nSFC) maps showed structural connections between conventional functional networks and the rest of the brain. Four examples of this analysis for controls are shown in Fig. 6, with the left column showing the resting state network (RSN), the middle column showing the nSFC, and the right column showing the difference. The basal ganglia network nSFC was greatest in the amygdala, precentral gyrus, and frontal areas. Similarly, the primary visual network was enhanced by nSFC along the pathway from V1, to the thalamus, and finally the orbitofrontal gyrus. The prefrontal network nSFC enhanced the precuneus, the cerebellum, and brainstem areas. An independent component that was localized to the cerebellum demonstrated structurofunctional connectivity throughout the brain at projection fiber endpoints. In general, nSFC demonstrated expected structurofunctional connectivity to the resting state network. Because this analysis expands the networks, we refer the nSFC maps as the SFC-enhanced networks.
Fig. 6.
3D visualization of structurofunctional resting-state networks in healthy individuals. Shown are four selected resting-state independent components from conventional MELODIC time-concatenated group ICA (RSN) (left column) and the same components enhanced with the structurofunctional correlation metric (nSFC) (middle column). Note that these maps show the mean voxel-wise mean across the control group. The right column shows the difference in RSN and nSFC. In general, voxels that were structurally connected to the resting state network were enhanced by the correlation. Isosurface colors for individual RSN and nSFC maps correspond to positive correlation coefficients of 0.3, 0.4, 0.5, 0.6, and 0.7. In the nSFC − RSN column (right), an isovalue of 0.3 was extracted, and the surface is colored by nSFC − RSN ranging from − 1 to 1, with Red: 1.0; Green: 0.0; Blue: − 1.0. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
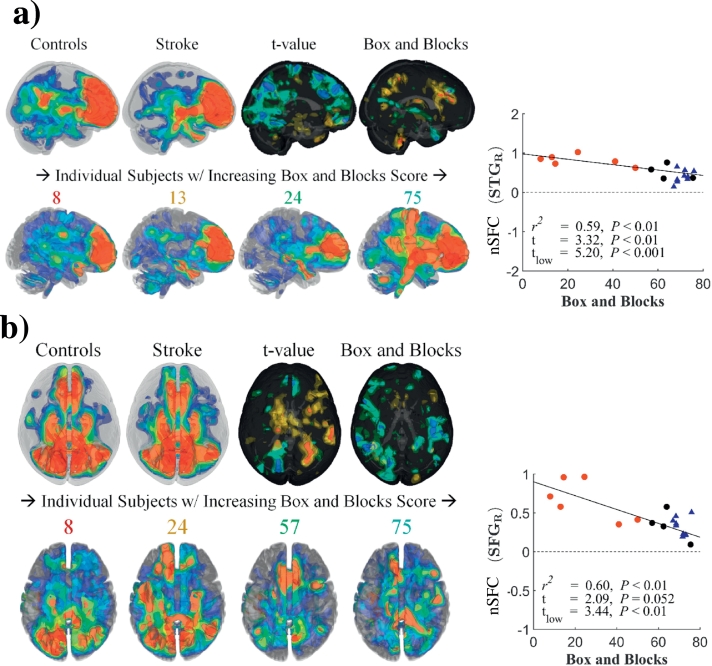
3.4. Reduced nSFC in motor networks correlated with impairment
Group differences in the SFC-enhanced (nSFC) ipsilesional sensorimotor network are shown in Fig. 7a. In control subjects, the enhanced network was spatially distributed in the left sensorimotor cortex and structurally connected regions. These regions included the prefrontal and posterior parietal areas of the left hemisphere and regions in the right hemisphere connected by commissural fibers (Fig. 7a, upper left). In stroke subjects, the nSFC in the contralesional superior temporal gyrus and inferior frontal gyrus was lower in subjects with greater impairments and was correlated with Box and Blocks score (R2 = 0.9, t = − 4.48) (Fig. 7a, right). This effect was observed across stroke subjects, with higher functioning subjects presenting with greater extent of enhanced sensorimotor networks (Fig. 7a, lower row). Although nSFC was similar for stroke and control subjects within the ipsilesional hemisphere, higher functioning stroke individuals had a greater level of contralesional enhancement.
Fig. 7.
Behavioral correlations of nSFC in sensorimotor and cerebellar networks.
Network-based structurofunctional correlations (nSFC) of the ipsilesional sensorimotor network (a) and cerebellar network (b) are shown for controls (upper left in each subfigure) stroke subjects (to the right of the controls) and individual stroke subjects (lower row in each subfigure). For t-value maps, blue and red surfaces indicate negative and positive t-values, where a negative t value indicates stroke subjects had lower nSFC than controls. Box and Blocks maps indicate clusters with significant correlations between nSFC and the Box and Blocks score (P < 0.05). The numbers over individual nSFC maps indicate the Box and Blocks scores for those subjects. Correlation plots for the Box and Blocks are shown for the right superior temporal gyrus (STG R) (a) and right cerebellum (Cbl R) (b). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Shown in Fig. 7b and Table 3, the enhanced cerebellar network in control subjects had structurofunctional correlations in frontal and parietal cortical areas, the bilateral cerebellum, brainstem, and thalamus. In this enhanced network, nSFC was reduced in the right cerebellum of stroke subjects, and these differences were larger in subjects with greater impairment (R2 = 0.72, t = − 3.8) (Fig. 7b, right). Similar to the sensorimotor network, the enhanced cerebellar network showed greater extent in higher functioning stroke subjects (Fig. 7b, lower row).
Table 3.
Voxel-based group differences in SFC-enhanced networks. Clusters of significantly different voxels between stroke and age-matched controls are reported for the SFC-enhancement resting-state networks. Acronyms: Crus: AG: angular gyrus, CGH: cingulate gyrus, Fu: fusiform gyrus, IFG: inferior frontal gyrus, Ins: insular cortex, ITG: inferior temporal gyrus, MFG: middle frontal gyrus, MFOG: middle fronto-orbital gyrus, MTG: middle temporal gyrus, PoCG: postcentral gyrus, PrCG: precentral gyrus, PreCu: precuneus, SFG: superior frontal gyrus, SMG: supramarginal gyrus, STG: superior temporal gyrus, Thal: thalamus.
| Network |
Cluster |
Stroke vs controls |
Box & Blocks |
|||||
|---|---|---|---|---|---|---|---|---|
| Independent component | ROI | x | y | z | nVox | t (all) | t (low) | R |
| Right insula | SS_L | − 42 | − 18 | − 16 | 31† | 1.472†† | 2.896†† | − 0.75†† |
| Prefrontal | Cbl_R | 11 | − 75 | − 40 | 57 | 2.024†† | 3.280†† | − 0.80†† |
| Left insula | STG_R | 27 | 14 | − 32 | 130 | 2.848†† | 4.281†† | − 0.77†† |
| STG_R | 55 | − 14 | 0 | 42 | 1.759†† | 3.933†† | − 0.67†† | |
| Default-mode (post) | SFG_R | 27 | − 2 | 65 | 378 | 2.086†† | 3.444†† | − 0.77†† |
| Cbl_R | 15 | − 51 | − 32 | 150 | 3.669†† | 4.954†† | − 0.64†† | |
| Cbl_L | − 34 | − 63 | − 52 | 114 | 2.704†† | 3.449†† | − 0.66†† | |
| Cbl_R | 31 | − 79 | − 48 | 46 | 3.741†† | 3.844†† | − 0.62†† | |
| Basal ganglia | Cbl_R | 23 | − 59 | − 48 | 226 | 2.049†† | 3.879†† | − 0.77†† |
| Caud_L | − 18 | 10 | 16 | 235 | 2.093†† | 4.060†† | − 0.90†† | |
| SFG_R | 7 | 6 | 57 | 266 | 2.187†† | 2.831†† | − 0.64†† | |
| Right sensorimotor | IFG_R | 27 | 14 | 25 | 55 | − 4.400†† | − 3.731†† | 0.48†† |
| PrCG_L | − 18 | − 18 | 53 | 39 | − 3.851†† | − 4.185†† | 0.44†† | |
| Left amygdala | Cbl_L | − 34 | − 39 | − 36 | 31 | 1.631†† | 2.773†† | − 0.73†† |
| Left sensorimotor | IFG_R | 35 | − 18 | 0 | 487 | − 2.485†† | − 6.045†† | 0.91†† |
| MFG_R | 51 | 14 | 41 | 175 | − 1.400†† | − 2.885†† | 0.88†† | |
Network acronyms: Salience: salience network nodes in anterior cingulate gyrus and bilateral insula, SM: sensorimotor network and contralateral cerebellum, Cerebellum: bilateral cerebellum network, DMN ant: default-mode network anterior half, Prefrontal: prefrontal cortex near anterior surface, aCingG: anterior cingulate gyrus.
p < 0.05.
p < 0.01.
3.5. Stroke-related nSFC increases in contralesional hemisphere
Group differences in the enhanced prefrontal network, shown in Fig. 8a and Table 3, had mixed correlations with Box and Blocks scores. The control group had greatest nSFC within the prefrontal cortex and precuneus (Fig. 8a, upper left), while nSFC in stroke subjects was highest in the prefrontal cortex and contralesional superior temporal gyrus (Fig. 8a, upper middle). The nSFC within the prefrontal resting-state network was correlated with Box and Blocks score in the precuneus (R2 = 0.72), although the stroke group was not significantly different from controls (t = − 0.20). However, nSFC was greater in the contralesional superior temporal gyrus and inferior frontal gyrus (t = 2.8) and was correlated with Box and Blocks score (R2 = 0.58) (Fig. 8a, right). In contrast, the prefrontal network in the low-functioning stroke subjects had lower nSFC in the ipsilesional middle temporal gyrus (R2 = 0.61, t = − 2.76).
Fig. 8.
Behavioral correlations of nSFC in prefrontal and default-mode networks. SFC-enhanced prefrontal network (a) and default-mode network (b) are shown in stroke subjects, age-matched controls, and individual stroke subjects. Blue and red surfaces indicate negative and positive t-values and correlations with significance level of P < 0.05. STG R: right superior temporal gyrus, PrCG R: right precentral gyrus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Structurofunctional correlations of the posterior default-mode network, which was distributed in the amygdala, anterior cingulate, precuneus, and posterior parietal cortex (Fig. 8b), generally had negative correlations with the Box and Blocks scores. The DMN had increased nSFC to the contralesional hemisphere and cerebellum (Table 3) in stroke subjects. Specifically, nSFC was greater within the precentral gyrus (t = 2.45, tlow = 3.70), postcentral gyrus (t = 1.56, tlow = 2.92), and right cerebellar crus IV (t = 3.0, tlow = 3.48) for stroke subjects. These differences were correlated with Box and Blocks score (P < 0.05), with all significant correlations being negative (i.e. higher connectivity in lower-functioning stroke subjects) (Fig. 8b, right; Table 3).
4. Discussion and conclusions
The results of this study indicated a decrease in structurofunctional connectivity to key integrative areas of the brain, including the cerebellum, midbrain and thalamus. These areas are critical to motor function and iSFC in the cerebellum was significantly correlated to hand motor function (i.e. the Box and Blocks score). Thus, connectivity of the cerebellum based on iSFC might be a key indicator of sensorimotor function in stroke subjects. This observation with voxel-based iSFC measures was supported by observations of changes in connectivity of the cerebellar network identified by an ICA analysis of resting state fMRI data. A comparison of the SFC-enhanced cerebellar network across stroke and control subjects demonstrated decreases in connections to the cerebellum that were correlated with hand function. These observations strongly suggest that connections to the cerebellum are important indicators of motor function in stroke subjects.
Although complex, a married structural-functional connectivity both complements and builds upon the studies that focus on either structural or functional connectivity. Enhancing well-documented resting-state networks from fMRI studies with established structural connectivity techniques may reveal network-specific mechanisms underlying pathology and recovery. This paper developed a novel metric that is not limited to stroke populations. Regions structurally connected to a damaged functional network may be central to cortical reorganization. These local areas may have direct structural connections to the perilesional tissue. SFC-enhancement is a novel way for investigators to focus in on specific RSNs to investigate the structure-function relationship at the voxel level.
Stroke subjects had reduced voxel-wise intrinsic structurofunctional connectivity, iSFC, in highly integrative cortical areas, while structurofunctional resting-state networks revealed changes in organization. The iplesional sensorimotor network and cerebellar network both had reduced structurofunctional connectivity that was correlated with Box and Blocks Score. The default-mode network and prefrontal network each had increased structurofunctional connectivity to the contralesional hemisphere. These group differences and functional correlations were often located in regions that were outside but structurally connected to the conventional resting-state networks.
Our novel analysis paradigm successfully married structural and functional connectivity. Indeed, the results indicate that intrinsic functional connectivity distributions can be expressed as a function of fiber length. Although resting-state functional connectivity in itself can detect differences in stroke subjects that are correlated with function, SFC may predict the broader impact of lesions on these same networks. For example, the posterior default-mode network was enhanced by SFC within the anterior cingulate gyrus, but this enhancement was lower in stroke subjects.
Stroke subjects had reduced intrinsic structurofunctional connectivity (e.g. iSFC) between areas connected at greater fiber path distances, as shown in Fig. 3. These differences were notably seen in regions that are associated with nodes of integrative cortical networks, such as the cerebellum and prefrontal cortex. The medial prefrontal cortex is densely connected to the precuneus and posterior parietal areas, forming the core of the default-mode network (DMN). It is possible that the nodes of integrative networks, such as the DMN or cerebellar networks, reorganize their connections after stroke. Whether these changes are compensatory or maladaptive remains speculation. This result is consistent with studies that have found reduced anterior-posterior DMN connectivity in aging (Vidal-Piñeiro et al., 2014). Vidal-Pineiro et al. found that white matter integrity of the cingulum was correlated with reduced DMN functional connectivity in the aging population. The results in this study suggest that anterior-posterior DMN connectivity is further reduced after stroke when compared to age-matched individuals.
We speculate that differences in the structurofunctional networks of the cerebellum and prefrontal cortex after stroke may be compensatory or maladaptive mechanisms of reorganization. Weakened sensorimotor and cerebellar networks may be shift to shorter distance connections to default-mode and prefrontal network nodes. The right cerebellum, which had reduced intrinsic SFC, also had lower SFC within the cerebellar and ipsilesional sensorimotor networks. This result complements other studies that have found decreased functional connectivity between the ipsilesional sensorimotor cortex and contralateral cerebellum (Tang et al., 2016). However, the default-mode network had increased structurofunctional connectivity to the right cerebellum. The contralesional superior temporal gyrus had increased SFC to the prefrontal network but decreased SFC to the ipsilesional sensorimotor network, suggesting an adaptive role of the prefrontal cortex. This result is consistent with stroke subjects having a reduced iSFC and product within the prefrontal cortex, as shown in Fig. 4, which suggested that the prefrontal cortex shifts its connectivity to shorter distances.
In the stroke group, there were differences in intrinsic structurofunctional connectivity at specific fiber path lengths. As shown in Fig. 5, a cluster within the ipsilesional precentral gyrus had increased iSFC at the fiber length range 7–10 cm. These differences were negatively correlated with Box and Blocks Score, suggesting that greater motor impairment is associated with the sensorimotor cortex increases its functional connectivity with nearby areas. Furthermore, we speculate that residual structural connectivity to perilesional cortical areas may guide cortical reorganization following a stroke.
Structurofunctional resting-state network analysis is a viable extension to conventional ICA methods when structural connectivity information is also available. Our technique may be the first to explicitly fuse structural and functional connectivity by combining BOLD signals across fiber endpoints. We believe that our technique is the first to express each voxel's intrinsic functional connectivity distribution as a function of fiber length. This work is also the first to produce a new fMRI time course at each voxel that is a combined signal of its structural connections. The structurofunctional correlation technique is different from others that combine structural and functional connectivity. Methods introduced in the past use structural connectivity to impose constraints on functional connectivity and dynamic causal models (Sporns et al., 2000, Stephan et al., 2009). The current approach attempts to enhance rather than constrain functional networks in order to estimate a broader network.
Lack of task-based fMRI data is another limitation when interpreting behavioral correlations with brain function metrics. Resting-state and task-based functional connectivity have shown different changes in the stroke population versus healthy individuals. However, since lower functioning stroke subjects often exhibit more head movement during a motor task, task-based fMRI data are more susceptible to motion artifacts. Moreover, these artifacts contaminate behavioral spatial maps.
Our approach shares many of the same limitations as functional MRI and diffusion MRI tractography. Functional connectivity MRI assumes that voxels with correlations in BOLD signal are co-active and thus functionally connected. Due to the course temporal resolution of fMRI and the slow nature of the hemodynamic response, this modeled connectivity is left undirected. The accuracy of structural connectivity estimated by diffusion MRI tractography approaches is limited by the fiber orientation model. Models of the orientation distribution function at each voxel make the assumption that axon organization is the prime contributor to diffusion directionality.
A common limitation of diffusion MRI tractography is that voxels with crossing white matter fibers can lead to inaccurate structural mappings. Most tractography fibers originating in the cerebellum, for example, fail to cross at the pons, thus leading to many positive connections with the ipsilateral hemisphere and false negative connections with the contralateral hemisphere. In this study, we chose to exclude fibers that did not cross at the pons in order to mitigate the connectivity bias introduced by this limitation of tractography. However, it is known that there are in fact fibers that synapse at the red nucleus and influence the ipsilateral hemisphere. Thus, our technique was not able to detect these connections that may indeed play a role in alternative pathways after stroke.
Structurofunctional connectivity analysis has additional limitations associated with its principle assumptions. First, the SFC model assumes that every reconstructed fiber endpoint contains neurons that are connected to the neurons located at the other endpoint. Since there are cases in which reconstructed fibers terminate within the white matter, our current implementation occasionally mixes the BOLD signals of gray matter voxels with white matter voxels. These violations of our assumption led to the enhancement of white matter pathways within our resulting maps. Another limitation was the usage of deterministic tractography, which does not provide a connectivity distribution for every voxel as do probabilistic approaches. Probabilistic approaches are capable of generating more fibers from the cerebellum that cross at the brainstem, which would have been beneficial for studying the cerebellum's structural-functional connectivity. In the case of a stroke, there are alterations of the vasculature and white matter microstructure within and nearby the lesion that may impact the SFC model's assumptions.
The SFC-metric results in this study may have been impacted by data artifacts in resting-state fMRI data caused by head motion. Head motion was corrected using affine image registration, and these motion regressors were treated as covariates in all functional connectivity analysis. However, recent resting-state fMRI studies have introduced a scrubbing technique that excludes frames based on frame-by-frame change in percent BOLD signal (Power et al., 2012). Strong spikes in the global BOLD signal may have then impacted the temporal correlations used to identify RSNs. The distribution of frame-by-frame global BOLD signal in each subject is shown in Fig. S4. There were no significant differences between stroke subjects and controls in framewise displacement (FD) nor DVARS. Additionally, these metrics were not correlated with the behavioral measures collected.
Another critical source of potential error in any voxel-based approach is intersubject and intermodality image registration. This study involved coregistration of functional, structural, and diffusion MRI within and between subjects. Each of these modalities has its own unique artifacts. Spatial distortions inherent to echo-planar imaging have different characteristics between diffusion and functional MRI, leading to a nonlinear mapping between voxels. An additional field-correction map can be acquired before the EPI scan in order to correct the distortions. This study did not include this field-map in its acquisition. In order to improve mappings between diffusion and functional MRI spaces, a nonlinear deformable registration was performed on mean BOLD images between subjects. Such a registration between functional images has been subject to criticism.
In this study, respiration and cardiac cycle were not treated as coregressors, which leaves them as potential confounds. These physiological processes have been shown to introduce artifacts into resting-state connectivity analyses. It has been recommended that these data be collected during the scan and be treated as coregressors during the ICA and/or seed-based connectivity analyses. However, other investigators have argued against this correction measure since that there may be important cortical activity that is highly correlated with respiration and heart rate. Independent components that were determined to be related to artifact or noise were regressed out of our data before applying our analyses.
The innovative significance of the structurofunctional correlation metrics is governed by the biological interpretation of BOLD signals averaged at fixed fiber distances. Transmission time of an action potential depends not only on physical distance, but also on conduction velocity. Conduction velocity increases with axon diameter and can scale tenfold in the presence of myelin (Caminiti et al., 2013, Hartline and Colman, 2007). On the other hand, the time-scale of the hemodynamic response and BOLD signal fluctuations is over multiple seconds. Thus, conduction velocity is not expected to have a significant impact on the SFC model.
Hemodynamic lag in resting-state data (Mitra et al., 2014) was not estimated in this study and may have contributed to the differences in functional connectivity. However, since differences in hemodynamic lag may be influenced by more than one fiber pathway, incorporating such delay estimates into our model is nontrivial. The impact of hemodynamic lag on SFC network enhancement will need to be investigated in future studies.
The residual structural and functional connectivity of the brain after stroke may be used to select a personalized treatment that maximizes functional outcomes. In such an approach, structurofunctional resting-state networks would allow for targeting specific functional systems while incorporating both types of connectivity information.
In conclusion, our novel analysis can detect changes in structure-function interactions in cortical networks after stroke. Future work will investigate changes in structurofunctional networks between acute and chronic phases of stroke, and potential applications for personalized rehabilitation will be considered. Additionally, SFC will be validated with a publicly available collection of young healthy adults.
Acknowledgments
Acknowledgements
This work was supported by the Marquette University Strategic Innovation Fund (grant number 37) and the Ralph and Marion C. Falk Medical Research Trust. The contents of this article were developed under a grant from the National Institute on Disability, Independent Living, and Rehabilitation Research (NIDILRR grant number 90RE5006, formerly NIDRR H133E100007). NIDILRR is a Center within the Administration for Community Living (ACL), Department of Health and Human Services (HHS). The contents of this article do not necessarily represent the policy of NIDILRR, ACL, HHS, and you should not assume endorsement by the Federal Government. The funding sources had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit this article for publication.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2017.07.002.
Appendix A. Supplementary data
Supplementary material
References
- Basser P.J., Mattiello J., Le Bihan D. MR diffusion tensor spectroscopy and imaging. Biophys. J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann Mackay, Filippini Smith. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. NeuroImage. 2009;47:S148. [Google Scholar]
- Beckmann C.F., Smith S.M. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Behrens T.E.J., Johansen-Berg H., Woolrich M.W., Smith S.M., Wheeler-Kingshott C.A.M., Boulby P.A., Barker G.J., Sillery E.L., Sheehan K., Ciccarelli O., Thompson A.J., Brady J.M., Matthews P.M. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat. Neurosci. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- Behrens T.E.J., Berg H.J., Jbabdi S., Rushworth M.F.S., Woolrich M.W. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? NeuroImage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B., Zerrin Yetkin F., Haughton V.M., Hyde J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Buch E.R., Modir Shanechi A., Fourkas A.D., Weber C., Birbaumer N., Cohen L.G. Parietofrontal integrity determines neural modulation associated with grasping imagery after stroke. Brain. 2012;135:596–614. doi: 10.1093/brain/awr331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminiti R., Carducci F., Piervincenzi C., Battaglia-Mayer A., Confalone G., Visco-Comandini F., Pantano P., Innocenti G.M. Diameter, length, speed, and conduction delay of callosal axons in macaque monkeys and humans: comparing data from histology and magnetic resonance imaging diffusion tractography. J. Neurosci. 2013;33:14501–14511. doi: 10.1523/JNEUROSCI.0761-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.R., Astafiev S.V., Lang C.E., Connor L.T., Strube M.J., Pope D.L.W., Shulman G.L., Corbetta M. Resting inter-hemispheric fMRI connectivity predicts performance after stroke. Ann. Neurol. 2010;67:365–375. doi: 10.1002/ana.21905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.R., Shulman G.L., Corbetta M. Why use a connectivity-based approach to study stroke and recovery of function? NeuroImage. 2012;62:2271–2280. doi: 10.1016/j.neuroimage.2012.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L., Schlaug G. Increased resting state connectivity between ipsilesional motor cortex and contralesional premotor cortex after transcranial direct current stimulation with physical therapy. Sci Rep. 2016;6:23271. doi: 10.1038/srep23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts J.J., Higham D.J., Bosnell R., Jbabdi S., Matthews P.M., Behrens T.E.J., Johansen-Berg H. Network analysis detects changes in the contralesional hemisphere following stroke. NeuroImage. 2011;54:161–169. doi: 10.1016/j.neuroimage.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux J.S., Greicius M.D. Greater than the sum of its parts: a review of studies combining structural connectivity and resting-state functional connectivity. Brain Struct. Funct. 2009;213:525–533. doi: 10.1007/s00429-009-0208-6. [DOI] [PubMed] [Google Scholar]
- Du Y., Fan Y. Group information guided ICA for fMRI data analysis. NeuroImage. 2013;69:157–197. doi: 10.1016/j.neuroimage.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Fjell A.M., Sneve M.H., Grydeland H., Storsve A.B., Amlien I.K., Yendiki A., Walhovd K.B. Relationship between structural and functional connectivity change across the adult lifespan: a longitudinal investigation. Hum. Brain Mapp. 2017;38:561–573. doi: 10.1002/hbm.23403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M.D. Clinical applications of resting state functional connectivity. Front. Syst. Neurosci. 2010;4 doi: 10.3389/fnsys.2010.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugl-Meyer A., Jaasko L., Leyman I., Olsson S., Steglind S. The post-stroke hemiplegic patient. Scand. J. Rehabil. Med. 1975;7:13–31. [PubMed] [Google Scholar]
- Hagmann P., Cammoun L., Gigandet X., Meuli R., Honey C.J., Van Wedeen J., Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:1479–1493. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartline D.K., Colman D.R. Rapid conduction and the evolution of giant axons and myelinated fibers. Curr. Biol. 2007;17:29–35. doi: 10.1016/j.cub.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Honey C.J., Honey C.J., Sporns O., Sporns O., Cammoun L., Cammoun L., Gigandet X., Gigandet X., Thiran J.P., Thiran J.P., Meuli R., Meuli R., Hagmann P., Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey C.J., Thivierge J.P., Sporns O. Can structure predict function in the human brain? NeuroImage. 2010;52:766–776. doi: 10.1016/j.neuroimage.2010.01.071. [DOI] [PubMed] [Google Scholar]
- Horn A., Ostwald D., Reisert M., Blankenburg F. The structural-functional connectome and the default mode network of the human brain. NeuroImage. 2014;102:142–151. doi: 10.1016/j.neuroimage.2013.09.069. [DOI] [PubMed] [Google Scholar]
- Hyvarinen A. Fast and robust fixed-point algorithms for independent component analysis. Neural Netw, IEEE Trans. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Smith S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Joel S.E., Caffo B.S., Van Zijl P.C.M., Pekar J.J. On the relationship between seed-based and ICA-based measures of functional connectivity. Magn. Reson. Med. 2011;66:644–657. doi: 10.1002/mrm.22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinosky B.T., Schindler-Ivens S., Schmit B.D. White matter structural connectivity is associated with sensorimotor function in stroke survivors. NeuroImage Clin. 2013:2. doi: 10.1016/j.nicl.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P., Schulz R., Hummel F.C. Structural connectivity analyses in motor recovery research after stroke. Ann. Clin. Transl. Neurol. 2016;3:233–244. doi: 10.1002/acn3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J.L., Ripley B.D. A new statistical approach to detecting significant activation in functional MRI. NeuroImage. 2000;12:366–380. doi: 10.1006/nimg.2000.0628. [DOI] [PubMed] [Google Scholar]
- Mathiowetz V., Weber K. 1985. Adult N o Drills for the Box and Block; p. 39. [DOI] [PubMed] [Google Scholar]
- Mathiowetz V., Volland G., Kashman N., Weber K. Adult norms for the Box and Block Test of manual dexterity. Am. J. Occup. Ther. Off. Publ. Am. Occup. Ther. Assoc. 1985 doi: 10.5014/ajot.39.6.386. [DOI] [PubMed] [Google Scholar]
- van Meer M.P.A., van der Marel K., Wang K., Otte W.M., el Bouazati S., Roeling T.A.P., Viergever M.A., Berkelbach van der Sprenkel J.W., Dijkhuizen R.M. Recovery of sensorimotor function after experimental stroke correlates with restoration of resting-state interhemispheric functional connectivity. J. Neurosci. 2010;30:3964–3972. doi: 10.1523/JNEUROSCI.5709-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minka T.P. Automatic choice of dimensionality for PCA. M.I.T. media aboratory percept. Comput. Sect. 2000;1–16 (10.1.1.19.9545) [Google Scholar]
- Mitra A., Snyder A.Z., Hacker C.D., Raichle M.E. Lag structure in resting-state fMRI. J. Neurophysiol. 2014;111:2374–2391. doi: 10.1152/jn.00804.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovadia-Caro S., Margulies D.S., Villringer A. The value of resting-state functional magnetic resonance imaging in stroke. Stroke. 2014:2818–2824. doi: 10.1161/STROKEAHA.114.003689. [DOI] [PubMed] [Google Scholar]
- Park B., Kim D.S., Park H.J. Graph independent component analysis reveals repertoires of intrinsic network components in the human brain. PLoS One. 2014;9:1–10. doi: 10.1371/journal.pone.0082873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehme A.K., Grefkes C. Cerebral network disorders after stroke: evidence from imaging-based connectivity analyses of active and resting brain states in humans. J. Physiol. 2013;591:17–31. doi: 10.1113/jphysiol.2012.243469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytty R., Nikkinen J., Paavola L., Abou Elseoud A., Moilanen V., Visuri A., Tervonen O., Renton A.E., Traynor B.J., Kiviniemi V., Remes A.M. GroupICA dual regression analysis of resting state networks in a behavioral variant of frontotemporal dementia. Front. Hum. Neurosci. 2013;7:461. doi: 10.3389/fnhum.2013.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. The human connectome: a complex network. Ann. N. Y. Acad. Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Sporns O., Tononi G., Edelman G.M. Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb. Cortex. 2000;10:127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- Stephan K.E., Tittgemeyer M., Knösche T.R., Moran R.J., Friston K.J. Tractography-based priors for dynamic causal models. NeuroImage. 2009;47:1628–1638. doi: 10.1016/j.neuroimage.2009.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C., Zhao Z., Chen C., Zheng X., Sun F., Zhang X., Tian J., Fan M., Wu Y., Jia J. Decreased functional connectivity of homotopic brain regions in chronic stroke patients: a resting state fMRI study. PLoS One. 2016;11:1–13. doi: 10.1371/journal.pone.0152875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion J.P. Image matching as a diffusion process: an analogy with Maxwell's demons. Med. Image Anal. 1998;2:243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- Tuch D.S. Q-ball imaging. Magn. Reson. Med. 2004;52:1358–1372. doi: 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- Tuch D.S., Reese T.G., Wiegell M.R., Makris N., Belliveau J.W., Van Wedeen J. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn. Reson. Med. 2002;48:577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- Urbin M.A., Hong X., Lang C.E., Carter A.R. Resting-state functional connectivity and its association with multiple domains of upper-extremity function in chronic stroke. Neurorehabil. Neural Repair. 2014;28:761–769. doi: 10.1177/1545968314522349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Piñeiro D., Valls-Pedret C., Fernández-Cabello S., Arenaza-Urquijo E.M., Sala-Llonch R., Solana E., Bargalló N., Junqué C., Ros E., Bartrés-Faz D. Decreased default mode network connectivity correlates with age-associated structural and cognitive changes. Front. Aging Neurosci. 2014;6:256. doi: 10.3389/fnagi.2014.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Benner T., Sorensen A.G., Wedeen V.J. Diffusion Toolkit: a software package for diffusion imaging data processing and tractography. Proc. Int. Soc. Magn. Reson. Med. 2007;15:3720. [Google Scholar]
- Wang L., Yu C., Chen H., Qin W., He Y., Fan F., Zhang Y., Wang M., Li K., Zang Y., Woodward T.S., Zhu C. Dynamic functional reorganization of the motor execution network after stroke. Brain. 2010;133:1224–1238. doi: 10.1093/brain/awq043. [DOI] [PubMed] [Google Scholar]
- Wolf S.L., Catlin P.A., Ellis M., Archer A.L., Morgan B., Piacentino A. Assessing Wolf motor function test as outcome measure for research in patients after stroke. Stroke. 2001;32:1635–1639. doi: 10.1161/01.str.32.7.1635. [DOI] [PubMed] [Google Scholar]
- Wolf S.L., Winstein C.J., Miller J.P., Thompson P.A., Taub E., Uswatte G. The excite trial: retention of improved upper extremity function among stroke survivors receiving CI Movement therapy. Lancet Neurol. 2008;7:33–40. doi: 10.1016/S1474-4422(07)70294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh F.C., Tang P.F., Tseng W.Y.I. Diffusion MRI connectometry automatically reveals affected fiber pathways in individuals with chronic stroke. NeuroImage Clin. 2013;2:912–921. doi: 10.1016/j.nicl.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Olivi A., Hertig S.J., Zijl P. Van. Automated fiber tracking of human brain white matter using diffusion tensor imaging. NeuroImage. 2009;42:771–777. doi: 10.1016/j.neuroimage.2008.04.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material