Abstract
Quantification of DNA lesions constitutes one of the main tasks in toxicology and in assessing health risks accompanied by exposure to carcinogens. Tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) can undergo metabolic transformation to give a reactive intermediate that pyridyloxobutylates nucleobases and phosphate backbone of DNA. Here, we reported a highly sensitive method, relying on the use of nanoflow liquid chromatography-nanoelectrospray ionization-tandem mass spectrometry (nLC-nESI-MS/MS), for the simultaneous quantifications of O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine (O6-POBdG) as well as O2-and O4-[4-(3-pyridyl)-4-oxobut-1-yl]-thymidine (O2-POBdT and O4-POBdT). By using this method, we measured the levels of the three DNA adducts with the use of 10 µg of DNA isolated from cultured mammalian cells exposed to a model pyridyloxobutylating agent, 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone (NNKOAc). Our results demonstrated, for the first time, the formation of O4-POBdT in naked DNA and in genomic DNA of cultured mammalian cells exposed with NNKOAc. We also revealed that the levels of the three lesions increased with the dose of NNKOAc and that O2-POBdT and O4-POBdT could be subjected to repair by the nucleotide excision repair (NER) pathway. The method reported here will be useful for investigations about the involvement of other DNA repair pathways in the removal of these lesions and for human toxicological studies in the future.
Graphical abstract
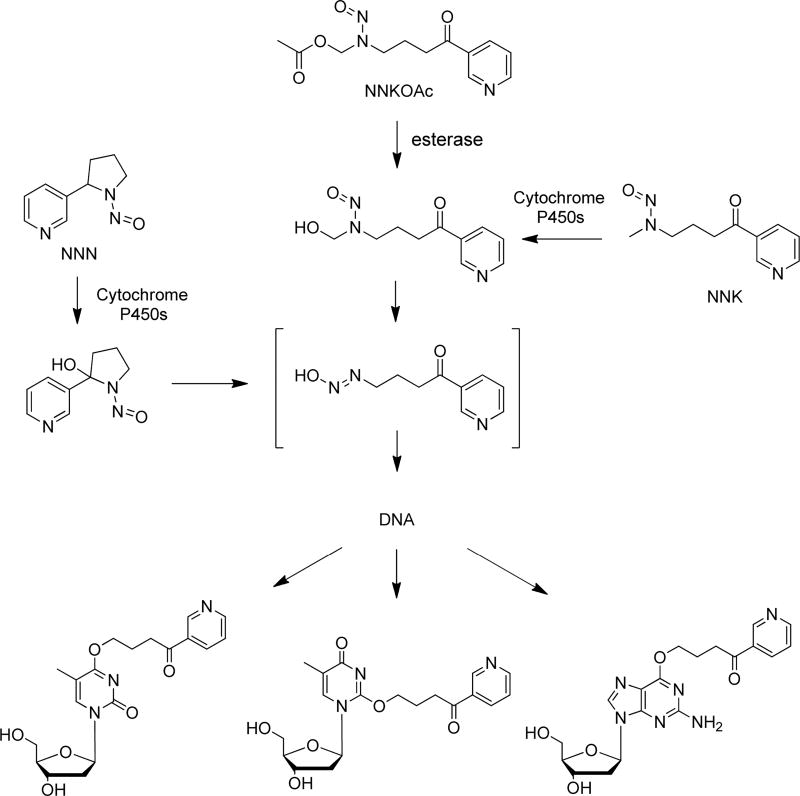
Human genome is constantly attacked by various toxic chemicals formed from endogenous metabolism and present in the environment, which can result in DNA damage and perturbation of genomic stability.1 Tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) are well-known carcinogens that induce cancer in rodents, and they are also considered as human carcinogens.2,3 The carcinogenic effects of NNK and NNN reside on their capability in inducing the formation of DNA adducts, which may give rise to mutations in DNA during DNA replication.2–4
Hecht and co-workers4,5 found that NNK and NNN, after metabolic activation by cytochrome P450 enzymes, can both give rise to a reactive intermediate that can pyridyloxobutylate DNA, and that of NNK can also lead to the methylation of DNA (Scheme 1). The contribution of the resulting O6-methylguanine (O6-mG) to the carcinogenic properties of NNK has been well documented,2 where a strong correlation between O6-mG levels measured at 96 h following NNK exposure and tumor multiplicity was observed.6 GC → AT transition is the major type of mutation induced by methylating agents, demonstrating the dominant role of O6-mG in the overall mutagenicity of these agents.7 Apart from methylation, the role of the pyridyloxobutylation pathway in carcinogenesis has also been investigated, where an increasing body of literature indicates that pyridyloxobutylated DNA lesions could contribute to the carcinogenic effects of NNK and NNN.8–14 In addition, 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone (NNKOAc), a model pyridyloxobutylating agent, is a lung carcinogen in A/J mice under chronic dosing conditions.6 Pyridyloxobutylated DNA adducts, 7-POBG, O2-POBdT, and O6-POBdG, persisted in lung DNA of A/J mice at significant levels for up to 96 h post-treatment.12 These DNA lesions also accumulate in normal lung tissues of lung cancer patients15 and in lung stem cells,22 indicating that the formation and persistence of these adducts may be important in tobacco-derived human lung cancer. Other types of pyridyloxobutylated DNA adducts were also discovered recently. For instance, Ma et al.23 characterized, by using liquid chromatography-nanoelectrospray ionization-high-resolution tandem mass spectrometry technique, pyridyloxobutyl phosphate adducts in NNKOAc-treated calf thymus DNA and in DNA isolated from tissues of rats exposed with NNK. More recently, Michel et al.24 identified O2-POBdC as the major, and N3-POBdC and N4-POBdC as the minor adducts of dC.
Scheme 1.
Activation of NNKOAc by Cellular Esterase and the Resulting Formation of O4-POBdT, O2-POBdT, and O6-POBdG
Repair studies have also been conducted for the pyridyloxobutylated DNA lesions. In this vein, O6-alkylguanine-DNA alkyltransferase (AGT) was found to be able to transfer the POB group from O6-POBdG in DNA to Cys145 in the protein.9,10,12,18 In addition, heightened AT → TA transversion mutations were observed for O2-POBdT in nucleotide excision repair (NER)-deficient cells, suggesting the involvement of NER in repairing this lesion.18
When not repaired, pyridyloxobutylated DNA adducts may perturb the transmission of genetic information by compromising the efficiency and fidelity of DNA replication and transcription. Previous studies have been conducted for assessing the cytotoxic and mutagenic properties of pyridyloxobutylated DNA lesions.16–20 Li et al.18 demonstrated that O6-POBdG is mutagenic in both bacterial and human cells. This adduct induced only GC → AT transition mutations in bacteria, whereas GC → TA transversions and more complex mutations were observed in the K-ras oncogene of NNKOAc-induced lung tumor in A/J mice.6 There were few reports about how O2-POBdT perturbs DNA replication,12,18,21 where the lesion was found to be strongly blocking to DNA replication, and direct significant frequencies of nucleotide misincorporations during replication in bacterial and human cells.21,25 Similar findings were made for other O2-alkylated thymidine lesions.26,27
Due to the increasing demand for the risk assessment of tobacco smoking and the resulting DNA pyridyloxobutylation in human carcinogenesis,28–30 it is important to establish reliable methods for the unequivocal identification and accurate quantification of pyridyloxobutylated DNA adducts. In this respect, 32P-postlabeling assay, immunoblot analysis and excision assay were used for measuring the levels and examining the repair of these DNA lesions in vitro and in vivo.16,17,20,21 Because of its high sensitivity and specificity, mass spectrometry has been extensively employed for the analyses of DNA adducts.31–36 There were several reports about the use of solid-phase extraction followed by LC-MS/MS analysis on a triple-quadrupole mass spectrometer to quantify different pyridyloxobutyl DNA adducts, directly or after conversion of the modified nucleosides to the respective modified nucleobases with acid hydrolysis.12,15,18
Here, we developed a highly sensitive nanoflow liquid chromatography-nanoelectrospray ionization tandem mass spectrometry (nLC-nESI-MS/MS) together with the stable isotope-dilution method for the measurements of O4-POBdT, O2-POBdT, and O6-POBdG, where O4-POBdT was identified here for the first time (Scheme 1). We also examined the dose-dependent formation and repair of these DNA lesions in repair-competent and NER-deficient human skin fibroblasts and Chinese hamster ovary cells.
EXPERIMENTAL SECTION
Materials
All enzymes and chemicals, if not specifically described, were obtained from Sigma-Aldrich (St. Louis, MO) or New England Biolabs (Ipswich, WA). NNKOAc was purchased from Toronto Research Chemicals Inc. (North York, Ontario). All stable isotope-labeled starting materials were purchased from Cambridge Isotope Laboratories (Cambridge, MA), and erythro-9-(2-hydroxy-3-nonyl)adenine (EHNA) hydrochloride was obtained from Tocris Bioscience (Ellisville, MO). Repair-competent AA8 Chinese hamster ovary (CHO) cells and the isogenic CHO cells depleted of excision repair cross-complementing rodent repair deficiency, complementation group 1 (ERCC1, CHO-7–27)37 were provided by M. M. Seidman (National Institute of Aging, Bethesda, MD). Human skin fibroblasts that are defective in xeroderma pigmentosum complementation group A (XPA, GM04429) or repair-proficient (GM00637) were kind gifts from G. P. Pfeifer (Van Andel Research Institute, Grand Rapids, MI).
Preparation of Standards
O2-POBdT, O6-POBdG, and their corresponding stable isotope-labeled derivatives were synthesized following previously described procedures (Scheme S1).38,39 Notably, O4-POBdT and its stable isotope-labeled counterpart were synthesized for the first time in the present study,41 and the synthetic route and the spectroscopic characterizations of this modified nucleoside are provided in the Supporting Information. In particular, exact mass measurement (Agilent 6210 ESI-TOF MS) yielded m/z 390.1689 and 394.1981 for the [M + H]+ ions of the O4-POBdT and [pyridine-D4]-O4-POBdT, respectively, which are in line with their calculated m/z of 390.1665 and 394.1916, respectively. The structure of O4-POBdT was also characterized by two-dimensional NMR spectroscopy, where the 1H–13C hetero-nuclear multiple bond correlation (HMBC) spectrum of the modified nucleoside revealed the correlation between the terminal hydrogen atoms of the POB moiety with the C4, but not the C2 of the pyrimidine ring, supporting that the POB moiety is attached to the O4, but not the O2 or N3 of dT (Figure S1).
Treatment of Calf Thymus DNA with NNKOAc and Esterase
Calf thymus DNA (50 µg) was incubated with 50 and 200 µg NNKOAc in the presence of porcine liver esterase (0.4 U) in 0.1 M phosphate buffer (400 µL, pH 7.0) at 37 °C for 1.5 h. The resulting solution was extracted sequentially with equal volumes of CHCl3/isoamyl alcohol (24:1) and ethyl acetate. The DNA in the aqueous layer was precipitated by adding cold ethanol, washed with 70% ethanol and then with pure ethanol, dried in air at room temperature, redissolved in water, and stored at −20 °C until enzymatic digestion and LC-MS/MS analysis.
Cell Culture and NNKOAc Treatment
Cells were maintained at 37 °C in a 5% CO2 atmosphere, where human skin fibroblasts were cultured in Dulbecco’s modified Eagle’s medium, and CHO cells were cultured in Alpha Minimum Essential Medium without ribonucleosides or 2′-deoxyribonucleosides. All culture media were supplemented with fetal bovine serum (10%, v/v) and penicillin (100 IU/mL). Cells (1–1.5 × 106) were seeded in 75 cm2 flasks in complete medium. At 24 h later, the cells were unexposed or exposed to 5, 10, or 25 µM of NNKOAc. After treatment for 24 h, the media were removed and the cells were washed with phosphate-buffered saline (1× PBS) for two times to remove residual medium and NNKOAc. For the repair study, the cells were subsequently cultured in the corresponding media at 37 °C for different time intervals to permit lesion repair. The cells were then detached by using trypsin-EDTA and harvested by centrifugation.
Extraction and Enzymatic Digestion of DNA
The experimental procedures for the extraction and enzymatic hydrolysis of genomic DNA, and HPLC enrichment of pyridyloxobutylated nucleosides were similar to those described previously.42,43 The details are provided in the online Supporting Information, and the HPLC enrichment trace is shown in Figure S2.
nLC-nESI-MS/MS Analysis
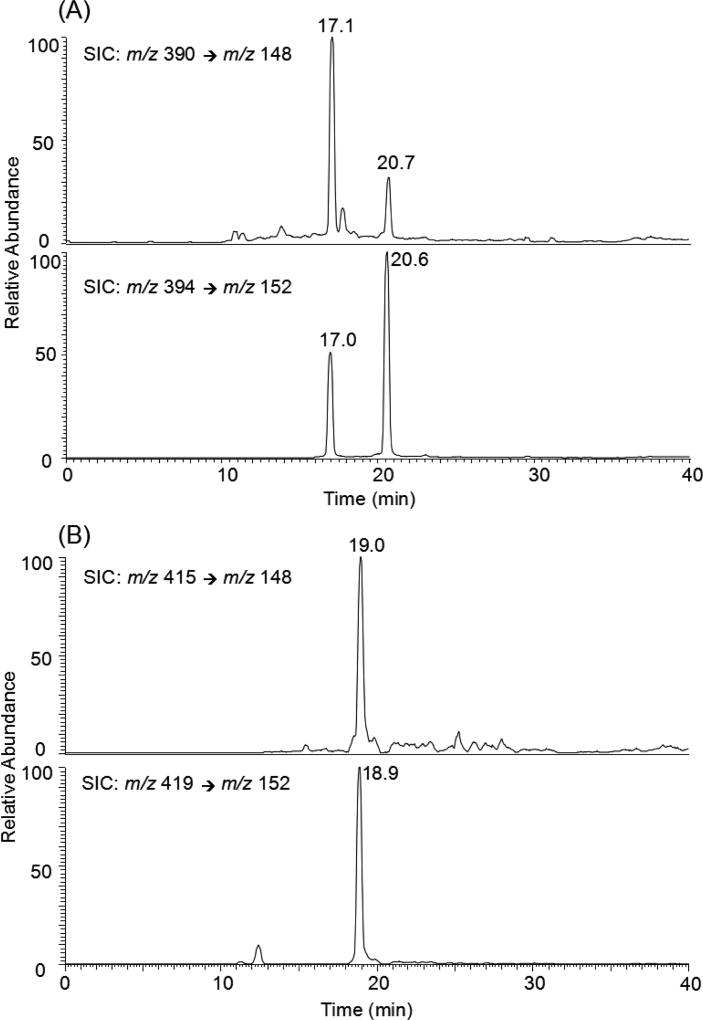
Online nLC-nESI-MS/MS analyses were performed on a TSQ-Vantage triple quadrupole mass spectrometer (Thermo Fisher Scientific) coupled with an EASY nLC II system (Thermo Fisher Scientific). The detailed experimental conditions for nLC separation are provided in the Supporting Information. The TSQ-Vantage mass spectrometer was set up in the multiple-reaction monitoring mode. We monitored the transitions corresponding to the neutral loss of an unmodified nucleoside (i.e., 242-Da for the modified dT derivatives, and 267 Da for the modified dG counterpart) from the [M + H]+ ions of the three modified nucleosides (i.e., m/z 390 → 148, 390 → 148, and 415 → 148 for O2-POBdT, O4-POBdT, and O6-POBdG, respectively) and their corresponding stable isotope-labeled derivatives (i.e., m/z 394 → 152, 394 → 152, and 419 → 152, Figure 1). The electrospray voltage was 2.0 kV and the temperature for the ion transfer tube was maintained at 275 °C. The width for parent ion isolation was 3 m/z units in MS/MS mode, and the collision energy was 15 V. The limit of quantitation (LOQ), reported as the amount of analyte giving a signal-to-noise ratio (S/N) of 10 in the selected-ion chromatograms (SICs) generated for the transitions employed for quantification, was obtained from three separate experiments.
Figure 1.
Representative selected-ion chromatograms (SICs) for monitoring the m/z 390 → 148 (A, top panel), 394 → 152 (A, bottom panel), 415 → 148 (B, top panel), and 419 → 152 (B, bottom panel) transitions for the [M + H]+ ions of the unlabeled and stable isotope-labeled O2- and O4-POBdT (A), and O6-POBdG (B), respectively, in the nucleoside mixture of DNA extracted from the CHO-7–27 cells treated with 10 µM NNKOAc for 24 h.
Method Development
The intra- and interday accuracy and precision were assessed by measuring quality control samples of O2-POBdT, O4-POBdT, and O6-POBdG at three different concentrations. The samples for calibration curve generation and quality control were prepared from 10 µg of calf thymus DNA mixed with standard solutions of the three modified oligodeoxyribonucleotides (ODNs) and the three stable isotope-labeled mononucleosides, following the same procedures of DNA digestion, HPLC enrichment, and LC-MS/MS analysis as described above for the cellular DNA samples. Each calibration curve was obtained from triplicate analyses, where the molar ratios of the unlabeled ODNs to their respective labeled mononucleoside adducts were 0.25, 0.50, 1.00, 1.25, 2.50, 5.00, and 10.0 for O2-POBdT and O6-POBdG, and 0.040, 0.080, 0.15, 0.30, 0.60, 1.00, and 2.00 for O4-POBdT. Data based on peak area ratios of responses of unlabeled/ labeled adduct standard versus the molar ratios of unlabeled/labeled adduct standard were then fitted to straight lines to yield the calibration curves (Figure S3). The quantities of the modified nucleosides (in moles) in the nucleoside mixtures were determined from the peak area ratios observed in the SICs for the analytes over their respective stable isotope-labeled standards, the number of moles of the labeled standards added, and the calibration curves. The final DNA lesion levels, reported as the numbers of lesions per 108 nucleosides, were determined by dividing the number of moles of the DNA adducts by the total number of moles of nucleosides in the DNA digestion mixture.
RESULTS
The primary goal of this study is to establish a robust nLC-nESI-MS/MS in combination with the stable isotope-dilution method for the measurements of O-pyridyloxobutylated dT and dG lesions in cellular DNA.
Preparations of Unlabeled and Stable Isotope-Labeled Standards
We first prepared the unlabeled O2-POBdT, O4-POBdT, and O6-POBdG, and their corresponding stable isotope-labeled derivatives (see Experimental Section). In this context, it is worth noting that O4-POBdT is a novel pyridyloxobutylated DNA adduct, and we synthesized this modified nucleoside from the reaction of 4-hydroxy-1-(pyridin-3-yl)butan-1-one with O4-(1,2,4-triazolyl)-substituted dT. For the chemical syntheses of stable isotope-labeled standards, we employed [D4]-4-hydroxy-1-(pyridin-3-yl)butan-1-one to react with the activated forms of O2-dT, O4-dT, and O6-dG, where we were not able to detect any appreciable H/D exchange for the D4-labeled nucleosides at room temperature over a month.
nLC-nESI-MS/MS for the Quantifications of O2-POBdT, O4-POBdT, and O6-POBdG
We next established an LC-MS/MS method for the sensitive and accurate measurements of O2-POBdT, O4-POBdT, and O6-POBdG. In this respect, we first digested cellular DNA so as to release these lesions as mononucleosides using a combination of four enzymes, as described in the Experimental Section. The stable isotope-labeled O2-POBdT, O4-POBdT, and O6-POBdG were added before DNA digestion, which corrects for potential loss of analytes in the following sample preparation. We then assessed the LOQs of the nLC-nESI-MS/MS method prior to the analyses of cellular DNA samples by using pure unlabeled standards. It turned out that the LOQs for O2-POBdT, O4-POBdT, and O6-POBdG were 6.1, 4.6, and 1.8 amol, respectively. We also examined the intra- and interday accuracy and precision (n = 3) by measuring calf thymus DNA samples doped with different amounts of lesion-containing ODNs. Our results showed that the method offers reasonably good precision (4.0–13.7%) and accuracy (86.2–96.1%) for measuring these three lesions (Table 1).
Table 1.
Intraday and Interday Precision and Accuracy for the Measurements of O2-POBdT, O4-POBdT, and O6-POBdG
| intraday | interday | |||
|---|---|---|---|---|
|
|
|
|||
| ODN amts (fmol) |
precision (%) |
accuracy (%) |
precision (%) |
accuracy (%) |
| O2-POBdT | ||||
| 5 | 7.7 | 94.4 | 11.8 | 91.7 |
| 15 | 6.5 | 88.6 | 10.5 | 92.0 |
| 50 | 5.9 | 90.0 | 12.3 | 87.2 |
| O4-POBdT | ||||
| 0.5 | 8.8 | 86.2 | 13.1 | 89.1 |
| 1.5 | 4.5 | 89.3 | 12.9 | 93.2 |
| 5 | 7.6 | 92.6 | 9.7 | 88.4 |
| O6-POBdG | ||||
| 5 | 7.4 | 92.8 | 8.5 | 90.3 |
| 15 | 5.3 | 93.6 | 12.4 | 96.1 |
| 50 | 4.0 | 88.7 | 13.7 | 90.9 |
The detection of low levels of DNA lesions are often significantly affected by relatively large amounts of unmodified nucleosides. To address this issue, we adopted offline HPLC to enrich O2-POBdT, O4-POBdT, and O6-POBdG from the nucleoside mixtures before analysis using nLC-nESI-MS/MS. In this aspect, owing the better sensitivity provided by the positive-than negative-ion mode, we measured the pyridyloxobutylated nucleosides by operating the mass spectrometer in the positive-ion mode, where the mobile phase contained 0.1% formic acid (v/v) for promoting analyte protonation. Figure S4 illustrates the proposed fragmentation pathways for the three modified nucleosides.
Dose-Dependent Formation of O2-POBdT, O4-POBdT, and O6-POBdG in Mammalian Cells
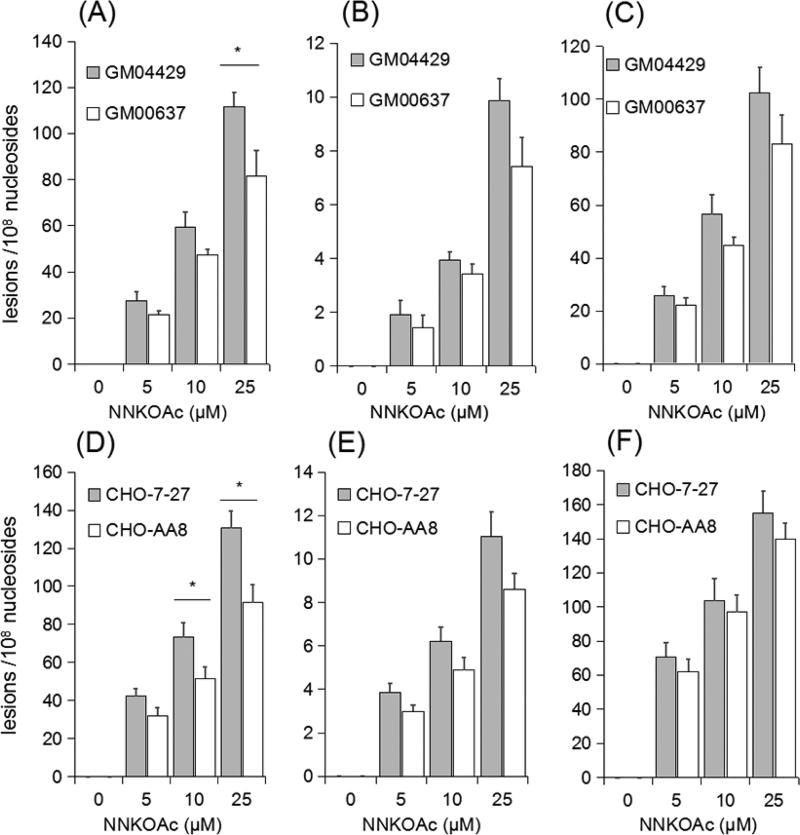
After establishing a robust nLC-nESI-MS/MS method, we subsequently measured the frequencies of O2-POBdT, O4-POBdT, and O6-POBdG in genomic DNA isolated from human skin fibroblasts and Chinese hamster ovary cells exposed with different concentrations of NNKOAc. The quantification data revealed a dose-dependent formation of the three modified nucleosides in these cells (Figure 2). For instance, as the dose of NNKOAc was elevated from 5 to 25 µM, the levels of O4-POBdT in the NER-deficient GM04429 cells and repair-proficient GM00637 cells increased from 1.3 to 10.9 and from 0.9 to 8.9 lesions per 108 nucleosides, respectively (Figure 2B). Similar dose-dependent elevation in the levels of O4-POBdT were found in the NER-deficient CHO (7–27) and repair-proficient CHO (AA8) cells, that is, from 3.3 to 12.4 and from 2.6 to 9.2 lesions per 108 nucleosides, respectively (Figure 2E). Furthermore, all three modified nucleosides were not detectable in control samples without NNKOAc exposure, indicating the absence of endogenous agents that can induce DNA pyridyloxobutylation.
Figure 2.
LC-MS/MS quantification results for O2-POBdT (A, D), O4-POBdT (B, E), and O6-POBdG (C, F) in DNA samples isolated from human skin fibroblast cells (A–C) that are repair-proficient (GM00637) or deficient in XPA (GM04429) and Chinese hamster ovary cells (D–F) that are repair-competent (CHO-AA8) or deficient in ERCC1 (CHO-7–27) exposed to different concentrations of NNKOAc for 24 h. The data represent mean ± S. D. (n = 3). *p < 0.05. Unpaired, two-tailed Student’s t-test was employed for calculating the p values.
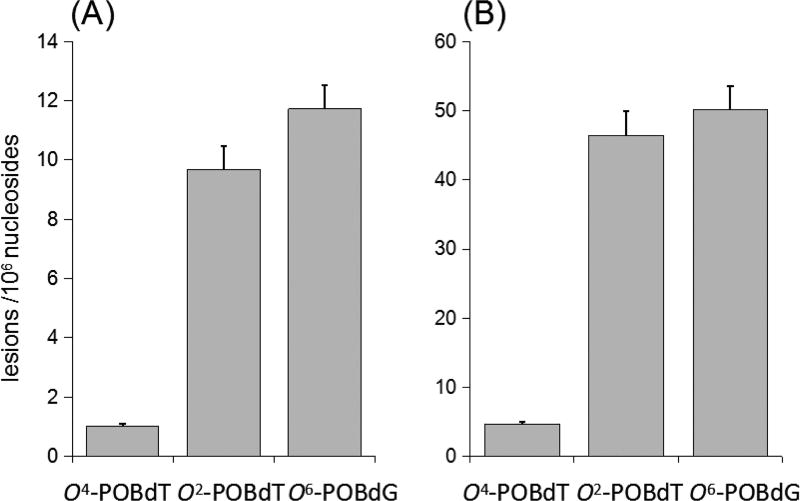
Our quantification data also revealed that, following NNKOAc exposure, O2-POBdT and O6-POBdG were accumulated at levels that are at least 10-fold higher than O4-POBdT in mammalian cells (Figure 2). The more pronounced accumulation of O2-POBdT over the regioisomeric O4-POBdT could be attributed to the preferential formation and/or less-efficient repair of the former lesion. To examine whether O2-POBdT could be induced more preferentially than O4-POBdT by the pyridyloxobutylating agent, we treated calf thymus DNA with NNKOAc and porcine liver esterase at 37 °C for 1.5 h, and measured the levels of the three lesions by the same LCMS/ MS method (Figure 3). Our results indeed revealed the preferential formation O2-POBdT over O4-POBdT, demonstrating that the higher level of accumulation of the former lesion arises, at least in part, from the its higher rate of formation than O4-POBdT. Additionally, the relative levels of O2-POBdT and O6-POBdG were in keeping with what was reported previously for calf thymus DNA exposed to NNKOAc.39,40
Figure 3.
Frequencies of formation of O4-POBdT, O2-POBdT, and O6-POBdG in calf thymus DNA treated with 50 (A) and 200 (B) µg NNKOAc, together with porcine liver esterase. The data represent mean ± SD (n = 3).
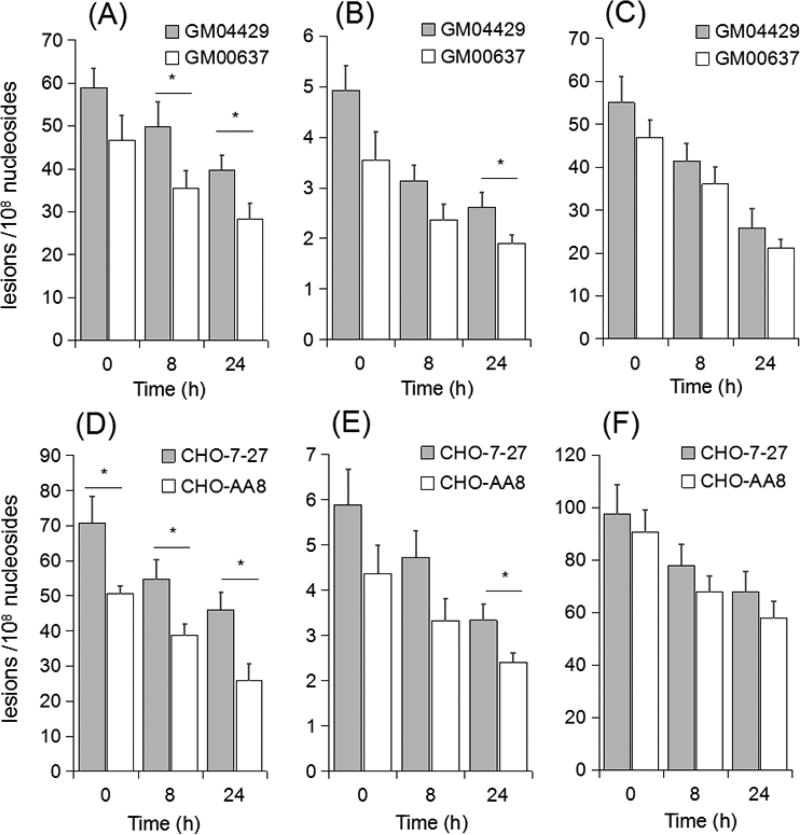
Repair of O2-POBdT, O4-POBdT, and O6-POBdG in Cells
Our above quantification data showed that the repair-proficient cells exhibited lower levels of the three lesions relative to XPA-deficient GM04429 cell and ERCC1-deficient CHO-7–27 cells, especially for O2-POBdT, suggesting that NER may play an important role in repairing these lesions. To examine this aspect further, we next monitored the removal of the three pyridyloxobutyl DNA adducts in the aforementioned cells at 0, 8, and 24 h after exposure to 10 µM NNKOAc (Figure 4). This concentration was employed since it was only slightly toxic and resulted in less than 25% of cell death. It turned out that deficiency in NER did not confer any statistically significant difference in the levels of O6-POBdG at 24 h following exposure to NNKOAc. The lesion was repaired in human skin fibroblast cells by 24 h but persisted in CHO cells, consistent with previous reports showing that O6-POBdG is readily repaired by AGT and there is no AGT activity in CHO cells.9,10,12,18
Figure 4.
LC-MS/MS for monitoring the repair of O2-POBdT (A, D), O4-POBdT (B, E), and O6-POBdG (C, F) in human skin fibroblast (A–C) and Chinese hamster ovary (D–F) cells following a 24 h treatment with 10 µM NNKOAc. The data represent mean ± S. D. (n = 3). *p < 0.05. The p values were calculated using unpaired, two-tailed Student’s t-test.
The levels of O2-POBdT, however, were significantly lower in NER-proficient GM00637 and CHO-AA8 cells than the corresponding NER-deficient cells (i.e., GM04429 and CHO-7–27) at all three time points (i.e., 0, 8 and 24 h) after NNKOAc treatment. In addition, the rates of removal of O2-POBdT were similar in the repair-competent GM00637 and CHO-AA8 cells. Whereas there was no apparent difference in the removal of O4-POBdT in human skin fibroblast cells and CHO cells at 0 and 8 h following NNKOAc exposure, we observed significantly different levels of O4-POBdT at 24 h following the exposure, suggesting the involvement of NER in repairing O4-POBdT.
DISCUSSION
Because of its specificity, accuracy and sensitivity, LC-MS/MS in combination with the stable isotope-dilution technique constitutes a reliable analytical method for the measurement of DNA lesions in complex biological matrices.31–36 Superior to traditional methods for DNA adduct measurements (e.g., immunoassay and 32P-postlabeling), this method not only enables accurate identification (i.e., by providing structural information), but also offers reliable quantification for various modified nucleosides.34,44 In addition, we added the stable isotope-labeled standards of the lesions to the nucleoside mixture before enzymatic digestion, and the levels of the lesions were measured from the molar ratios of the analytes over their respective stable isotope-labeled standards. Hence, the quantifications of these lesions are not influenced by variations in experimental conditions of enzymatic digestion, HPLC enrichment, and LC–MS/MS measurement. It is also worth noting that, different from previously reported methods for quantifying the relevant DNA adducts,45,46 the calibration curves reported in the present study were constructed by spiking calf thymus DNA with lesion-carrying ODNs. This allows for the correction of potential incomplete release of the modified nucleosides from DNA. Furthermore, when compared to the previous solid-phase extraction method for DNA adduct enrichment,45,46 the off-line HPLC enrichment used in the this study provides much better elimination of unmodified nucleosides and buffer salts added during the enzymatic digestion, thereby providing better sensitivity for measuring these modified nucleosides.
Mammalian cells are equipped with a battery of DNA repair mechanisms to remove various types of DNA lesions from genomic DNA, thereby maintaining genomic stability. In this study, we carefully compared the adduct levels in four lines of mammalian cells that are deficient or proficient in NER pathway. Our results revealed that the formation of O2-POBdT, O4-POBdT, and O6-POBdG in genomic DNA of mammalian cells increase with the dose of NNKOAc. In addition, we found that the levels of O6-POBdG of human skin fibroblast cells were much lower than those of Chinese hamster ovary (CHO) cells, which is in keeping with the known role of AGT in repairing this lesion and the lack of AGT in CHO cells.9,10,12,18 We also observed that the extents of the removal of O6-POBdG following NNKOAc exposure were not altered by deficiency in NER. By contrast, the NER repair pathway plays a significant role in repairing O2-POBdT. The levels of O2-POBdT were significantly lower in NER-proficient GM00637 and CHO-AA8 cells than the corresponding NER-deficient cells (i.e., GM04429 and CHO-7–27). Moreover, O4-POBdT was produced at much lower levels than O2-POBdT and O6-POBdG in all four lines of mammalian cells exposed to NNKOAc. This finding, along with the measurement of these three lesions in calf thymus DNA showed that O4-POBdT is less preferentially formed than O2-POBdT and O6-POBdG. Lastly, our results support that the NER pathway is involved in the repair of the minor-groove O2-POBdT lesion and, to a lesser degree, the major-groove O4-POBdT lesion.
In summary, we reported, for the first time, the formation of O4-POBdT in mammalian cells upon exposure to NNKOAc, a model pyridyloxobutylating agent as well as the simultaneous quantifications of O2-POBdT, O4-POBdT, and O6-POBdG in mammalian cells exposed to NNKOAc with the use of off-line HPLC enrichment in combination with nLC-nESI-MS/MS with the stable isotope-dilution method. The robust analytical method reported here may serve as a powerful tool for studying the repair of these lesions and for exploring the use of pyridyloxobutylated DNA lesions as biomarkers for tobacco smoking-induced cancer in the future.
Supplementary Material
Acknowledgments
The authors thank the National Institutes of Health for supporting this research (R01 ES025121).
Footnotes
ASSOCIATED CONTENT
- Details for the chemical synthesis of O4-POBdT, its spectroscopic characterizations, and detailed experimental procedures for the extraction and enzymatic digestion of DNA, HPLC enrichment of modified nucleosides, and online nLC separation. Figure S1 (selected region of 1H–13C HMBC spectrum of O4-POBdT), Figure S2 (HPLC trace for the enrichment of O4-POBdT, O2-POBdT, and O6-POBdG from the enzymatic digestion mixture of genomic DNA isolated from NNKOAc-treated cells), Figure S3 (calibration curves for the quantifications of O4-POBdT, O2-POBdT, and O6-POBdG), Figure S4 (proposed fragmentation pathways for the observed fragment ions in MS/MS; PDF).
The authors declare no competing financial interest.
References
- 1.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS. Chem. Res. Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. J. Natl. Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS. Mutat. Res., Fundam. Mol. Mech. Mutagen. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 5.Hecht SS, Villalta PW, Sturla SJ, Cheng G, Yu N, Upadhyaya P, Wang M. Chem. Res. Toxicol. 2004;17:588–597. doi: 10.1021/tx034263t. [DOI] [PubMed] [Google Scholar]
- 6.Ronai ZA, Gradia S, Peterson LA, Hecht SS. Carcinogenesis. 1993;14:2419–2422. doi: 10.1093/carcin/14.11.2419. [DOI] [PubMed] [Google Scholar]
- 7.Horsfall MJ, Gordon AJ, Burns PA, Zielenska M, van der Vliet GM, Glickman BW. Environ. Mol. Mutagen. 1990;15:107–122. doi: 10.1002/em.2850150208. [DOI] [PubMed] [Google Scholar]
- 8.Hecht SS. Mutat. Res., Fundam. Mol. Mech. Mutagen. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 9.Peterson LA, Liu XK, Hecht SS. Cancer Res. 1993;53:2780–2785. [PubMed] [Google Scholar]
- 10.Mijal RS, Thomson NM, Fleischer NL, Pauly GT, Moschel RC, Kanugula S, Fang Q, Pegg AE, Peterson LA. Chem. Res. Toxicol. 2004;17:424–434. doi: 10.1021/tx0342417. [DOI] [PubMed] [Google Scholar]
- 11.Staretz ME, Foiles PG, Miglietta LM, Hecht SS. Cancer Res. 1997;57:259–266. [PubMed] [Google Scholar]
- 12.Urban AM, Upadhyaya P, Cao Q, Peterson LA. Chem. Res. Toxicol. 2012;25:2167–2178. doi: 10.1021/tx300245w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, Wang M, Villalta PW, Lindgren BR, Lao Y, Hecht SS. Chem. Res. Toxicol. 2009;22:949–956. doi: 10.1021/tx900040j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L, Balbo S, Wang M, Upadhyaya P, Khariwala SS, Villalta PW, Hecht SS. Chem. Res. Toxicol. 2013;26:1526–1535. doi: 10.1021/tx400235x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holzle D, Schlobe D, Tricker AR, Richter E. Toxicology. 2007;232:277–285. doi: 10.1016/j.tox.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Brown PJ, Bedard LL, Massey TE. Cancer Lett. 2008;260:48–55. doi: 10.1016/j.canlet.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Brown PJ, Massey TE. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2009;663:15–21. doi: 10.1016/j.mrfmmm.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Perdigao J, Pegg AE, Lao Y, Hecht SS, Lindgren BR, Reardon JT, Sancar A, Wattenberg EV, Peterson LA. Chem. Res. Toxicol. 2009;22:1464–1472. doi: 10.1021/tx9001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotandeniya D, Murphy D, Yan S, Park S, Seneviratne U, Koopmeiners JS, Pegg A, Kanugula S, Kassie F, Tretyakova N. Biochemistry. 2013;52:4075–4088. doi: 10.1021/bi4004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi JY, Guengerich FP. J. Biol. Chem. 2008;283:23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weerasooriya S, Jasti VP, Bose A, Spratt TE, Basu AK. DNA Repair. 2015;35:63–70. doi: 10.1016/j.dnarep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balbo S, Upadhyaya P, Villalta PW, Qian X, Kassie F. Chem. Res. Toxicol. 2013;26:511–513. doi: 10.1021/tx400054s. [DOI] [PubMed] [Google Scholar]
- 23.Ma B, Villalta PW, Zarth AT, Kotandeniya D, Upadhyaya P, Stepanov I, Hecht SS. Chem. Res. Toxicol. 2015;28:2151–2159. doi: 10.1021/acs.chemrestox.5b00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel AK, Zarth AT, Upadhyaya P, Hecht SS. ACS Omega. 2017;2:1180–1190. doi: 10.1021/acsomega.7b00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jasti VP, Spratt TE, Basu AK. Chem. Res. Toxicol. 2011;24:1833–1835. doi: 10.1021/tx200435d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai Q, Wang P, Wang Y. Carcinogenesis. 2014;35:2002–2006. doi: 10.1093/carcin/bgu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhai Q, Wang P, Cai Q, Wang Y. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson LA, Hecht SS. Curr. Opin. Pediatr. 2017;29:225–230. doi: 10.1097/MOP.0000000000000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht SS. Int. J. Cancer. 2012;131:2724–2732. doi: 10.1002/ijc.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecht SS. Chem. Res. Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balbo S, Turesky RJ, Villalta PW. Chem. Res. Toxicol. 2014;27:356–366. doi: 10.1021/tx4004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J, Yun BH, Upadhyaya P, Yao L, Krishnamachari S, Rosenquist TA, Grollman AP, Turesky RJ. Anal. Chem. 2016;88:4780–4787. doi: 10.1021/acs.analchem.6b00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao S, Guo J, Yun BH, Villalta PW, Krishna S, Tejpaul R, Murugan P, Weight CJ, Turesky RJ. Anal. Chem. 2016;88:12508–12515. doi: 10.1021/acs.analchem.6b04157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Wang Y. Chem. Soc. Rev. 2015;44:7829–7854. doi: 10.1039/c5cs00316d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumpathong W, Chan W, Taghizadeh K, Babu IR, Dedon PC. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E4845–4853. doi: 10.1073/pnas.1503945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monien BH, Schumacher F, Herrmann K, Glatt H, Turesky RJ, Chesne C. Anal. Chem. 2015;87:641–648. doi: 10.1021/ac503803m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolig RL, Layher SK, Santi B, Adair GM, Gu F, Rainbow AJ, Nairn RS. Mutagenesis. 1997;12:277–283. doi: 10.1093/mutage/12.4.277. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Spratt TE, Pegg AE, Peterson LA. Chem. Res. Toxicol. 1999;12:127–131. doi: 10.1021/tx980251+. [DOI] [PubMed] [Google Scholar]
- 39.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Chem. Res. Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturla SJ, Scott J, Lao Y, Hecht SS, Villalta PW. Chem. Res. Toxicol. 2005;18:1048–1055. doi: 10.1021/tx050028u. [DOI] [PubMed] [Google Scholar]
- 41.Xu YZ, Swann PF. Nucleic Acids Res. 1990;18:4061–4065. doi: 10.1093/nar/18.14.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Yuan B, Guerrero C, Bahde R, Gupta S, Wang Y. Anal. Chem. 2011;83:2201–2209. doi: 10.1021/ac103099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y, Wang J, Wang P, Wang Y. Anal. Chem. 2016;88:8036–8042. doi: 10.1021/acs.analchem.6b01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Y, Cui Y, Niedernhofer LJ, Wang Y. Chem. Res. Toxicol. 2016;29:2008–2039. doi: 10.1021/acs.chemrestox.6b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanden Bussche J, Moore SA, Pasmans F, Kuhnle GG, Vanhaecke L. J. Chromatogr. A. 2012;1257:25–33. doi: 10.1016/j.chroma.2012.07.040. [DOI] [PubMed] [Google Scholar]
- 46.Da Pieve C, Sahgal N, Moore SA, Velasco-Garcia MN. Rapid Commun. Mass Spectrom. 2013;27:2493–2503. doi: 10.1002/rcm.6709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.