Abstract
Here, we employed a bioinformatics approach to identify novel molecular determinants to predict tumor progression and overall survival in gastric cancer patients. In particular, we directly assessed whether nuclear-derived mRNA species encoding proteins involved in mitochondrial protein translation and OXPHOS are able to successfully predict clinical outcome in gastric cancer. As such, using in silico validation, we have now established the prognostic value of these mitochondrial biomarkers, in a defined population of gastric cancer patients. In this context, we interrogated 5 year follow-up data collected from a group of N = 359 gastric cancer patients. Importantly, in this group of cancer patients, Ki67 and PCNA (conventional markers of cell proliferation) were associated with tumor progression, as might be expected. Using this simplified informatics approach, we identified ∼75 new individual mitochondrial gene probes that effectively predicted tumor progression, with hazard-ratios (HR) of up to 2.22 (p < 2.1e-10). These mitochondrial mRNA transcripts included heat shock proteins/chaperones, membrane proteins, anti-oxidants, enzymes involved in genome maintenance, as well as mitochondrial ribosomal proteins (MRPs) and numerous members of the OXPHOS complexes. In addition, we combined 8 mitochondrial protein transcripts (NDUFS5, VDAC3, ATP5O, IMMT, MRPL28, COX5B, MRPL52, PRKDC), to generate a compact gastric mitochondrial gene signature, associated with a HR of 2.77 (p = 1.4e-14). As a result of this analysis and validation, we strongly suggest that proteins involved in mitochondrial protein translation and OXPHOS should be considered as targets for new drug discovery, for the treatment of gastric cancers. The mitochondrial markers we identified here could also be used as companion diagnostics, to predict clinical outcomes, as well as the patient response to therapy. This should allow a more successful and personalized approach to gastric cancer diagnosis and therapy.
Keywords: gastric cancer, mitochondrial biomarkers, overall survival, tumor progression
INTRODUCTION
Gastric cancer, also known as stomach cancer, starts as an infectious disease that drives the development of chronic inflammation and ulcerated lesions, located within the stomach wall lining. In most cases of gastric cancer, these lesions are morphologically defined as epithelial carcinomas [1–3]. During tumor progression, the gastric cancer cells migrate and metastasize to the lymph nodes, the abdomen, bones, lungs and the liver. The most common etiology of gastric cancer is chronic infection with Helicobacter pylori, a bacterium, which represents ∼50-70% of the cases [4–6]. Classic symptoms consist of “heartburn”, abdominal pain, nausea and vomiting, as well as anemia and occult blood in the feces. The diagnosis of gastric cancer is confirmed by an endoscopic biopsy, revealing an invasive carcinoma. The disease is most common in Asian countries, especially in South Korea and Japan, among others [1–6].
Unfortunately, the 5-year survival rates for gastric cancer patients vary widely, from as low as 15% to as high as 60%. As such, more effective biomarkers are urgently needed for the early stratification of gastric cancer patients, into low-risk and high-risk groups at diagnosis. Here, we directly tested the hypothesis that markers of mitochondrial biogenesis and respiratory function may have significant prognostic value in the early identification of high-risk gastric cancer patients, with increased tumor progression and poor overall survival.
More specifically, we employed a bioinformatics approach to determine the prognostic utility of nuclear-encoded mitochondrial mRNA species in predicting clinical outcome. Our results indicate that ∼75 individual mitochondrial gene probes can be effectively used, to predict progression and survival in gastric cancer patients. As a result, we propose the idea that mitochondria are novel therapeutic targets for drug discovery, aimed at optimizing the effectiveness of gastric cancer therapy [7–9].
RESULTS
Prognostic value of proliferative and inflammatory markers in gastric cancer patients
Here, we used publically available transcriptional profiling data from the tumors of gastric cancer patients, with 5 years of follow-up data, to identify new potential biomarkers (Figure 1). As proliferative biomarkers are often used as clinical endpoints in patient trials, we first assessed the prognostic value of Ki67 and PCNA, in this patient population. Tables 1, 2 and Figure 2 both show the prognostic value of these markers. The hazard-ratios for Ki67 and PCNA were 1.79 and 1.86, respectively, for time to first progression (FP).
Figure 1. A bio-informatics approach to gastric cancer marker discovery.
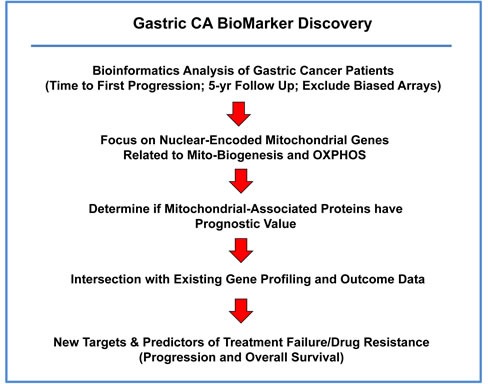
We chose to focus on gastric cancer patients, with 5-years of follow-up data (N = 359). In this context, we evaluated the prognostic value of mitochondrial markers for predicting time to first progression and overall survival.
Table 1. Prognostic value of Ki67 in gastric cancer.
| Gene Probe ID | Symbol | Hazard-Ratio | Log-Rank Test |
|---|---|---|---|
| 212020_s_at | MKI67 | 1.92 | 1e-06 |
| 212023_s_at | MKI67 | 1.92 | 2.7e-07 |
| 212022_s_at | MKI67 | 1.83 | 1.9e-06 |
| 212021_s_at | MKI67 | 1.60 | 0.002 |
| Combined | 1.79 | 6.2e-06 |
Table 2. Prognostic value of PCNA and CD163 in gastric cancer.
| Gene Probe ID | Symbol | Hazard-Ratio | Log-Rank Test |
|---|---|---|---|
| 217400_at | PCNA | 1.86 | 6.5e-06 |
| 216233_at | CD163 | 1.46 | 0.003 |
Figure 2. Markers of proliferation and inflammation predict poor clinical outcome in gastric cancer patients.
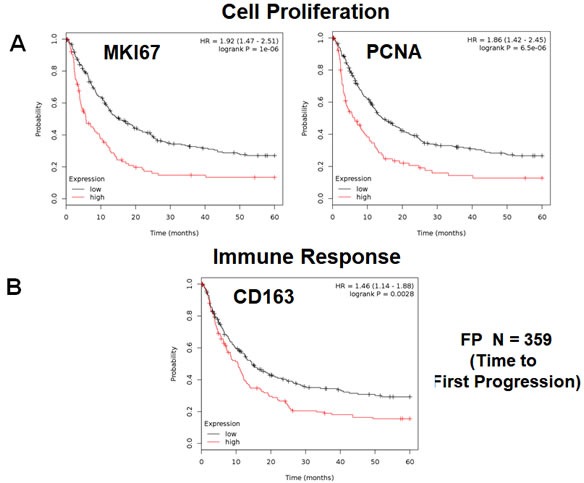
We assessed the predictive value of Ki67 and PCNA in N = 359 gastric cancer patients. A. Note that high transcript levels of Ki67 and PCNA are associated with significantly reduced time to first progression. Note that the official gene name for the Ki67 protein is MKI67. B. Note that that high transcript levels of CD163 are associated with significantly reduced time to first progression.
We also assessed the prognostic value of a macrophage-specific marker of inflammation, namely CD163. Table 2 and Figure 2 show that CD163 effectively predicted time to first progression, with a hazard-ratio of 1.46. Thus, conventional markers of proliferation and inflammation predict overall survival in gastric cancer patients.
Prognostic value of individual mitochondrial markers
To evaluate the hypothesis that elevated mitochondrial mass, biogenesis and function somehow contributes towards poor clinical outcome in gastric cancer patients, we also determined the prognostic value of individual mitochondrial markers.
First, we examined the behavior of mitochondrial membrane proteins and HSPs/chaperones. Table 3 and Figure 3 both show that VDAC3 and IMMT had the best prognostic value, with a hazard-ratio of up to 2.22 (p<1.4e-09). HSPD1 and members of the TIMM and TOMM gene families also had prognostic value; SOD2 also had significant value. Positive results were also obtained with DNA-PK (PRKDC), a kinase that maintains the integrity and copy number of the mitochondrial genome (mt-DNA).
Table 3. Prognostic value of mitochondrial HSPs and other mitochondrial proteins.
| Gene Probe ID | Symbol | Hazard-Ratio | Log-Rank Test |
|---|---|---|---|
| Heat Shock Proteins and Chaperones (4 probes) | |||
| 200807_s_at | HSPD1 | 1.83 | 1.9e-06 |
| 200806_s_at | HSPD1 | 1.56 | 0.003 |
| 200691_s_at | HSPA9 | 1.61 | 0.0002 |
| 205565_s_at | FXN | 1.38 | 0.01 |
| Membrane Proteins (9 probes) | |||
| 208844_at | VDAC3 | 2.22 | 1.4e-09 |
| 211662_s_at | VDAC2 | 1.51 | 0.002 |
| 200955_at | IMMT | 2.20 | 2.4e-09 |
| 218118_s_at | TIMM23 | 1.91 | 4.2e-07 |
| 218408_at | TIMM10 | 1.88 | 1.4e-06 |
| 218357_s_at | TIMM8B | 1.49 | 0.002 |
| 201821_s_at | TIMM17A | 1.33 | 0.025 |
| 201870_at | TOMM34 | 1.95 | 5.1e-07 |
| 202264_s_at | TOMM40 | 1.44 | 0.009 |
| Mitochondrial Anti-Oxidants (2 probes) | |||
| 215223_s_at | SOD2 | 1.72 | 2.1e-05 |
| 215078_at | SOD2 | 1.70 | 2.9e-05 |
| Mitochondrial Genome Maintenance (3 probes) | |||
| 208694_at | PRKDC | 2.05 | 1.2e-07 |
| 210543_s_at | PRKDC | 1.78 | 6.9e-06 |
| 215757_at | PRKDC | 1.47 | 0.003 |
Figure 3. Mitochondrial membrane proteins and PRKDC are associated with poor clinical outcome in gastric cancer patients.
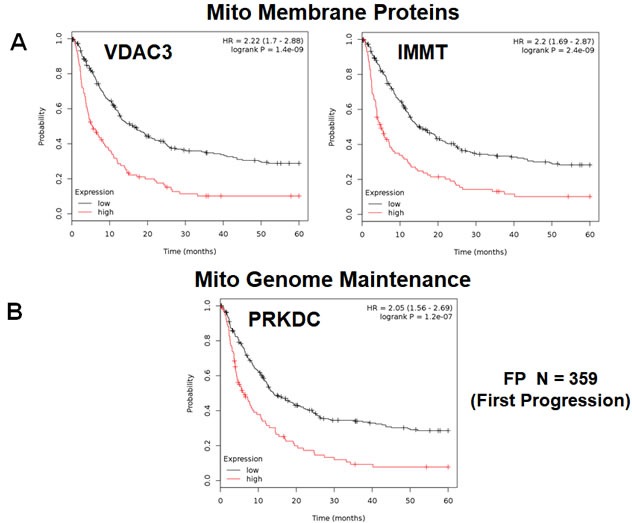
A. Note that that high transcript levels of VDAC3 and IMMT are associated with significantly reduced time to first progression. B. Note that that high transcript levels of PRKDC are associated with significantly reduced time to first progression.
Next, we determined the prognostic value of mitochondrial ribosomal proteins (MRPs), which are essential for mitochondrial biogenesis (Table 4). Twelve components of the large subunit (MRPLs) showed significant prognostic value, with hazard-ratios between 2.17 and 1.29. Notably, MRPL28 had the best prognostic value. Eight different components of the small subunit (MRPSs) showed significant prognostic value, with hazard-ratios between 1.89 and 1.36. As a result, twenty different MRPs all predicted reduced time to first progression. Representative examples of Kaplan-Meier curves are shown in Figure 4, panels A & B.
Table 4. Prognostic value of mitochondrial ribosomal proteins.
| Gene Probe ID | Symbol | Hazard-Ratio | Log-Rank Test |
|---|---|---|---|
| Large Ribosomal Subunit (12 probes) | |||
| 204599_s_at | MRPL28 | 2.17 | 1.2e-08 |
| 221997_s_at | MRPL52 | 2.12 | 3.2e-09 |
| 222216_s_at | MRPL17 | 1.68 | 0.0001 |
| 220527_at | MRPL20 | 1.67 | 0.0002 |
| 217907_at | MRPL18 | 1.62 | 0.0004 |
| 218887_at | MRPL2 | 1.60 | 0.0002 |
| 203931_s_at | MRPL12 | 1.56 | 0.001 |
| 208787_at | MRPL3 | 1.53 | 0.0007 |
| 217919_s_at | MRPL42 | 1.52 | 0.002 |
| 218049_s_at | MRPL13 | 1.47 | 0.008 |
| 218281_at | MRPL48 | 1.40 | 0.009 |
| 213897_s_at | MRPL23 | 1.29 | 0.049 |
| Small Ribosomal Subunit (8 probes) | |||
| 215919_s_at | MRPS11 | 1.89 | 5.1e-07 |
| 213840_s_at | MRPS12 | 1.84 | 1.5e-06 |
| 210008_s_at | MRPS12 | 1.47 | 0.004 |
| 204330_s_at | MRPS12 | 1.37 | 0.015 |
| 204331_s_at | MRPS12 | 1.37 | 0.037 |
| 203800_s_at | MRPS14 | 1.53 | 0.002 |
| 219220_x_at | MRPS22 | 1.44 | 0.005 |
| 219819_s_at | MRPS28 | 1.42 | 0.01 |
| 218112_at | MRPS34 | 1.36 | 0.02 |
Figure 4. Mitochondrial ribosomal proteins (MRPs) are associated with poor clinical outcome in gastric cancer patients.
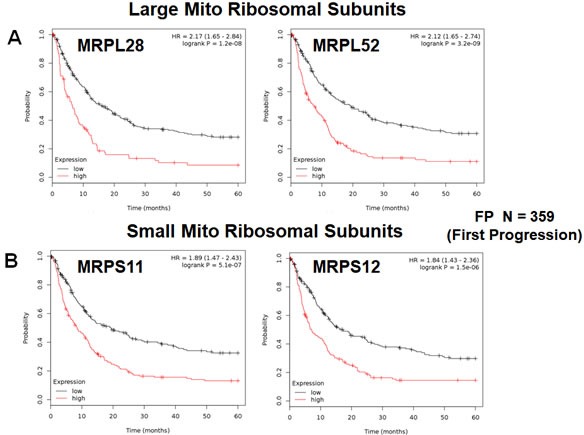
A. Note that high transcript levels of MRPL28 and MRPL52 predict significantly reduced time to first progression. B. Similarly, high transcript levels of MRPS11 and MRPS12 predict significantly reduced time to first progression.
In addition, we evaluated the prognostic value of members of the OXPHOS complexes I-V. These results are summarized in Table 5. Interestingly, 37 different gene probes for the OXPHOS complexes showed hazard-ratios between 2.27 and 1.30. NDUFS5 (complex I) had the best prognostic value (HR=2.27; p=6e-10). COX5B (complex IV) and ATP5O (complex V) also showed significant prognostic value (HR=2.14; p=1.4e-08 versus HR=2.22; p=2.1e-10). Kaplan-Meier analysis for components of complex I and II are shown in Figure 5A, 5B, while results with complex III and IV are shown in Figure 6A, 6B. Results with complex V are highlighted in Figure 7.
Table 5. Prognostic value of mitochondrial OXPHOS complexes.
| Gene Probe ID | Symbol | Hazard-Ratio | Log-Rank Test |
|---|---|---|---|
| Complex I (11 probes) | |||
| 201757_at | NDUFS5 | 2.27 | 6e-10 |
| 215850_s_at | NDUFA5 | 1.93 | 2.1e-07 |
| 208969_at | NDUFA9 | 1.92 | 1.5e-06 |
| 203606_at | NDUFS6 | 1.74 | 7.9e-05 |
| 214241_at | NDUFB8 | 1.67 | 5.7e-05 |
| 203371_s_at | NDUFB3 | 1.51 | 0.002 |
| 218226_s_at | NDUFB4 | 1.49 | 0.003 |
| 202001_s_at | NDUFA6 | 1.37 | 0.02 |
| 218160_at | NDUFA8 | 1.31 | 0.04 |
| 202785_at | NDUFA7 | 1.31 | 0.04 |
| 218563_at | NDUFA3 | 1.30 | 0.04 |
| Complex II (1 probe) | |||
| 214166_at | SDHB | 1.40 | 0.009 |
| Complex III (2 probes) | |||
| 207618_s_at | BCS1L | 1.76 | 7.1e-06 |
| 202233_s_at | UQCR8 | 1.51 | 0.001 |
| Complex IV (10 probes) | |||
| 213736_at | COX5B | 2.14 | 1.4e-08 |
| 218057_x_at | COX4NB | 1.94 | 7.7e-07 |
| 201754_at | COX6C | 1.74 | 7.1e-05 |
| 201441_at | COX6B1 | 1.67 | 0.0001 |
| 200925_at | COX6A1 | 1.64 | 8.8e-05 |
| 203880_at | COX17 | 1.60 | 0.0003 |
| 217451_at | COX5A | 1.49 | 0.006 |
| 202110_at | COX7B | 1.42 | 0.01 |
| 217249_x_at | COX7A2 | 1.33 | 0.035 |
| 216003_at | COX10 | 1.33 | 0.046 |
| Complex V (13 probes) | |||
| 221677_s_at | ATP5O | 2.22 | 2.1e-10 |
| 207552_at | ATP5G2 | 1.90 | 7.5e-06 |
| 207335_x_at | ATP5I | 1.84 | 8.4e-06 |
| 217801_at | ATP5E | 1.64 | 0.0002 |
| 208972_s_at | ATP5G1 | 1.51 | 0.002 |
| 210149_s_at | ATP5H | 1.49 | 0.003 |
| 202961_s_at | ATP5J2 | 1.47 | 0.004 |
| 210453_x_at | ATP5L | 1.45 | 0.006 |
| 207573_x_at | ATP5L | 1.44 | 0.01 |
| 208746_x_at | ATP5L | 1.40 | 0.009 |
| 213366_x_at | ATP5C | 1.33 | 0.03 |
| 206993_at | ATP5S | 1.29 | 0.04 |
| 213366_x_at | ATP5C1 | 1.33 | 0.03 |
Figure 5. Mitochondrial complex I and II proteins are associated with poor clinical outcome in gastric cancer patients.
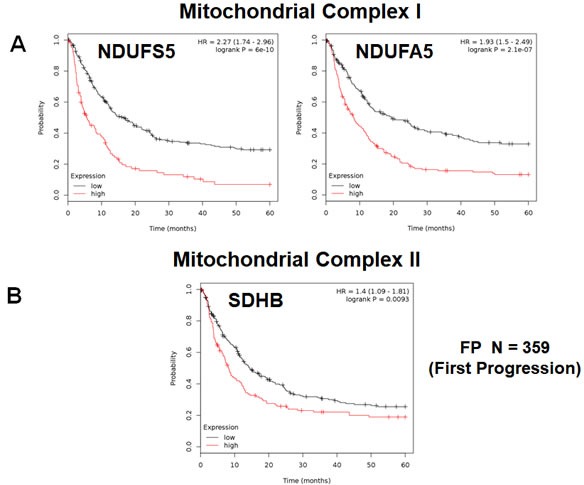
A. Note that high levels of NDUFS5 and NDUFA5 predict significantly reduced time to first progression. B. Similarly, high levels of SDHB predict significantly reduced time to first progression.
Figure 6. Mitochondrial complex III and IV proteins are associated with poor clinical outcome in gastric cancer patients.
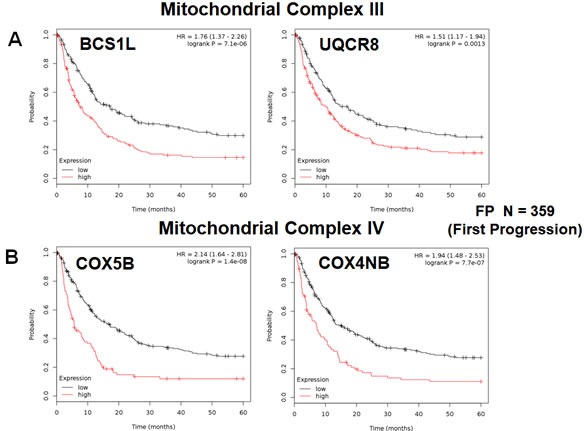
A. Note that high levels of BCS1L and UQCR8 predict significantly reduced time to first progression. B. Similarly, high levels of COX5B and COX4NB predict reduced time to first progression.
Figure 7. Mitochondrial complex V proteins are associated with poor clinical outcome in gastric cancer patients.
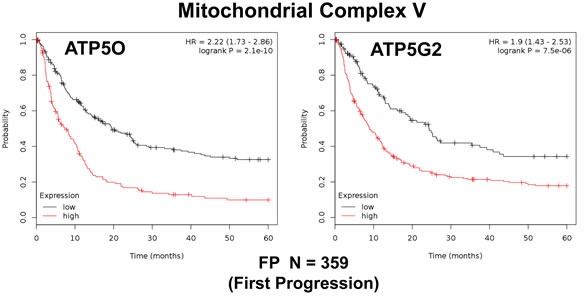
Note that high levels of ATP5O and ATP5G2 predict significantly reduced time to first progression.
Prognostic value of a short gastric (Ga) mito-signature
To increase the prognostic significance of these unique mitochondrial markers, we then combined the most promising markers to create a new gastric mitochondrial gene signature.
More specifically, Ga-Mito-Signature-1 contains 8 genes (NDUFS5, VDAC3, ATP5O, IMMT, MRPL28, COX5B, MRPL52, PRKDC). Importantly, Ga-Mito-Signature-1 yielded a significantly improved hazard-ratio for time to first progression (FP) (HR=2.77; p=1.4e-14), as well as for overall survival (OS) (HR=2.01; p=6e-11). These results are summarized in Table 6 and highlighted in Figure 8.
Table 6. Compact gastric CA mito-signature for predicting clinical outcome.
| Gene Probe ID | Symbol | Hazard-Ratio | Log-Rank Test |
|---|---|---|---|
| 201757_at | NDUFS5 | 2.27 | 6e-10 |
| 208844_at | VDAC3 | 2.22 | 1.4e-09 |
| 221677_s_at | ATP5O | 2.22 | 2.1e-10 |
| 200955_at | IMMT | 2.20 | 2.4e-09 |
| 204599_s_at | MRPL28 | 2.17 | 1.2e-08 |
| 213736_at | COX5B | 2.14 | 1.4e-08 |
| 221997_s_at | MRPL52 | 2.12 | 3.2e-09 |
| 208694_at | PRKDC | 2.05 | 1.2e-07 |
| Combined | 2.77 | 1.4e-14 |
Figure 8. A short mitochondrial signature predicts tumor progression and overall survival in gastric cancer patients.
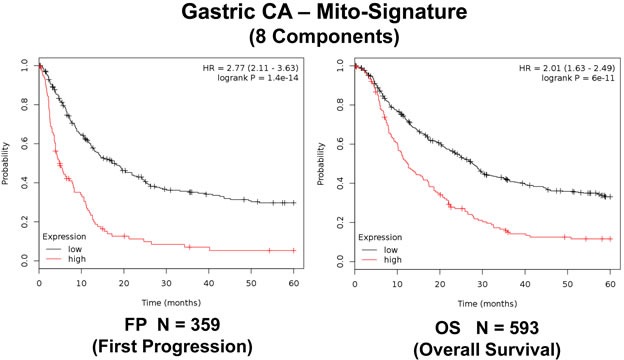
We combined 8 mitochondrial proteins (NDUFS5, VDAC3, ATP5O, IMMT, MRPL28, COX5B, MRPL52, PRKDC), to generate a compact gastric mitochondrial gene signature.
As such, the Ga-Mito-Signature-1 represents a significant improvement over individual mitochondrial biomarkers, as well as Ki67 and PCNA.
DISCUSSION
New “druggable” targets for inhibiting mitochondrial biogenesis and blocking oxidative metabolism in cancer cells
Here, we suggest that the new mitochondrial markers that we developed here may also be useful for selecting new “druggable” targets for innovative drug development, to prevent treatment failure and improve overall survival. As a consequence of our K-M analyses, we believe that the mitochondrial ribosome would be an attractive new target for developing novel inhibitors of mitochondrial protein translation in cancer cells; similarly, mitochondrial chaperones, the OXPHOS complexes and the mitochondrial ATP-synthase may also be suitable drug targets. Particular components of these multi-subunit protein complexes show significant prognostic value, suggesting that modulation of their intrinsic activity may provide therapeutic benefits. Targeting of these large protein complexes would be predicted to suppress tumor recurrence and prevent disease progression in these gastric cancer patients.
Similarly, these mitochondrial markers could also be employed as companion diagnostics for novel therapies targeting either mitochondrial protein translation or cellular respiration, to select the high-risk sub-population of gastric cancer patients, resulting in the proper treatment stratification (Figure 9). In direct support of this assertion, we showed here that ∼75 different mitochondrial probes could be used to successfully identify the sub-population of high-risk gastric cancer patients. As such, these results indicate that mitochondrial markers could be used to monitor and/or predict the response to therapy, identifying patients at high-risk for treatment failure at diagnosis, several years in advance, even prior to the initiation of therapy.
Figure 9. Gastric cancer: Mitochondrial-based diagnostics for personalized therapy.
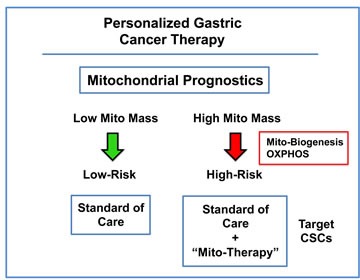
In this proposed scheme, mitochondrial markers could be used to separate gastric cancer patients into high-risk and low-risk groups. Then, patients with high levels of mitochondrial markers in their primary tumor (“bad prognosis”) would be treated with mitochondrial-based therapies, to prevent progression and increase survival.
Cancer stem cells (CSCs) have increased mitochondrial power: diagnostic and therapeutic implications
Recently, we identified increased mitochondrial function as a new feature of cancer stem cells (CSCs), based on unbiased proteomics analysis [7–9,11]. This increase is mitochondrial function in CSCs may be secondary to increased mitochondrial biogenesis and/or elevated mitochondrial protein translation, resulting in the steady-state amplification of mitochondrial mass [7–9,11]. These findings have important implications for designing new approaches for identifying and eradicating CSCs.
Importantly, our previous studies directly show that MitoTracker (a fluorescent mitochondrial vital dye) can be used, together with flow cytometry, to purify CSCs from heterogeneous populations of cancer cells [12,13]. Importantly, the “Mito-high” cells showed the greatest functional capacity for i) anchorage-independent growth and ii) tumor-initiating activity in vivo [12–14]. Quantitatively similar results were also obtained with other mitochondrial probes (for detecting ROS and H2O2) and/or NAD(P)H auto-fluorescence, all surrogate markers of increased mitochondrial respiratory activity [14].
In support of these functional studies, we have also shown that FDA-approved antibiotics can be used to effectively target mitochondria in CSCs [15–21]. Because mitochondria evolved from aerobic bacteria, over a period >1.5 billion years, certain types of antibiotics functionally inhibit mitochondrial biogenesis, as a manageable side-effect. In this context, tetracycline antibiotics, such as Doxycycline, appeared to be the most promising. For example, Doxcycline effectively eradicates CSCs in 12 different cell lines, representing 8 different cancer types, as well as in primary tissue samples derived from patients with advanced metastatic disease (collected from pleural effusions or ascites fluid) [15,16].
Interestingly, these functional observations could mechanistically explain the high prognostic value of mitochondrial markers in predicting progression and survival in gastric cancer patients. Patients with high levels of mitochondrial markers may have increased levels of CSCs (Figure 9). As a result, these mitochondrial biomarkers could also be used as companion diagnostics, to identify which gastric cancer patients would benefit most from anti-cancer therapy with Doxycycline (or perhaps with other inhibitors of mitochondrial biogenesis and/or drugs metabolically targeting respiratory function, such as Metformin).
METHOD OF ANALYSIS
Kaplan-Meier (K-M) Analyses. To perform K-M analysis on nuclear mitochondrial gene transcripts, we used an open-access, online survival, data analysis tool to interrogate publically available microarray data from up to 1,065 gastric cancer patients [10]. This allowed us to determine their overall prognostic value. For this purpose, we primarily analyzed 5-year follow-up data from gastric cancer patients (N = 359). Biased array data were excluded from the analysis. This allowed us to identify ∼75 nuclear mitochondrial gene probes, with significant prognostic value. Hazard-ratios for time to first progression (FP) and overall survival (OS) were calculated, at the best auto-selected cut-off, and p-values were calculated using the logrank test and plotted in R. K-M curves were also generated online using the K-M-plotter (as high-resolution TIFF files) [10], using univariate analysis:
http://kmplot.com/analysis/index.php?p=service&cancer=gastric
This allowed us to directly perform in silico validation of these mitochondrial biomarker candidates.
Abbreviations
- CSCs
cancer stem-like cells
- HR
hazard ratio
- K-M
Kaplan-Meier
- MRPL
mitochondrial ribosomal proteins, large subunit
- MRPS
mitochondrial ribosomal proteins, small subunit
- N
number of patients in a given data set
- OS
overall survival
- OXPHOS
oxidative phosphorylation (mitochondrial respiration)
- FP
time to first progression
Footnotes
Author contributions
Professor Lisanti and Dr. Sotgia conceived and initiated this project. Professor Lisanti and Dr. Sotgia both performed the bioinformatics analysis, and wrote the manuscript.
FUNDING
It should be noted that this bioinformatics analysis, focused on nuclear-encoded mitochondrial-related gene transcripts, was not funded by a specific grant and did not require any research expenditures, since no “wet” laboratory experiments were performed.
CONFLICTS OF INTEREST
MPL and FS hold a minority interest in Lunella, Inc.
REFERENCES
- 1.Luo G, Hu Y, Zhang Z, Wang P, Luo Z, Lin J, Cheng C, Yang Y. Clinicopathologic significance and prognostic value of Ki-67 expression in patients with gastric cancer: a meta-analysis. Oncotarget. 2017;8:50273–50283. doi: 10.18632/oncotarget.17305. https://doi.org/10.18632/oncotarget.17305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machairas N, Charalampoudis P, Molmenti EP, Kykalos S, Tsaparas P, Stamopoulos P, Sotiropoulos GC. The value of staging laparoscopy in gastric cancer. Ann Gastroenterol. 2017;30:287–294. doi: 10.20524/aog.2017.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid BJ. Genomics, Endoscopy, and Control of Gastroesophageal Cancers: A Perspective. Cell Mol Gastroenterol Hepatol. 2017(3):359–366. doi: 10.1016/j.jcmgh.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Meltzer SJ. Gastric Cancer in the Era of Precision Medicine. Cell Mol Gastroenterol Hepatol. 2017;3:348–358. doi: 10.1016/j.jcmgh.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayakawa Y, Fox JG, Wang TC. The Origins of Gastric Cancer From Gastric Stem Cells: Lessons From Mouse Models. Cell Mol Gastroenterol Hepatol. 2017;3:331–338. doi: 10.1016/j.jcmgh.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu D, Kanda M, Kodera Y. Review of recent molecular landscape knowledge of gastric cancer. Histol Histopathol. 2017;27:11898. doi: 10.14670/HH-11-898. [DOI] [PubMed] [Google Scholar]
- 7.Peiris-Pagès M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18:55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 9.Killock D. Drug therapy: Can the mitochondrial adverse effects of antibiotics be exploited to target cancer metabolism? Nat Rev Clin Oncol. 2015;12:190. doi: 10.1038/nrclinonc.2015.24. [DOI] [PubMed] [Google Scholar]
- 10.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–33. doi: 10.18632/oncotarget.10337. https://doi.org/10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb R, Harrison H, Hulit J, Smith DL, Lisanti MP, Sotgia F. Mitochondria as new therapeutic targets for eradicating cancer stem cells: Quantitative proteomics and functional validation via MCT1/2 inhibition. Oncotarget. 2014;5:11029–37. doi: 10.18632/oncotarget.2789. https://doi.org/10.18632/oncotarget.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnie G, Sotgia F, Lisanti MP. High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget. 2015;6:30472–86. doi: 10.18632/oncotarget.5401. https://doi.org/10.18632/oncotarget.5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb R, Bonuccelli G, Ozsvári B, Peiris-Pagès M, Fiorillo M, Smith DL, Bevilacqua G, Mazzanti CM, McDonnell LA, Naccarato AG, Chiu M, Wynne L, Martinez-Outschoorn UE, et al. Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: Understanding WNT/FGF-driven anabolic signaling. Oncotarget. 2015;6:30453–71. doi: 10.18632/oncotarget.5852. https://doi.org/10.18632/oncotarget.5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonuccelli G, De Francesco EM, de Boer R, Tanowitz HB, Lisanti MP. NADH autofluorescence, a new metabolic biomarker for cancer stem cells: Identification of Vitamin C and CAPE as natural products targeting “stemness”. Oncotarget. 2017;8:20667–20678. doi: 10.18632/oncotarget.15400. https://doi.org/10.18632/oncotarget.15400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget. 2015;6:4569–84. doi: 10.18632/oncotarget.3174. https://doi.org/10.18632/oncotarget.3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb R, Fiorillo M, Chadwick A, Ozsvari B, Reeves KJ, Smith DL, Clarke RB, Howell SJ, Cappello AR, Martinez-Outschoorn UE, Peiris-Pagès M, Sotgia F, Lisanti MP. Doxycycline down-regulates DNA-PK and radiosensitizes tumor initiating cells: Implications for more effective radiation therapy. Oncotarget. 2015;6:14005–25. doi: 10.18632/oncotarget.4159. https://doi.org/10.18632/oncotarget.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonuccelli G, Peiris-Pages M, Ozsvari B, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Targeting cancer stem cell propagation with palbociclib, a CDK4/6 inhibitor: Telomerase drives tumor cell heterogeneity. Oncotarget. 2017;8:9868–84. doi: 10.18632/oncotarget.14196. https://doi.org/10.18632/oncotarget.14196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Luca A, Fiorillo M, Peiris-Pagès M, Ozsvari B, Smith DL, Sanchez-Alvarez R, Martinez-Outschoorn UE, Cappello AR, Pezzi V, Lisanti MP, Sotgia F. Mitochondrial biogenesis is required for the anchorage-independent survival and propagation of stem-like cancer cells. Oncotarget. 2015;6:14777–95. doi: 10.18632/oncotarget.4401. https://doi.org/10.18632/oncotarget.4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiorillo M, Lamb R, Tanowitz HB, Cappello AR, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Bedaquiline, an FDA-approved antibiotic, inhibits mitochondrial function and potently blocks the proliferative expansion of stem-like cancer cells (CSCs) Aging (Albany NY) 2016;8:1593–607. doi: 10.18632/aging.100983. https://doi.org/10.18632/aging.100983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorillo M, Lamb R, Tanowitz HB, Mutti L, Krstic-Demonacos M, Cappello AR, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Repurposing Atovaquone: Targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget. 2016;7:34084–99. doi: 10.18632/oncotarget.9122. https://doi.org/10.18632/oncotarget.9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiorillo M, Sotgia F, Sisci D, Cappello AR, Lisanti MP. Mitochondrial “power” drives tamoxifen resistance: NQO1 and GCLC are new therapeutic targets in breast cancer. Oncotarget. 2017;8:20309–327. doi: 10.18632/oncotarget.15852. https://doi.org/10.18632/oncotarget.15852 [DOI] [PMC free article] [PubMed] [Google Scholar]


