Abstract
Maintenance of mammary functional capacity during cycles of proliferation and regression depend on appropriate cell fate decisions of mammary progenitor cells to populate an epithelium consisting of secretory luminal cells and contractile myoepithelial cells. It is well established that transforming growth factor β (TGFβ) restricts mammary epithelial cell proliferation and that sensitivity to TGFβ is decreased in breast cancer. Here we show that TGFβ also exerts control of mammary progenitor self-renewal and lineage commitment decisions by stringent regulation of breast cancer early onset 1 (BRCA1), which controls stem cell self-renewal and lineage commitment. Either genetic depletion of Tgfb1or transient blockade of TGFβ increased self-renewal of mammary progenitor cells in mice and cultured primary mammary epithelial cells, and also skewed lineage commitment towards the myoepithelial fate. TGFβ stabilized the expression of BRCA1 by suppressing miR-182. Ectopic expression of BRCA1 or antagonism of miR-182 in cultured TGFβ-deficient mammary epithelial cells restored lineage commitment. These findings reveal that TGFβ modulation of BRCA1 directs mammary epithelial cell fate and, since stem/progenitor cells are thought to be the cell of origin for aggressive breast cancer subtypes, suggest that TGFβdysregulation during carcinogenesis may promote distinct breast cancer subtypes.
Introduction
Transforming growth factor β (TGFβ) is a pluripotent cytokine that plays multiple roles in mammary development and breast carcinogenesis. (1)Daniel and Silberstein first showed that pellets containing TGFβ profoundly inhibit mammary epithelial proliferation (2). Subsequent genetic depletion of Tgfb1 in mice confirmed this finding by demonstrating significant acceleration of ductal morphogenesis during puberty and increased proliferation of both hormone receptor positive and negative luminal cells as a function of ovarian hormones during proliferative stages of the estrus cycle (3, 4). In contrast, overexpression of active TGFβ alters differentiation and induces replicative senescence (5, 6). Studies in mammary gland and other tissues suggest that TGFβ is involved in stem cell functions, including cell fate decisions, cell cycle entry, self-renewal and niche formation (1, 7-9).
The development of the mouse mammary anlage in utero and ductal morphogenesis during puberty give rise to luminal cells that respond to hormonal stimulation to proliferate and undergo secretory differentiation during pregnancy, followed by dramatic involution upon weaning. Luminal cells are supported by a basal layer consisting of myoepithelial cells that perform contractile function during lactation. Thus, this dynamic organ engages multiple endocrine systems to orchestrate cycles of differentiation, functional maturation to produce milk, and regression to a quiescent state. Delineation of the mammary epithelial hierarchy is essential for understanding the complex development, maturation, and cyclical secretory differentiation of the breast (10).
The epithelium harbors mammary stem cells (MaSC) that provide the remarkable regenerative capacity of the organ. The regenerative capability of MaSC is astounding; a single mammary gland stem cell is predicted to be capable of generating 1012-1013 progeny (11). Although a single MaSC can give rise to an entire mammary epithelial tree in mice, this capability appears to be restricted in situ. Two fundamental properties define MaSC: the ability to self-renew through symmetric division to maintain a pool of stem cells, and multipotency that confers the potential by asymmetric division to generate differentiated daughter cells. Analysis of self-renewal and lineage commitment using mammary repopulating activity (12) (13) and lineage tracing (14) (15) provides strong evidence for MaSC, albeit with varying degrees of functional capacity to recapitulate the mammary hierarchy. Although most current research focuses on identifying features of discrete MaSC, the functional potential of the tissue depends on appropriate cell fate decisions to maintain homeostasis of a relatively quiescent ductal system consisting of a bilayer of luminal and myoepithelial cells during repeated ovarian cycles.
Unraveling the mechanisms that regulate the mammary epithelial lineage is important for not only elucidating how homeostasis is maintained but is also thought to be important in the etiology of breast cancer since the cell of origin hypothesis posits that certain breast cancers arise from transformation of stem or progenitor cells (16). The expression profiles of basal-like and claudin-low intrinsic breast cancer subtypes are enriched in the signatures of MaSC and progenitor cells (17) and mutations in the gene for breast cancer associated 1 (BRCA1) cause dysregulation of MaSC and luminal progenitors (18-20), and predisposes the gland to develop aggressive hormone receptor negative breast cancer. Interestingly, TGFβ biology appears to discriminate between these two subtypes (21) (16, 22). Here we show that TGFβ controls mammary self-renewal and lineage commitment during homeostasis through stringent regulation of BRCA1 through a miRNA-mediated mechanism.
Results
TGFβ1 depletion promotes self-renewal in mammary gland
Overexpression of constitutively active TGFβ1 in the mammary gland decreases mammary repopulating activity (7). We first sought to investigate the effect of TGFβ depletion on this process. Hence, we evaluated serial transplantation efficiency of the mammary glands of BALB/c Tgfb1 wild-type and heterozygous (HT) littermates. TGFβ1 protein is reduced by 70-90% in tissue extracts from Tgfb1 HT mice relative to that in tissue extracts from wild-type mice (3). Transplants of fragments from Tgfb1 HT generated more outgrowths and had greater serial transplantation capacity than did wild-type MEC (fig. S1, A and B), which indicates greater self-renewal capacity. The repopulation capacity of isolated mammary epithelial cells (MEC) from Tgfb1 HT and wild-type mice was then assessed with a limiting dilution assay that determines the frequency of MEC capable of recapitulating mammary morphogenesis when inoculated into fat pads divested of endogenous epithelium (23). The repopulating capacity of Tgfb1 HT MEC was 3-fold greater than that of wild-type MEC (1/1500 and 1/4712, respectively; Fig. 1A). Next, we used flow cytometry to evaluate Lin-/CD24med/CD49fhigh (12) or CD29hiCD24hiCD61+ (17), two sets of cell surface markers of MEC populations shown to be enriched in mammary repopulating activity (fig. S2). Compared to those from wild-type mice, the MEC prepared from adult Tgfb1 HT mice had approximately 2-fold more cells that exhibited MaSC cell surface markers (Fig. 1, B and C). Consistent with this, qRT-PCR revealed that stem cell genes NOTCH1 and PROCR were increased 3.4-fold and PRC2 was increased 7-fold in RNA from Tgfb1 HT MEC compared to wild-type MEC (Fig. 1D). These data support the idea that TGFβ inhibits MaSC expansion.
Figure 1. TGFβ1 suppresses MaSC expansion.
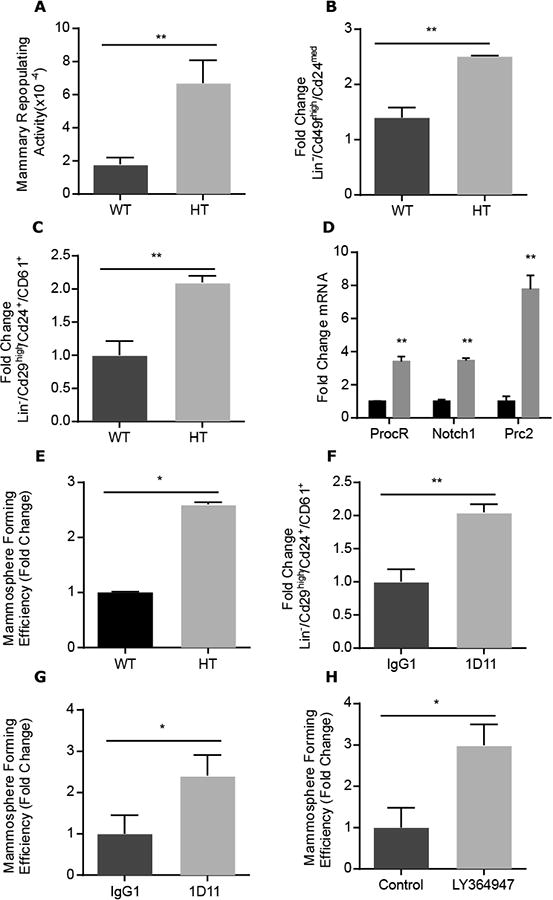
(A) Frequency of mammary repopulating cells from 15-week-old adult Tgfb1 HT mice compared to WT mice as measured by limiting dilution mammary repopulation capacity. Data are mean ±95% confidence intervals from 3 biological replicates (N=50 transplants per group). (B) Change in Lin-/Cd24med/Cd49fhi population lineage markers enriched for MaSC in HT relative to WT MEC (C) The Lin-/Cd29high/Cd24+/CD61+ lineage markers enriched for MaSC is significantly increased in HT versus WT MEC. (D) Expression of stem cell-associated genes, Notch1, ProcRand Prc2, analyzed by qPCR in Tgfb1 HT relative to WT MEC.(E) MFE of Tgfb1 HT MEC relative to that of WT MEC. (F) The difference in MaSC-enriched Lin-/Cd29high/Cd24+/Cd61+ population in MEC isolated from 10-week old BALB/c mice treated with TGFβ neutralizing antibody 1D11 (10 mg/kg) twice a week for 4 weeks compared to those treated with control antibodies (IgG1). (G) Mammosphere generation by WT MEC treated in vitro with 1D11 relative to those treated with control antibody. (H) Mammosphere generation by WT MEC treated in vitro with TGFβ type I receptor inhibitor LY364947 relative to those treated with vehicle control. Data in (B-H) are mean fold change ± S.E.M. from three biological replicates; *P<0.05, **P<0.01, by Student's t-test
Mammosphere formation by cells cultured in restricted growth factor conditions is a surrogate of repopulating capacity (24). Tgfb1 HT and wild-type MEC were sorted using cell surface markers CD29, CD24 and CD61 to define populations enriched in MaSC (CD29highCD24+CD61+), luminal progenitors (CD29lowCD24+CD61+), and stromal cells (CD29lowCD24-) and seeded into mammosphere culture conditions (24). After 7 days, the characteristics of each sorted population were assessed. As described, cells sorted from the CD29lowCD24-fraction did not form multicellular colonies. Acinar structures evidenced by hollow lumen were obtained from cells sorted from the CD29lowCD24+CD61+ fraction, whereas solid-appearing mammospheres, thought to represent MaSC, were obtained from the CD29highCD24+CD61+ fraction. Similar to the increase in in vivo repopulating activity, we conducted a limiting dilution assay based on the formation of solid mammospheres (fig. S3). The mammosphere forming efficiency (MFE) increased 2.5-fold from Tgfb1 HT MEC compared to wild-type MEC cultured (Fig. 1E). Consistent with TGFβ's role inhibiting proliferation (3), Tgfb1 HT MEC-derived mammospheres were significantly larger compared to those derived from wild-type MEC (97.0 ± 5.5, n=12; versus 76.3 ± 4.8, n=10; p<0.05).
MaSC established during development are susceptible to physiological mediators of expansion, as is evident during pregnancy (10). To test whether decreased TGFβ activity during development alters the frequency of MaSC or constitutively inhibits MaSC self-renewal, we treated BALB/c mice with TGFβ pan-isoform neutralizing monoclonal antibodies, 1D11, or isotypematched IgG1 control antibodies twice a week for 4 weeks. Compared to control-treated mice, the mammary epithelium of mice treated with neutralizing antibodies exhibited significantly less phosphorylation of the TGFβ effector SMAD2 (p-SMAD2; fig. S4). The fraction of CD29hiCD24hiCD61+ MEC isolated from mice treated with 1D11 was increased 2-fold, similar to the difference seen between wild-type and Tgfb1 HT MEC (Fig. 1F). Because neutralizing antibody captures ligand released from extracellular stores, these data suggest that TGFβ activity dynamically restricts MaSC self-renewal.
To confirm the idea that TGFβ is necessary to restrain self-renewal, mammospheres from primary MEC were treated for one week with 1D11 or a small-molecule inhibitor of TGFβ type I receptor kinase, LY364947. Dissociated mammospheres from treated or control cultures were placed into secondary passage to test MFE without further treatment. Both ligand capture by 1D11 (Fig. 1G) and receptor signaling blockade by LY364947 (Fig. 1H) resulted in a more than 2-fold increase in mammosphere formation (Fig. S3). Together these data indicate that TGFβ activity inhibits MaSC self-renewal.
TGFβ1 depletion skews lineage commitment
We noticed that the CD24+/CD49fmed population, which is enriched in myoepithelial cells, was significantly increased in MEC isolated from Tgfb1 HT mice (Fig. 2A). To confirm this, we conducted flow cytometry analysis using cytokeratin 14 (K14) and 18 (K18), which mark myoepithelial and luminal populations, respectively. More K14-positive cells were obtained from Tgfb1 HT MEC (Fig. 2B). This expansion was at the expense of luminal cells in MEC from Tgfb1 HT compared to wild-type mice (Fig. 2C). The gene encoding the protein p63 is expressed only in basal mammary epithelial cells, which are primarily myoepithelial. The percentage of p63-positive myoepithelial cells was significantly increased in tissue from Tgfb1 HT compared to wild-type mice (Fig. 2D). This was also evident in the mammary glands of mice treated with the TGFβ neutralizing antibody 1D11 (Fig. 2E). Because proliferation is increased in the mammary glands of Tgfb1 HT mice (4), we investigated whether proliferation was increased specifically in myoepithelial cells by assessing the co-localization of the proliferation marker Ki-67 and p63 by immunostaining. The frequency of Ki-67 and p63 double-positive cells was not significantly increased in tissue from either Tgfb1 HT mice or 1D11-treated mice (Fig. 2, D and E). Together these data suggested that TGFβ depletion skews lineage commitment toward basal cells.
Figure 2. TGFβ1 regulates mammary lineage commitment.
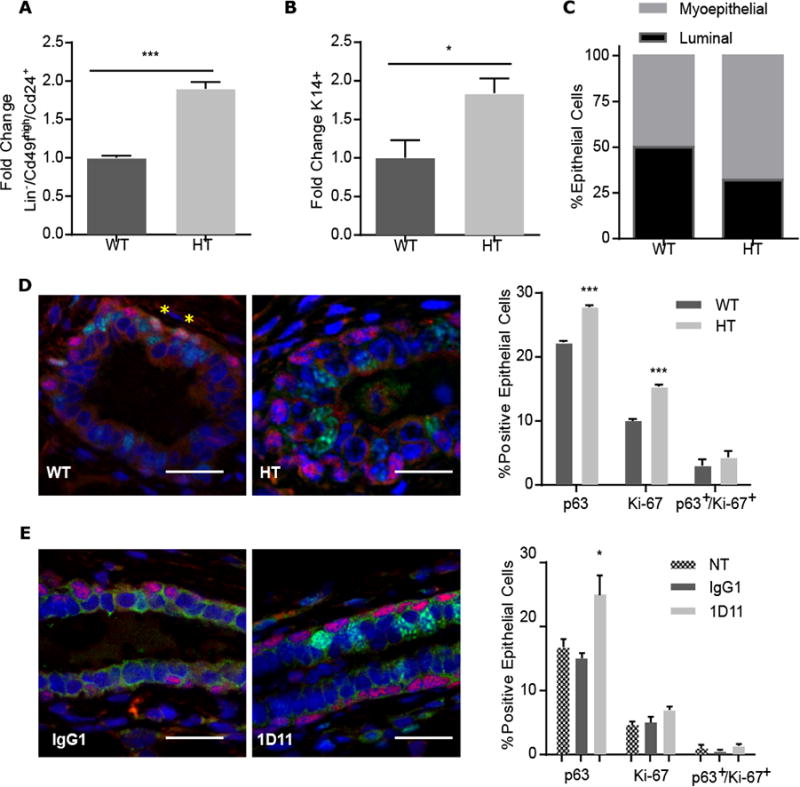
(A) FACS analysis of the Lin/Cd24+/Cd49fhimyoepithelial population in MEC isolated from 15 week-old Tgfb1 HT or WT mice.(B)Proportion of cells positive for myoepithelial marker cytokeratin 14 (K14) in Tgfb1 HT MEC relative to WT MEC. (C) The proportion of myoepithelial (grey) to luminal (black) cells in Tgfb1 HT compared to WT MEC cultures on the basis of FACS analysis. (D) Representative images of mammary glands isolated from 15-week-old Tgfb1 WT or HT mice and immunostained for p63 (red) and Ki-67 (green). Nuclei were counterstained blue. Yellow asterisks indicate rare co-localizing cells. Percentage of epithelial cells was quantified from 10 random fields per mouse. (E) Representative images of p63 (red) and Ki-67 (green) immunostaining in mammary glands from adult mice treated as in Figure 1 with control (IgG1) TGFβ neutralizing antibody (1D11). Percentage of epithelial cells was quantified from 10 random fields per mouse. NT, non-treated control. Data in (A-E) are means ± S.E.M. from three biological replicates,;*P<0.05, ***P<0.001, by Student's t-test. Scale bars, 50 μm.
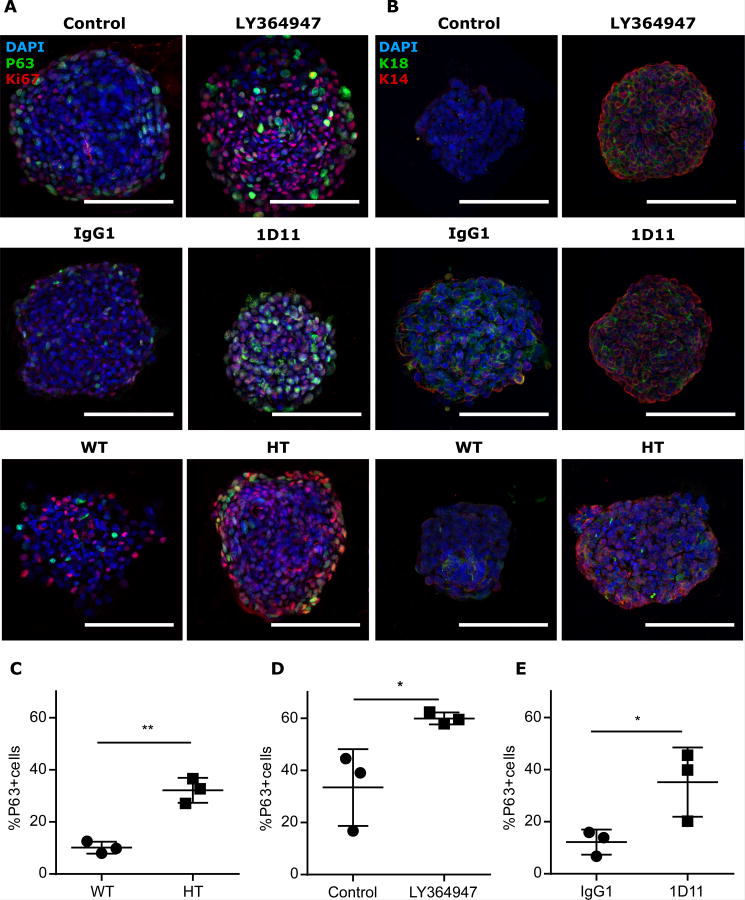
We next examined mammosphere composition as a function of TGFβ depletion by immunostaining for myoepithelial marker, p63 (Fig. 3A). Relative to their respective controls, the p63-positive population was increased in mammospheres derived from Tgfb1 HT MEC (Fig. 3C), those treated with 1D11 TGFβ neutralizing antibody (Fig. 3D), and those treated with a small-molecule TGFβ type I receptor kinase inhibitor LY364947 (Fig. 3E). The proportion of K14-positive cells in mammospheres also appeared to increase when TGFβ was compromised in these three models (Fig. 3B). Thus, we concluded that TGFβ depletion also skews epithelial lineage commitment toward myoepithelial differentiation.
Figure 3. TGFβ1 depletion skews lineage commitment in mammospheres.
(A and B) Representative images of immunostaining for (A) p63 (red) and Ki-67 (green) or (B) K14 (red) and K18 (green) in WT and Tgfb1 HT mammospheres, treated as indicated. Scale bars, 100 μm.(C)Frequency of p63+ cells in Tgfb1 HT or WT primary mammospheres. (D) Frequency of p63+ cells in WT secondary mammospheres treated with LY364947 in culture compared to vehicle control. (E) Frequency of p63+ cells in WT mammospheres treated with 1D11 or antibody control. Scale bar, 100 μm. Data in (C-E) are means ± S.E.M. from three biological replicates; *P<0.05, **P<0.01, by Student's t-test.
The TGFβ1-BRCA1 axis restricts MaSC self-renewal
BRCA1 mutations expand MaSC and luminal progenitors in human cells and tissues (18-20). We have previously reported that Brca1 mRNA is decreased in murine Tgfb1 null keratinocytes compared to HT keratinocytes, whereas BRCA1 abundance is decreased in cultured human epithelial cells treated with a small-molecule TGFβ type 1 receptor kinase inhibitor (25); moreover Brca1 mRNA is also significantly decreased in gene expression microarrays from mammary glands from Tgfb1 HT mice compared to WT mice (21).
Thus, we examined the abundance of BRCA1 protein and mRNA in MEC from Tgfb1 HT mice compared to those from WT mice. Both protein and message abundance were decreased in Tgfb1 HT MEC (Fig. 4, A and B). BRCA1 expression is tightly regulated during cell cycle progression in many cell types and is increased during S-phase (26, 27). To examine this, we used MCF10A cells, a non-malignant human mammary epithelial cell line, treated with LY364947 to inhibit TGFβ signaling. BRCA1 protein rapidly increased in response to TGFβ; concomitant treatment with LY364957 blocked this effect. Effective suppression of TGFβ signaling by LY364947 was evident by inhibition of TGFβ-induced p-SMAD2 (fig. S5). Both protein and mRNA abundance were further decreased with prolonged inhibition (Fig. 4C). The amount of BRCA1 mRNA in LY364947-treated MCF10A cells was comparable to that in SUM1315 cells, a human BRCA1 haplo-insufficient breast cancer cell line (Fig. 4D). Likewise, treatment of MCF10A with TGFβ neutralizing antibody similarly decreased BRCA1 protein and mRNA (Fig. 4, E and F). As BRCA1 is increased during S-phase and inhibition of TGFβ signaling increases proliferation in MCF10A cells, BRCA1 would be expected to increase if due to cell cycle rather than decrease as observed. Moreover, TGFβ treatment of MCF10A increased BRCA1 mRNA (Fig. 4G). Together these data indicate discrete regulation of BRCA1 by TGFβ.
Figure 4. Inhibition of TGFβ1 suppresses the abundance of BRCA1 protein and mRNA.
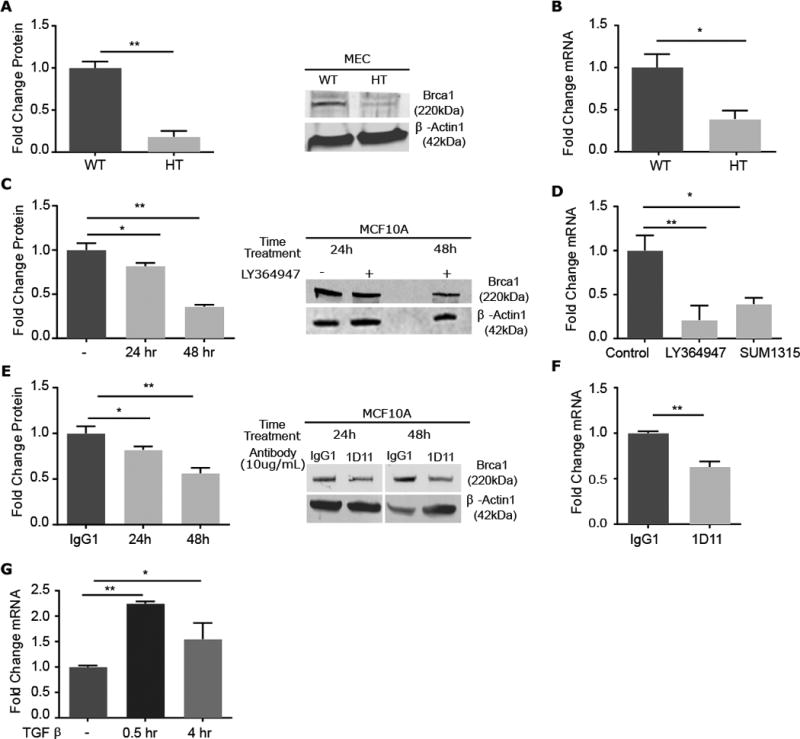
(A)Quantitation of Brca1 abundance in Tgfb1 WT and HT MEC and representative immunoblot.(B) Corresponding Brca1 mRNA expression in specimens in (A). (C) Quantitation of BRCA1 protein abundance in MCF10A cells treated with small molecule inhibitor LY364947 for 24 hours and a representative immunoblot. (D) Corresponding Brca1 mRNA expression in MCF10A cells, with comparison to that in BRCA1-mutant SUM1315 cells. (E) Quantitation of BRCA1 protein abundance in MCF10A cells treated with TGFβ neutralizing antibody 1D11 for 24 or 48 hours and a representative immunoblot. (F) Corresponding mRNA for specimens in (E). (G) BRCA1 mRNA measured by qRT-PCR in MCF10A cells treated with TGFβ for 0.5 or 4 hours. Graphed data are means ± S.E.M. of biological triplicates,;*P<0.05, **P<0.01, by Students t- test in (A, B and F) and ANOVA in (C, D, E and G).
Given that BRCA1 mutations alter human mammary lineage commitment (20), we next tested whether overexpressing Brca1 in Tgfb1 HT MEC could reverse effect of TGFβ on mammary lineage. BRCA1 protein abundance increased in Tgfb1 HT MEC infected with the Brca1-expressing lentivirus compared to those infected with an empty vector (Fig. 5A). Functional evaluation of lentiviral BRCA1 overexpression in Tgfb1 HT MEC compared to Tgfb1 HT MEC expressing the empty vector resulted in more than 3-fold decrease in mammosphere forming capacity, which was similar to that of WT mammospheres (Fig. 5B). Although Tgfb1 HT mammospheres contain more p63-positive cells than Tgfb1 WT mammospheres, their frequency in mammospheres from Tgfb1 HT MEC overexpressing BRCA1 was similar to that of wild-type MEC mammospheres (Fig. 5C). These data indicate that BRCA1 mediates TGFβ effects on mammary self-renewal and lineage commitment.
Figure 5. TGFβ promotes expression of BRCA1.
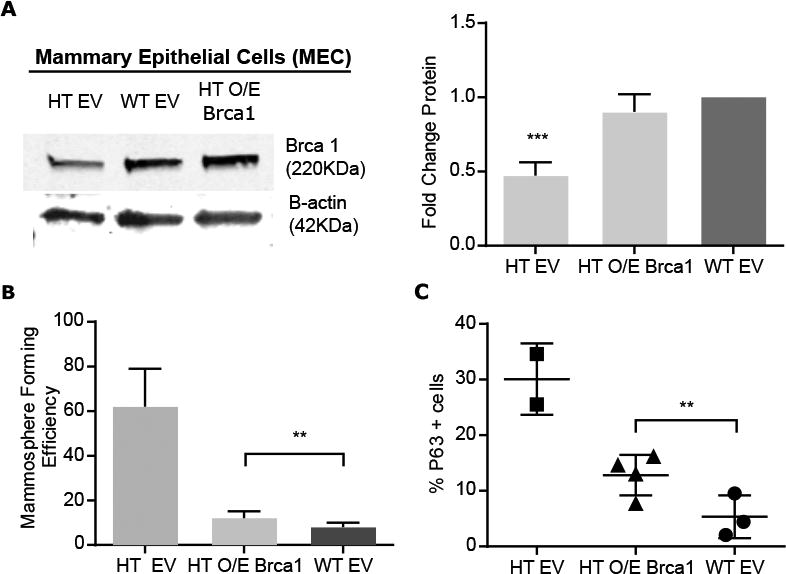
(A) Representative immunoblot and quantitation of Brca1 in Tgfb1 HT MEC expressing lentiviral-encoded BRCA1(O/E) or empty vector (EV) and WT MEC expressing EV. (B) Mammosphere formation by Tgfb1 HT MEC overexpressing BRCA1 compared to Tgfb1 WT or HT MEC expressing EV. (C) p63+ cells in mammospheres formed by Tgfb1 HT MEC overexpressing BRCA1 compared to those from Tgfb1 WT or HT MEC expressing EV. Data are means ± S.E.M. from three biological replicates; **P<0.01, ***P<0.001, by ANOVA.
TGFβ1 increases BRCA1 by inhibiting miR-182
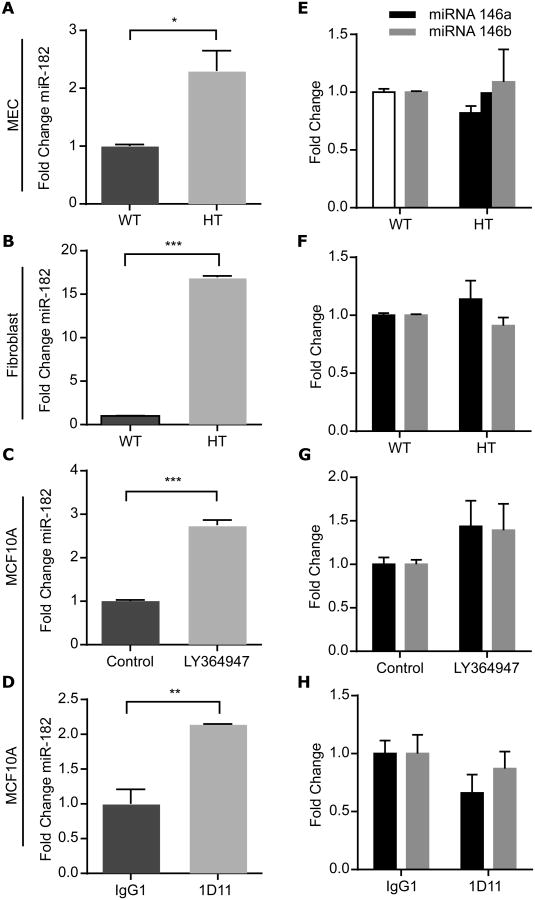
BRCA1 is post-transcriptionally suppressed by several microRNAs (miR) that include miR-182 (28) and miR-146a and 146b-5p (28, 29). We analyzed the expression of these miRNAs as a function of TGFβ depletion in MEC, MCF10A cells, and mammary fibroblasts. Notably, inhibition of TGFβ ligand or signaling in all cells significantly increased the abundance of miR-182 (Fig. 6, A to D) but did not affect that of miR-146a or miR-146b (Fig. 6, E to H), nor that of miR-181a and miR-181b (fig. S6), which have been reported to be modulated by TGFβ (30). In contrast, TGFβ treatment of MCF10A cells for as little as 30 min significantly decreased the abundance of miR-182 (fig. S7A), concordant with the observed increase in BRCA1 mRNA abundance (Fig. 4G), which persisted for 4 hours, whereas the abundance of neither miR-146a nor miR-146b was affected (fig. S7B).
Figure 6. Genetic or transient depletion of TGFβ1 increases miR182, but not miR-146a or miR-146b. (A-H).
qRT-PCR of miR-182 (A-D) or miR-146a and miR-146b-5p (black and grey, respectively; E-H) abundance in MEC (A and E) and fibroblasts (B and F) from WT or Tgfb1 HT mice, and MCF10A cells treated with LY364947 (C and G) or 1D11 (D and H). Data are means ± S.E.M. from three biological replicates,; *P<0.05, **P<0.01, ***P<0.001, by Student's t-test.
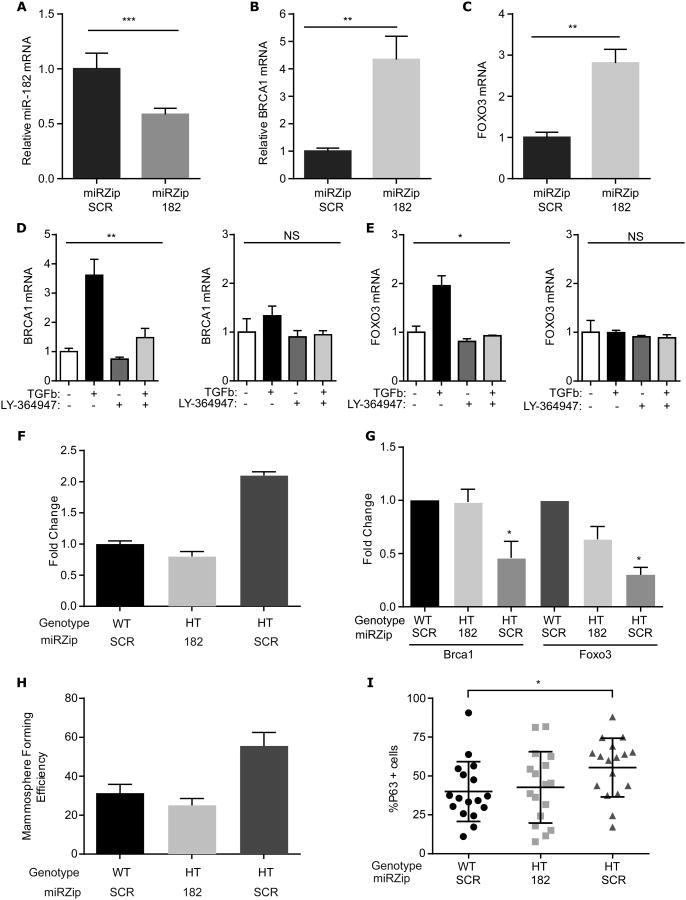
To test whether miR-182 indeed mediated BRCA1 mRNA abundance, we used a lentiviral miRZip to inhibit miR-182 function in MCF10A cells. Stable MCF10A cell lines were established that expressed a miRZip-182 or a scrambled (SCR) miRZip. Expression of miR-182 mRNA was reduced by about half in cells transfected with miRZip-182 (Fig. 7A), whereas the abundance of BRCA1 and FOXO3, another validated miR-182 target, increased several-fold (Fig. 7B,C). Infection of Tgfb1 WT and HT MEC with miRZip-182, but not the scrambled mirZip (SCR), abrogated TGFβ induction and LY364957 suppression of both BRCA1 (Fig. 7D) and FOXO3 mRNA abundance (Fig. 7E). These data confirm that miR-182 targets both mRNAs as reported (31), and demonstrate that the requirement for TGFβ control of miR-182 in its regulation of BRCA1.
Figure 7. TGFβ inhibition of miR-182 mediates BRCA1 and mammary phenotype.
(A-C) MCF10A cell lines engineered to stably express mirZip-182 or miRZip-SCR were compared using qRT-PCR to measure (A) miR-182(B) BRCA1 and (C) FOXO3 mRNA. The abundance of each in MCF10 cells expressing miRZip-182 was normalized to that in cells expressing miRZip-SCR. Data are means ±S.E.M. of 4 or more independent experiments analyzed using Student's t-test. (D and E) qRT-PCR (D) BRCA1 or (E) FOXO3 mRNA abundance measured in MCF10A cell lines engineered to stably express miRZip-SCR (left) or mirZip-182 (right) in response to TGFβ and/or LY364947 normalized to their respective untreated controls. Data are means ± S.E.M from 4 independent experiments, analyzed using ANOVA. (F) Abundance of miR-182 in Tgfb1 HT MEC infected with miRZIP-182 lentivirus compared to HT MEC and WT MEC transfected with miR-ZIP SCR. (G) Quantitation of immunoblots for BRCA1 and FOXO3 in (F). (H) Mammosphere forming efficiency of HT and WT MEC expressing miRZip-182 or mirZip-SCR. Data are means ± S.D. from 2 biological replicates. (I) Mammospheres from (H) analyzed for percentage of cells staining positively for the basal/myoepithelial marker p63. Data are means ± S.D. from 17 mammospheres, pooled from two independent experiments; *P<0.05, **P<0.01, ***P<0.001, by Student's t-test.
We next used HT MEC expression of miRZIP-182 or miRZip-SCR to compare to Tgfb1 WT expressing miRZip-SCR. As expected the miRZip-182 decreased mir-182 to that similar to WT and 2-fold less than compared to HT MEC expressing miRZip-SCR (Fig.7F). Antagonism of miR-182 also restored Brca1 and Foxo3 protein as assayed by immunoblotting of lysates (fig. S8A).; their abundance increased in Tgfb1 HT MEC expressing miRZip-182 to levels similar to miRZip-SCR expressing WT MEC, and were nearly twice that of miRZip-SCR infected Tgfb1 HT MEC (Fig. 7G). These results indicated that TGFβ suppression of miR-182 is necessary for BRCA1 mRNA and protein accumulation.
If TGFβ depletion decreased BRCA1 abundance, which in turn increases MaSC self-renewal, by increasing miR-182, then antagonizing miR-182 should suppress this phenotype. To test this prediction, self-renewal was measured in MEC by mammosphere formation. The Tgfb1 HT cells expressing mirZip-182 generated half as many mammospheres as Tgfb1 HT cells expressing the control SCR vector (Fig. 7H). Relative to SCR control mammospheres, miR-182 was reduced in mammospheres of Tgfb1 HT cells expressing miRZip-182 (fig. S8B). Moreover, the basal-skewed lineage commitment of Tgfb1 HT mammospheres was reversed (Fig. 7I), but proliferation was not significantly affected (fig. S8C). Together these studies indicate that TGFβ maintains MaSC and lineage commitment by suppressing miR-182 that targets BRCA1 mRNA for degradation. TGFβ depletion releases miR-182 to repress BRCA1 mRNA stability or translation, which leads to increased mammary self-renewal and skews lineage commitment to the myoepithelial cell fate.
Discussion
Our studies demonstrate that TGFβ restricts mammary stem cell expansion and directs mammary lineage commitment to luminal cell fate, which together enforces mammary gland homeostasis. The molecular mechanism underpinning this shift involves stringent control of BRCA1 by TGFβ. We determined that TGFβ regulates BRCA1 by inhibiting miR-182; miR-182 typically functions to repress BRCA1 protein translation and target its mRNA for degradation. The Brca1 protein level in mammary glands from Tgfb1 HT mice was half that of TGFβ competent mice and was accompanied by a doubling of mammary repopulating cells. We used in vitro mammosphere formation as a surrogate to evaluate the underlying mechanisms of mammary repopulation, which allowed us to ascertain that either Brca1 overexpression or miR-182 antagonism reversed the consequences of genetic depletion of TGFβ on self-renewal and lineage commitment. Thus, TGFpTGFβ, miR-182 and BRCA1 axis regulates self-renewal and epithelial lineage commitment during mammary homeostasis.
The biology of TGFβ in the mammary gland is complex (reviewed in (32)]. TGFβ1 is arguably the most abundant isoform during pubertal morphogenesis, but all three TGFβ isoform-encoding mRNAs are expressed in the mammary epithelium across all phases of mammary development (33). The product of each gene is a latent TGFβ complex that, when activated extracellularly, release ligands that bind the same set of receptors. Thus, the key determinant is biological activity that is controlled by the latent complex and latency associated peptide, which are distinctly different for each isoform. We cannot exclude the potential for other isoforms to regulate BRCA1 as we did not use reagents to selectively block one isoform, but various studies show that any active TGFβ would result in comparable signaling.
The TGFβ pathway intersects with multiple miRNAs (34-37). Our data in human mammary cell line MCF10A shows that TGFβ inhibits miR-182, and, conversely, TGFβ inhibition increases miR-182, while neither TGFβ addition nor inhibition affects the abundance of miRNAs 146a and 146b (29) or miRNAs 181a and 181b (38), which also target BRCA1 mRNA for degradation. The amount of miR-182 is increased in breast tumors compared with normal breast tissue (39) and in serum of patients with breast cancer (40). High miR-182 is associated with worse outcome in a meta-analysis of 10 studies (41). Overexpression of miR-182 increases the sensitivity of breast cancer cell lines to poly(ADP-ribose) polymerase 1 (PARP1) inhibitors (28, 42), similar to that observed in BRCA1-deficient cells and tumors (42).
The luminal progenitor population is expanded in the haploinsufficient non-malignant tissue of BRCA1 mutation carriers (19, 20), while engineered deletion of BRCA1 in human cells leads to stem cell expansion (18). Interestingly, miR-182 is increased in human luminal progenitors compared to MaSC (43), which our data suggests might be due to reduced TGFβ activity. TGFβ inhibition also significantly increases colony formation of basal cells, which appear to have mammary repopulating capacity (44). A paracrine mechanism by which hormone-responsive luminal epithelial cells restrict the expansion of MaSC (45) may also be reinforced by TGFβ because TGFβ activation in estrogen receptor-positive luminal epithelial cells at estrus inhibits their proliferation and thus, the proportion of estrogen-receptor cells (3, 4).
The frequency of MaSC and the subsequent development of aggressive breast cancer are thought to be functionally linked by the cell-of-origin hypothesis, which posits that malignant transformation of long-lived MaSC or progenitors underlie the heterogeneity of tumor subtypes [reviewed in (16)]. For example, women with germline mutations of BRCA1, which was the first genetic locus associated with familial breast cancer risk (46), have an extremely high risk of developing breast carcinomas negative for estrogen receptor, progesterone receptor and HER2 amplification; these so-called triple-negative breast cancer occur at a young age and generally have a very poor prognosis. BRCA1 knockdown in primary breast epithelial cells increases the proportion of stem or undifferentiated cells and decreases the proportion of cells expressing luminal epithelial markers and estrogen receptor (18). Even before cancer onset, the breast tissue of BRCA1 mutation carriers contains a disproportionate number of luminal progenitor cells (19) and exhibits aberrant progenitor cell lineage commitment (20). Thus, the prevalence of triple-negative breast cancer per se is linked to aberrant mammary lineage commitment (18-20). The effect of radiation on MaSC and the development of aggressive cancer are also highly correlated as a function of TGFβ in murine mammary carcinogenesis studies (21, 47, 48). These data provide substantial support for the idea that dysregulation of stem and progenitor cell during development is involved in the etiology of breast carcinogenesis.
Material and Methods
Mice
All animal experiments were performed at New York University School of Medicine with institutional review and approval. BALB/c mice were purchased from TACONIC and housed four per cage with Lab Diet 5008 chow and water provided ad libidum. Tgfb1 heterozygote (HT) BALB/c mice were bred in-house under similar conditions. For assays assessing mammary stem cell activity, cells were isolated from 5-8 15-week-old wild-type (WT) or Tgfb1 HT BALB/c mice. Neutralizing antibody 1D11 (R&D Systems, Inc.) or isotype control antibody IgG1 (R&D Systems, Inc.), was administered by intraperitoneal injection (10 mg/kg) twice a week for 4 weeks. Mammary repopulation capacity was assayed by limiting dilution (23)using 5 cell doses and 58 mice as recipients per treatment (23). The repopulating capacity was calculated using L-Calc V1.1.1 (StemCell Technologies).
Flow Cytometry
Mammary gland tissue was enzymatically digested to single-cell suspensions, depleted of non-epithelial cells by using the MACS lineage cell depletion kit (Militenyi Biotec GmbH, Bergisch Gladbach, Germany), and sorted in three experiments with technical triplicates based on the expression of CD24, CD29 and CD61. Phycoerythrin (PE)-conjugated CD24 antibody (BD Pharmingen, San Diego, CA), fluorescein (FITC)-conjugated CD29 antibody (EMD Millipore, Billerica, MA), and allophycocyanin (APC)-conjugated CD61 antibody (Invitrogen, Carlsbad, CA) were used to detect the cell surface markers. Stem cell-enriched, subpopulations were identified by the CD24+CD29hi, CD24+CD29loCD61+ and CD24+CD29loCD61- phenotypes, respectively, as previously reported (17). Cells were also isolated using CD49f antibody (BD Pharmingen, San Diego, CA) and CD24 antibody as previously described (49).
Mammary epithelial cells (MEC) were also sorted after staining with rabbit-raised antibody against K14 at 1/100 dilution (Covance) and guinea pig-raised antibody against K8/18 at 1/500 dilution (Fitzgerald) overnight. Prior to staining cells were fixed in cold 80% methanol. MEC were incubated for 1 hour at room temperature with secondary antibodies Alexa 488-conjugated donkey anti-rabbit IgG and Alexa 594-conjugated donkey an antibody recognizing mouse IgG (Invitrogen, Carlsbad, CA), each at 1/200 dilution. Cells were stained with propidium iodide (PI) to analyze for cell cycle by flow cytometry. Flow cytometry was performed with MO-FLO (Becton Dickinson) and analyzed with Flow Jo software (version 7.6.4).
Cell culture
MEC and mammary fibroblasts were isolated from 15-week-old nulliparous Tgfb1 WT and HT BALB/c female mice as previously described (23). Fibroblasts were cultured in DMEM/F12 supplemented with 10% fetal calf serum, penicillin (100 IU/ml), streptomycin (100 μg/ml), 4 mM L-glutamine, 5 μg/ml bovine pancreatic insulin, 10 ng/ml cholera toxin and 10 ng/ml epidermal growth factor (EGF). Cells were passaged by differential trypsinization until fibroblasts spontaneously immortalized. Fibroblasts were serum-starved and then cultured in serum replacement medium for 1 day for cell cycle synchronization, then qRT-PCR and immunostaining assays were performed.
MCF10A cells (ATCC, Manassas, VA) were cultured in serum free medium as previously described in the presence or absence of recombinant human TGFβ1 (2 ng/ml; R&D Systems, Minneapolis, MN), TGFβ type I receptor kinase inhibitor LY364947 (2 μM; Calbiochem), TGFβ neutralizing antibody 1D11 (10 μg/ml), or IgG1 isotype control (R&D Systems Inc) added at the time of plating and with medium change every other day thereafter (50). The number of cells plated under each condition was selected so that all cultures reached similar degrees of confluence at experiment termination.
Mammosphere Culture
MEC were dissociated into single cells according to standard protocol and cultured in mammosphere media consisting of Epicult-B medium (Stem Cell Technology) containing 5% Matrigel, 10 ng/ml EGF (Sigma), 20 ng/ml bFGF (BD Biosciences), 4 μg/ml Heparin (Sigma), 5% heat inactivated FBS, 5 μM Rock inhibitor Y-27632 (Tocris) (24). Cells were seeded at 1,000-2,000 per well in 96-well ultralow attachment plates (Corning). The number of mammospheres (solid appearance, >50 um in diameter) was counted 7 days after seeding. Mammospheres were treated in vitro with 10 μg/ml TGFβ neutralizing antibody 1D11, IgG1 isotype control antibody, or 2 μM LY364947. Mammospheres pretreated for one week were dissociated and limiting dilutions were seeded into secondary passages without further treatment to measure self-renewal using L-Calc V1.1.1 (StemCell Technologies).
Immunofluorescence and image acquisition
3 or 5 μm sections of formalin fixed, paraffin embedded mammary gland tissue were deparaffinized, incubated with antigen unmasking solution (Vector, Burlingame, CA), blocked with 0.5% casein and incubated with primary antibodies over night at 4°C using mouse p63 antibody (1:100, Lab Vision, Kalamazoo, MI), rabbit Ki-67 antibody (1:1000, Novocastra NCL-ki67p), rabbit K14 antibody (1;1500, Covance, NJ), guinea pig K8/18 antibody (1:200, Fitzgerald, MA), or mouse phosphorylated Ser465/467 SMAD2 (1:50, pSMAD) antibody (clone 138D4, CAT#3108, Cell Signaling). Slides were washed 3 times with PBS and incubated for 1 hour at room temperature with Alexa 488-conjugated donkey an antibody recognizing rabbit and Alexa 594-conjugated donkey an antibody recognizing mouse antibodies (Invitrogen, Carlsbad, CA). DAPI was used for nuclear counterstaining, and samples where mounted with Vectashield (Vector, Burlingame, CA).
Ten mammospheres per treatment in each experiment were randomly imaged using 20X and 40X objectives with 0.95 numerical aperture Zeiss Plan-Apochromat objective on a Zeiss Axiovert microscope equipped with epifluorescence. For image analysis, 12-bit images for each color channel were used with macros developed in house from the open source platform Fiji-ImageJ (NIH, Bethesda, MA) (51). The DAPI channel was used to generate region of interest (ROI) containing the whole area of each mammosphere. The total number of cells per ROI was estimated dividing the area of the ROI by an estimate of the nuclei area, a calculated mean of 150 measured nuclei areas. Finally an estimated frequency was calculated using the following formula: p63 counted cells per ROI/total estimated cells per ROI x 100.
RT-PCR
RNA was extracted from epithelial preps by using the TRIzol reagent (Invitrogen, Carlsbad, California). cDNA was synthesized from 1 μg of RNA by means the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, California). Real-time quantitative PCR was performed using SYBR Green Mix (Applied Biosystems, Foster City, CA), according to the manufacturer's instructions. Primers (table S1) were purchased from Sigma-Aldrich (St. Louis, MO). The amount of genes of interest were normalized against expression of SPARC and analyzed with the comparative Ct method for quantitation of transcripts.
Western blots
Lysates were obtained from MCF10A cells, log phase murine mammary fibroblasts, snap-frozen murine mammary glands cleared of muscle and lymph nodes, or MEC transferred into modified RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP40) with protease and phosphatase inhibitor cocktails (Sigma Aldrich, St. Louis, MO). Protein extracts (100 μg) were electrophoresed and transferred to membrane. BRCA1 was detected using antibodies that target its N-terminus (BRCA1-N; a gift from Dr. L. A. Chodosh, University of Pennsylvania) or C-terminus (C20, Santa Cruz Biotechnology), and phosphorylated SMAD2 at 1:500 dilution (clone 138D4, CAT#3108, Cell Signaling Technology). Relative densitometry of the bands was normalized to the densitometry of actin(the loading control), detected using the LICOR Odyssey system.
miRNA qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, California) or the miRNAeasy Mini Kit (Qiagen). qRT-PCR analysis of mir-182, mir-146a, and mir-146b-5p, was performed by using miRNA-specific TaqMan MicroRNA Assay Kit (Applied Biosystems); 12.5 ng of total RNA was reversed transcribed using the corresponding RT Primer and the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). PCR was performed on 1.33 μL of RT products by adding the TaqMan PCR primers and the TaqMan Universal PCR Master Mix (Applied Biosystems). SNO and RNU44 small RNAs were used for normalization of input RNA/cDNA amounts.
Stable overexpression in cultured cells
The BRCA1 (Ex-H0047-Lv105) and control (EX-NEG-Lv105) vectors were purchased from GeneCopoeia, and lentiviruses were prepared according to the manufacturer's protocol. Virus-containing media was centrifuged, filtered (0.45 μM PES low protein filter) and stored at -80°C. WT or Tgfb1 HT MEC were diluted into low attachment plates to obtain mammospheres that were then dissociated and seeded as single cells in mammosphere media containing 5% Matrigel as described above. Cells in mammosphere media were infected with concentrated virus containing media and polybrene (6 μg/mL). Twenty-four hours after transfection, media containing virus was removed and replaced with mammosphere medium. Then cells were selected with 1 μM puromycin for 2 days to generate mammospheres with stable overexpression of wild-type BRCA1. After selection cells were dissociated and seeded by limiting dilution in 96-well low attachment plates to assess mammosphere forming efficiency.
Lentiviral anti-sense miR-182 (miRZip-182), control vectors and packaging plasmids were generated as described (52). Stable expression of miRZip-182 in MCF10A was achieved through lentiviral infection. Cells were growing exponentially before transduction and infected at 70-80% confluent state. Stable expression was achieved through selection in 1 μg/ml puromycin (Fisher GibcoTM, A1113803). Infection of lentiviral vectors were undertaken in the presence of 6 μg/ml Polybrene (Sigma, H9268). Fresh media with 1 μg/ml puromycin was replaced every two days. Media without puromycin was used for experiments. For RT-qPCR assay, 2 μg/ml of TGFBR1 kinase inhibitor LY364947 was given 20 hours before TGFβ treatment (2 ng/ml, R&D, 240-B-002). Cells were then lysed by QIAzol (QIAGEN, 79306) 4h post TGFβ treatment. Total RNA was extracted with miRNeasy Mini Kit (QIAGEN, 217004).
Oligonucleotide miRZip Transfection
Tgfb1 WT and HT MEC were seeded in 12-well plates and transfected with 150nM of miRZIP-182 or scrambled miRZIP negative control oligonucleotides (miRIDIAN, Applied Biosystems) using Lipofectamine 2000 (Invitrogen) according to suggested manufacturer procedures in triplicate. Transfection efficiency was monitored using Renilla Luciferase.
Statistics
Data are presented as mean ± SEM from 3 biological replicates conducted with 3 technical triplicates unless noted otherwise. Differences between treatment groups were determined using the Chi-square test (FACS), two-tailed Student's t-test (comparison between two group/treatments) or one-way ANOVA (comparison between three groups/treatments). Results were considered statistically significant at p<0.05; *p<0.05, **p<0.01, ***p<0.001.
Supplementary Material
Acknowledgments
The authors thankfully acknowledge technical assistance from William Chou, Xiaolu Li, Shiva Boloruchi and Manan Patel, and helpful discussions with Drs. Ann Lazar, Pamela Cowin, David Kleinberg and Sangeetha Vijayakumar. We are grateful to Dr. L. A. Chodosh, University of Pennsylvania, for the gift of BRCA1-N antibody. Funding: This research was supported by the NIH Graduate Program Training grant 5T32GM007238 (HMR), R01CA155234-03 (E.H.), DOE grant DE-FG02-08ER64654 (M.H.B.-H.) and NIH/NCI grant K08CA172722 (C.O.). The authors acknowledge the Histopathology and Flow Cytometry Cores that are partially supported by the Cancer Center Support Grant, P30CA016087 at the Laura and Isaac Perlmutter Cancer Center.
Footnotes
“This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at https://urldefense.proofpoint.com/v2/url?u=http-3A__www.sciencesignaling.org_&d=DQIFCQ&c=j5oPpO0eBH1iio48DtsedbOBGmuw5jHLjgvtN2r4ehE&r=LOVGNbw7ucmQf7Dhzd9TxPTQtywATsPyfcYiQ1Y_1U&m=KpGle5LdZ5wpgMyH2z6pmJmEq8lisXbG1-LuPTJzJuo&s=LEbFoKNQe8uc7nQU3mnN7B33oZDFE1mJO02ZfXOEqIU&e=. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Author Contributions: M. H. B.-H. and H. M.-R. conceived and designed the study. H. M.-R., I. I.-B., C. O., Q.L. and M. H. B.-H. collected and/or assembled the data. H. M.-R., I. I.-B., D. H., E. H., and M. H. B.-H. analyzed and interpreted the data. E.H. and M.H.B.-H. wrote the manuscript, with contributions from H. M.-R., I. I.-B., and C.O.
Competing interests: The authors have no competing interests regarding the data reported herein.
Figure S1: Tgfb1 WT and HT mammary serial transplantation capacity.
Figure S2: Representative FACS profiles of Tgfb1 WT and HT MEC.
Figure S3: Representative phase contrast images of mammospheres.
Figure S4: Phosphorylated SMAD2 (pSMAD2) intensity in mammary epithelial in mice treated with TGFβ neutralizing antibody.
Figure S6: Analysis of miR-181.
Figure S7: Analysis of miR-146.
Figure S8: Effects of miRZIP-182 expression in MEC.
Table S1: Primers.
References
- 1.Oshimori N, Fuchs E. Paracrine TGF-β Signaling Counterbalances BMP-Mediated Repression in Hair Follicle Stem Cell Activation. Cell stem cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silberstein GB, Daniel CW. Reversible inhibition of mammary gland growth by transforming growth factor-β. Science (New York, NY) 1987;237:291–293. doi: 10.1126/science.3474783. [DOI] [PubMed] [Google Scholar]
- 3.Ewan KB, et al. Latent TGF-β activation in mammary gland: Regulation by ovarian hormones affects ductal and alveolar proliferation. Am J Path. 2002;160:2081–2093. doi: 10.1016/s0002-9440(10)61158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewan KBR, et al. Proliferation of estrogen receptor-alpha-positive mammary epithelial cells is restrained by transforming growth factor-beta 1 in adult mice. Am J Pathol. 2005;167:409–417. doi: 10.1016/s0002-9440(10)62985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierce DFJ, et al. Inhibition of mammary duct development but not alveolar outgrowth during pregnancy in transgenic mice expressing active TGF-β 1. Genes Dev. 1993;7:2308–2317. doi: 10.1101/gad.7.12a.2308. [DOI] [PubMed] [Google Scholar]
- 6.Jhappan C, et al. Targeting expression of a transforming growth factor p1 transgene to the pregnant mammary gland inhibits alveolar development and lactation. EMBO J. 1993;12:1835–1845. doi: 10.1002/j.1460-2075.1993.tb05832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kordon EC, et al. Ectopic TGF beta 1 expression in the secretory mammary epithelium induces early senescence of the epithelial stem cell population. Dev Biol. 1995;168:47–61. doi: 10.1006/dbio.1995.1060. [DOI] [PubMed] [Google Scholar]
- 8.Salm SN, et al. TGF-{beta} maintains dormancy of prostatic stem cells in the proximal region of ducts. J Cell Biol. 2005;170:81–90. doi: 10.1083/jcb.200412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falk S, et al. Brain area-specific effect of TGF-beta signaling on Wnt-dependent neural stem cell expansion. Cell stem cell. 2008;2:472–483. doi: 10.1016/j.stem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 12.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 13.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 14.Van Keymeulen A, et al. Nature. advance online publication; 2011. Distinct stem cells contribute to mammary gland development and maintenance. [DOI] [PubMed] [Google Scholar]
- 15.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 16.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 17.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast cancer research: BCR. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nature medicine. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 20.Proia TA, et al. Genetic Predisposition Directs Breast Cancer Phenotype by Dictating Progenitor Cell Fate. Cell stem cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen DH, et al. Radiation Acts on the Microenvironment to Affect Breast Carcinogenesis by Distinct Mechanisms that Decrease Cancer Latency and Affect Tumor Type. Cancer Cell. 2011;19:640–651. doi: 10.1016/j.ccr.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie W, et al. Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: A tissue microarray study. Cancer Res. 2002;62:497–505. [PubMed] [Google Scholar]
- 23.Illa-Bochaca I, et al. Limiting-dilution transplantation assays in mammary stem cell studies. Methods Mol Biol. 2010;621:29–47. doi: 10.1007/978-1-60761-063-2_2. [DOI] [PubMed] [Google Scholar]
- 24.Guo W, et al. Slug and Sox9 Cooperatively Determine the Mammary Stem Cell State. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxwell CA, et al. Targeted and nontargeted effects of ionizing radiation that impact genomic instability. Cancer research. 2008;68:8304–8311. doi: 10.1158/0008-5472.CAN-08-1212. [DOI] [PubMed] [Google Scholar]
- 26.Ruffner H, Verma IM. BRCA1 is a cell cycle-regulated nuclear phosphoprotein. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7138–7143. doi: 10.1073/pnas.94.14.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaughn JP, et al. BRCA1 expression is induced before DNA synthesis in both normal and tumor-derived breast cells. Cell Growth Differ. 1996;7:711–715. [PubMed] [Google Scholar]
- 28.Moskwa P, et al. miR-182-Mediated Downregulation of BRCA1 Impacts DNA Repair and Sensitivity to PARP Inhibitors. Molecular cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia AI, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO molecular medicine. 2011;3:279–290. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisso A, et al. Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle. 2013;12:1679–1687. doi: 10.4161/cc.24757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segura MF, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moses H, Barcellos-Hoff MH. TGF-β Biology in Mammary Development and Breast Cancer. Cold Spring Harbor Perspectives in Biology. 2011;3:a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson SD, Silberstein GB, Roberts AB, Flanders KC, Daniel CD. Regulated expression and growth inhibitory effects of transforming growth factor-β isoforms in mouse mamary gland development. Development. 1991;113:867–878. doi: 10.1242/dev.113.3.867. [DOI] [PubMed] [Google Scholar]
- 34.Kong W, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature;2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda N, et al. TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. Journal of immunology. 2012;188:3323–3331. doi: 10.4049/jimmunol.1100876. [DOI] [PubMed] [Google Scholar]
- 37.Qin W, et al. TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephro. 2011;l22:1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, et al. TGFβ Induces “BRCAness” and Sensitivity to PARP Inhibition in Breast Cancer by Regulating DNA-Repair Genes. Molecular Cancer Research. 2014;12:1597–1609. doi: 10.1158/1541-7786.MCR-14-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li P, et al. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Research. 2014;16:473. doi: 10.1186/s13058-014-0473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang PY, et al. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol Lett. 2013;6:1681–1686. doi: 10.3892/ol.2013.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, et al. Prognostic Value of MicroRNA-182 in Cancers: A Meta-Analysis. Dis Markers. 2015;2015:482146. doi: 10.1155/2015/482146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rottenberg S, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pal B, et al. Integration of microRNA signatures of distinct mammary epithelial cell types with their gene expression and epigenetic portraits. Breast Cancer Research. 2015;17:85. doi: 10.1186/s13058-015-0585-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prater MD, et al. Mammary stem cells have myoepithelial cell properties. Nat Cell Biol. 2014;16:942–950. doi: 10.1038/ncb3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asselin-Labat ML, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 46.Bowcock AM, et al. THRA1 and D17S183 flank an interval of < 4 cM for the breast-ovarian cancer gene (BRCA1) on chromosome 17q21. Am J Hum Genet. 1993;52:718–722. [PMC free article] [PubMed] [Google Scholar]
- 47.Tang J, et al. Irradiation of juvenile, but not adult, mammary gland increases stem cell self-renewal and estrogen receptor negative tumors. Stem Cells. 2013;32:649–661. doi: 10.1002/stem.1533. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen DH, et al. Murine Microenvironment Metaprofiles Associate with Human Cancer Etiology and Intrinsic Subtypes. Clin Cancer Research. 2013;19:1353–1362. doi: 10.1158/1078-0432.CCR-12-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shelton DN, et al. Use of stem cell markers in dissociated mammary populations. Methods Mol Biol. 2010;621:49–55. doi: 10.1007/978-1-60761-063-2_3. [DOI] [PubMed] [Google Scholar]
- 50.Andarawewa KL, et al. Ionizing radiation predisposes nonmalignant human mammary epithelial cells to undergo transforming growth factor (3 induced epithelial to mesenchymal transition. Cancer research. 2007;67:8662–8670. doi: 10.1158/0008-5472.CAN-07-1294. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Z, et al. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. The Journal of pathology. 2012;228:204–215. doi: 10.1002/path.4000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.