Abstract
MicroRNAs contribute to the development of kidney disease. Previous analyses of microRNA expression in human kidneys, however, were limited by tissue heterogeneity or the inclusion of only one pathologic type. In this study, we used laser-capture microdissection to obtain glomeruli and proximal tubules from 98 human needle kidney biopsy specimens for microRNA expression analysis using deep sequencing. We analyzed specimens from patients with diabetic nephropathy (DN), FSGS, IgA nephropathy (IgAN), membranoproliferative GN (MPGN) (n=19–23 for each disease), and a control group (n=14). Compared with control glomeruli, DN, FSGS, IgAN, and MPGN glomeruli exhibited differential expression of 18, 12, two, and 17 known microRNAs, respectively. The expression of several microRNAs also differed between disease conditions. Specifically, compared with control or FSGS glomeruli, IgAN glomeruli exhibited downregulated expression of hsa-miR-3182. Furthermore, in combination, the expression levels of hsa-miR-146a-5p and hsa-miR-30a-5p distinguished DN from all other conditions except IgAN. Compared with control proximal tubules, DN, FSGS, IgAN, and MPGN proximal tubules had differential expression of 13, 14, eight, and eight microRNAs, respectively, but expression of microRNAs did not differ significantly between the disease conditions. The abundance of several microRNAs correlated with indexes of renal function. Finally, we validated the differential glomerular expression of select microRNAs in a second cohort of patients with DN (n=19) and FSGS (n=21). In conclusion, we identified tissue-specific microRNA expression patterns associated with several kidney pathologies. The identified microRNAs could be developed as biomarkers of kidney diseases and might be involved in disease mechanisms.
Keywords: microRNA, glomerulus, proximal tubule, chronic kidney disease, kidney, biopsy
MicroRNAs are conserved, endogenous, small RNA molecules. The canonical role of microRNAs is to regulate the expression of their target proteins by mRNA degradation or translation repression.1–4 More than 2000 human microRNAs have been discovered,5 and it is estimated that they regulate more than 60% of protein-coding genes.6 microRNAs are known to regulate many cellular processes, including cell type differentiation, proliferation, migration, and survival, and play an active role in cellular physiology and pathophysiology.2,7,8
Many microRNAs are involved in the development or progression of chronic or acute kidney disease in patients or animal models.9 For example, miR-29 attenuates renal interstitial fibrosis.10–13 miR-382 and miR-21 contribute to renal interstitial fibrosis.14–16 miR-192 mediates the effect of TGF-β1 on glomerular mesangial cells and glomerular injury in diabetes.17,18 The miR-30 family protects against podocyte injury and proteinuria.19,20 miR-21 contributes to the protective effect of ischemic or xenon preconditioning on renal ischemia-reperfusion injury.21,22
In addition to playing mechanistic roles, microRNAs could serve as potential disease markers, which may contribute to the development of precision medicine.23 For example, a study of 112 patients with nonsmall-cell lung cancer showed a signature of five microRNAs that predicted the disease progression. Patients with a high risk score on the five-microRNA signature had poor overall and disease-free survival compared with patients with a low risk score.24 Another study of polyps from 109 patients undergoing colonoscopy screenings found microRNA signatures for four different histopathologies compared with normal mucosa.25
Several studies have searched for microRNA signatures in kidney disease. Kidney diseases can involve several types of pathologic changes in glomerular, tubulointerstitial, and/or vascular compartments. A study of several progressive kidney diseases involving 72 patients concluded that specific microRNAs were associated with kidney disease progression.26 In this study, patients were separated by disease progression rather than types of kidney disease. The study was done on mixed tissue containing all compartments of renal biopsy samples. Numerous studies have been done on allografts looking for microRNA signatures for transplant rejection, AKI, and tubulointerstitial fibrosis.27–29 These studies analyzed mixed tissue samples from up to 12 patients.
In this study, laser-capture microdissection (LCM) was used to obtain glomeruli and proximal tubules from human needle kidney biopsy specimens for microRNA expression analysis using deep sequencing. Nearly 100 patients with four different types of kidney diseases including diabetic nephropathy (DN), FSGS, IgA nephropathy (IgAN), and membranoproliferative GN (MPGN), and a control group with minimal kidney injury were analyzed. The analysis identified several microRNAs that were differentially expressed specifically in the glomeruli or proximal tubules in the four types of kidney disease. Select findings were validated in a second cohort of 40 patients.
Results
General Characteristics of MicroRNA Expression in Human Glomeruli and Proximal Tubules
LCM was used to separately collect glomeruli and proximal tubules from archived kidney biopsy specimens of 84 patients with DN, FSGS, IgAN, or MPGN, and 14 controls. Supplemental Figure 1 shows representative pictures of the structures captured. The purity of the glomerular and proximal tubule sections was verified by quantitative PCR (qPCR) analysis of marker genes of different nephron segments (Supplemental Figure 2).
A total of 191 samples were used for small RNA deep sequencing, 94 samples from glomeruli and 97 samples from proximal tubules (Supplemental Table 1). Samples with fewer than 10,000 reads mapped to microRNAs were excluded from the data analysis (Supplemental Table 1). On average, each sample had 1.1 million mapped reads and a 46% mapping rate (Supplemental Table 2). Demographic and clinical characteristics of the patients analyzed are summarized in Supplemental Table 3.
We detected 205 known unique mature human microRNAs from the glomeruli and proximal tubules. In addition, 223 novel unique human microRNA candidates were identified. Finally, 66 unique mature homolog microRNAs corresponding to known microRNAs in nonhuman species were detected. The original read count for each microRNA in all samples are available in Supplemental Material.
MicroRNA abundance profiles detected by the deep sequencing analysis of laser-captured formalin-fixed, paraffin-embedded (FFPE) tissues were reproducible across samples of the same tissue type (Supplemental Figure 3). The correlation of microRNA profiles in glomerular and proximal tubular samples was lower, as expected (Supplemental Figure 3).
The top 5% most abundant known human microRNAs detected in glomeruli or proximal tubules on the basis of the average of all samples analyzed are shown in Supplemental Table 4. These microRNAs were largely consistent with the most abundant microRNAs found in our previous laser-capture study of rat glomeruli (Supplemental Table 4).30 The top 5% most abundant microRNAs in pathologically normal glomeruli or proximal tubules on the basis of the average of control patients or in each individual control patient are shown in Supplemental Table 5.
MicroRNAs Differentially Expressed in the Glomeruli
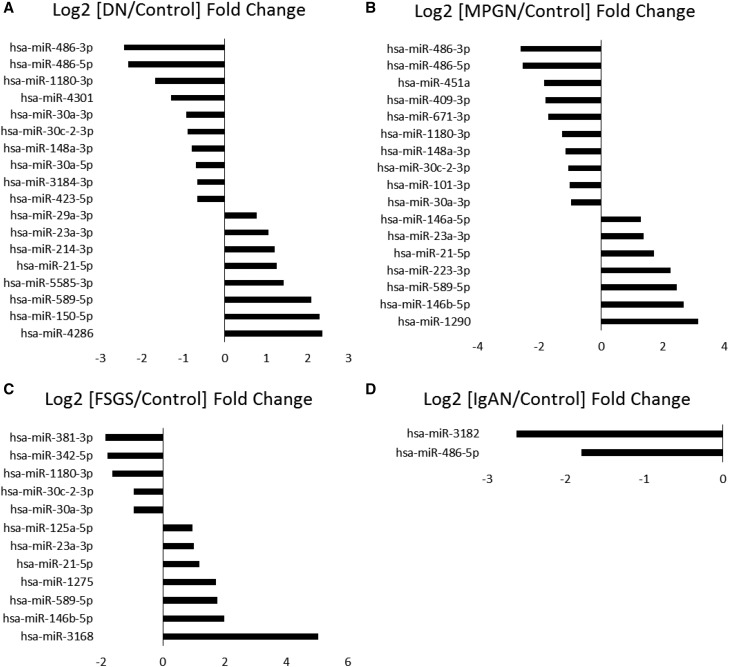
Adjusted P values of <0.05 from an edgeR2 analysis were used to identify differentially expressed microRNAs. In the glomeruli, 21 microRNAs were differentially expressed between control and DN groups, 18 of which were known human microRNAs. Seventeen microRNAs (including 12 known human microRNAs) were differentially expressed between control and FSGS groups. The comparison between control and IgAN groups yielded only five differentially expressed microRNAs, two of which were human microRNAs. Lastly, 25 microRNAs (including 17 known human microRNAs) were differentially expressed between the control group and the MPGN group. The known human microRNAs differentially expressed in the glomeruli are shown in Figure 1. The upregulation of miR-21-5p in the DN group and miR-146b-5p in the MPGN group compared with controls was verified by qPCR analysis of the original RNA samples (Supplemental Figure 4).
Figure 1.
Several microRNAs are differentially expressed in the glomeruli between controls and each type of kidney disease. (A) DN. (B) MPGN. (C) FSGS. (D) IgAN. Log2 fold changes of each disease over controls for microRNAs with FDR<0.05 are shown.
The sample size of the control group is not sufficient for full stratification by sex or race. However, we recalculated the statistical significance of the control versus DN group using only male or white patients. As one would expect from smaller sample sizes, fewer microRNAs reached significance compared with the analysis of entire groups. Nevertheless, all microRNAs identified in men only or whites only as differentially expressed between control and DN groups (Supplemental Figure 5) overlapped with microRNAs identified from entire groups.
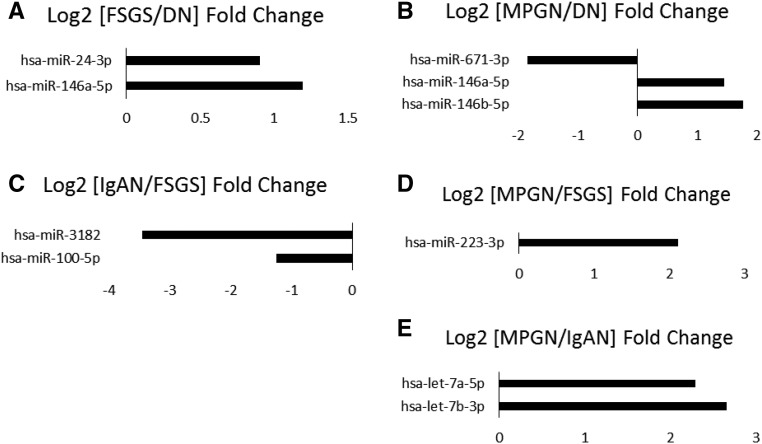
The comparison between disease conditions yielded fewer differentially expressed microRNAs, with only two known human microRNAs differentially expressed between DN and FSGS, FSGS and IgAN, and IgAN and MPGN groups, one differentially expressed between FSGS and MPGN groups, and none between DN and IgAN groups (Figure 2).
Figure 2.
Several microRNAs are differentially expressed in the glomeruli between different types of kidney disease. (A) FSGS compared with DN. (B) MPGN compared with DN. (C) IgAN compared with FSGS. (D) MPGN compared with FSGS. (E) MPGN compared with IgAN. Log2 fold changes for microRNAs with FDR<0.05 are shown.
MicroRNAs from the miR-30-3p family were significantly downregulated in the glomeruli of patients with DN, MPGN, and FSGS but not in patients with IgAN. hsa-miR-30a-5p was only significantly downregulated in DN patients compared with controls. hsa-miR-146a-5p was upregulated in patients with FSGS and MPGN compared with patients with DN. hsa-miR-3182 was significantly downregulated only in patients with IgAN compared with controls and patients with FSGS. hsa-miR-21-5p was upregulated in patients with DN, FSGS, and MPGN but not IgAN compared with controls. hsa-miR-486-5p and hsa-miR486-3p were downregulated in patients with DN and MPGN, whereas patients with IgAN showed only a downregulation of hsa-miR486-5p. Neither hsa-miR-486-5p nor hsa-miR486-3p was downregulated in FSGS. Lastly, there was a five-fold upregulation of hsa-miR-3168 in patients with FSGS compared with controls.
MicroRNAs Differentially Expressed in the Proximal Tubules
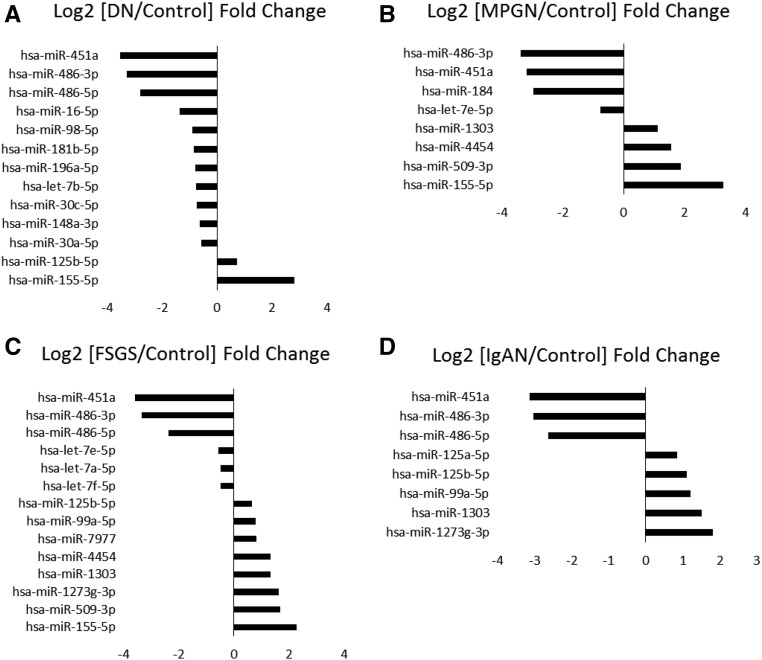
The proximal tubule samples showed several microRNAs differentially expressed when comparing controls with each disease condition. A total of 18 microRNAs, 13 of which were known human microRNAs, were differentially expressed between the control group and the DN group. A total of 23 microRNAs were differentially expressed between control and FSGS groups, 14 of which were known human microRNAs. Eleven microRNAs (including eight known human microRNAs) were differentially expressed between control and IgAN groups. Finally, all eight differentially expressed microRNAs between control and MPGN groups in the proximal tubules samples were known human microRNAs. The known human microRNAs differentially expressed in the proximal tubules are shown in Figure 3.
Figure 3.
Several microRNAs are differentially expressed in the proximal tubules between controls and each type of kidney disease. (A) DN. (B) MPGN. (C) FSGS. (D) IgAN. Log2 fold changes of each disease over controls for microRNAs with FDR<0.05 are shown.
hsa-miR-486-3p in the proximal tubules was downregulated by about three-fold in all diseases compared with controls, and hsa-miR486-5p was downregulated in all diseases compared with controls with the exception of MPGN. Another microRNA downregulated by nearly three-fold in all disease conditions compared with controls was hsa-miR-451a. MicroRNAs of the hsa-let-7 family were downregulated in patients with FSGS compared with controls, and hsa-let-7e-5p was downregulated in patients with MPGN compared with controls. The hsa-miR-30-5p family was only downregulated in DN compared with controls. Lastly, hsa-miR-125b-5p was upregulated in DN, FSGS, and IgAN compared with controls. IgAN also exhibited upregulation of hsa-miR-125a-5p compared with controls.
We recalculated the statistical significance of the control versus DN groups using only male patients and only white patients. Similar to the findings in glomeruli, fewer microRNAs reached significance compared with the analysis of entire groups. Nevertheless, four of six microRNAs identified in proximal tubules of men only (Supplemental Figure 5) overlapped with microRNAs identified from entire groups.
In contrast with the findings in the glomeruli, none of the microRNAs detected in the proximal tubules was differentially expressed between any two of the four types of kidney disease analyzed.
Correlation of MicroRNAs with Clinical Phenotypes
The correlation with microRNA abundance was examined for the following clinical parameters at the time of the biopsy, with all groups of patients combined: BP (systolic and diastolic), BUN, serum creatinine, eGFR, and urine protein excretion. The coefficients of regression are shown in Supplemental Figure 6. At the significance level of false discovery rate (FDR) <0.05, miR-148a-3p in proximal tubules was inversely correlated with BUN, and miR-155-5p and miR-182-5p were positively correlated with serum creatinine. At FDR<0.10, miR-21-5p in both glomeruli and proximal tubules was inversely correlated with eGFR. Several additional microRNAs were correlated with renal function or diastolic BP at FDR<0.20.
Inverse Correlations between MicroRNAs and mRNAs in the Glomeruli
mRNA expression profiles, assessed by microarray analysis, have been reported for glomeruli and tubulointerstitial fragments in 157 patients representing nine kidney diseases.31 Comparison of the microRNA data obtained from the glomeruli in this study with the previously reported mRNA data indicated several pairs of inversely and significantly differentially expressed microRNAs and predicted target mRNAs (Supplemental Figure 7). miR-21-5p was upregulated in the glomeruli of patients with FSGS, whereas its predicted target gene, KIF11, was downregulated in FSGS. miR-381-3p was downregulated in FSGS, whereas 18 of its predicted target genes were upregulated in FSGS. miR-24-3p was upregulated in FSGS compared with DN, whereas nine of its predicted target genes were downregulated.
We tested multiple qPCR primer pairs for several mRNAs, shown in Supplemental Figure 7. Only one pair of primers for SERPINH1 allowed amplification without dimerization. SERPINH1 tended to be downregulated in FSGS compared with DN in both the original sample set and validation sample set (Supplemental Figure 8), which was consistent with what was shown in Supplemental Figure 7. The downregulation, however, did not reach statistical significance and the data variability was large. The difficulty of mRNA analysis in these samples was likely because mRNA was fragmented in FFPE tissues, which was compounded by the small amount of RNA available from LCM samples.
Validation in a Second Cohort of Patients
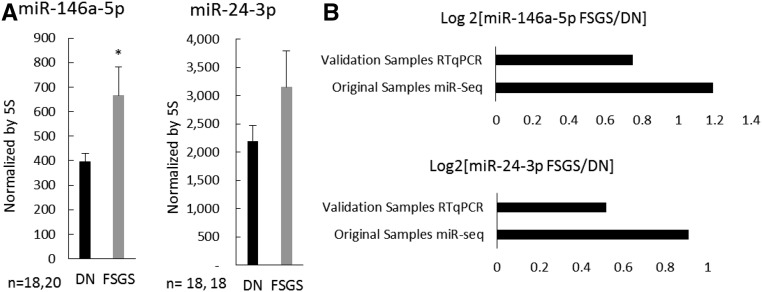
We carried out a study to examine if the microRNA differential expression identified in this study could be validated in another cohort of patients. We obtained biopsy samples from 19 additional patients with DN and 21 additional patients with FSGS, and collected glomeruli by LCM as we did in the original study. We performed qPCR analysis for miR-146a-5p and miR-24-3p on this set of validation samples. The results indicated miR-146a-5p abundance was significantly higher in FSGS compared with DN in the validation samples (Figure 4), which was consistent with findings from microRNA-sequencing analysis of the original samples. miR-24-3p abundance tended to be higher in FSGS (Figure 4), which again, was consistent with findings from the original samples, although the difference did not reach statistical significance in the validation sample set.
Figure 4.
Differential expression of select miRNAs was validated in a second cohort of patients. (A) Abundance of miR-146a-5p and miR-24-3p in glomerular samples from additional patients with DN and FSGS on the basis of qPCR analysis. (B) Log2 fold differences of FSGS/DN from microRNA-sequencing analysis of original samples and qPCR analysis of new validation samples. *P<0.05; P=0.17 for miR-24-3p.
Discussion
In this study, we performed microRNA deep sequencing in glomeruli and proximal tubules obtained by LCM from kidney biopsy specimens of nearly 100 patients, including four different types of kidney pathology. We identified several microRNAs that were differentially expressed in DN, FSGS, IgAN, and MPGN, respectively, compared with controls in the glomeruli or the proximal tubules. A small number of microRNAs in the glomeruli, but none in the proximal tubules, were differentially expressed between types of disease.
Our study is the first to use a tissue type–specific approach to analyze microRNA expression profiles in a large cohort of patient samples with different kidney diseases. The tissue specificity allows us to differentiate microRNAs that are altered specifically in the glomeruli or the proximal tubules for each disease. MicroRNA expression is highly tissue specific. Analysis of whole kidney biopsy specimens risks missing differential expression of microRNAs that might occur in a specific tissue region. The small quantity of the tissues microdissected from kidney biopsy specimens posed a technical challenge for genome-scale next-generation sequencing analysis, as did the FFPE present in archived biopsy specimens. We were able to modify the small RNA deep sequencing methods to obtain microRNA expression profile data from glomeruli and proximal tubules laser-captured from FFPE kidney biopsy specimens. The data appear to be of high quality, as indicated by the consistency between samples of the same tissue type and with animal model studies, verification of differential expression by qPCR in original samples, and validation of select findings in a second cohort of patients.
The microRNAs identified in this study could be developed as biomarkers of specific kidney disease in future studies utilizing additional, larger cohorts of patients. In that regard, it is interesting to note that we observed several microRNAs as differentially expressed between different types of kidney disease in the glomeruli, but not in the proximal tubules. We did observe differences in the proximal tubules between the control and each of the four diseases. The findings suggest these diseases might have specific effects on microRNAs in the glomeruli, which could be developed as disease-specific biomarkers. On the other hand, although the tubules were affected in these diseases, the molecular changes in the tubules at the stage of disease progression when the biopsy was performed might primarily result from pathologic changes common to all four diseases, such as increased filtration of proteins. The lack of differentially expressed microRNAs between diseases in the proximal tubule, therefore, could be paradoxical support for the quality of our data as it is consistent with the fact that the kidney diseases studied are pathologically distinct primarily in the glomeruli.
Several findings of this study are consistent with previous findings. miR-21 is known to be upregulated by TGF-β1 and suppress PTEN, leading to collagen deposition and fibrosis.9,32–34 We observed upregulation of miR-21-5p in the glomeruli from patients with DN, FSGS, or MPGN, and apparently an inverse correlation between miR-21-5p in both glomeruli and proximal tubules with eGFR. Numerous studies have reported miR-30 family microRNAs, which are highly abundant in the glomeruli and particularly the podocytes, to be downregulated in glomerular diseases.19,20,35,36 We also found downregulation of miR-30 family microRNAs in the glomeruli of patients with DN, FSGS, or MPGN. An mRNA profiling study in nine types of kidney diseases suggested a close interconnection between lupus nephritis, IgAN, and DN.31 Our study is consistent with this finding as we did not find any microRNAs differentially expressed between IgAN and DN.
This study found a significant downregulation of miR-486 in either the glomeruli, proximal tubules, or both, in all types of kidney pathologies compared with controls, particularly miR-486-3p. miR-486 has been reported to protect against ischemia-reperfusion injury in mouse kidneys. miR-486 targets PTEN and was found to be enriched in exosomes extracted from human cord blood. Protection against ischemic injury was achieved by infusion of exosomes.37 miR-486 has also been found to be protective against different types of cancer,38–41 including renal carcinoma, for which it has been proposed as a prognostic marker after nephrectomy in advanced renal cell carcinoma.38
Several of the microRNAs identified in this study have not been examined in kidney disease and could represent new disease mechanisms worthy of further investigation. For example, hsa-miR-589-5p was upregulated in the glomeruli of patients with DN, MPGN, or FSGS. A study in peritoneal mesothelial cells reported that miR-589 mediates TGF-β1-induced epithelial-mesenchymal transition.42 The functional role of miR-589 in the kidney is unknown. miR-3182 was downregulated only in the glomeruli of IgAN. miR-3168 was the most abundant microRNA we found in the glomeruli and proximal tubules, and it was upregulated by five-fold in the glomeruli of patients with FSGS compared with controls. The functions of these microRNAs are unknown and warrant further investigation.
In summary, we have identified several microRNAs that are differentially expressed specifically in glomeruli or proximal tubules in four types of kidney diseases in a cohort of nearly 100 patients. Select findings were validated in a second cohort of 40 patients. The identified microRNAs could be developed as biomarkers of kidney diseases and might be involved in the disease mechanisms.
Concise Methods
Selection of Biopsy Specimens
The Institutional Review Board at the Medical College of Wisconsin approved the use of human samples from kidney biopsies for the experiments outlined in this study. The kidney biopsy archive at the Medical College of Wisconsin was searched for patients with DN, FSGS, MPGN, and IgAN diagnosed between 1980 and 2014. Samples from patients who had previously received a kidney transplant were excluded. Each case was reviewed by a research pathologist to confirm that the biopsy specimen demonstrated the appropriate pathologic changes for the disease category and did not demonstrate significant changes characteristic of other diseases. The control group consisted of pathologically normal kidney tissues. Most of the controls were uninvolved tissues from partial nephrectomies of patients with renal masses. Other patient types included were a patient with proteinuria, a patient with a gunshot wound, and a transplant donor kidney. Again, all control samples were without any significant pathologic findings.
LCM
Laser capture was performed with a Zeiss P.A.L.M. Microbeam III LCM system (Carl Zeiss Microscopy) housed at the Children’s Research Institute Imaging Core. FFPE samples were cut into 10-μm sections and lightly stained with eosin. The laser capture microscope is equipped with an infrared laser and a computer. Glomeruli or proximal tubules were outlined with the drawing tool on the computer view of the field. The laser cut the selected structures, following the drawn outline, and blew the cut sections off the slide by a higher energy laser pulse, to be captured onto a 200-μl AdhesiveCap tube (Carl Zeiss Microscopy) placed over the sample area. Approximately 200,000–400,000 μm2 of each tissue type were collected from each sample.30
Small RNA Deep Sequencing
Qiagen’s AllPrep DNA/RNA FFPE kit was used following the manufacturer’s instructions for extraction of total RNA from laser-captured samples. The manufacturer’s modifications for extraction of small RNA were followed and 20 µl DEPC water were used for final elution of RNA. The RNA concentration was too low to quantify by nanodrop. A standard curve for qPCR analysis of 5S rRNA was used to confirm the presence of RNA in a few samples. A total of 5 µl extracted RNA from each sample was used for library preparation. Illumina’s TruSeq Small RNA Library Preparation Kits were used following the manufacturer’s instructions, with modifications for preparation of sequencing libraries. Optimization of library preparation was performed with two samples, and 19 cycles of PCR were selected and subsequently used for amplification in all samples. Four groups of 48 samples each were made, utilizing all 48 unique adapters in the library kits (one sample from a different study was sequenced along with this study to complete the fourth 48 sample group, maintaining the number of samples multiplexed each time in this study). The quantity and size distribution of the libraries were confirmed with a DNA 1000 chip in an Agilent 2000 Bioanalyser. Sequencing was performed on an Illumina HiSEquation 2000 sequencer.30
Analysis of Small RNA Deep Sequencing Data
Sequencing data analysis was performed using an in-house analytical pipeline essentially as described previously.30,43 Briefly, trimmed sequencing reads were mapped to miRbase for identification of known microRNAs using Bowtie. Remaining sequences were mapped against the mRNA database, Rfamand RepBase, to remove sequences not corresponding to microRNAs. miRanalyser was then used to predict new microRNAs. To normalize and test differential expression, we used number of reads of known and newly identified microRNAs as input for the Bioconductor edgeR package. edgeR uses a negative binomial distribution to model reads of microRNAs and to test for differential expression in deep sequencing datasets. The Benjamini–Hochberg method was used to control FDR in all statistical tests.
Real-Time PCR
An aliquot of the glomerular RNA samples used for microRNA library preparation was used to validate the sequencing results by qPCR. TaqMan microRNA assays specific for hsa-miR-21-5p, miR-146a-5p, miR-146b-5p, miR-24-3p, and SERPINH1 primers were used in an Applied Biosystems real-time PCR instrument, using 5S rRNA as normalizer.44 The purity of the glomerular and proximal tubule samples was verified by qPCR using specific primers for marker genes of different nephron segments. We used GLUT2 as a marker for proximal tubules, NPHS2 (podocin) as a glomerular marker, SLC8A1 (Na+/Ca++ exchanger) for the distal tubules, and AQP2 for the collecting ducts. SYBR Green dye was used for detection and 18S rRNA was used as a normalizer.
Correlation with Clinical Phenotypes
Clinical data were obtained by review of patients’ charts from the time of biopsy and pathology reports archived with the biopsy specimens. Clinical data were first log transformed followed by regression analysis to estimate relationships between microRNAs and clinical observations. The Benjamini–Hochberg procedure was used for controlling FDRs in multiple comparisons. A regression coefficient was obtained, which indicated how much a certain risk factor was expected to increase (if the coefficient was positive) or decrease (if the coefficient was negative) when the abundance of the significant microRNA increased by one.
Correlation with mRNA Data
Microarray-based mRNA expression data in glomeruli for three conditions that overlapped with this study (control, DN and FSGS) were downloaded from the GEO datasets (GSE47183).31 mRNAs differentially expressed between control and FSGS or DN and FSGS groups were detected by using two-sample t tests. We only compared control and FSGS or DN and FSGS groups because these were the only comparisons for which comparable microarray data were available. The Benjamini–Hochberg procedure was used for controlling FDRs in multiple comparisons (FDR<0.05). TargetScanHuman (v7.1) was used to identify predicted targets of microRNAs in glomeruli found to be differentially expressed (FDR<0.05) between control and FSGS or DN and FSGS groups in this study. Overlaps between differentially expressed mRNAs and predicted targets of differentially expressed microRNAs were identified. Finally, we determined whether the abundance of the identified mRNAs and microRNAs changed in opposite directions between the two conditions compared.
Disclosures
None.
Supplementary Material
Acknowledgments
M.A.B., S.J.D., A.M.W., K.A.I., S.T.G., K.B., K.R.R., and Y.L. performed the experiments and collected the data. P.L., X.P., and M.A.B. analyzed the data. M.L. designed and led the study. M.A.B. and M.L. drafted the paper. All authors edited and approved the paper.
This study was supported by National Institutes of Health grants HL121233, HL082798-6186, HL125409, GM066730, and DK098104.
An abstract of this work was presented at the American Society of Nephrology Kidney Week 2016 in Chicago, IL, November 15–20, 2016.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016121280/-/DCSupplemental.
References
- 1.Couzin J: Breakthrough of the year. Small RNAs make big splash. Science 298: 2296–2297, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Meister G: miRNAs get an early start on translational silencing. Cell 131: 25–28, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Couzin J: MicroRNAs make big impression in disease after disease. Science 319: 1782–1784, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Liang M, Liu Y, Mladinov D, Cowley AW Jr, Trivedi H, Fang Y, Xu X, Ding X, Tian Z: MicroRNA: A new frontier in kidney and blood pressure research. Am J Physiol Renal Physiol 297: F553–F558, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozomara A, Griffiths-Jones S: miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomes CP, Cho JH, Hood L, Franco OL, Pereira RW, Wang K: A review of computational tools in microRNA discovery. Front Genet 4: 81, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makeyev EV, Maniatis T: Multilevel regulation of gene expression by microRNAs. Science 319: 1789–1790, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M: The miR-29 family: Genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 44: 237–244, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trionfini P, Benigni A, Remuzzi G: MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 11: 23–33, 2015 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW Jr, Ferreri NR, Yeo NC, Liang M: Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension 55: 974–982, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin W, Chung AC, Huang XR, Meng XM, Hui DS, Yu CM, Sung JJ, Lan HY: TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol 22: 1462–1474, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, Koh P, Thomas M, Jandeleit-Dahm K, Gregorevic P, Cooper ME, Kantharidis P: Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol 23: 252–265, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Yu X, Liu Y, Kriegel AJ, Heng Y, Xu X, Liang M, Ding X: miR-29c is downregulated in renal interstitial fibrosis in humans and rats and restored by HIF-α activation. Am J Physiol Renal Physiol 304: F1274–F1282, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M: MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: A novel role of miR-382. Nucleic Acids Res 38: 8338–8347, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, Li J, Tran PT, Kaimal V, Huang X, Chang AN, Li S, Kalra A, Grafals M, Portilla D, MacKenna DA, Orkin SH, Duffield JS: MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 4: 121ra18, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kriegel AJ, Liu Y, Cohen B, Usa K, Liu Y, Liang M: MiR-382 targeting of kallikrein 5 contributes to renal inner medullary interstitial fibrosis. Physiol Genomics 44: 259–267, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R: MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A 104: 3432–3437, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R: TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11: 881–889, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramezani A, Devaney JM, Cohen S, Wing MR, Scott R, Knoblach S, Singhal R, Howard L, Kopp JB, Raj DS: Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: A pilot study. Eur J Clin Invest 45: 394–404, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Zheng C, Wang X, Yun S, Zhao Y, Liu L, Lu Y, Ye Y, Zhu X, Zhang C, Shi S, Liu Z: MicroRNA-30 family members regulate calcium/calcineurin signaling in podocytes. J Clin Invest 125: 4091–4106, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Kriegel AJ, Liu Y, Usa K, Mladinov D, Liu H, Fang Y, Ding X, Liang M: Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int 82: 1167–1175, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia P, Teng J, Zou J, Fang Y, Zhang X, Bosnjak ZJ, Liang M, Ding X: miR-21 contributes to xenon-conferred amelioration of renal ischemia-reperfusion injury in mice. Anesthesiology 119: 621–630, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotchen TA, Cowley AW Jr, Liang M: Ushering hypertension into a new era of precision medicine. JAMA 315: 343–344, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC: MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13: 48–57, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Tsikitis VL, Potter A, Mori M, Buckmeier JA, Preece CR, Harrington CA, Bartley AN, Bhattacharyya A, Hamilton SR, Lance P, Thompson PA: MicroRNA signatures of colonic adenomas according to histology. Cancer Prev Res (Phila) 9: 942–949, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudnicki M, Perco P, D Haene B, Leierer J, Heinzel A, Mühlberger I, Schweibert N, Sunzenauer J, Regele H, Kronbichler A, Mestdagh P, Vandesompele J, Mayer B, Mayer G: Renal microRNA- and RNA-profiles in progressive chronic kidney disease. Eur J Clin Invest 46: 213–226, 2016 [DOI] [PubMed] [Google Scholar]
- 27.Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M: MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A 106: 5330–5335, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Dov IZ, Muthukumar T, Morozov P, Mueller FB, Tuschl T, Suthanthiran M: MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation 94: 1086–1094, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilflingseder J, Sunzenauer J, Toronyi E, Heinzel A, Kainz A, Mayer B, Perco P, Telkes G, Langer RM, Oberbauer R: Molecular pathogenesis of post-transplant acute kidney injury: Assessment of whole-genome mRNA and miRNA profiles. PLoS One 9: e104164, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kriegel AJ, Liu Y, Liu P, Baker MA, Hodges MR, Hua X, Liang M: Characteristics of microRNAs enriched in specific cell types and primary tissue types in solid organs. Physiol Genomics 45: 1144–1156, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martini S, Nair V, Keller BJ, Eichinger F, Hawkins JJ, Randolph A, Böger CA, Gadegbeku CA, Fox CS, Cohen CD, Kretzler M; European Renal cDNA Bank; C-PROBE Cohort; CKDGen Consortium : Integrative biology identifies shared transcriptional networks in CKD. J Am Soc Nephrol 25: 2559–2572, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato M, Arce L, Natarajan R: MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol 4: 1255–1266, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudnicki M, Beckers A, Neuwirt H, Vandesompele J: RNA expression signatures and posttranscriptional regulation in diabetic nephropathy. Nephrol Dial Transplant 30[Suppl 4]: iv35–iv42, 2015 [DOI] [PubMed] [Google Scholar]
- 34.Bhatt K, Kato M, Natarajan R: Mini-review: Emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol 310: F109–F118, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP: Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol 19: 2159–2169, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Zheng C, Fan Y, Zeng C, Chen Z, Qin W, Zhang C, Zhang W, Wang X, Zhu X, Zhang M, Zen K, Liu Z: Downregulation of microRNA-30 facilitates podocyte injury and is prevented by glucocorticoids. J Am Soc Nephrol 25: 92–104, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viñas JL, Burger D, Zimpelmann J, Haneef R, Knoll W, Campbell P, Gutsol A, Carter A, Allan DS, Burns KD: Transfer of microRNA-486-5p from human endothelial colony forming cell-derived exosomes reduces ischemic kidney injury. Kidney Int 90: 1238–1250, 2016 [DOI] [PubMed] [Google Scholar]
- 38.Goto K, Oue N, Shinmei S, Sentani K, Sakamoto N, Naito Y, Hayashi T, Teishima J, Matsubara A, Yasui W: Expression of miR-486 is a potential prognostic factor after nephrectomy in advanced renal cell carcinoma. Mol Clin Oncol 1: 235–240, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma X, Wei J, Zhang L, Deng D, Liu L, Mei X, He X, Tian J: miR-486-5p inhibits cell growth of papillary thyroid carcinoma by targeting fibrillin-1. Biomed Pharmacother 80: 220–226, 2016 [DOI] [PubMed] [Google Scholar]
- 40.Ye H, Yu X, Xia J, Tang X, Tang L, Chen F: MiR-486-3p targeting ECM1 represses cell proliferation and metastasis in cervical cancer. Biomed Pharmacother 80: 109–114, 2016 [DOI] [PubMed] [Google Scholar]
- 41.Youness RA, El-Tayebi HM, Assal RA, Hosny K, Esmat G, Abdelaziz AI: MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol Lett 12: 2567–2573, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K, Zhang H, Zhou X, Tang WB, Xiao L, Liu YH, Liu H, Peng YM, Sun L, Liu FY: miRNA589 regulates epithelial-mesenchymal transition in human peritoneal mesothelial cells. J Biomed Biotechnol 2012: 673096, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kriegel AJ, Baker MA, Liu Y, Liu P, Cowley AW Jr, Liang M: Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension 66: 793–799, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang M, Pietrusz JL: Thiol-related genes in diabetic complications: A novel protective role for endogenous thioredoxin 2. Arterioscler Thromb Vasc Biol 27: 77–83, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.