Abstract
The SNP variant rs2943650 near IRS1 gene locus was previously associated with decreased body fat and IRS1 gene expression as well as an adverse metabolic profile in humans. Here, we hypothesize that these effects may be mediated by an interplay with epigenetic alterations. We measured IRS1 promoter DNA methylation and mRNA expression in paired human subcutaneous and omental visceral adipose tissue samples (SAT and OVAT) from 146 and 41 individuals, respectively. Genotyping of rs2943650 was performed in all individuals (N = 146). We observed a significantly higher IRS1 promoter DNA methylation in OVAT compared to SAT (N = 146, P = 8.0 × 10−6), while expression levels show the opposite effect direction (N = 41, P = 0.011). OVAT and SAT methylation correlated negatively with IRS1 gene expression in obese subjects (N = 16, P = 0.007 and P = 0.010). The major T-allele is related to increased DNA methylation in OVAT (N = 146, P = 0.019). Finally, DNA methylation and gene expression in OVAT correlated with anthropometric traits (waist- circumference waist-to-hip ratio) and parameters of glucose metabolism in obese individuals. Our data suggest that the association between rs2943650 near the IRS1 gene locus with clinically relevant variables may at least be modulated by changes in DNA methylation that translates into altered IRS1 gene expression.
Introduction
Body fat composition is a more precise parameter than body mass index (BMI) to analyze fat distribution. To identify genetic factors influencing body fat mass and body fat distribution large, genome wide association studies (GWAS) have been conducted1–3 Genes in close vicinity of the identified genetic variants and conferring important pathophysiological functions in metabolism are plausible candidate genes for further analysis on the link between adverse fat distribution, metabolic and cardiovascular diseases4. One such example is the previously reported single nucleotide polymorphism (SNP) rs2943650 near the insulin receptor substrate 1 (IRS1) gene locus. T-allele carriers showed decreased body fat percentage along with an unfavorable metabolic profile including decreased high density lipoprotein- (HDL) cholesterol, increased triglyceride concentrations and insulin resistance1. IRS1 is an important player in insulin signaling being one of the most relevant proteins able to bind the phosphorylated insulin receptor to activate downstream cascades5,6. IRS1 knock out mouse models further suggests that IRS1 plays a role in adipocyte differentiation7. Interestingly, the SNP variant rs2943650 was further shown to be associated with reduced subcutaneous adipose tissue in men1. Kilpeläinen et al. hypothesized that the SNP may lead to a predominant deposition of fat into visceral depots thereby contributing to insulin resistance and dyslipidemia, despite an overall decrease in body fat percentage. Furthermore, the body fat decreasing T-allele was shown the be associated with reduced IRS1 mRNA expression in human subcutaneous and omental adipose tissue (SAT and OVAT)1,8.
However, it is still an open question by which mechanisms rs2943650 contributes to regulation of IRS1 expression and subsequent alterations in metabolic variables.
Changes in DNA methylation pattern were described as highly dynamic in response to weight loss after bariatric surgery9, exercise10,11 and between different adipose tissue compartments12. Previously, we identified differences in DNA methylation between SAT and OVAT13,14. It has been demonstrated that genetic variants influence the DNA methylation of nearby CpG sites by introducing or deleting CpG sites (CpG-SNPs)15–17. Several CpG-SNPs, which partially change the methylation at the corresponding and/or surrounding CpG sites associate with alternative splicing events in human pancreatic islets16. A recent study identified a haplotype within TOMM20 which is related to changes in the methylation state of a core CpG site matching a relevant transcription factor binding site and to altered LDL-cholesterol levels in patients with metabolic syndrome18. These and several further studies provide evidence for triangular relationships between genetic variants, DNA methylation and gene expression and/or diseases state19–21. Hence, we hypothesized a potential interplay between the genetic variant rs2943650 near IRS1 and epigenetic variation, such as DNA methylation within its promoter region.
To test whether the observed effects of rs2943650 near IRS1 on expression and metabolic profiles can be mediated by an interplay with DNA methylation, we measured i) differential IRS1 DNA methylation and mRNA expression in paired samples of human SAT vs. OVAT. We tested for ii) correlation of DNA methylation with IRS1 gene expression levels and analyzed for association of rs2943650 effect allele (T) with both methylation and expression. iii) Finally, we performed correlation analysis to assess potential relationships between DNA methylation, mRNA expression and rs2943650 genotypes with metabolic and anthropometric variables.
Methods
Study population
A total of 146 individuals (men N = 55; women N = 91) from a Caucasian cohort (Germany) were included for DNA methylation analysis and SNP genotyping. Main characteristics of the study cohort are reported in Table 1. Paired samples of SAT and OVAT were obtained during open abdominal surgery for e.g. sleeve gastrectomy, Roux-en-Y gastric bypass, gastric banding, cholecystectomy, abdominal injuries or explorative laparotomy. Individuals with body weight fluctuations (partially self-reported) in the last 3 months before surgery were excluded (>2% fluctuations of body weight). mRNA expression data from either SAT or OVAT was available for 63 individuals14. Paired expression data from OVAT and SAT was available from 41 individuals and was used for phenotype-association analysis per adipose tissue depot. For subsequent analyses comparing both tissue depots we used 28 of these subjects for which overlapping paired methylation and gene expression data from SAT and OVAT were available. All analyses were performed in obesity-subgroups (lean, overweight and obese) if not described otherwise. Phenotyping of the study participants was performed as previously described22 and included anthropometric measurements (body weight, height, waist-to-hip-ratio (WHR)), body fat mass using bioimpedance analysis or dual-energy X-ray absorptiometry. Furthermore, metabolic parameters such as fasting plasma glucose (FPG) and insulin (FPI), a 75 g oral glucose tolerance test (OGTT), glycated haemoglobin (HbA1c), lipoprotein-, triglyceride-, free fatty acid- and cytokine- serum concentrations were obtained. Insulin sensitivity was assessed with hyperinsulinemic-euglycemic clamps. Determination of visceral- and subcutaneous fat areas was performed based on computed tomography or MRI scans. By setting a ratio of visceral/subcutaneous fat area of 0.5 obese individuals were further divided being either viscerally or subcutaneously obese (<0.5 = subcutaneous obesity; >0.5 = visceral obesity; 0.5 = obese (not specified)). The study was approved by the Ethics Committee of the University of Leipzig (approval no: 159-12-21052012), and performed in accordance to the declaration of Helsinki. All subjects gave written informed consent before taking part in this study.
Table 1.
Characteristics of the study cohort.
| Total | Lean | Overweight | Obese | P-valuelean vs obese | |
|---|---|---|---|---|---|
| N | 146 | 50 | 28 | 68 | 50/68 |
| Sex (m/f) | 55/91 | 20/30 | 14/14 | 21/47 | — |
| T2D (yes/no) | 42/104 | 7/43 | 7/21 | 28/40 | — |
| Age (years) | 58 ± 16 | 66 ± 12 | 67 ± 12 | 49 ± 15 | n.s. |
| Weight (kg) | 100 ± 43 | 63 ± 10 | 78 ± 11 | 137 ± 36 | <0.001 |
| Height (m) | 1.68 ± 0.90 | 1.67 ± 0.8 | 1.70 ± 0.12 | 1.68 ± 0.81 | n.s. |
| BMI (kg/m2) | 35.3 ± 14.7 | 22.4 ± 2.5 | 26.9 ± 1.4 | 48.3 ± 11.7 | <0.001 |
| Visceral fat area (cm²) | 178 ± 160 | 54 ± 41.5 | 134 ± 76 | 296 ± 161 | <0.001 |
| Subcutanoues fat area (cm²) | 720 ± 775 | 55.5 ± 27.5 | 271 ± 173 | 1438 ± 601 | <0.001 |
| CT-ratio (OVAT/SAT fat area) | 0.66 ± 0.89 | 1.21 ± 1.25 | 0.63 ± 0.40 | 0.22 ± 0.11 | <0.001 |
| Waist circumference (cm) | 106 ± 32 | 79 ± 17 | 94.5 ± 15 | 134 ± 21 | 0.022 |
| Hip circumference (cm) | 113 ± 30 | 87.5 ± 11 | 100 ± 12 | 140 ± 24 | <0.001 |
| Waist-to-hip ratio | 0.94 ± 0.12 | 0.89 ± 0.11 | 0.95 ± 0.12 | 0.96 ± 0.13 | n.s. |
| Body fat % | 30.9 ± 12.8 | 20.8 ± 4.7 | 25.9 ± 4.6 | 43.3 ± 10.2 | <0.001 |
| Fasting plasma glucose (mmol/l) | 6.03 ± 1.76 | 5.57 ± 1.00 | 5.95 ± 1.33 | 6.40 ± 2.23 | 0.011 |
| 2 hr OGTT plasma glucose (mmol/l) | 6.91 ± 2.76 | 6.18 ± 1.49 | 6.2 ± 1.02 | 7.85 ± 3.69 | n.s. |
| Fasting plasma Insulin (mmol/l) | 73.12 ± 120.87 | 11.77 ± 22.18 | 49.30 ± 57.68 | 138.83 ± 157.71 | <0.001 |
| HbA1c % | 5.80 ± 0.78 | 5.43 ± 0.54 | 5.79 ± 0.67 | 6.10 ± 0.86 | <0.001 |
| HDL cholesterol (mmol/l) | 1.36 ± 0.43 | 1.48 ± 0.49 | 1.39 ± 0.33 | 1.19 ± 0.37 | n.s. |
| LDL cholesterol (mmol/l) | 3.20 ± 1.09 | 3.08 ± 1.13 | 3.42 ± 0.93 | 3.21 ± 1.13 | n.s. |
| Triglycerides (mmol/l) | 1.39 ± 0.74 | 1.29 ± 0.67 | 1.21 ± 0.43 | 1.57 ± 0.88 | n.s. |
| Adiponectin (ug/ml) | 9.32 ± 5.74 | 13.74 ± 6.78 | 8.94 ± 3.86 | 6.59 ± 3.36 | <0.001 |
N = Number of subjects; T2D = diagnosed Type 2 Diabetes; BMI = Body mass index (WHO classification: lean ≥ 18; <25 kg/m²; <30 kg/m²obese ≥ 30 kg/m²); SAT = subcutaneous adipose tissue; OVAT = omental visceral adipose tissue; OGTT = oral glucose tolerance test; HDL = high density lipoprotein cholesterol; LDL = low density lipoprotein cholesterol. P-values were generated using unpaired t-tests.
DNA extraction and bisulfite conversion
DNA extraction and bisulfite conversion was carried out in the entire cohort (N = 146). After collecting adipose tissue, samples were frozen in liquid nitrogen and stored at −80 °C. Genomic DNA was extracted using GenEluteTM Mammalian Genomic DNA Miniprep Kit (SIGMA-ALDRICH, USA) and were bisulfite converted using Qiagen EpiTect Fast DNA Bisulfite Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols.
Analysing CpG methylation within IRS1 promoter region
To analyse DNA methylation at CpG sites we used the Hs_IRS1_05_PM PyroMark CpG assay (PM00011711) provided by Qiagen (Qiagen, Hilden, Germany) including 5 CpG sites in the promoter region of IRS1 (Fig. 1). Due to technical reasons (optimization of dispensation order in the PyroMark run) only 4 out of 5 CpG sites were analysed. Pyrosequencing was run on a PyroMark Q24 with subsequent analysis of the obtained results via PyroMark Q24 software, version 2.0.6 (Qiagen, Hilden, Germany). For replication purposes each DNA sample was PCR amplified and analysed twice on different plates. Calculating the interday coefficients of variance (CVs) for replicates results in 0.355 for SAT and 0.298 for OVAT. For statistical analysis the mean percentage of replicates for methylation at every CpG site was determined as well as the mean methylation level across all four analysed CpG sites. Two none template controls per plate containing water were included in PCR reactions and subsequent pyrosequencing analysis.
Figure 1.
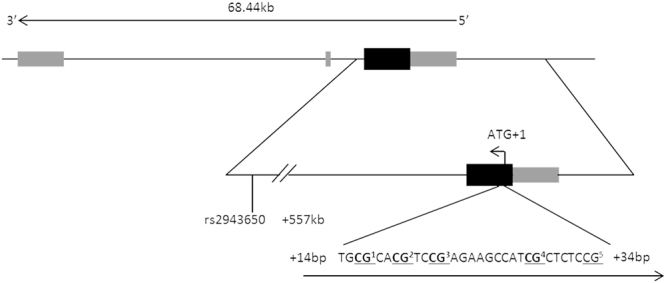
Schematic representation of IRS1 gene locus, analyzed CpG locus and SNP location. IRS1 exons are shown as filled boxes (black = coding exons; grey = non-coding exons). The CpG locus and analyzed SNP variant rs2943650 (C/T) are shown relative to the translation start site (ATG + 1). Analyzed CpGs are highlighted in bold and numbered according to order of analysis (CpG5 was excluded from analysisdue to optimization of dispensation order in the PyroMark run). Figure not scaled. kb = kilobases; bp = base pairs.
Genotyping of rs2943650 near IRS1
Genotyping of rs2943650 near the IRS1 locus was performed in the same 146 individuals for whom methylation analyses were applied (Fig. 1). Genomic DNA was extracted from adipose tissue using GenEluteTM Mammalian Genomic DNA Miniprep Kit (SIGMA-ALDRICH, USA) according to manufacturer,s instructions. Genotypes of rs2943650 were generated by using the KASPar genotyping system (KBioscience allele specific PCR Genotyping System; KBioscience, Teddington, Middlesex, UK). Fluorescence measurement was done with an ABI 7500 Real-Time PCR system. All genotypes were in Hardy-Weinberg equilibrium (P > 0.05). The minor allele frequency (MAF) of rs2943650 is 0.42. As we analysed a single SNP missingness per individual is negligible. Errors of genotyping were excluded by random re-genotyping (~5%) of the samples; all genotypes matched the initially designated genotypes. Genotypes of four subjects could not be examined due to technical reasons (uncertainty in allelic discrimination due to low DNA content), which results in a SNP call rate of 97.3%. Water was used as a no template control (NTC).
Gene expression analysis
mRNA expression data (IRS1 transcript: ILMN_1874569) was available for 63 subjects (total N = 63, SAT = 56, OVAT = 48) and was extracted from genome wide data achieved by Illumina human HT-12 expression arrays14. Corresponding methylation data were generated by pyrosequencing for a subset of these subjects (total N = 60, SAT = 40, OVAT = 32). Paired SAT and OVAT expression data per sample were available for 41 subjects. Complete methylation and expression data in both adipose tissue depots per sample were available for 28 subjects.
Statistical analysis
We used non-parametric tests as well as bivariate correlation analysis using residuals to adjust for potential confounders such as age, sex and BMI. DNA methylation levels at four individual CpG sites and mean methylation across the entire CpG locus were used as continuous variables. To analyse for differences in methylation/expression levels between OVAT and SAT in the entire cohort and also gender specific, Wilcoxon rank-sum tests were used. Mann-Whitney-U tests were performed to test for differences between obesity subgroups (lean, overweight and obese subgroup). Spearman correlation coefficients were used to assess bivariate correlation (ß = correlation coefficient) with measurements for anthropometric and metabolic traits using residuals to control for covariates sex, age and BMI. Genetic association of rs2943650 with DNA methylation, mRNA expression as well as metabolic and anthropometric traits was tested by using additive (MM vs Mm vs mm; M = major allele, m = minor allele), dominant (MM + Mm vs mm) and recessive modes (MM vs Mm + mm) of inheritance during linear regression analysis adjusted for study specific covariates (sex, age, BMI). All non-normally distributed variables have been log transformed prior to linear regression analysis. Bonferroni correction was used to take into account multiple testing (number of clinical variables*2 adipose tissue depots*3 inheritance models). We lowered the significance threshold to P = 4.9 × 10−4 (0.05/(17 * 2 * 3) = 4.9 × 10−4). Moreover, all P-values > 4.9 × 10−4 but ≤ 0.05 were considered nominally significant. All P-values are provided uncorrected for multiple testing. Statistical analysis were performed using SPSS statistics version 20.0.1 (SPSS, Inc.; Chicago, IL).
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the University of Leipzig, and performed in accordance to the declaration of Helsinki. All subjects gave written informed consent before taking part in this study.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Results
rs2943650 near IRS1 and relationship to clinical variables, IRS1 mRNA expression and DNA methylation
To dissect whether the reported IRS1 genetic variant relates to clinically important variables in our cohort we applied genetic association analysis in the total cohort (N = 142; 146 minus four drop outs). Although non-significant, we observed among T allele carriers an unfavourable metabolic profile (such as increased triglyceride levels, increased plasma glucose and insulin levels) along with lower body weight, BMI, waist, hip and visceral and subcutaneous fat area (Table 2) which is in line with previously reported data1. We also tested for association between rs2943650 and methylation/expression levels from SAT and OVAT. We observed significantly increased DNA methylation in OVAT among T-allele carriers which withstands adjustment for the covariates sex, age, BMI (Table 2) and also T2D, while IRS1 mRNA expression is, albeit non-significant, decreased in the same individuals.
Table 2.
Association between IRS1 variant rs2943650 and IRS1 methylation- and expression levels as well as quantitative traits.
| IRS1 rs2943650 | |||
|---|---|---|---|
| Genotype (N) | TT (49) | TC (67) | CC (26) |
| Association analysis with methylation level | |||
| Mean_4CpGs_SAT | 3.23 ± 1.72 | 3.07 ± 1.54 | 2.78 ± 1.43 |
| P-value | °0.278 #0.249 *0.512 | ||
| Mean_4CpGs_OVAT | 4.31 ± 2.22 | 4.26 ± 1.95 | 3.05 ± 1.4 |
| P-value | °0.096 #0.019 *0.592 | ||
| Association analysis with expression level | |||
| mRNA expression_SAT | −0.062 ± 0.139 | 0.057 ± 0.181 | 0.12 ± 0.126 |
| P-value | °0.270 #0.489 *0.303 | ||
| mRNA expression OVAT | −0.85 ± 0.123 | 0.003 ± 0.11 | 0.023 ± 0.229 |
| P-value | °0.101 #0.323 *0.144 | ||
| Association analysis with anthropometric traits | |||
| Weight (kg) | 97.9 ± 43.6 | 100.4 ± 43.5 | 111.5 ± 42.9 |
| P-value | °0.893 #0.398 *0.621 | ||
| Visceral fat area (cm²) | 163 ± 142 | 182 ± 176 | 215 ± 152 |
| P-value | °0.840 #0.298 *0.557 | ||
| Subcutaneous fat area (cm²) | 564 ± 687 | 751 ± 801 | 991 ± 829 |
| P-value | °0.482 #0.295 *0.868 | ||
| CT-ratio (OVAT/SAT fat area) | 0.96 ± 1.36 | 0.52 ± 0.51 | 0.47 ± 0.51 |
| P-value | °0.669 #0.942 *0.559 | ||
| BMI (kg/m²) | 34 ± 15 | 36 ± 15 | 38 ± 15 |
| P-value | °0.459 #0.564 *0.534 | ||
| Waist circumference (cm) | 104.8 ± 29.5 | 104.6 ± 33.8 | 118.3 ± 30.2 |
| P-value | °0.996 #0.128 *0.197 | ||
| Hip circumference (cm) | 109.2 ± 25.7 | 113.0 ± 31.2 | 124.9 ± 33.6 |
| P-value | °0.620 #0.446 *0.165 | ||
| Waist-to-hip ratio | 0.95 ± 0.13 | 0.92 ± 0.12 | 0.95 ± 0.11 |
| P-value | °0.707 #0.342 *0.813 | ||
| body fat (%) | 30.2 ± 13.5 | 32.3 ± 13.2 | 29.9 ± 9.1 |
| P-value | °0.445 #0.346 *0.675 | ||
| Association analysis with glucose/insulin metabolism | |||
| Fasting plasma glucose (mmol/l) | 6.41 ± 1.77 | 5.84 ± 1.93 | 5.85 ± 1.33 |
| P-value | °0.457 #0.813 *0.195 | ||
| 2 h OGTT plasma glucose (mmol/l) | 6.80 ± 2.17 | 7.18 ± 3.47 | 6.56 ± 1.43 |
| P-value | °0.975 #0.714 *0.798 | ||
| HbA1c (%) | 5.98 ± 0.92 | 5.68 ± 0.64 | 5.83 ± 0.77 |
| P-value | °0.824 #0.406 *0.319 | ||
| Fasting plasma Insulin (pmol/l) | 97.74 ± 164.59 | 62.45 ± 105.66 | 70.46 ± 68.3 |
| P-value | °0.988 #0.119 *0.166 | ||
| Association analysis with lipid metabolism | |||
| HDL cholesterol (mmol/l) | 1.33 ± 0.46 | 1.4 ± 0.47 | 1.31 ± 0.31 |
| P-value | °0.846 #0.810 *0.923 | ||
| LDL cholesterol (mmol/l) | 3.39 ± 1.04 | 3.19 ± 1.17 | 3.13 ± 0.85 |
| P-value | °0.694 #0.955 *0.532 | ||
| Triglycerides (mmol/l) | 1.49 ± 0.72 | 1.35 ± 0.85 | 1.30 ± 0.47 |
| P-value | °0.659 #0.961 *0.544 | ||
| Adiponectin (µg/ml) | 7.96 ± 4.54 | 10.17 ± 6.18 | 8.39 ± 5.17 |
| P-value | °0.541 #0.672 *0.229 | ||
N = number f subjects (total cohort N = 142) (T2D: N = 42 with TT = 19; TC = 16; CC = 7); P-values were calculated using °additive (MM vs Mm vs mm), #dominant (MM + Mm vs mm) and *recessive (MM vs Mm + mm) modes of inheritance by linear regression analysis adjusted for age, sex and logBMI (except for BMI). Nominal significant P-values are highlighted in bold. SAT = subcutaneous adipose tissue, OVAT = omental visceral adipose tissue; BMI = body mass index; OGTT = oral glucose tolerance test; HDL = high density lipoprotein cholesterol; LDL = low density lipoprotein cholesterol.
Inter-depot specific DNA methylation within IRS1 promoter region
We measured IRS1 DNA methylation at one CpG-locus including 4 CpG sites within the promoter region of IRS1 in 146 paired samples of SAT and OVAT. In the entire cohort, mean SAT methylation (average of all 4CpGs = 3.08 ± 1.57%) was significantly lower than OVAT methylation (average of all 4CpGs = 4.05 ± 1.98%, P < 0.0001, Fig. 2) which was also nominally significant for all 4 CpG sites separately (all P < 0.05). Similar directions were observed in BMI stratified subgroups of lean, overweight and obese individuals. CpG2 in OVAT is the only site representing slightly significant higher methylation levels in obese than in lean subjects (P = 0.003). Results are shown in Fig. 2.
Figure 2.
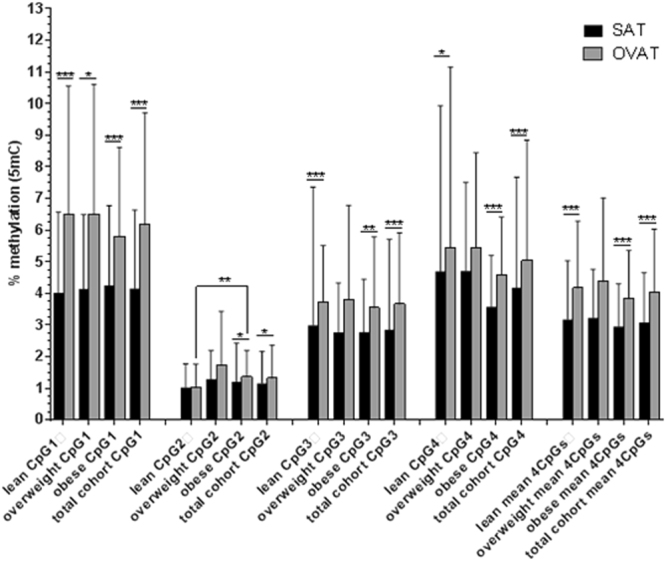
DNA methylation at one CpG locus within IRS1 promoter in SAT vs OVAT in BMI subgroups. Data are presented as mean ± standard deviation (S.D.). Number of participants: lean = 50; overweight = 28; obese = 68; total cohort = 146. P-values were calculated using non-parametric tests; significance of p-values are indicated as following *<0.05; **<0.001; ***<0.0001. 5mC = 5methyl cytosine; SAT = subcutaneous adipose tissue; OVAT = omental visceral adipose tissue.
Furthermore, no significant methylation differences were observed between subjects with (N = 42) and without type 2 diabetes (N = 104; data not shown). In the entire cohort (N = 146) the described inter-depot specific differences in IRS1 DNA methylation levels between SAT and OVAT are stronger in women (men P = 0.001; women P = 3.8 × 10−5, data not shown) which might be due to differences in sample sizes (men N = 55; women N = 91).
DNA methylation correlates with measures of fat distribution and glucose/lipid metabolism
To assess potential relationships of IRS1 promoter methylation and metabolic and anthropometric variables, we performed bivariate Spearman correlation analysis in all samples (N = 146). By analysing BMI stratified subgroups we observed in obese individuals that OVAT methylation positively correlates with variables of fat distribution such as waist (P = 0.006; ß = 0.349), and WHR (P = 0.038; ß = 0.266) and markers of glucose metabolism such as fasting plasma glucose (P = 0.026; ß = 0.271). HDL-cholesterol levels are negatively related to OVAT methylation (P = 0.013; ß = −0.415).
Further, in lean subjects, SAT methylation positively correlates with HDL cholesterol (P = 0.031; ß = 0.341) while OVAT methylation is related negatively to body fat percentage (P = 0.032; ß = −0.373).
In the overweight subgroup, HDL-cholesterol is negatively related to SAT methylation (P = 0.044; ß = −0.444) and subcutaneous fat area is negatively correlated with OVAT methylation (P = 0.041; ß = −0.412). All data are adjusted for sex, age and BMI and are summarized in Table 3. Additional adjustment for T2D results in the loss of association to FPG (data not shown). However, none of the here reported results withstands correction for multiple testing.
Table 3.
Correlation analysis of mean CpG locus methylation with quantitative phenotypes.
| Quantitative trait | N* | Total cohort ( N = 146) | Lean (N = 50) | Overweight (N = 28) | Obese (N = 68) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean CpG locus methylation in | |||||||||
| SAT | OVAT | SAT | OVAT | SAT | OVAT | SAT | OVAT | ||
| Weight | 146/50/28/68 | 0.813a [0.020]b | 0.467 [−0.061] | 0.061 [−0.267] | 0.846 [0.028] | 0.175 [0.264] | 0.130 [−0.293] | 0.490 [0.085] | 0.717 [0.045] |
| Visceral fat area (cm²) | 135/49/25/61 | 0.928 [−0.008] | 0.716 [0.032] | 0.502[0098] | 0.435 [0.114] | 0.060 [−0.382] | 0.216 [−0.256] | 0.871 [0.021] | 0.222 [0.159] |
| Subcutanoues fat area (cm²) | 135/49/25/61 | 0.181 [0.116] | 0.948 [−0.006] | 0.134 [0.217] | 0.862 [−0.026] | 0.875 [−0.033] | 0.041 [−0.412] | 0.411 [0.107] | 0.665 [0.057] |
| CT−ratio (OVAT/SAT fat area) | 135/49/25/61 | 0.633 [0.042] | 0.272 [0.095] | 0.959 [0.008] | 0.723 [−0.052] | 0.852 [0.039] | 0.230 [0.249] | 0.247 [0.151] | 0.595 [−0.069] |
| BMI (kg/m2) | 146/50/28/68 | 0.469 [0.060] | 0.731 [0.029] | 0.790 [−0.039] | 0.145 [−0.209] | 0.301 [0.203] | 0.312 [0.198] | 0.318 [0.123] | 0.210 [−0.154] |
| Waist circumference (cm) | 139/50/28/61 | 0.372 [0.076] | 0.280 [0.092] | 0.736 [0.049] | 0.536 [−0.090] | 0.595 [0.105] | 0.327 [−0.192] | 0.636 [0.062] | 0.006 [0.349] |
| Hip circumference (cm) | 139/50/28/61 | 0.088 [0.145] | 0.603 [−0.044] | 0.512 [0.095] | 0.259 [−0.163] | 0.353 [0.182] | 0.816 [−0.046] | 0.294 [0.136] | 0.847 [−0.025] |
| Waist-to-hip ratio | 139/50/28/61 | 0.873 [0.014] | 0.574 [0.048] | 0.878 [0.022] | 0.467 [−0.105] | 0.857 [−0.036] | 0.446 [−0.150] | 0.863 [−0.023] | 0.038 [0.266] |
| Body fat (%) | 81/33/15/33 | 0.161 [−0.157] | 0.384 [−0.098] | 0.370 [0.161] | 0.032 [−0.373] | 0.071 [−0.479] | 0.830 [−0.061] | 0.074 [−0.315] | 0.875 [0.028] |
| Fasting plasma glucose (mmol/l) | 146/50/28/68 | 0.590 [0.045] | 0.168 [0.115] | 0.419 [0.117] | 0.671 [−0.062] | 0.551 [0.118] | 0.914 [0.021] | 0.943 [−0.009] | 0.026 [0.271] |
| 2 hr OGTT plasma glucose (mmol/l) | 75/32/10/33 | 0.510 [0.077] | 0.980 [−0.003] | 0.465 [0.134] | 0.595 [−0.098] | 0.533 [−0.224] | 0.777 [−0.103] | 0.519 [0.116] | 0.856 [0.033] |
| Fasting plasma Insulin (pmol/l) | 123/46/25/52 | 0.838 [0.019] | 0.757 [0.028] | 0.371 [0.135] | 0.595 [−0.097] | 0.606 [−0.108] | 0.679 [−0.087] | 0.981 [−0.003] | 0.179 [0.189] |
| HbA1c (%) | 137/49/27/61 | 0.798 [0.022] | 0.775 [0.025] | 0.386 [0.127] | 0.774 [−0.042] | 0.626 [0.098] | 0.417 [−0.163] | 0.585 [−0.071] | 0.114 [0.204] |
| HDL cholesterol (mmol/l) | 96/40/21/35 | 0.697 [0.040] | 0.083 [−0.178] | 0.031 [0.341] | 0.631 [−0.078] | 0.044 [−0.444] | 0.695 [0.091] | 0.799 [−0.045] | 0.013 [−0.415] |
| LDL cholesterol (mmol/l) | 93/41/21/31 | 0.931 [0.009] | 0.310 [−0.106] | 0.241 [0.187] | 0.121 [−0.246] | 0.427 [−0.183] | 0.729 [−0.081] | 0.660 [−0.082] | 0.236 [0.219] |
| Triglycerides (mmol/l) | 86/32/18/36 | 0.806 [0.027] | 0.696 [−0.043] | 0.973 [0.006] | 0.130 [−0.273] | 0.179 [0.331] | 0.345 [−0.236] | 0.621 [−0.085] | 0.293 [0.180] |
| Adiponectin (µg/ml) | 98/32/17/49 | 0.777 [−0.029] | 0.642 [0.048] | 0.068 [−0.326] | 0.938 [−0.014] | 0.256 [0.292] | 0.765 [0.078] | 0.729 [0.051] | 0.429 [0.116] |
WHO classification: lean ≥ 18; <25 kg/m²; overweight ≥ 25 kg/m²; <30 kg/m²; obese ≥ 30 kg/m²; SAT (subcutaneous adipose tissue); OVAT (omental visceral adipose tissue), N = subjects (N* in total cohort/lean/overweight/obese individuals, respectively); a = P-value. P-values were calculated using bivariate Spearman correlation analysis (adjusted for age, sex and BMI (except for BMI) by calculating standardized residuals); nominal significant P-values are highlighted in bold. b = beta (ß; effect size and direction), BMI = body mass index; OGTT = oral glucose tolerance test; HDL = high density lipoprotein cholesterol; LDL = low density lipoprotein cholesterol.
IRS1 mRNA expression is adipose tissue depot specific
Expression data either from SAT or OVAT were extracted from genome wide expression data reported previously14 for 63 subjects. To compare SAT and OVAT expression level, we lowered the sample size to N = 41 – subjects representing paired SAT and OVAT expression data. We observed significantly higher IRS1 mRNA expression in SAT compared to OVAT in the entire cohort (N = 41; P = 0.0004) and obese individuals (N = 28; P = 0.001; Fig. 3) while this relationship is non-significant in lean subjects. No significant differences in IRS1 mRNA expression were detected in men vs women while T2D patients (N = 10) show lower IRS1 expression ( = mean −0.04 ± SD 0.14) than subjects without T2D (N = 31; = mean 0.08 ± SD 0.15) in SAT (P = 0.032).
Figure 3.
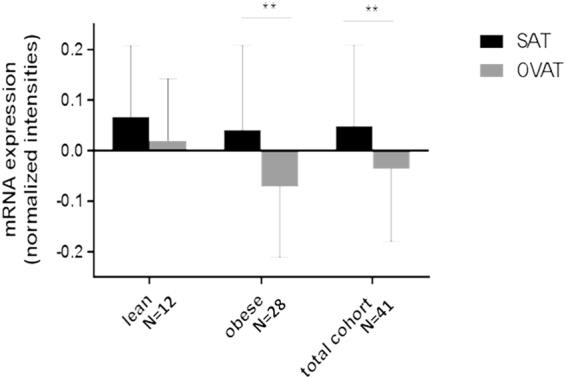
IRS1 mRNA expression in SAT and OVAT. Total N = 41 represent paired SAT and OVAT expression data per sample. Overweight subgroup was excluded due to small sample size (N = 1). Data are presented as mean ± standard deviation. P-values were calculated using non-parametric t-tests. SAT = subcutaneous adipose tissue; OVAT = omental visceral adipose tissue.
IRS1 mRNA expression and DNA methylation are correlated in obese subjects
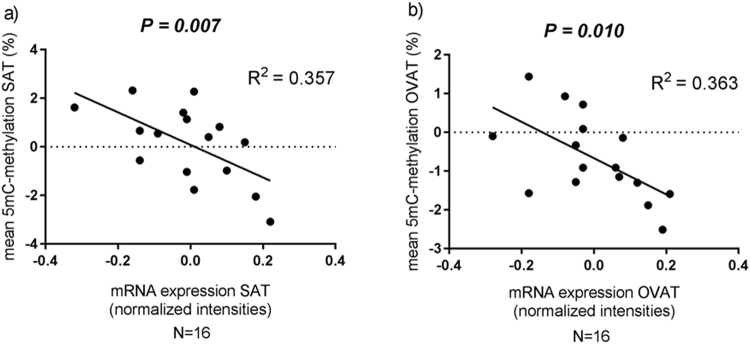
In order to better understand whether DNA methylation may impact on gene expression we performed correlation analysis in all individuals for whom DNA methylation and expression were available in both SAT and OVAT (N = 28). Among obese individuals (N = 16), DNA methylation of IRS1 and its gene expression are negatively correlated in both SAT and OVAT (Fig. 4) and withstand adjustment for T2D (data not shown).
Figure 4.
Correlation analysis of IRS1 DNA methylation and mRNA gene expression in SAT and OVAT among obese subjects. (a) correlation analysis in SAT; (b) correlation analysis in OVAT. Data were calculated by using Spearman correlation analysis by using standardized residuals to adjust for age, sex and BMI. 5mC = 5methyl cytosine; SAT = subcutaneous adipose tissue; OVAT = omental visceral adipose tissue.
IRS1 mRNA expression is related to variables of glucose metabolism
To analyse whether the IRS1 mRNA expression relates to clinically relevant variables, we conducted bivariate Spearman correlation analysis in the total cohort and in BMI stratified subgroups. In order to compare results, these analysis were performed among all 60 individuals from whom overlapping methylation and expression data in either SAT or OVAT were available. All data were adjusted for age, sex and BMI (summarized in Table 3). Albeit not statistically significant we observed in the entire cohort and in subgroups nominal significant relationships of IRS1 expression levels and variables of glucose metabolism such as fasting and/or 2 h plasma glucose levels (total cohort: fasting plasma glucose (P = 0.046; ß = −0.317); lean: fasting plasma glucose (P = 0.046; ß = −0.463) and 2 h plasma glucose levels (P = 0.023; ß = −0.582); obese: 2 h plasma glucose levels (P = 0.047; ß = 0.714). Only the association between SAT IRS1 mRNA expression and FPG in lean individuals survives additional adjusting for T2D (P = 0.021; ß = −0.589, data not shown). However, none of these associations would withstand correction for multiple testing. Moreover, they confer different effect directions between obese individuals and the remainder (Table 4 ).
Table 4.
Correlation analysis between IRS1 mRNA expression and quantitative phenotypes.
| Quantitative trait | N* | Total cohort (N = 60) | Lean (N = 23) | Obese (N = 34) | |||
|---|---|---|---|---|---|---|---|
| mean IRS1 mRNA expression in | |||||||
| SAT | OVAT | SAT | OVAT | SAT | OVAT | ||
| SAT/OVAT | |||||||
| total – lean – obese | |||||||
| Weight | 40/32 – 19/14 – 19/17 | 0.270a [0.179]b | 0.239 [−0.214] | 0.663 [0.107] | 0.464 [−0.213] | 0.473 [0.175] | 0.451 [−0.196] |
| Visceral fat area (cm²) | 34/27 – 19/14 – 13/12 | 0.260 [−0.198] | 0.058 [0.370] | 0.229 [−0.289] | 0.474 [0.209] | 0.201 [−0.379] | 0.255 [0.357] |
| Subcutanoues fat area (cm²) | 34/27 – 19/14 – 13/12 | 0.890 [0.025] | 0.249 [−0.230] | 0.842 [0.049] | 0.899 [−0.037] | 0.494 [0.209] | 0.208 [−0.392] |
| CT-ratio (OVAT/SAT fat area) | 34/27 – 19/14 – 13/12 | 0.713 [−0.066] | 0.866 [0.034] | 0.943 [0.018] | 0.455 [0.218] | 0.803 [0.077] | 0.779 [0.091] |
| BMI kg/m2 | 40/32 – 19/14 – 19/17 | 0.274 [−0.177] | 0.236 [−0.216] | 0.065 [0.432] | 0.605 [0.152] | 0.344 [−0.230] | 0.248 [−0.297] |
| Waist circumference (cm) | 34/27 – 19/14 – 13/12 | 0.701 [−0.068] | 0.696 [−0.079] | 0.391 [−0.209] | 0.681 [−0.121] | 0.873 [0.049] | 0.795 [−0.084] |
| Hip circumference (cm) | 34/27 – 19/14 – 13/12 | 0.926 [0.017] | 0.583 [−0.111] | 0.586 [0.133] | 0.543 [−0.178] | 0.845 [0.060] | 0.430 [−0.252] |
| Waist-to-hip ratio | 34/27 – 19/14 – 13/12 | 0.598 [−0.094] | 0.854 [−0.037] | 0.057 [−0.444] | 0.899 [0.037] | 0.721 [0.110] | 0.914 [−0.035] |
| Body fat (%) | 23/18 – 11/8 – 11/10 | 0.511 [−0.144] | 0.829 [−0.055] | 0.484 [0.236] | 0.823 [−0.095] | 0.650 [0.155] | 0.881 [0.055] |
| Fasting plasma glucose (mmol/l) | 40/32 – 19/14 – 19/17 | 0.046 [−0.317] | 0.655[−0.082] | 0.046 [−0.463] | 0.840 [−0.059] | 0.088 [−0.402] | 0.866 [−0.044] |
| 2 hr OGTT plasma glucose (mmol/l) | 25/17 – 15/8 – 8/8 | 0.303 [−0.215] | 0.135 [0.377] | 0.023 [−0.582] | 0.911 [0.048] | 0.352 [0.381] | 0.047 [0.714] |
| Fasting plasma Insulin (pmol/l) | 38/29 – 19/13 – 17/15 | 0.282 [−0.179] | 0.190 [0.251] | 0.367 [−0.219] | 0.972 [0.011] | 0.593 [−0.140] | 0.405 [0.232] |
| HbA1c (%) | 39/31 – 19/14 – 18/16 | 0.382 [−0.144] | 0.763 [0.056] | 0.178 [−0.323] | 0.615 [0.147] | 0.576 [−0.141] | 0.854 [0.050] |
| HDL cholesterol (mmol/l) | 27/22 – 15/11 – 10/10 | 0.765 [0.060] | 0.334 [−0.216] | 0.820 [0.064] | 0.160 [−0.455] | 0.651 [−0.164] | 0.934 [−0.030] |
| LDL cholesterol (mmol/l) | 27/23 – 15/12 – 10/10 | 0.947 [0.013] | 0.477 [0.156] | 1.000 [0.000] | 0.245 [0.364] | 0.726 [−0.127] | 0.446 [0.273] |
| Triglycerides (mmol/l) | 22/18 – 11/8 – 10/9 | 0.393 [−0.191] | 0.705 [0.096] | 0.937 [0.027] | 0.823 [−0.095] | 0.934 [0.030] | 0.668 [0.167] |
| Adiponectin (µg/ml) | 26/21 – 12/9 – 13/12 | 0.350 [0.191] | 0.475 [−0.165] | 0.880 [−0.049] | 0.460 [−0.283] | 0.061 [0.533] | 0.319 [−0.315] |
all subjects representing methylation and expression data in either SAT (N = 40) or OVAT (N = 32) have been included while N* = subjects in SAT/OVAT per group (total – lean –obese); overweight subgroup was excluded due to N = 3; WHO classification: lean≥18; <25 kg/m²; <30 kg/m²obese≥ 30 kg/m²; SAT (subcutaneous adipose tissue); OVAT (omental visceral adipose tissue), a = P-value. P-values were calculated using bivariate Spearman correlation analysis (adjusted for age, sex and BMI (except for BMI) by calculating standardized residuals); nominal significant P-values are highlighted in bold. b = beta (ß; effect size and direction) BMI = body mass index; OGTT = oral glucose tolerance test; HDL = high density lipoprotein cholesterol; LDL = low density lipoprotein cholesterol.
Discussion
Body fat distribution is strongly related to obesity associated co-morbidities4. While large GWAS studies have identified numerous SNPs associated with WHR and body fat percentage, it is poorly understood how these variants influence metabolic traits2,23. Interestingly, many GWAS variants were identified as being methylation quantitative trait loci (meQTL) indicating that the interplay between genetics and epigenetics is important for disease development24. Moreover, nearly half of the identified T2D risk variants, including one variant of IRS1 (rs7578326), are CpG-SNPs with an influence on methylation status of these SNP sites and/or surrounding CpG sites, hence might have an impact on gene expression16.
Here, we tested whether genotypes at a previously identified body fat associated SNP variant near IRS1 rs2943650 interplay with IRS1 DNA methylation and gene expression, thereby potentially mediating the observed effects on individual metabolic profiles and body fat distribution. We genotyped rs2943650 and measured DNA methylation in 146 subjects. Additionally, IRS1 mRNA expression data were extracted from a previous data set14. Although non-significant, which is most likely due to the small sample size for genetic association studies, our data are in line with others1,25. We found among T allele carriers (major allele) a favourable anthropometric pattern, which is however combined with more unhealthy metabolic variables compared to C allele carriers. These results largely support the previously reported hypothesis that a relative decrease in subcutaneous to visceral adipose tissue mass is related to e.g. insulin resistance, dyslipidemia and lower adiponectin levels1. Our data further suggest that IRS1 DNA methylation at several CpG sites within the promoter is strongly fat depot specific with significantly higher levels in OVAT. Interestingly, we found that rs2943650 T-allele is related to increased methylation in OVAT while OVAT DNA methylation levels correlate with variables of fat distribution and glucose/lipid metabolism. Interestingly, this relationship may be obesity specific as we observed opposite effect directions of OVAT DNA methylation on e.g. body fat in obese compared to lean individuals; while in general obesity-specific methylation effects were reported in many studies26. In the light of these results one could speculate, that the observed effects on fat distribution are, at least partially and tissue specifically, driven by a potential interplay between genetic variation and IRS1 DNA methylation.
Future studies could focus on the use of both, a patient’s SNP and DNA methylation data as diagnostic tool for the development of adiposity related co-morbidities like T2D. Hence, since OVAT isn’t easy accessible it would be interesting to know whether DNA methylation level of IRS1 in blood reflect those derived in OVAT. However, larger studies are warranted which address these questions but also include further SNP markers, their potential role in predicting DNA methylation and the use as diagnostic tool.
Since both, DNA methylation and SNP variants can influence gene expression we tested for associations with IRS1 mRNA expression. IRS1 mRNA is differentially expressed in SAT and OVAT with lower levels in visceral adipose tissue depot while T-allele carriers show lower IRS1 mRNA expression in OVAT. Further, among obese individuals IRS1 DNA methylation is negatively related to IRS1 gene expression in SAT and OVAT. We therefore conclude that both SNP and IRS1 DNA methylation are involved in regulating depot-specific IRS1 mRNA expression which might be more prominent among obese subjects. Taken together, our data suggest that the mechanisms underlying the association between rs2943650 near IRS1 and body composition measures may be mediated by promoter hyper-methylation among risk allele carriers in a tissue specific manner. Further, DNA methylation and gene expression are most strongly correlated among obese individuals underlining that this relationship may be obesity-specific. Possible mechanisms underlying the observed results may also include microRNA expression which was previously described to be adipose tissue depot-specific27–29. This can result in depot-specific IRS1 mRNA translation in SAT and OVAT which consequently affects protein content and subsequently metabolic traits tissue specifically.
Our study has several limitations including the small sample size. We included 146 individuals in the genetic association study for whom we measured DNA methylation data. Although the number of individuals is sufficient for studying epigenetic effects it is too small to identify genetic associations. This is most likely the reason for the lack of statistically significant results in SNP analysis, although they show the same trend and effect directions as larger previously published studies. In addition, expression data were only available for a subgroup of subjects which can lead to false positive or false negative results. We only measured DNA methylation at 4 CpG sites which may not be representative for the entire promoter region. Moreover, we used adipose tissue biopsies which naturally contain several cell types. Therefore, it cannot be ruled out that effects from other cell types such as macrophages may have influenced our results. Finally, we do not have information about nutrition, smoking and other environmental factors that may impact on epigenetic mechanisms. However, T2D and medication do not affect the shown associations.
In conclusion, our results suggest, that the previously reported body fat associated variant rs2943650 (T) interacts with DNA hypermethylation in OVAT at several CpG sites within the IRS1 promoter linking epigenetic and genetic effects. Since DNA methylation and gene expression are negatively correlated in both SAT and OVAT among obese individuals, the observed findings might be obesity-specific. However, the interplay of genetic and epigenetic factors at the IRS1 locus seems do not sufficiently explain the overall variability of metabolic alterations which clearly indicates that other mechanisms need to be taken into account.
Perspectives
Individual adipose tissue distribution is a critical parameter for developing obesity-related co-morbidities such as type 2 diabetes mellitus. Many studies identified genetic risk variants related to fat distribution. However, underlying mechanisms on how these variants influence the risk for co-morbidities still needs to be explored and will add substantial clinical relevance for the treatment of patients.
This study identifies IRS1 to be differentially methylated and expressed in human SAT and OVAT which correlates to metabolic and anthropometric variables. Furthermore, higher OVAT methylation in obese individuals associates with the previously identified risk variant for fat distribution rs2943650 providing insights into genetic and epigenetic interactions on human fat distribution.
IRS1 is a major player in insulin signalling. Hence, understanding on how genetic variants near or within its gene influence the metabolism alone or in concert with epigenetic alterations will help us identifying clinically relevant underlying mechanisms related to metabolic co-morbidities.
Acknowledgements
We are grateful to all individuals who participated in the study. We appreciate the excellent technical assistance of Ines Müller. This work was supported by a research grant from the IFB AdiposityDiseases ADI-K50D and K7–45; The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7-PEOPLE-2013-COFUND) under grant agreement n° 609020 - Scientia Fellows. M.K. was funded by ADI-K7–39 (to Y.B.). K.R. was funded by ADI-K6e-96 (to Y.B.) and by an EU-Scientia Fellowship from the University of Oslo (to K.R.). IFB AdiposityDiseases is supported by the Federal Ministry of Education and Research (BMBF), Germany, FKZ: 01EO1501. This work was further supported by the Kompetenznetz Adipositas to M.B. (Competence network for Obesity) funded by the Federal Ministry of Education and Research (German Obesity Biomaterial Bank; FKZ 01GI1128). Further support was obtained from the Deutsche Forschungsgemeinschaft for a Collaborative Research Center (CRC 1052/2): “Obesity mechanisms” (B1 to MB; B3 to P.K.; and A1 to M.S.). M.K. was funded by a research fellowship from the German Research Foundation “Deutsche Forschungsgemeinschaft” (KE 2182/1-1 to M.K.).
Author Contributions
KR performed experiments and wrote the first draft of the manuscript. MKlös and MK contributed in sample preparation. LH and XL supported by statistical analysis. AD, MS, DG, TL and MD provided adipose tissue samples. MS, HB, PK and MB contributed with data analysing, interpretation and edited the final manuscript. YB designed and conceived the study and overlooked the experiments. KR and YB wrote the final manuscript while all authors contributed by editing and proof reading.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kilpeläinen TO, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Y, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Comms. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schleinitz D, Böttcher Y, Kovacs P, Blühr M. The genetics of fat distribution. Diabetologia. 2014;57:1276–86. doi: 10.1007/s00125-014-3214-z. [DOI] [PubMed] [Google Scholar]
- 5.Lipes MA, et al. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–90. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 6.Brüning JC, et al. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;88:561–72. doi: 10.1016/S0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 7.Miki H, et al. Essential Role of Insulin Receptor Substrate 1 (IRS-1) and IRS-2 in Adipocyte Differentiation. Molecular and Cellular Biology. 2001;21:2521–2532. doi: 10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rung J, Cauchi S, Albrechtsen A, Shen L. RG, C-PC, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 9.Barres R, et al. Weight Loss after Gastric Bypass Surgery in Human Obesity Remodels Promoter Methylation. Cell Reports. 2013;3:1020–1027. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Ling C, Rönn T. Epigenetic adaptation to regular exercise in human. Drug Discovery Today. 2014;19:1015–1018. doi: 10.1016/j.drudis.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Rönn T, et al. A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehrke S, et al. Epigenetic Regulation of Depot-Specific Gene Expression in Adipose Tissue. PLoS ONE. 2013;8:e82516. doi: 10.1371/journal.pone.0082516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller M, et al. Global DNA methylation levels in human adipose tissue are related to fat distribution and glucose homeostasi. Diabetologia. 2014;57:2374–2383. doi: 10.1007/s00125-014-3356-z. [DOI] [PubMed] [Google Scholar]
- 14.Keller M, et al. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol Metab. 2016;6:86–100. doi: 10.1016/j.molmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Putku M, et al. CDH13 promoter SNPs with pleiotropic effect on cardiometabolic parameters represent methylation QTLs. Hum Genet. 2015;134:291–303. doi: 10.1007/s00439-014-1521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dayeh TA, et al. Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia. 2013;56:1036–1046. doi: 10.1007/s00125-012-2815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansego ML, Milagro FI, Zulet MA, Martinez JA. SH2B1 CpG-SNP Is Associated with Body Weight Reduction in Obese Subjects Following a Dietary Restriction Program. Ann Nutr Metab. 2015;66:1–9. doi: 10.1159/000368425. [DOI] [PubMed] [Google Scholar]
- 18.Toro-Martín J et al. Methylation quantitative trait loci within the TOMM20 gene are associated with metabolic syndrome-related lipid alterations in severely obese subjects. Diabetol Metab Syndr8, doi:10.1186/s13098-016-0171-3 (2016). [DOI] [PMC free article] [PubMed]
- 19.Volkov P, et al. A Genome-Wide mQTL Analysis in Human Adipose Tissue Identifies Genetic Variants Associated with DNA Methylation, Gene Expression and Metabolic Traits. PLoS ONE. 2016;11:e0157776. doi: 10.1371/journal.pone.0157776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaghlool SB, et al. Mendelian inheritance of trimodal CpG methylation sites suggests distal cis-acting genetic effects. Clin Epigenet. 2016;8:124. doi: 10.1186/s13148-016-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemire M, et al. Long-range epigenetic regulation is conferred by genetic variation located at thousands of independent loci. Nat Comms. 2015;6:6326. doi: 10.1038/ncomms7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klöting N, et al. Insulin-sensitive obesity. Am J Physiol Endocrinol Metab. 2010;299:E506–E515. doi: 10.1152/ajpendo.00586.2009. [DOI] [PubMed] [Google Scholar]
- 23.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsson AH, et al. Genome-Wide Associations between Genetic and Epigenetic Variation Influence mRNA Expression and Insulin Secretion in Human Pancreatic Islet. PLoS Genet. 2014;10:e1004735. doi: 10.1371/journal.pgen.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Q, et al. Genetic variation near IRS1 is associated with adiposity and a favorable metabolic profile in U.S. Hispanics/Latinos. Obesity. 2016;11:2407–2413. doi: 10.1002/oby.21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS. and members of EpiSCOPE. Epigenetics and human obesity. Int J Obes. 2015;39:85–97. doi: 10.1038/ijo.2014.34. [DOI] [PubMed] [Google Scholar]
- 27.Arias N et al. MicroRNAs involved in the browning process of adipocytes. J Physiol Biochem72, 509-521 (2015). [DOI] [PubMed]
- 28.Klöting N, et al. MicroRNA Expression in Human Omental and Subcutaneous Adipose Tissue. PLoS ONE. 2009;4:e4699. doi: 10.1371/journal.pone.0004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Development20, 515–524 (2006). [DOI] [PubMed]