Abstract
The transcription factor hepatocyte nuclear factor–1β (HNF-1β) is essential for normal kidney development and function. Inactivation of HNF-1β in mouse kidney tubules leads to early-onset cyst formation and postnatal lethality. Here, we used Pkhd1/Cre mice to delete HNF-1β specifically in renal collecting ducts (CDs). CD-specific HNF-1β mutant mice survived long term and developed slowly progressive cystic kidney disease, renal fibrosis, and hydronephrosis. Compared with wild-type littermates, HNF-1β mutant mice exhibited polyuria and polydipsia. Before the development of significant renal structural abnormalities, mutant mice exhibited low urine osmolality at baseline and after water restriction and administration of desmopressin. However, mutant and wild-type mice had similar plasma vasopressin and solute excretion levels. HNF-1β mutant kidneys showed increased expression of aquaporin-2 mRNA but mislocalized expression of aquaporin-2 protein in the cytoplasm of CD cells. Mutant kidneys also had decreased expression of the UT-A urea transporter and collectrin, which is involved in apical membrane vesicle trafficking. Treatment of HNF-1β mutant mIMCD3 cells with hypertonic NaCl inhibited the induction of osmoregulated genes, including Nr1h4, which encodes the transcription factor FXR that is required for maximal urinary concentration. Chromatin immunoprecipitation and sequencing experiments revealed HNF-1β binding to the Nr1h4 promoter in wild-type kidneys, and immunoblot analysis revealed downregulated expression of FXR in HNF-1β mutant kidneys. These findings reveal a novel role of HNF-1β in osmoregulation and identify multiple mechanisms, whereby mutations of HNF-1β produce defects in urinary concentration.
Keywords: transcription factors, collecting ducts, urea, cystic kidney, water-electrolyte balance, osmolality
Hepatocyte nuclear factor–1β (HNF-1β) is an essential transcription factor that regulates tissue-specific gene expression in the epithelium of the kidney, liver, pancreas, and genitourinary tract.1 HNF-1β contains an N-terminal dimerization domain, a POU homeodomain that mediates binding to the consensus sequence (5′-RGTTAATNATTAACM-3′), and a C-terminal transactivation domain.2 HNF-1β binds to target gene promoters and activates transcription, although transcriptional repression has also been reported.3,4 In the adult kidney, HNF-1β is expressed in tubular epithelial cells in all segments of the nephron and renal collecting ducts (CDs), where it regulates the expression of kidney-specific genes, such as Slc12a1 and Cdh16.5 In the developing kidney (metanephros), HNF-1β is expressed in the ureteric bud, renal vesicles, comma- and S-shaped bodies, and maturing nephrons.6 Inactivation of HNF-1β in the ureteric bud leads to abnormal branching and defective mesenchymal-epithelial transition,7 whereas inactivation in the metanephric mesenchyme prevents the proper segmentation of the nephron.6
Consistent with its essential role in kidney development, humans with heterozygous mutations of HNF1B develop congenital renal abnormalities, including renal cysts and diabetes, congenital anomalies of the kidney and urinary tract, and autosomal dominant tubulointerstitial kidney disease.8 The spectrum of renal structural anomalies includes renal hypoplasia/agenesis, renal dysplasia, horseshoe kidney, glomerulocystic kidney disease, and multicystic dysplastic kidney.9,10 Functional abnormalities include hyperuricemia and hypomagnesemia.11
HNF-1β also plays a central role in a transcriptional network that is involved in polycystic kidney disease. Transgenic mice expressing dominant negative mutant HNF-1β under the control of a kidney-specific promoter develop kidney cysts and renal failure, which is similar to the phenotype of humans with HNF1B mutations.12 Similarly, kidney-specific deletion of Hnf1β using Cre/loxP recombination results in renal cyst formation in mice.13 Characterization of the cystic kidneys has revealed that HNF-1β regulates the transcription of numerous cystic disease genes, including Pkhd1, Pkd2, Umod, Glis2, and Kif12,14,15 suggesting that mutations of HNF-1β produce kidney cysts by downregulating the expression of these genes. HNF-1β may also control tubular diameter through regulation of planar cell polarity.16
Using ChIP-chip in combination with gene expression profiling, we have described additional roles for HNF-1β in the mouse kidney. HNF-1β regulates renal cholesterol metabolism by controlling the expression of cholesterol biosynthetic genes and Pcsk9, a key gene involved in cholesterol uptake.17 HNF-1β also regulates the expression of microRNAs, and we have discovered a novel lncRNA under regulatory control of HNF-1β that gives rise to the miR-200 family of miRNAs.18
Kidney-specific inactivation of HNF-1β using a tubule-specific Cre line (Ksp/Cre) results in the rapid formation of kidney cysts, renal failure, and early postnatal lethality.13 To assess the functions of HNF-1β specifically in renal CDs, we inactivated HNF-1β using a CD-specific Cre line (Pkhd1/Cre). Characterization of CD-specific mutant mice revealed a novel role of HNF-1β in urinary concentration and osmoregulation.
Results
CD-Specific Deletion of Hnf1β
To examine the role of HNF-1β in the CD, we generated Pkhd1/Cre;Hnf1βf/f (HNF-1β mutant) mice. Pkhd1/Cre transgenic mice carry a 4.7-kb genomic fragment that contains the promoter of Pkhd1 and directs expression of Cre recombinase in renal CD beginning at embryonic day 14.15 Hnf1βf/f mice contain two loxP sites flanking the essential first exon of Hnf1β.13 Generation of bitransgenic Pkhd1/Cre;Hnf1βf/f mice allowed targeted deletion of HNF-1β in CD without affecting other nephron segments. PCR analysis detected the recombined allele of Hnf1β in genomic DNA from kidneys of HNF-1β mutant mice, confirming Cre-mediated recombination (Supplemental Figure 1A). To identify the cells in which recombination occurred, an enhanced yellow fluorescent protein (EYFP) reporter that is activated by Cre/loxP recombination (R26R-EYFP) was introduced into the genetic cross. Kidney sections from control mice (Pkhd1/Cre;Hnf1βf/+;R26R-EYFP) and HNF-1β mutant mice (Pkhd1/Cre;Hnf1βf/f;R26R-EYFP) were costained with an antibody against HNF-1β and an antibody against green fluorescent protein that also recognizes EYFP. In Cre-positive control mice that were heterozygous for the floxed Hnf1β allele, expression of HNF-1β was maintained in EYFP-positive renal tubules (Supplemental Figure 1, B–E). In Cre-positive homozygous Hnf1βf/f mutant mice, HNF-1β was absent in EYFP-positive tubular epithelial cells at postnatal day 7 (P7) and P14 (Supplemental Figure 1, F–M), confirming the deletion of Hnf1β at two different time points.
Histologic Characterization of HNF-1β Mutant Mice
Previous studies showed that kidney-specific knockout of HNF-1β with the Ksp/Cre transgene produced 75% mortality by age P21.15 In contrast, CD-specific HNF-1β knockout mice showed similar survival as wild-type littermates up to 420 days of age (P=0.90; Mantel–Cox test; n=9). Hematoxylin and eosin staining of kidney sections from CD-specific HNF-1β mutant mice revealed slowly progressive cyst formation (Figure 1A). At P7, HNF-1β mutant mice had relatively normal medullary architecture (Supplemental Figure 1F). A few dilated medullary tubules and small cysts started to develop at P14 (Figure 1, B and C). Thereafter, medullary cyst number and size increased significantly with age, such that, by P35, all mutant mice exhibited medullary cysts (Figure 1D). Cortical cysts were first noted at P21 and became more prominent over time (Figure 1G). In contrast to kidney-specific Ksp/Cre;Hnf1βf/f mice that developed azotemia as early as P8,15 serum creatinine at P35 was only mildly elevated in CD-specific Pkhd1/Cre;Hnf1βf/f mice compared with control littermates (0.110±0.005 versus 0.083±0.004 mg/dl; P<0.05; n=9).
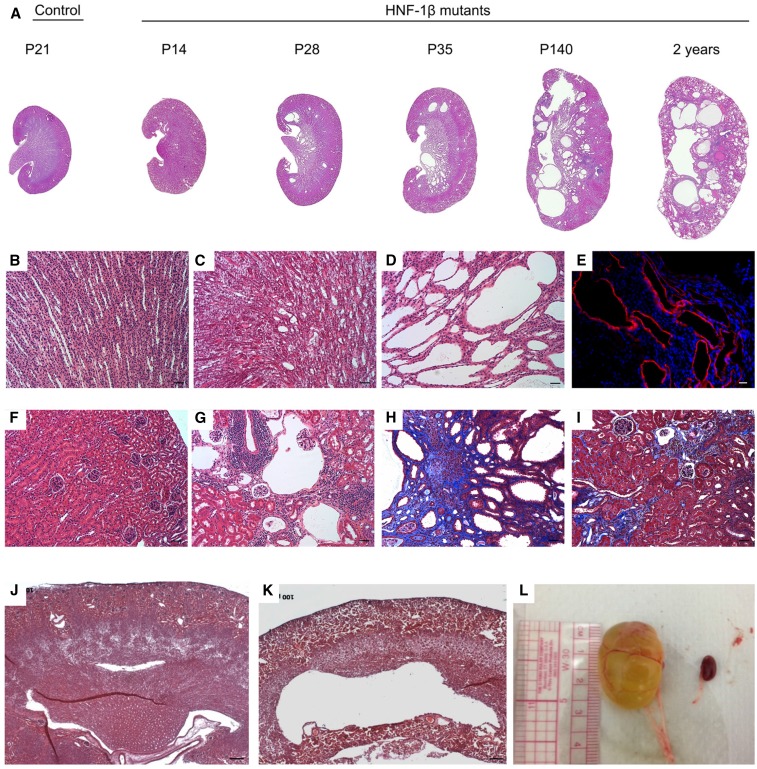
Figure 1.
Characterization of CD-specific HNF-1β mutant mice. (A) Hematoxylin and eosin (H&E) staining of kidney sections from control mice and HNF-1β mutant mice at the indicated ages. All images were acquired at the same magnification. (B and F) Higher magnification images of control kidneys at P14 showed normal histology of (B) the medulla and (F) the cortex. (C and D) Higher magnification images of HNF-1β mutant kidneys showed (C) a few dilated tubules at P14 and (D) significant cyst formation at P35. (E) Staining with an antibody against aquaporin-2 (red) showed that the cysts originated from the CD. Nuclei were counterstained with 4′6-diamidino-2-phenylindol (blue). (G) At 20 weeks of age (P140), the majority of HNF-1β mutant mice also exhibited glomerular cysts. (H and I) Trichrome staining of kidneys from mutant mice at 20 weeks revealed interstitial fibrosis (blue; H) surrounding medullary cysts and (I) in the renal cortex. (J and K) H&E staining showed that hydronephrosis was (K) present in some mutant mice at P7 and (J) absent in control littermates. (L) Hydronephrosis was more severe and frequent in mutant mice at later ages (P140). A control kidney is shown to the right. Scale bars, 50 μm in B–D and F–I; 20 μm in E; 100 μm in J–K.
Immunostaining with an antibody against aquaporin-2 (AQP2) confirmed that the medullary cysts in Pkhd1/Cre;Hnf1βf/f mutant kidneys originated from the CD (Figure 1E, Supplemental Figure 1N). NKCC2-positive thick ascending limbs of loops of Henle (TALH) were unaffected (Supplemental Figure 1O). Interstitial fibrosis developed beginning around P35 in both cystic and noncystic regions of the kidney (Figure 1, H and I). Mild hydronephrosis was present as early as P7 (Figure 1, J and K) and progressed to severe hydronephrosis by the age of 40 weeks old (Figure 1L). The complete spectrum of phenotypes observed in HNF-1β mutant mice is depicted in Supplemental Table 1. Mutant phenotypes were observed in both male and female mice.
HNF-1β Mutant Mice Develop Polyuria and Impaired Urinary Concentration
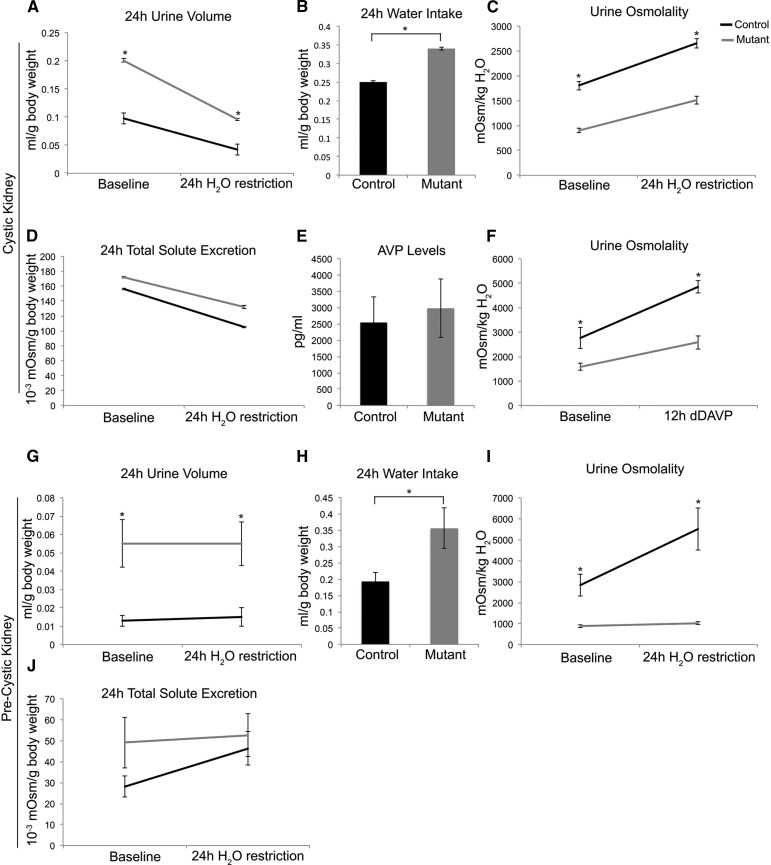
HNF-1β mutant and control mice were placed in metabolic cages and acclimated for 3 days; then, water intake and urine output were measured. HNF-1β mutant mice (n=11) produced approximately twice the volume of urine compared with control mice both at baseline and after 24 hours of water restriction, indicating the presence of polyuria (Figure 2A). Mutant mice had higher water intake and lower urine osmolality at baseline compared with controls (Figure 2, B and C). Although urine osmolality increased after water restriction, the maximal urine concentrating ability was still impaired in mutant mice (Figure 2C). Baseline plasma sodium concentration was not significantly different in mutant mice (139±1.5 mEq/L) and control mice (136±1.3 mEq/L; P=0.13; n=9). When hydronephrosis was excluded, mutant mice (n=7) were persistently polyuric (4.10 versus 1.81 ml; P=0.001) with defective urinary concentrating ability (852 versus 1821 mosmol/kg H2O; P=0.001). The urinary concentrations of sodium, potassium, and urea were all reduced by a similar magnitude in HNF-1β mutant mice (Supplemental Table 2). Total 24-hour urinary solute excretion was similar to control mice, further indicating a water diuresis rather than a solute diuresis (Figure 2D). Plasma arginine vasopressin levels were similar between control and mutant mice after 24 hours of water restriction (Figure 2E). The V2 vasopressin receptor agonist desmopressin was administered after 24 hours of water restriction, and urine was collected over a 12-hour period. Urine osmolality increased in HNF-1β mutant mice but remained lower than in control mice (2579 versus 4854 mosmol/kg H2O; n=4) (Figure 2F).
Figure 2.
CD-specific HNF-1β mutant mice develop polyuria and urine concentration defects. (A) Adult (P41) HNF-1β mutant mice produced higher urine volumes than controls both at baseline and after 24 hours of water restriction (n=11). (B and C) Mutant mice had (B) higher water intake and (C) lower urine osmolality than controls at baseline and after 24 hours of water restriction. (D) Total solute excretion was not significantly different between control and mutant mice. (E) Control and HNF-1β mutant mice had similar plasma vasopressin (AVP) levels 24 hours after water restriction (n=6). (F) Urine osmolality increased after desmopressin (dDAVP) injection but remained lower in HNF-1β mutant mice compared with controls (n=4). (G–J) At P17, a precystic time point, HNF-1β mutant mice (n=7) produced (G) higher urine volumes than controls both at baseline and after 24 hours of water restriction. Mutant mice had (H) higher water intake and (I) lower urine osmolality than controls at baseline and after 24 hours of water restriction. (J) No significant differences were observed in total solute excretion between control and mutant mice. Error bars indicate SEM. *P<0.05.
To determine if polyuria was present before the development of significant renal structural abnormalities, we analyzed mice at an earlier time point (P17) and excluded those with visible hydronephrosis. Similar to adult mice, 17-day-old HNF-1β mutant mice (n=7) exhibited polyuria and produced approximately three times the volume of urine compared with control mice both at baseline and after water restriction (Figure 2G). Baseline water intake was also higher in P17 mutant mice (Figure 2H). HNF-1β mutant mice at P17 had lower urine osmolality at baseline compared with controls and also failed to maximally concentrate their urine after water restriction (Figure 2I). Mutant mice had similar 24-hour total solute excretion as control littermates (Figure 2J). The effects of exogenous desmopressin could not be determined, because young mutant mice did not tolerate administration of desmopressin after water restriction. Collectively, these data indicate that the development of polyuria and impaired urinary concentration in HNF-1β mutant mice is not simply secondary to cyst formation or hydronephrosis.
Next, we performed quantitative real-time PCR (qPCR) to measure the expression of genes that are known to be involved in urinary concentration. Two genes, Aqp2 and Uta (Slc14a2), encoding the aquaporin-2 water channel and urea transporters UT-A1–UT-A4, respectively, were differentially expressed between HNF-1β mutant and control kidneys. Uta1/3 mRNA levels were decreased by 54%, whereas Aqp2 mRNA was increased by 72% (Supplemental Figure 2A). Western blot analysis using an antibody against AQP2 and an antibody that recognizes UT-A1 and UT-A2 showed that HNF-1β mutant kidneys had increased levels of AQP2 (Supplemental Figure 3A) and reduced levels of UT-A1/2 (Supplemental Figure 2J). Immunostaining of control kidneys showed that AQP2 was colocalized with UT-A1, the isoform that is expressed in renal CD (Supplemental Figure 2, B–E). Consistent with the changes in mRNA expression, antibody staining for UT-A1 was reduced, and staining for AQP2 was increased in the cystic CD of HNF-1β mutant mice (Supplemental Figure 2, F–I). The urea transporter gene Uta has been shown to be required for urinary concentration.19 However, ChIP-seq and ChIP-PCR showed that the Uta (Slc14a2) region did not contain binding sites for HNF-1β, indicating that the downregulation of UT-A1 was indirect (not shown).
Kidneys from CD-specific HNF-1β mutant mice contained elevated levels of the second messenger cAMP (Supplemental Figure 3B). The increased levels of cAMP may explain the increased expression of AQP2, because Aqp2 transcription is stimulated by cAMP response element binding protein (CREB).20 Expression of another cAMP-regulated gene, V2 vasopressin receptor,21 was also increased in CD-specific HNF-1β mutant kidneys (Supplemental Figure 3C). Because defects in urinary concentrating ability would not be expected in mice with increased expression of AQP2, we performed confocal laser scanning microscopy to study the subcellular localization of AQP2. In control kidneys, AQP2 was localized exclusively in the apical membrane and subapical region of cells in the CD (Supplemental Figure 3D). In contrast, HNF-1β mutant CD cells showed accumulation of AQP2 in the cytoplasm and basolateral region, suggesting a defect in apical protein trafficking (Supplemental Figure 3D). Consistent with this mechanism, mutant kidneys showed decreased expression of Tmem27, an HNF-1β–regulated gene that encodes collectrin, a protein involved in apical vesicle trafficking (Supplemental Figure 3E).
HNF-1β Plays a Novel Role in the Transcriptional Response to Hypertonicity
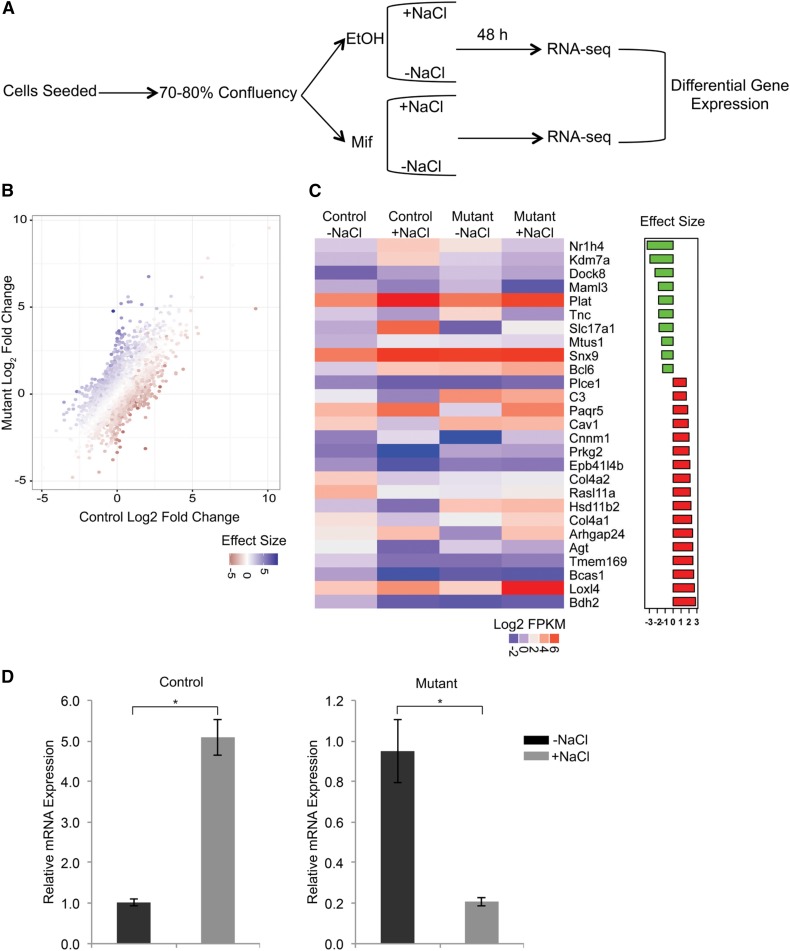
To further understand the mechanism of the urinary concentrating defect in HNF-1β mutant mice, we treated HNF-1β mutant mIMCD3 cells with hypertonic NaCl. mIMCD3 cells exposed to hypertonic NaCl upregulate expression of osmosensitive genes and mimic some of the changes that occur in the CDs of dehydrated mice.22 To assess the dependency on HNF-1β, we studied mIMCD3 cells that express dominant negative mutant HNF-1β (53A cells).3 53A cells were treated with mifepristone to induce expression of mutant HNF-1β and then incubated in medium supplemented with 100 mM NaCl for 48 hours. Uninduced cells were used as a control (Figure 3A). RNA was extracted, and gene expression profiles were obtained by RNA-seq. Treatment of control cells with hypertonic medium resulted in downregulation of 567 genes and upregulation of 1242 genes (−1> log2 fold change >1 and a Benjamini–Hochberg corrected P value <0.05) (Figure 3B). As validation, we observed upregulation of several genes that are known to be induced by chronic hypertonicity, including Nfat5, Slc6a12, Akr1b3, Sgk1, and Aqp1.23–26 In HNF-1β mutant cells, 648 genes were downregulated by hypertonicity, and 2022 were upregulated. Of the 1809 genes that were differentially regulated in control cells, 52 genes with size effect >1 or <−1 failed to respond in mutant cells. To identify genes that were directly regulated by HNF-1β, we overlapped the RNA-seq data with our previous ChIP-seq data obtained from mIMCD3 cells.17 Twenty-seven of 52 genes contained nearby HNF-1β binding sites and were, therefore, considered direct HNF-1β targets (Figure 3C). The remaining 25 genes were considered indirect targets (Supplemental Table 3).
Figure 3.
HNF-1β mutant cells display impaired response to hypertonicity. (A) mIMCD3 cells that can be induced to express dominant negative mutant HNF-1β (53A cells) were cultured for 24 hours until they reached 70%–80% confluency; then, they were treated with mifepristone (Mif) to induce mutant HNF-1β or vehicle (EtOH) as a control. After 6 hours of induction, NaCl was added to the medium (100 mM), and cells were cultured for another 48 hours followed by RNA extraction and sequencing. (B) Scatter plot showing differential gene regulation in response to NaCl treatment in control cells (x axis) and HNF-1β mutant cells (y axis). The color scale bar indicates the degree of interaction (effect size) between the dominant negative mutant effect and the hypertonicity effect in the statistical model for each gene. (C) Heat map (left panel) showing the changes in gene expression in response to hypertonicity in control and HNF-1β mutant cells for a subset of genes (wild-type log2 fold change >1 or <−1; effect size >1 or <−1; P<0.05) that are directly bound by HNF-1β. The scale of expression measured in fragment per kilobase of transcript per million mapped reads (log2 FPKM) is indicated. Effect size (right panel) shows top up- (green) and downregulated (red) genes in control cells compared with mutant cells. (D) Changes in expression of Nr1h4 (Fxr) in response to hypertonicity in mutant cells (right panel) compared with control cells (left panel). Error bars indicate SEM. *P<0.05.
Of the 27 direct targets, Fxr (Nr1h4) showed the greatest difference in osmoregulation between HNF-1β mutant and control cells. Fxr (Nr1h4) encodes the farnesoid X receptor, FXR, a member of the nuclear hormone receptor superfamily. Genetic deletion of FXR has been shown to impair urinary concentration in mice,27 similar to the phenotype of HNF-1β mutant mice. Quantitative RT-PCR confirmed that the expression of Fxr (Nr1h4) was fivefold higher in control cells treated with hypertonic medium compared with those treated with isotonic medium (Figure 3D). In contrast, expression was downregulated by 80% in HNF-1β mutant cells under hypertonic conditions. We conclude that HNF-1β plays an important role in regulating genes that are induced by hypertonicity, including Fxr (Nr1h4).
HNF-1β Binds to the Fxr Promoter and Regulates Its Expression In Vivo
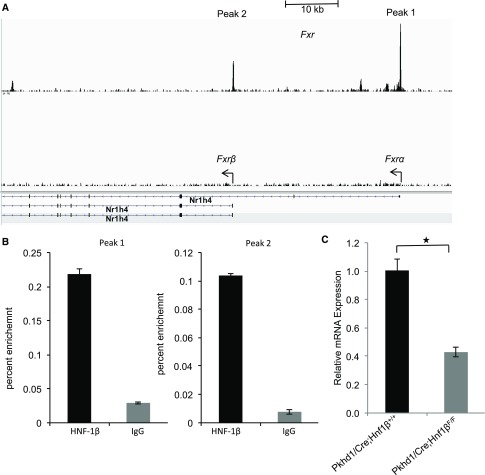
FXR (NR1H4) encodes four isoforms in both humans and mice (FXRα1, FXRα2, FXRα3, and FXRα4) as a result of alternative splicing of exon 5 and the use of two distinct promoters that initiate transcription from either exon 1 or exon 3.28,29 ChIP-seq experiments in mIMCD3 cells identified HNF-1β binding sites within the two known Fxr promoters (Figure 4A).17 We confirmed HNF-1β binding at the respective sites in vivo by performing ChIP-qPCR on chromatin isolated from mouse kidney. Relative to control IgG, HNF-1β was significantly enriched by >14-fold at both Fxr promoters (Figure 4B). To determine if the binding of HNF-1β correlated with changes in Fxr expression, we measured Fxr mRNA in wild-type and HNF-1β mutant kidneys by qPCR. The expression of Fxr was reduced by 60% in mutant kidneys compared with controls (Figure 4C). Collectively, these experiments indicate that HNF-1β transcriptionally regulates Fxr in vivo by binding to its promoter region.
Figure 4.
HNF-1β binds to the Fxr (Nr1h4) promoter and regulates its expression in vivo. (A) ChIP-seq showing occupancy of HNF-1β on the promoter regions of Fxrα and Fxrβ (arrows) in chromatin from mIMCD3 cells. Black boxes indicate exons, and arrowheads indicate the direction of transcription. Data were visualized using IGV software. (B) Quantitative ChIP-qPCR confirming binding of HNF-1β to the respective sites on the Fxr (Nr1h4) promoter in the adult kidney. Data shown are mean±SEM of three independent experiments. (C) Expression of Fxr (Nr1h4) mRNA transcripts in control kidneys and HNF-1β mutant cystic kidneys (n=3). *P<0.05.
FXR Protein Is Downregulated in Cyst Epithelium of HNF-1β Mutant Kidney
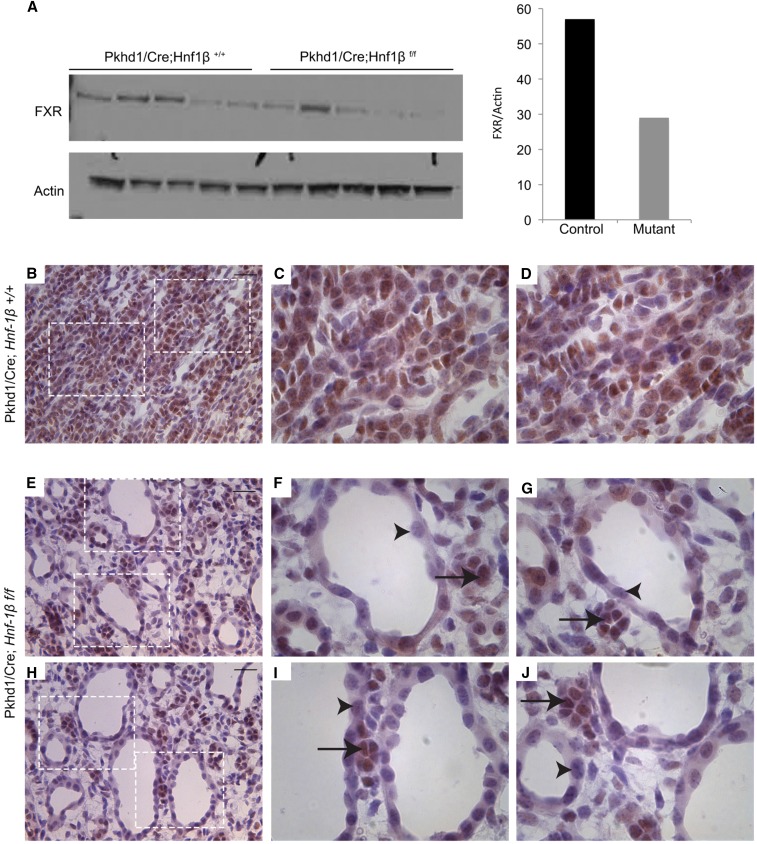
To determine whether the changes in Fxr (Nr1h4) mRNA were associated with corresponding changes in FXR protein, we performed western blot analysis and antibody staining. As shown previously,27 FXR was expressed in renal tubules in both the cortex and the medulla (Supplemental Figure 4, A–F). Minimal FXR was detected in glomeruli (Supplemental Figure 4, B and C). Western blot analysis showed that the expression of FXR protein was reduced in HNF-1β mutant kidneys compared with controls (Figure 5A). Antibody staining in control mice showed that FXR was located in the nuclei of tubular epithelial cells in the renal medulla (Figure 5, B–D). In mutant mice, FXR was almost undetectable in the epithelia lining the cysts but present in the surrounding noncystic tubules (Figure 5, E–J). FXR expression did not change in the proximal tubules of mutant kidneys, where HNF-1β expression was still maintained (Supplemental Figure 4, G–L). These data indicate that HNF-1β is required for the expression of FXR mRNA and protein in the CD.
Figure 5.
Loss of FXR expression in the cyst epithelium in HNF-1β mutant kidney. (A) Western blot analysis showing the expression of FXR in control kidneys (Pkhd1/Cre;Hnf1β+/+) and cystic kidneys (Pkhd1/Cre;Hnf1βf/f) at P35 (n=5). Right panel shows densitometric analysis of the western blots. FXR levels were normalized to the levels of actin. (B–J) Immunohistochemical staining for FXR on (B–D) kidney sections from control mice and (E–J) cystic kidneys from HNF-1β mutant mice. FXR staining is indicated in brown, and sections were counterstained with hematoxylin. All images were taken from the medullary region of the kidney. (F, G, I, and J) Higher magnification images show absence of staining in the cyst epithelium (arrowheads) and presence in surrounding noncystic tubules (arrows). Scale bars, 20 μm.
Discussion
In this study, we describe a new mouse model, in which HNF-1β was deleted from the CD of the mouse kidney. Unlike removal of HNF-1β throughout the nephron, which results in early cyst formation, renal failure, and postnatal lethality,13 CD-specific deletion leads to more slowly progressive cyst formation and long-term survival. HNF-1β mutant mice developed hydronephrosis as early as the first week of life. Hydronephrosis can be caused by obstruction of the urinary tract or excessive urine production that results in the dilation of the renal pelvis. When dye was injected into the kidneys of mutant mice, it appeared in the bladder, indicating that hydronephrosis was not due to complete obstruction of the urinary tract (data not shown). Consistent with a functional etiology, HNF-1β mutant mice exhibited polyuria due to defects in the ability to maximally concentrate urine. Even in the absence of overt hydronephrosis, mutant mice were persistently polyuric. In addition, polyuria and impaired urinary concentration persisted after 24 hours of water restriction, which suggests that polydipsia was not the cause of polyuria. Both control and mutant adult mice had similar plasma vasopressin levels, indicating that polyuria is not due to the complete absence of vasopressin. Administration of exogenous desmopressin raised urine osmolality; however, mutant mice had persistently more dilute urine compared with control mice. Collectively, these results indicate that HNF-1β mutant mice have partial nephrogenic diabetes insipidus and develop hydronephrosis as a consequence of polyuria. Hydronephrosis has previously been observed in polyuric mice with mutations in other genes affecting the urinary concentration mechanism (e.g., Taz and Nkcc2).30,31
Expression of Slc14a2, which encodes the UT-A family of renal urea transporters, was decreased in HNF-1β mutant kidneys. Inactivation of UT-A1 and UT-A3, two members of the family that are expressed in renal CD, produces urinary concentration defects.19 Thus, inhibition of Slc14a2 may contribute to the HNF-1β mutant phenotype. However, the mechanism of downregulation is not known, because transcription of Slc14a2 does not seem to be directly regulated by HNF-1β. Expression of another renal urea transporter, UT-B (Slc14a1), has been correlated with the miR-200 family of microRNAs, members of which are regulated by HNF-1β.18 The 3′ untranslated region of Slc14a1 contains a sequence that is complementary to the seed sequence of miR-200c, and UT-B and miR-200c show reciprocal expression in dehydrated mice, suggesting that miR-200c regulates levels of UT-B.32 HNF-1β regulates expression of miR-200b and miR-429, which contain the same seed sequence as miR-200c; however, it is unlikely that HNF-1β regulates tonicity-responsive expression of UT-B in the kidney, because HNF-1β is exclusively expressed in renal tubules whereas UT-B is primarily expressed in vasa recta.
We previously showed that the levels of cAMP and activity of CREB were elevated in HNF-1β mutant kidneys and cells.33 Here, we found that cAMP content was also increased in mice with CD-specific ablation of HNF-1β. Elevated cAMP would explain the increased expression of Aqp2 observed in HNF-1β mutant kidneys, because activated CREB stimulates transcription of Aqp2.20 Although the expression of Aqp2 was increased, confocal laser scanning microscopy revealed that AQP2 protein accumulated in the cytoplasm and basolateral region of HNF-1β mutant CD cells rather than the exclusively apical localization found in wild-type cells. Similar cytoplasmic and basolateral mislocalization has been observed in mice with nephrogenic diabetes insipidus due to missense mutations of AQP2 that affect protein trafficking.34 These findings suggest that urinary concentrating defects are present, despite increased abundance of AQP2 owing to impaired trafficking to the apical membrane. As a potential mechanism, we found decreased expression of Tmem27, an HNF-1β–regulated gene that encodes collectrin, a protein involved in apical vesicle exocytosis in the kidney.35 Collectrin directly binds to snapin and thereby, regulates soluble N-ethylmaleimide–sensitive factor attachment protein receptor complexes that mediate vesicle fusion with the plasma membrane.35,36 Collectrin also interacts with actin-myosin proteins that are involved in vesicle trafficking. Inhibiting collectrin reduces the plasma membrane expression of apical transport proteins in renal epithelial cells.37–39 Because vesicle fusion represents the final step in the insertion of AQP2 in the apical membrane, inhibition at this step could reduce the apical localization of AQP2, despite increased cAMP levels. It is possible that other HNF-1β–dependent genes that have not yet been identified also contribute to the observed defect in AQP2 trafficking.
In addition to the CD, abnormalities in the loop of Henle can produce defects in urinary concentration. We have previously identified an HNF-1 binding site in the promoter of the Slc12a1 gene, which encodes the apical NKCC2 in the TALH.40 Inactivation of Slc12a1 in mice produces polyuria and hydronephrosis,30 which is similar to the phenotype of HNF-1β mutant mice. However, Cre activity in HNF-1β mutant mice was absent from TALH, expression of Nkcc2 mRNA transcripts was unchanged, and immunostaining showed apical localization of NKCC2 protein in noncystic TALH. These findings indicate that polyuria observed in CD-specific HNF-1β mutant mice is not due to alterations in TALH structure or NKCC2 expression.
To further understand the mechanism underlying the defects in urinary concentration in HNF-1β mutant mice, we performed RNA-seq on HNF-1β mutant mIMCD3 cells treated with hypertonic NaCl. We identified 52 genes, 27 direct targets and 25 indirect targets, with regulation in response to hypertonicity that was dependent on HNF-1β. By comparing the lists of HNF-1β targets with the National Institutes of Health (NIH) Database of Osmoregulated Proteins in Mammalian Cells,41 we identified three direct gene targets (Plat, Cav1, and Loxl4) and two indirect targets (Bsnd and Zscan4c) that encode known osmoregulated proteins, further supporting the validity of the experimental approach. These studies are the first to implicate HNF-1β in the transcriptional response to hypertonicity. The levels of expression of HNF-1β itself were not altered in mIMCD3 cells subjected to hypertonic stress (data not shown), which suggests that HNF-1β might have a permissive effect on osmoregulatory gene expression. The transcription factor NFAT5/TonEBP is a key regulator of gene expression in response to osmotic stress.42 Although ChIP-seq identified a small HNF-1β binding peak near the Nfat5 gene, we were unable to confirm this result by ChIP-qPCR. Moreover, NFAT5 and several of its known targets (Slc6a12, Akr1b3, Sgk1, and Aqp1) were appropriately upregulated in both control cells and HNF-1β mutant cells in response to hypertonicity. In contrast, expression of another NFAT5-regulated gene, Slc14a2,43 was decreased in HNF-1β mutant mice. Two osmosensitive genes that we identified as direct targets of HNF-1β, Paqr5 and Cnnm1, are also known targets of NFAT5.44 Further studies will be needed to determine whether HNF-1β is needed for NFAT5-dependent gene regulation.
Of the 27 osmosensitive genes that were directly regulated by HNF-1β, Fxr (Nr1h4) showed the greatest difference in osmoregulation between wild-type and mutant cells. We presented multiple lines of evidence establishing that HNF-1β directly regulates the transcription of Fxr in renal CD. Previous studies have shown that the expression of FXR in the liver is regulated by HNF-1α45; however, regulation by HNF-1β has not been shown previously. The principal known function of FXR is to regulate bile acid metabolism in the liver and intestine.46 Although the functions of FXR in the kidney are less clear, a recent study showed that deficiency of FXR impairs urinary concentration.27 This phenotype was attributed to decreased expression of Aqp2, which was identified as an FXR target. In contrast, we found that the expression of Aqp2 was increased in HNF-1β mutant mice, despite downregulation of FXR, which suggests that additional FXR targets other than Aqp2 underlie the urinary concentrating defects in HNF-1β mutant mice.
We examined the published lists of genes that are known to be regulated by FXR47,48 and identified several genes other than Aqp2 that have been implicated in the response to hyperosmotic stress, including Aqp4, Il1b, Mapk14, and Eno1.49 Several of the osmosensitive genes that we identified as targets of HNF-1β are also known targets of FXR, including C3, Agt, Loxl4, P2rx7, Mgst1, and Plat. Comparison of the lists of known FXR-regulated genes with the NIH Database of Osmoregulated Proteins in Mammalian Cells identified additional genes that encode osmoregulated proteins (Supplemental Table 4). These results suggest that FXR regulates the expression of multiple osmosensitive genes that could contribute to the urinary concentrating defect in HNF-1β knockout mice.
Hydronephrosis and polyuria have been observed in humans with mutations of HNF1B.11 Proposed etiologies of polyuria include glycosuria due to early-onset diabetes mellitus and renal sodium and potassium wasting causing intrauterine polyuria and polyhydramnios. Here, we identify another possible mechanism involving impaired HNF-1β–dependent expression of FXR and UTA-1. In addition, patients with sporadic renal dysplasia frequently present with polyuria, nocturia, and enuresis, reflecting an impairment in urinary concentration. Because mutations of HNF-1β are among the more common genetic abnormalities seen in renal dysplasia, one possibility is that the downregulation of HNF-1β targets underlies the observed urinary abnormalities.
In summary, loss of HNF-1β in the renal CD produces defects in urinary concentration through multiple mechanisms, including directly inhibiting the transcription of osmosensitive genes, such as FXR; indirectly downregulating genes, such as the urea transporter UT-A1; and altering trafficking of AQP2. These findings reveal a novel role of HNF-1β in osmoregulation and expand the spectrum of renal disorders caused by inherited mutations in HNF-1β.
Concise Methods
Transgenic Mice
Pkhd1/Cre mice that express Cre recombinase under the control of the Pkhd1 promoter and Hnf1βf/f mice containing loxP sites flanking exon 1 of Hnf1β have been described previously.15,50 R26R-EYFP mice that express EYFP after Cre/loxP recombination were provided by Frank Costantini (Columbia University).51 Pkhd1/Cre mice were crossed with Hnf1βf/+ mice, and the bitransgenic progeny were intercrossed to generate Pkhd1/Cre;Hnf1βf/f mice (HNF-1β mutant mice). Cre-negative or Pkhd1/Cre;Hnf1βf/+ littermates were used as negative controls. The genetic background of the mice was C57BL/6, and animals of both sexes were studied. All experiments involving animals were performed under the auspices of the Institutional Animal Care and Research Advisory Committees at the University of Texas Southwestern and the University of Minnesota.
Cell Culture
mIMCD3 cells were obtained from ATCC and maintained in DMEM supplemented with 10% FCS. A cell line derived from mIMCD3 (53A cells) that expresses dominant negative mutant HNF-1β has been described previously3; 53A cells were seeded in triplicate and cultured for 24 hours to 70%–80% confluency. Cells were then treated with mifepristone to induce expression of dominant negative mutant HNF-1β or vehicle as a negative control. For chronic hypertonicity experiments, the medium was supplemented with 100 mM NaCl, and cells were cultured for an additional 48 hours before analysis.
Metabolic Studies
Mice were housed in a controlled 12-hour light/dark cycle in individual metabolic cages (Tecniplast) with free access to water and food. After 3 days of acclimation, urine was collected for 24 hours as a baseline. Mice were then water restricted for 24 hours with ad libitum access to food, and urine was collected for osmolality and solute measurements. Urine osmolality was measured using an osmometer (model 3D3; Advanced Instruments, Norwood, MA). Urine sodium, potassium, and urea were measured using a Vitros 250 chemistry analyzer. Blood was obtained by cardiac puncture, and plasma vasopressin was measured by RIA as described previously.52 Measurements of serum creatinine by capillary electrophoresis were performed by the University of Texas Southwestern O’Brien Kidney Research Core Center. In some experiments, desmopressin (0.4 μg/kg) was injected intraperitoneally after 24-hour water restriction, and urine was collected for 12 hours after injection. Animals with grossly apparent hydronephrosis were excluded from the analysis.
qPCR
Total RNA was extracted from cells or kidneys from adult wild-type or HNF-1β mutant mice using the RNeasy Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad), and qPCR was performed with the iTAG Universal SYBER Green Supermix (Bio-Rad) using the CFX Connect Real-Time System (Bio-Rad). Gene expression levels were normalized to 18S rRNA. Primers used for qRT-PCR are listed in Supplemental Table 5.
RNA Sequencing
TruSeq stranded mRNA libraries were made for each sample and sequenced in a 50-bp PE run on the HiSEq 2500 instrument using v4 chemistry by the University of Minnesota Genomics Center. Sequence data were trimmed (Trimmomatic, version 0.32), mapped to the GRCm38 build of mouse genome guided by ENSEMBL 84 annotations (STAR, version STAR_2.4.2a_modified), and sorted (Samtools version 1.0) using resources at the Minnesota Supercomputing Institute. Gene quantification and deferential expression were performed using the GenomicAlignments53 and DESeq254 packages in Bioconductor. Raw and processed data have deposited with NCBI GEO under accession number GSE85976.
Quantitative ChIP-qPCR
ChIP assays were performed using the EZ ChIP kit (Millipore) according to the manufacturer’s protocol. Briefly, mouse kidney tissue was crosslinked with 1% formaldehyde for 15 minutes at room temperature. Crosslinked tissues were homogenized into a single-cell suspension, and chromatin samples were extracted from the nuclei and sonicated. Immunoprecipitation was performed with 5 μg rabbit anti–HNF-1β (sc-22840; Santa Cruz) or rabbit IgG (sc-2027; Santa Cruz) as a negative control. Immunoprecipitated DNA or 1% of the input was diluted 1:20 in dH2O, and real-time PCR was performed in triplicate using gene-specific primers and SOSadvanced cyber green supermix (Bio Rad). ChIP-qPCR data were normalized using the percentage input method according to the following formula: 100×2((Ct(input) −6.644)− Ct(HNF-1β)). Primers used for ChIP-qPCR were Peak 1 (5′-AACCCTGAAACTGCATCACC-3′, 5′-GAAAGGGTTCCCCAAAATGT-3′) and Peak 2 (5′-TTCTCTGGGGTGAGATGAGG-3′, 5′-ACAAGACAGCCAGCAGACCT-3′).
Western Blot
Whole kidneys from control and mutant mice (n=5 each group) were homogenized, and proteins were extracted and separated by SDS-PAGE (25 μg protein per lane). Proteins were transferred to nitrocellulose membranes using the wet transfer method. Immunoblotting was performed using primary antibodies against FXR (1:200; Santa Cruz), AQP2 (1:500; Sigma Aldrich), UT-A1/2 (1:1000; Stressmarq Biosciences Inc.), and β-actin (1:5000; Santa Cruz) or GAPDH (1:10,000; Santa Cruz) as an internal control. Densitometric analysis was performed using a Bio-Rad gel analyzer and Quantity One 1-D analysis software. Band densities were normalized to β-actin or GAPDH.
Antibody Staining
Mice were euthanized, and kidneys were fixed by perfusion and incubation in 4% paraformaldehyde. Kidney tissue was embedded in paraffin, sectioned, and stained with hematoxylin and eosin or incubated overnight in 30% sucrose, frozen in OCT, and sectioned at 10 μm. For immunohistochemistry, cryosections were heated in heat-induced antigen retrieval buffer (RV100M; Biocare Medical) for 30 minutes, treated with 3% hydrogen peroxide, and blocked in 100% Rodent Block M (RBM961; Biocare Medical) for 1 hour. FXR primary antibody (sc-13063; 1:4000; Santa Cruz) was added to the slides in 10% Rodent Block and incubated overnight at 4°C. Slides were incubated in Rabbit-on-Rodent HRP-Polymer (RMR622; Biocare Medical) for 20 minutes and then treated with DAB (926603-buffer A; 926503-buffer B; Biolegend) for 5 minutes. Sections were counterstained with hematoxylin. For immunofluorescence, the following antibodies and lectins were used: HNF-1β (1:500; Santa Cruz), green fluorescent protein (1:400; Rockland), AQP2 (1:400; Sigma Aldrich), UT-A1/2 (1:500; Stressmarq Biosciences Inc.), NKCC2 (1:500; LifeSpan BioSciences), and fluorescein-labeled Lotus tetragonolobus agglutinin (FL-1321; 1:200; LTA; Vector Laboratories). Secondary antibodies were conjugated to Alexa Fluor 488 or Alexa Fluor 594 (1:400; Molecular Probes). Costaining of AQP2 and UT-A1/2 was performed using a tyramide amplification kit (HRP goat anti-rabbit IgG and Alexa Fluor 488 tyramide; Life Technologies). Nuclei were counterstained with 4′6-diamidino-2-phenylindol. Images were captured using a Leica DM5500 B upright microscope and DFC7000 T camera. Confocal laser scanning microscopy was performed by the Imaging Core of the University of Minnesota Center for Immunology with an Olympus FV1000 Confocal Microscope.
cAMP Assays
Content of AMP was measured using the Direct cAMP ELISA Kit from Enzo Life Sciences according to the manufacturer’s directions. Briefly, kidneys from control and mutant mice were snap frozen in liquid nitrogen and pulverized, and 0.1 mg tissue was resuspended in 1 ml 0.1 M HCl. Absorbance at 405 nm was measured using a plate reader. cAMP was measured in triplicate for each sample and normalized to protein content.
Statistical Analyses
Statistical analysis was performed using GraphPad Prism software. Two-tailed unpaired t test was used for pairwise comparisons. The Mantel–Cox test was used for survival analysis. P<0.05 was considered to be significant.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Jeff Sands (Emory University) and Chou-Long Huang (University of Texas Southwestern) for helpful discussions and Steffan Nye and Alan Mickelson for expert technical assistance. We also thank Jason Mitchell and Brian Fife of the University of Minnesota Center for Immunology for assistance with confocal laser scanning microscopy.
This work was supported by National Institutes of Health (NIH) grants K08DK084311 (to V.P.), R37DK049291 (to P.I.), and P30DK079328 (University of Texas Southwestern O’Brien Kidney Research Core Center). K.A. and L.N. were supported by NIH training grant T32DK007257.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016101095/-/DCSupplemental.
References
- 1.Igarashi P, Shao X, McNally BT, Hiesberger T: Roles of HNF-1beta in kidney development and congenital cystic diseases. Kidney Int 68: 1944–1947, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Hiesberger T, Shao X, Gourley E, Reimann A, Pontoglio M, Igarashi P: Role of the hepatocyte nuclear factor-1beta (HNF-1beta) C-terminal domain in Pkhd1 (ARPKD) gene transcription and renal cystogenesis. J Biol Chem 280: 10578–10586, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Gong Y, Ma Z, Patel V, Fischer E, Hiesberger T, Pontoglio M, Igarashi P: HNF-1beta regulates transcription of the PKD modifier gene Kif12. J Am Soc Nephrol 20: 41–47, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Z, Gong Y, Patel V, Karner CM, Fischer E, Hiesberger T, Carroll TJ, Pontoglio M, Igarashi P: Mutations of HNF-1beta inhibit epithelial morphogenesis through dysregulation of SOCS-3. Proc Natl Acad Sci USA 104: 20386–20391, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Pontoglio M, Hiesberger T, Sinclair AM, Igarashi P: Regulation of kidney-specific Ksp-cadherin gene promoter by hepatocyte nuclear factor-1beta. Am J Physiol Renal Physiol 283: F839–F851, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Massa F, Garbay S, Bouvier R, Sugitani Y, Noda T, Gubler MC, Heidet L, Pontoglio M, Fischer E: Hepatocyte nuclear factor 1β controls nephron tubular development. Development 140: 886–896, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Lokmane L, Heliot C, Garcia-Villalba P, Fabre M, Cereghini S: vHNF1 functions in distinct regulatory circuits to control ureteric bud branching and early nephrogenesis. Development 137: 347–357, 2010 [DOI] [PubMed] [Google Scholar]
- 8.Bellanné-Chantelot C, Chauveau D, Gautier JF, Dubois-Laforgue D, Clauin S, Beaufils S, Wilhelm JM, Boitard C, Noël LH, Velho G, Timsit J: Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann Intern Med 140: 510–517, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Decramer S, Parant O, Beaufils S, Clauin S, Guillou C, Kessler S, Aziza J, Bandin F, Schanstra JP, Bellanné-Chantelot C: Anomalies of the TCF2 gene are the main cause of fetal bilateral hyperechogenic kidneys. J Am Soc Nephrol 18: 923–933, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Heidet L, Decramer S, Pawtowski A, Morinière V, Bandin F, Knebelmann B, Lebre AS, Faguer S, Guigonis V, Antignac C, Salomon R: Spectrum of HNF1B mutations in a large cohort of patients who harbor renal diseases. Clin J Am Soc Nephrol 5: 1079–1090, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adalat S, Woolf AS, Johnstone KA, Wirsing A, Harries LW, Long DA, Hennekam RC, Ledermann SE, Rees L, van’t Hoff W, Marks SD, Trompeter RS, Tullus K, Winyard PJ, Cansick J, Mushtaq I, Dhillon HK, Bingham C, Edghill EL, Shroff R, Stanescu H, Ryffel GU, Ellard S, Bockenhauer D: HNF1B mutations associate with hypomagnesemia and renal magnesium wasting. J Am Soc Nephrol 20: 1123–1131, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiesberger T, Bai Y, Shao X, McNally BT, Sinclair AM, Tian X, Somlo S, Igarashi P: Mutation of hepatocyte nuclear factor-1beta inhibits Pkhd1 gene expression and produces renal cysts in mice. J Clin Invest 113: 814–825, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gresh L, Fischer E, Reimann A, Tanguy M, Garbay S, Shao X, Hiesberger T, Fiette L, Igarashi P, Yaniv M, Pontoglio M: A transcriptional network in polycystic kidney disease. EMBO J 23: 1657–1668, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verdeguer F, Le Corre S, Fischer E, Callens C, Garbay S, Doyen A, Igarashi P, Terzi F, Pontoglio M: A mitotic transcriptional switch in polycystic kidney disease. Nat Med 16: 106–110, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams SS, Cobo-Stark P, Hajarnis S, Aboudehen K, Shao X, Richardson JA, Patel V, Igarashi P: Tissue-specific regulation of the mouse Pkhd1 (ARPKD) gene promoter. Am J Physiol Renal Physiol 307: F356–F368, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer E, Legue E, Doyen A, Nato F, Nicolas JF, Torres V, Yaniv M, Pontoglio M: Defective planar cell polarity in polycystic kidney disease. Nat Genet 38: 21–23, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Aboudehen K, Kim MS, Mitsche M, Garland K, Anderson N, Noureddine L, Pontoglio M, Patel V, Xie Y, DeBose-Boyd R, Igarashi P: Transcription factor hepatocyte nuclear factor-1β regulates renal cholesterol metabolism. J Am Soc Nephrol 27: 2408–2421, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajarnis SS, Patel V, Aboudehen K, Attanasio M, Cobo-Stark P, Pontoglio M, Igarashi P: Transcription factor hepatocyte nuclear factor-1β (HNF-1β) regulates MicroRNA-200 expression through a long noncoding RNA. J Biol Chem 290: 24793–24805, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenton RA, Chou CL, Stewart GS, Smith CP, Knepper MA: Urinary concentrating defect in mice with selective deletion of phloretin-sensitive urea transporters in the renal collecting duct. Proc Natl Acad Sci USA 101: 7469–7474, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yasui M, Zelenin SM, Celsi G, Aperia A: Adenylate cyclase-coupled vasopressin receptor activates AQP2 promoter via a dual effect on CRE and AP1 elements. Am J Physiol 272: F443–F450, 1997 [DOI] [PubMed] [Google Scholar]
- 21.Machida K, Wakamatsu S, Izumi Y, Yosifovska T, Matsuzaki T, Nakayama Y, Kohda Y, Inoue T, Saito H, Tomita K, Nonoguchi H: Downregulation of the V2 vasopressin receptor in dehydration: Mechanisms and role of renal prostaglandin synthesis. Am J Physiol Renal Physiol 292: F1274–F1282, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Lanaspa MA, Andres-Hernando A, Rivard CJ, Dai Y, Berl T: Hypertonic stress increases claudin-4 expression and tight junction integrity in association with MUPP1 in IMCD3 cells. Proc Natl Acad Sci USA 105: 15797–15802, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Rodríguez C, Antos CL, Shelton JM, Richardson JA, Lin F, Novobrantseva TI, Bronson RT, Igarashi P, Rao A, Olson EN: Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc Natl Acad Sci USA 101: 2392–2397, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko BC, Ruepp B, Bohren KM, Gabbay KH, Chung SS: Identification and characterization of multiple osmotic response sequences in the human aldose reductase gene. J Biol Chem 272: 16431–16437, 1997 [DOI] [PubMed] [Google Scholar]
- 25.Ortells MC, Morancho B, Drews-Elger K, Viollet B, Laderoute KR, López-Rodríguez C, Aramburu J: Transcriptional regulation of gene expression during osmotic stress responses by the mammalian target of rapamycin. Nucleic Acids Res 40: 4368–4384, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umenishi F, Narikiyo T, Schrier RW: Effect on stability, degradation, expression, and targeting of aquaporin-2 water channel by hyperosmolality in renal epithelial cells. Biochem Biophys Res Commun 338: 1593–1599, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Huang S, Gao M, Liu J, Jia X, Han Q, Zheng S, Miao Y, Li S, Weng H, Xia X, Du S, Wu W, Gustafsson JA, Guan Y: Farnesoid X receptor (FXR) gene deficiency impairs urine concentration in mice. Proc Natl Acad Sci USA 111: 2277–2282, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber RM, Murphy K, Miao B, Link JR, Cunningham MR, Rupar MJ, Gunyuzlu PL, Haws TF, Kassam A, Powell F, Hollis GF, Young PR, Mukherjee R, Burn TC: Generation of multiple farnesoid-X-receptor isoforms through the use of alternative promoters. Gene 290: 35–43, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Kast-Woelbern HR, Edwards PA: Natural structural variants of the nuclear receptor farnesoid X receptor affect transcriptional activation. J Biol Chem 278: 104–110, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Takahashi N, Chernavvsky DR, Gomez RA, Igarashi P, Gitelman HJ, Smithies O: Uncompensated polyuria in a mouse model of Bartter’s syndrome. Proc Natl Acad Sci USA 97: 5434–5439, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, Nakagawa O, Small EV, Cobo-Stark P, Igarashi P, Murakami M, Tominaga J, Sato T, Asano T, Kurihara Y, Kurihara H: Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol 294: F542–F553, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Wang XH, Wang H, Chen L, Klein JD, Sands JM: Urea transporter B and MicroRNA-200c differ in kidney outer versus inner medulla following dehydration. Am J Med Sci 352: 296–301, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi YH, Suzuki A, Hajarnis S, Ma Z, Chapin HC, Caplan MJ, Pontoglio M, Somlo S, Igarashi P: Polycystin-2 and phosphodiesterase 4C are components of a ciliary A-kinase anchoring protein complex that is disrupted in cystic kidney diseases. Proc Natl Acad Sci USA 108: 10679–10684, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDill BW, Li SZ, Kovach PA, Ding L, Chen F: Congenital progressive hydronephrosis (cph) is caused by an S256L mutation in aquaporin-2 that affects its phosphorylation and apical membrane accumulation. Proc Natl Acad Sci USA 103: 6952–6957, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Wada J, Yasuhara A, Iseda I, Eguchi J, Fukui K, Yang Q, Yamagata K, Hiesberger T, Igarashi P, Zhang H, Wang H, Akagi S, Kanwar YS, Makino H: The role for HNF-1beta-targeted collectrin in maintenance of primary cilia and cell polarity in collecting duct cells. PLoS One 2: e414, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Fukui K, Yang Q, Cao Y, Takahashi N, Hatakeyama H, Wang H, Wada J, Zhang Y, Marselli L, Nammo T, Yoneda K, Onishi M, Higashiyama S, Matsuzawa Y, Gonzalez FJ, Weir GC, Kasai H, Shimomura I, Miyagawa J, Wollheim CB, Yamagata K: The HNF-1 target collectrin controls insulin exocytosis by SNARE complex formation. Cell Metab 2: 373–384, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Yasuhara A, Wada J, Malakauskas SM, Zhang Y, Eguchi J, Nakatsuka A, Murakami K, Kanzaki M, Teshigawara S, Yamagata K, Le TH, Makino H: Collectrin is involved in the development of salt-sensitive hypertension by facilitating the membrane trafficking of apical membrane proteins via interaction with soluble N-ethylmaleiamide-sensitive factor attachment protein receptor complex. Circulation 118: 2146–2155, 2008 [DOI] [PubMed] [Google Scholar]
- 38.Malakauskas SM, Quan H, Fields TA, McCall SJ, Yu MJ, Kourany WM, Frey CW, Le TH: Aminoaciduria and altered renal expression of luminal amino acid transporters in mice lacking novel gene collectrin. Am J Physiol Renal Physiol 292: F533–F544, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Danilczyk U, Sarao R, Remy C, Benabbas C, Stange G, Richter A, Arya S, Pospisilik JA, Singer D, Camargo SM, Makrides V, Ramadan T, Verrey F, Wagner CA, Penninger JM: Essential role for collectrin in renal amino acid transport. Nature 444: 1088–1091, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Igarashi P, Whyte DA, Li K, Nagami GT: Cloning and kidney cell-specific activity of the promoter of the murine renal Na-K-C1 cotransporter gene. J Biol Chem 271: 9666–9674, 1996 [DOI] [PubMed] [Google Scholar]
- 41.Grady CR, Knepper MA, Burg MB, Ferraris JD: Database of osmoregulated proteins in mammalian cells. Physiol Rep 2: e12180, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM: Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA 96: 2538–2542, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama Y, Peng T, Sands JM, Bagnasco SM: The TonE/TonEBP pathway mediates tonicity-responsive regulation of UT-A urea transporter expression. J Biol Chem 275: 38275–38280, 2000 [DOI] [PubMed] [Google Scholar]
- 44.Izumi Y, Yang W, Zhu J, Burg MB, Ferraris JD: RNA-Seq analysis of high NaCl-induced gene expression. Physiol Genomics 47: 500–513, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, Stoffel M: Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet 27: 375–382, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA: FXR regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res 47: 201–214, 2006 [DOI] [PubMed] [Google Scholar]
- 47.Zhan L, Liu HX, Fang Y, Kong B, He Y, Zhong XB, Fang J, Wan YJ, Guo GL: Genome-wide binding and transcriptome analysis of human farnesoid X receptor in primary human hepatocytes. PLoS One 9: e105930, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gai Z, Gui T, Hiller C, Kullak-Ublick GA: Farnesoid X receptor protects against kidney injury in uninephrectomized obese mice. J Biol Chem 291: 2397–2411, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brocker C, Thompson DC, Vasiliou V: The role of hyperosmotic stress in inflammation and disease. Biomol Concepts 3: 345–364, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coffinier C, Gresh L, Fiette L, Tronche F, Schütz G, Babinet C, Pontoglio M, Yaniv M, Barra J: Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1β. Development 129: 1829–1838, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F: Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bichet DG, Arthus MF, Barjon JN, Lonergan M, Kortas C: Human platelet fraction arginine-vasopressin. Potential physiological role. J Clin Invest 79: 881–887, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M: Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods 12: 115–121, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.