Abstract
Lung cancer, with 80–85% being non-small cell lung cancer (NSCLC), is the leading cause of cancer-related death in both men and women. Long non-coding RNAs (lncRNAs), always defined as non-protein-coding RNA molecules longer than 200 nucleotides, are now thought as a new frontier in the study of human malignant diseases including NSCLC. As researches continue, increasing number of roles that lncRNAs play in NSCLC has been found, and more and more evidences show lncRNAs have a close relationship with patients’ response to radiochemotherapy or molecular therapy. The aim of this review is to disclose the roles that lncRNAs play in NSCLC and how lncRANs influence the treatment of NSCLC.
Keywords: NSCLC, lncRNAs, chemotherapy, radiotherapy, molecular therapy
INTRODUCTION
Lung and bronchus cancer leads to most cancer-related death, with an estimation about 85,920 and 72,160 death in men and women respectively in America, and it was also reported that there were totally 224,390 American people being diagnosed with lung and bronchus cancer in 2016 [1]. Looking around the world, greater than one-third of all newly diagnosed lung cancers occurred in China, resulting in a large social and economic burden. According to the annual report on the status of cancer in China, in total, 651,053 patients were newly diagnosed with lung cancer in 2011, including 441,364 men and 209,689 women [2]. Lung and bronchus cancer is usually classified into NSCLC and small cell lung cancer (SCLC) accounting for approximately 80% and 20% respectively, the former of which is traditionally treated with surgery combined with or without radiochemotherapy [3].
Long non-coding RNAs (lncRNAs), always defined as non-protein-coding RNA molecules longer than 200 nucleotides, are not thought as transcriptional “noise” any more, and have been regarded as a new frontier in the study of human malignant diseases such as brain, breast, prostate, liver, ovary, esophagus tumors and other kinds of diseases like Fragile X syndrome, Alzheimer's disease and etc [4, 5]. LncRNAs, on the one hand, can regulate the expression of nearby protein-coding genes, and on the other hand, they themselves can serve key regulatory roles. In Jeremy's review, there are basically eight models for lncRNAs to influence the gene expressions, and eventually play the biological roles [6]. As researches continue, it has been increasingly recognized that its dysregulations contribute to the development and progression of lung and bronchus cancer [7]. We conduct a comprehensive review of the published literatures focusing on the roles that lncRNAs play in the presence and development of NSCLC, retrospect the relationship between lncRNAs and radiochemotherapy as well as molecular targeted therapy of NSCLC, and discuss future directions about lncRNAs in the researches of NSCLC.
Roles of LncRNAs in NSCLC
It has been proved that the abnormal expression of lncRNAs has a close relationship to NSCLC. Here is a quick review of some popular and well-studied lncRNAs related to NSCLC.
MALAT1 and NSCLC
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear-enriched abundant transcript 2 (NEAT2), was firstly identified in 2003 by subtractive hybridization as prognostic parameters for patient survival in stage I of NSCLC. MALAT1, more than 8000 nt expressed from chromosome 11q13, was detected not only in NSCLC, but also in some normal tissues such as pancreas, lung, prostate, ovary, colon, placenta, spleen, small intestine, kidney, heart, liver, testis and brain [8]. It was believed that MALAT1 expression levels were associated with patient survival by affecting genes involved in cancer like cellular growth, movement, proliferation, signaling, and immunoregulation [9]. Q-PCR was performed to confirm that the MALAT1 were upregulated in cancerous tissues than that in adjacent normal tissues [10]. And in vitro studies, migration and clonogenic growth could be suppressed by RNA-interference-mediated suppression of MALAT1 in A549 cells, while forced expression of MALAT1 in NIH 3T3 cells significantly increased migration [9]. What's more, the level of MALAT1 was higher in brain metastasis and it was increased in highly invasive subline of brain metastasis lung cancer cells, which was speculated on account for epithelial-mesenchymal transition (EMT) [11]. For the mechanism, there is no final conclusion until now. Some scholars thought it was regulated by DNA methylation [12] and some suspected MALAT1 of contributing to NSCLC by upregulating the expression of Bcl-2 and its interacting proteins [13]. Besides, it is reported that MALAT1 regulates alternative splicing (AS) of pre-mRNAs, which is a key step in the regulation and diversification of gene function, by controlling the levels of active serine/arginine (SR) proteins and the distributions to nuclear speckles [14]. Above all were recognized as modes of action for MALAT1: regulation of gene expression or alternative splicing [15].
HOTAIR and NSCLC
HOX antisense intergenic RNA (HOTAIR), a 2.2 kilobase noncoding RNA residing in the HOXC locus, was firstly identified in 2007. Rinn et al. proved in that paper that it might regulate gene expression in HOX loci in cis or trans; alternatively, it might be the act of antisense transcription in the HOXC locus [16]. And it was widely accepted that HOTAIR regulated gene expression by EZH2 (a subunit of PRC2), which led to histone H3 lysine 27 trimethylation of the HOXD locus, and it also could mediate chromosomal remodeling [17, 18]. In addition, it was confirmed that HOTAIR was highly expressed in both NSCLC samples and cell lines compared with adjacent tissues and it indicated a poor prognosis [19]. In the mechanism of how HOTAIR contributed to NSCLC, it was thought that HOTAIR might facilitate the tumor development but not the carcinogenesis of NSCLC [20]. In the meantime, some scholars found that HOTAIR modified the promoter of p53 and enhanced histone H3 lysine 27 trimethylation, which showed a negative relationship between HOTAIR and p53 in NSCLC cells [21]. What's more, it was reported that HOTAIR can activate Wnt/β-catenin signaling pathway in esophageal squamous cell carcinoma [22]. In addition, HOTAIR could involve in EMT, and also worked as competitive endogenous RNAs (ceRNAs) [23].
HOTTIP and NSCLC
HOXA distal transcript antisense RNA (HOTTIP) is an antisense non-coding transcript located at the distal end of HOXA gene cluster. It was regarded as key intermediates to transmit information from higher order chromosomal looping into chromatin modifications, and thus coordinated long range gene activation, which was associated with the WDR5/MLL complex to drive the H3 lysine 4 trimethylation and gene transcription [24, 25]. And it was identified as the most significantly up-regulated lncRNAs in human primary hepatocellular carcinoma even in early stage [26]. In NSCLC, HOTTIP expression was higher than corresponding adjacent normal tissues and contributed to cell proliferation and migration, which was by regulating HOXA13 and functioning as oncogene [27].
Besides these relatively common and popular lncRNAs mentioned above, there were still some other lncRNAs proved having close relationship to NSCLC which will be presented in Table 1 and Table 2.
Table 1. Overexpressed or upregulated lncRNAs in NSCLC tissues or cell lines and their functions and probable mechanism.
| LncRNA | Function in NSCLC | probable mechanism | cition |
|---|---|---|---|
| AGAP2-AS1 | negatively correlated with poor prognostic outcomes | repressed tumor-suppressor LATS2 and KLF2 transcription | [28] |
| ATB | presented a lower survival probability | [29] | |
| TCF7 | promoted invasion and self-renewal | TCF7 upregulated EpCAM expression through functioning as a competitive endogenous RNA (ceRNA) | [30] |
| SBF2-AS1 | increased the proliferation of NSCLC cells | negatively regulated P21 | [31] |
| FOXD2-AS1 | promoted NSCLC cell growth and NSCLC tumor progression | Wnt/β-catenin signaling | [32] |
| HOXA11-AS | promoted development and progression of NSCLC | regulated the expression of various pathways and genes, especially DOCK8 and TGF-beta pathway. | [33] |
| PCAT-1 | played an oncogenic role in NSCLC progression | [34] | |
| BCAR4 | associated with poorer 5-year overall survival rate of NSCLC patients | [35] | |
| CCAT2 | promoted tumorigenesis | over-expression of Pokemon | [36] |
| 00511 | functioned as an oncogene | acted as a modular scaffold of EZH2/PRC2 complexes | [37] |
| XIST | associated with shorter survival and poorer prognosis | by epigenetically repressing KLF2 expression | [38] |
| NEAT1 | correlated with poor prognosis | inhibition of miR-377-3p/ E2F3 axis. | [39] |
| ANRIL | correlated with advanced tumor–node–metastasis stage and greater tumor diameter | [40] | |
| ZFAS1 | an independent prognostic factor for poor survival of NSCLC patients | [41] | |
| SNHG1 | associated with a poor overall survival | inhibited miR-101-3p and activated of Wnt/β-catenin signaling pathway | [42] |
| RGMB-AS1 | correlated with differentiation, TNM stage, and lymph node metastasis | by regulating RGMB expression though exon2 of RGMB | [43] |
Table 2. Lower expressed or downregulated lncRNAs in NSCLC tissues or cell lines and their functions and probable mechanism.
| LncRNA | Function in NSCLC | probable mechanism | cition |
|---|---|---|---|
| TUSC7 | associated with worse overall survival | [44] | |
| CASC2 | independent predictor for overall survival of NSCLC | [45] | |
| GAS5 | indicated a poor prognosis and regulated cell proliferation | [46] | |
| TUG1 | related to the proliferation of NSCLC cells | TUG1 RNA could bind to PRC2 in the promotor region of CELF1 and negatively regulated CELF1 expressions | [47] |
| AK126698 | inhibited the proliferation and migration | inhibited the activation of Wnt/β-catenin pathway | [48] |
| GAS5-AS1 | regulated NSCLC cell migration and invasion | through regulation of EMT | [49] |
LncRNAs associated with radiochemotherapy of NSCLC
LncRNAs and chemotherapy
As is well-known, DNA damage repair (DDR) mechanisms, such as single-strand break, double-strand break, bulky adducts, base mismatches and base alkylation, are playing important roles to maintain genomic stability. Thanks to these precise modulations, cells could maintain genomic integrity confronted with numerous physical or chemical or even deadly strikes [50–52]. Platinum, a kind of chemical elements, could also cause DNA damage, especially in tumor cells, where the DDR is not complete.
Since found by Rosenbery in 1969, platinum was widely used in clinical practice including chemotherapeutics of NSCLC, which benefited a lot of incipient or advanced patients [53, 54]. Tumor cells, which proliferated more rapidly than normal ones, could be influenced directly by anticarcinogen, thus stopping equal division of DNA to next generation [55]. Binding of platinum and genomic DNAs in cell nucleus was thought to play important roles in cancer therapy, which influenced transcription and DNA replication and finally led to death of tumor cells [56]. However, the use of platinum was limited due to its toxicity, drug resistance, and some other side effects, which was demonstrated closely to lncRNAs [57, 58]. The polymorphisms of lncRNAs such as HOTTIP, CCAT2, H19, HOTAIR, MALATI, ANRIL and CASC8 were proved significantly associated with lung cancer risk or platinum-based chemotherapy response [58, 59]. It was reported that HOTAIR was significantly upregulated in cisplatin-resistant NSCLC cells both in vitro and in vivo, and it could enhance tumor cell proliferation, influence G0/G1 cell-cycle arrest, and decrease tumor cell apoptosis. Further studies showed that overexpression of HOTAIR could promote tumor sphere formation, which upregulated expression of the tumor stem cell-related biomarkers such as Nanog, Oct3/4, Sox2, c-Myc, β-catenin, and Klf4 [60, 61]. It was found inverse correlation between HOTAIR and p21 [62], the latter of which was proved as a negative regulator of the cell cycle [63]. What's more, lncRNA H19 had a negative relationship with cisplatin-based chemotherapy response, the enhancement of which associated with metastasis, induction of G0/G1 cell-cycle arrest, cell proliferation, and increased apoptosis [64]. Other lncRNAs were reported to relate to response to chemotherapy such as HOTTIP in osteosarcoma [65], MALAT1 in laryngeal squamous cell carcinoma [66], and ANRIL in nasopharyngeal carcinoma [67].
LncRNAs and radiotherapy
Radiotherapy is essential in most patients especially with advanced stage NSCLC, usually sequentially or simultaneously combined with surgery, chemotherapy and molecular therapy [68, 69]. Radioactive rays could cause a series of physical, chemical and biological damages to both tumor cells and normal cells, of which doctors make use, to cure cancer by reducing radiological dose of normal tissues and increasing that of tumor cells [70]. LncRNAs were proved to involve in the DNA damage response after radioactive rays, which might lead to the failure of radiotherapy [71]. However, the detailed mechanisms about lncRNA and resistance to radioterapy haven't been found, but it is certain that miRNAs do involve in the radioresistance of head and neck cancer [72]. And it is deserved to investigate whether there are some relationship between lncRNAs and radioresistance in NSCLC.
LncRNAs associated with molecular targeted therapy of NSCLC
Basically speaking, there are three kinds of targeted therapies for NSCLC so far, namely EGFR tyrosine kinase inhibitors (EGFR-TKIs), antiangiogenic agents and Programmed cell death protein 1 inhibitors, which brings hopes and prospects to patients suffering NSCLC.
EGFR and LncRNAs
Epidermal growth factor receptor (EGFR) super-family has been regarded as a therapeutic target to NSCLC. It was firstly reported in 2004 that the mutations of patients who were sensitive to gefitinib were around the ATP-binding pocket of the tyrosine kinase domain, which were small, in-frame deletions or amino acid substitutions [73]. Adenocarcinomas from never smokers were easier to acquire this kind of gene mutation, which meant they were more sensible to gefitinib or erlotinib [74]. According to statistics from Ohashi K, patients whose metastatic tumors identified EGFR mutations were expected to live longer than 2 years [75]. Following the use of these drugs, most patients who were initially sensitive to EGFR TKIs eventually acquired inevitable resistance after long-term therapy. And the EGFR T790M secondary mutation, which substituted methionine for threonine at residue 790, was firstly reported only a year later after the discovery of EGFR-TKIs [76]. The EGFR T790M secondary mutation was identified from patients who were not sensitive to EGFR-TKIs up to 68%. Besides, there were some other rare mutation such as D761Y, L747S, and T854A discovered in 2006, 2007, 2008 respectively [77–79]. In addition, mechanisms such as MET amplification, PTEN downregulation, CRKL amplification, High-level expression of HGF, FAS–NFƙB activation, EMT, and transformation to small cell lung cancer, were also working in the EGFR-TKIs resistance [80].
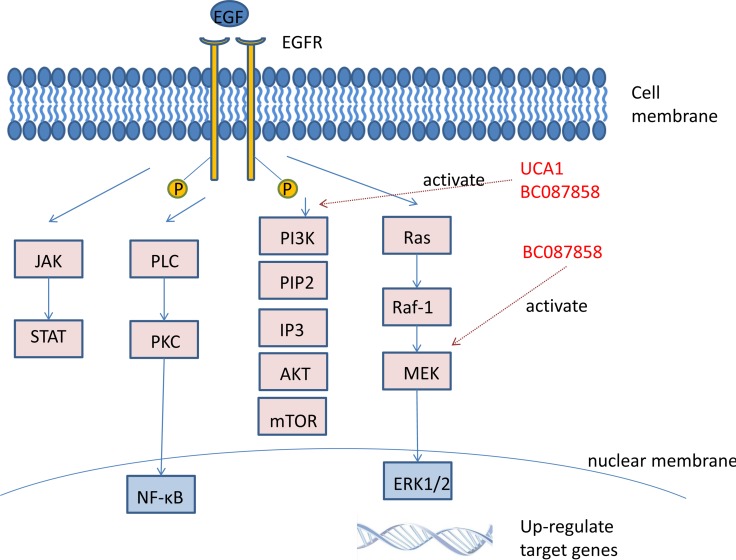
Few papers mentioned the relationship between lncRNAs and EGFR mutations in NSCLC. Urothelial cancer-associated 1 (UCA1), a long non-coding RNA highly specific to Bladder transitional cell carcinoma (TCC), was proved to have a close relationship to colorectal, gastric, ovarian cancer and NSCLC [81, 82]. UCA1 acted as an oncogenic role in NSCLC and it was proved that the expression of UCA1 in NSCLC samples was significantly higher compared with adjacent tissues partly through competitively ‘sponging’ miR-506-3p [83]. UCA1 was also highly expressed in patients with acquired resistance to EGFR-TKIs, and further studies showed that it was related to non-T790M by activating AKT/mTOR pathway and EMT [84].
LncRNA BC087858 could stimulate acquired resistance to EGFR-TKIs in NSCLC and might contribute to a shorter progression-free survival (PFS) [85]. Further study showed that lncRNA BC087858 could induce non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT via upregulating ZEB1 and Snail and eventually promoted cell invasion [86].
Not only single lncRNA could influence EGFR-TKIs resistance, but also lncRNAs could interact each other to affect the sensitivity to targeted therapy. The over and lower expression of CASC9 and lncRNA 00277 were respectively negative to sensitivity to gefitinib in PC9G2 cells, and taken together, it was reported that they contributed to NSCLC cells EGFR-TKI resistance through interacting with their co-expressed gene, namely PcGs, and affected different biological pathways [87] (Figure 1).
Figure 1. LncRNAs associated non-T790M mutation of NSCLC.
UCA1 activated AKT/mTOR pathway and related to non-T790M mutation. LncRNA BC087858 induced non-T790M mutation by activating PI3K/AKT and MEK/ERK pathways.
PD-1, PDL-1 and LncRNAs
The microenvironment of malignant cells were gaining highlight to the treatment of tumors and many labs were concentrating on finding ways to make immune cells kill cancer cells. T cells were the major cells to fight or kill malignant cells, and the activation of T cells was partly depending on immune checkpoints [88, 89]. Programmed cell death protein 1 (PD-1) and programmed cell death protein ligand 1 (PDL-1) are two of key components of immune checkpoints. It was widely accepted that the engagement of PD-1 by PDL-1 could suppress immune responses and consequently led to immune evasion [90, 91]. Therefore the study of PD-1 and PDL-1 is now offering new important opportunities for the therapy of cancer.
PDL-1 expression of tumor was significantly associated with a shorter PFS [92], and for the researches of its receptor, namely PD-1, showed that cumulative response rates to anti-PD-1 antibody were 18% among NSCLC patients according to a clinical trial in America (14 of 76 patients), which provided a kind of new method to NSCLC treatment [93]. Some scholars suggested the combination of EGFR-TKIs and immune checkpoints inhibitors, but due to the toxicity of this kind of combination, it aroused a lot of controversy [94, 95].
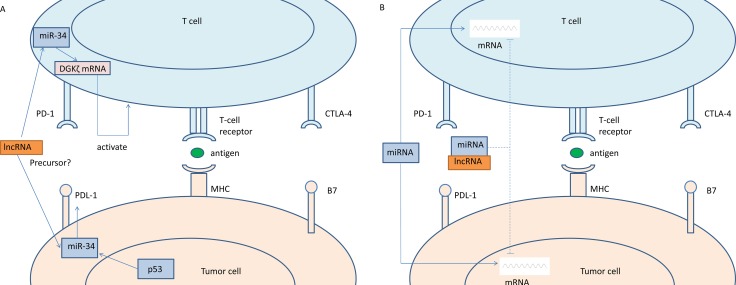
Although many evidences showed that PD-1/PDL-1 had a promising future to treat with NSCLC, we knew little about the regulation about expression of PD-1/PDL-1. Previous studies demonstrated that PD-1.5 C/T significantly increased advanced NSCLC risk and potentially related to NSCLC susceptibility in Chinese Han population [96]. However, to our knowledge, there was no report about the relationship between PD-1/PDL-1 and lncRNAs in NSCLC, and only one paper was found to reveal that the co-expression of lncRNA AFAP1-AS1 and PD-1 predicted poor prognosis of nasopharyngeal carcinoma (NPC) [97]. At present, it was found that p53 regulated PDL-1 via miR-34, which directly binded to the PDL-1 3′untranslated region in models of NSCLC [98]. What's more, it was also reported that miR-34 enhanced T cell activation via targeting diacylglycerol kinase ζ [99] (Figure 2A). Because the expression of miRNAs was quite specific to distinct tumors, and they could affect early regulation of immune responses, they were regarded as suitable molecules for cancer therapy [100]. In addition, lncRNAs could be precursors of miRNAs and act as ceRNAs to alter the distribution of miRNA molecules on their targets [6, 101] (Figure 2B), for example, it was found that lncRNA ARSR acted as a ceRNA for miR-34 and miR-449 and finally promoted Sunitinib resistance in renal cancer [102]. Thereby we can speculate that lncRNAs probably influence the patients’ response to anti-PD-1 or anti-PDL-1 treatment.
Figure 2. Hypothesis: LncRNAs associated the patients’ response to anti-PD-1 or anti-PDL-1 treatment.
(A) LncRNAs could be precursors of miRNAs. P53 regulated PDL-1 via miR-34, and miR-34 enhanced T cell activation via targeting diacylglycerol kinase ζ. (B) LncRNAs could act as ceRNAs to alter the distribution of miRNA molecules on their targets.
Antiangiogenic agents and LncRNAs
Researches showed that tumor growth and metastatic potential partly related to tumor angiogenesis. Vascular endothelial growth factor (VEGF), inducing angiogenesis in vivo, was expressed in most solid cancers including NSCLC [103]. Bevacizumab, a humanized monoclonal antibody, could block the binding of VEGF-A isoforms to VEGF receptors and therefore against tumors [104]. The Food and Drug Administration approved bevacizumab for the treatment in first-line metastatic setting of colorectal cancer, non-small cell lung cancer and breast cancer, and randomized controlled trials (RCTs) showed that bevacizumab-based regimens revealed significantly increased overall survival (OS) [105, 106].
For the relationship between VEGF and lncRNAs, it was proved that MALAT1 could promote angiogenesis and immunosuppressive properties of mesenchymal stem cells by inducing VEFG in preeclampsia [107]. More direct evidence is that when lincRNA p21 was inhibited, the expressions of angiogenesis-related genes were downregulated and lincRNA-p21-inhibited cells were observed to secrete less VEGFA than controls did [108].
CONCLUSIONS AND FUTURE DIRECTIONS
With the deep research, lncRNAs are not regarded as transcriptional “noise” any more, and they are thought to be a new frontier for many diseases including malignant tumors. Based on existing evidences, lncRNAs are playing important roles in the presence and development of NSCLC, which leads to most cancer-related death. What's more, over or lower expression of lncRNAs could alter the ability of cellular growth, movement, proliferation, signaling, immunoregulation and invasion, consequently to influence the prognostication of cancer. Besides, it has a close relationship between lncRNAs and the response to radiochemotherapy or molecular targeted therapy, by which ulteriorly affect the prognostication of NSCLC. Following the development of body fluid detection, lncRNAs test will not only be applied into operative tissues, but also in blood, urine and other body fluid and will have a better predictive and diagnostic function [109].
In the future, further studies would be concentrated on the following aspects: (1) identifying new lncRNAs (2) discovering more functions of lncRNAs (3) detecting more relationships with miRNAs and other non-coding RNAs (4) seeking more probable pathways that lncRNAs influence the gene transcript or protein expression (5) looking for possibility of lncRNAs as therapeutic targets (6) developing more precise and reliable ways to detect lncRNAs in body floods.
Ethics approval and consent to participate
Not applicable.
Acknowledgments
Not applicable.
Abbreviations
- NSCLC
non-small cell lung cancer
- lncRNA
long non-coding RNA
- SCLC
small cell lung cancer
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- NEAT2
nuclear-enriched abundant transcript 2
- EMT
epithelial-mesenchymal transition
- AS
alternative splicing
- SR
serine/arginine
- HOTAIR
HOX antisense intergenic RNA
- ceRNA
competing endogenous RNA
- HOTTIP
HOXA distal transcript antisense RNA
- SBF2-AS1
SBF2 antisense RNA 1
- HOXA11-AS
HOXA11 antisense RNA
- PCAT-1
Prostate cancer-associated transcript 1
- BCAR4
breast cancer anti-estrogen resistance 4
- CCAT2
colon cancer-associated transcript 2
- XIST
X inactivate-specific transcript
- NEAT1
nuclear enriched abundant transcript 1
- SNHG1
small nucleolar RNA host gene 1
- TUSC7
Tumor suppressor candidate 7
- CASC2
cancer susceptibility candidate 2
- GAS5
growth arrest-specific transcript 5
- UCA1
Urothelial cancer-associated 1
- AK126698
lncRNA AK126698
- DDR
DNA damage repair
- CCAT2
Colon Cancer Associated Transcript 2
- EGFR-TKIs
Epidermal growth factor receptor tyrosine kinase inhibitors
- EGFR
Epidermal growth factor receptor
- MET
Mesenchymal-epithelial transition
- TCC
transitional cell carcinoma
- PFS
progression-free survival
- AFAP1-AS1
actin filament-associated protein 1 antisense RNA
- NPC
nasopharyngeal carcinoma
- VEGF
Vascular endothelial growth factor
- RCT
randomized controlled trials
- OS
overall survival
Footnotes
Author contributions
Songqing Fan designed and revised the manuscript. Yuting Zhan wrote the manuscript and drew figures. Hongjing Zang collected related papers and created the tables. Juan Feng, Junmi Lu, Lingjiao Chen participated in the design and revise of the review. All the authors read and approved the final version of the review.
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
FUNDING
This work was supported by the National Natural Science Foundations of China (No: 81272566; 81472773).
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi: 10.3978/j.issn.1000-9604.2015.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pikin OV, Ryabov AB, Glushko VA, Kolbanov KI, Amiraliev AM, Vursol DA, Bagrov VA, Barmin VV. Surgery for non-small cell lung carcinoma after previous chemoradiotherapy alone. Khirurgiia (Mosk) 2016:28–31. doi: 10.17116/hirurgia201611228-31. [DOI] [PubMed] [Google Scholar]
- 4.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricciuti B, Mencaroni C, Paglialunga L, Paciullo F, Crinò L, Chiari R, Metro G. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33:18. doi: 10.1007/s12032-016-0731-2. [DOI] [PubMed] [Google Scholar]
- 8.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 10.Zhang CG, Yin DD, Sun SY, Han L. The use of lncRNA analysis for stratification management of prognostic risk in patients with NSCLC. Eur Rev Med Pharmacol Sci. 2017;21:115–119. [PubMed] [Google Scholar]
- 11.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 12.Guo F, Guo L, Li Y, Zhou Q, Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol. 2015;8:15903–15910. [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt LH, Görlich D, Spieker T, Rohde C, Schuler M, Mohr M, Humberg J, Sauer T, Thoenissen NH, Huge A, Voss R, Marra A, Faldum A, et al. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol. 2014;9:1294–1304. doi: 10.1097/JTO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 14.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 16.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y, Li J, Wang L. Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int J Mol Sci. 2014;15:18985–18999. doi: 10.3390/ijms151018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai) 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 21.Zhai N, Xia Y, Yin R, Liu J, Gao F. A negative regulation loop of long noncoding RNA HOTAIR and p53 in non-small-cell lung cancer. Onco Targets Ther. 2016;9:5713–5720. doi: 10.2147/OTT.S110219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng F, Zhou K, Cui W, Liu D, Ma Y. Clinicopathological significance of wnt/β-catenin signaling pathway in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:3045–3053. [PMC free article] [PubMed] [Google Scholar]
- 23.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess DJ. Non-coding RNA: HOTTIP goes the distance. Nat Rev Genet. 2011(12):300. doi: 10.1038/nrg2992. [DOI] [PubMed] [Google Scholar]
- 26.Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, Wong CM. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35:1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 27.Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao Z, Li Q, Zhang W. Up-regulation of long non-coding HOTTIP functions as an oncogene by regulating HOXA13 in non-small cell lung cancer. Am J Transl Res. 2016;8:2022–2032. [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Sun M, Zang C, Ma P, He J, Zhang M, Huang Z, Ding Y, Shu Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke L, Xu SB, Wang J, Jiang XL, Xu MQ. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clin Transl Oncol. 2016 doi: 10.1007/s12094-016-1572-3. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Wang D. Long noncoding RNA TCF7 promotes invasiveness and self-renewal of human non-small cell lung cancer cells. Hum Cell. 2017;30:23–29. doi: 10.1007/s13577-016-0147-5. [DOI] [PubMed] [Google Scholar]
- 31.Lv J, Qiu M, Xia W, Liu C, Xu Y, Wang J, Leng X, Huang S, Zhu R, Zhao M, Ji F, Xu L, Xu K, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75. doi: 10.1186/s13046-016-0352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rong L, Zhao R, Lu J. Highly expressed long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer progression via Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2017;484:586–591. doi: 10.1016/j.bbrc.2017.01.141. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, He RQ, Dang YW, Zhang XL, Wang X, Huang SN, Huang WT, Jiang MT, Gan XN, Xie Y, Li P, Luo DZ, Chen G, et al. Comprehensive analysis of the long noncoding RNA HOXA11-AS gene interaction regulatory network in NSCLC cells. Cancer Cell Int. 2016;16:89. doi: 10.1186/s12935-016-0366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao B, Hou X, Zhan H. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in non-small cell lung cancer cells. Int J Clin Exp Med. 2015;8:18482–18487. [PMC free article] [PubMed] [Google Scholar]
- 35.Gong J, Zhang H, He L, Wang L, Wang J. Increased Expression of Long Non-Coding RNA BCAR4 Is Predictive of Poor Prognosis in Patients with Non-Small Cell Lung Cancer. Tohoku J Exp Med. 2017;241:29–34. doi: 10.1620/tjem.241.29. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Z, Wang J, Wang S, Chang H, Zhang T, Qu J. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother. 2017;87:692–697. doi: 10.1016/j.biopha.2016.12.122. [DOI] [PubMed] [Google Scholar]
- 37.Sun CC, Li SJ, Li G, Hua RX, Zhou XH, Li DJ. Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non-small-cell Lung Cancer by Binding to EZH2 and Suppressing p57. Mol Ther Nucleic Acids. 2016;5:e385. doi: 10.1038/mtna.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 39.Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. https://doi.org/10.18632/oncotarget.10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Zhou X, Xu L, Rong C, Shen C, Bian W. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther. 2016;9:3077–3084. doi: 10.2147/OTT.S102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian FM, Meng FQ, Wang XB. Overexpression of long-noncoding RNA ZFAS1 decreases survival in human NSCLC patients. Eur Rev Med Pharmacol Sci. 2016;20:5126–5131. [PubMed] [Google Scholar]
- 42.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition ofmiR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. https://doi.org/10.18632/oncotarget.14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P, Zhang G, Li J, Yang R, Chen S, Wu S, Zhang F, Bai Y, Zhao H, Wang Y, Dun S, Chen X, Sun Q, et al. Long Noncoding RNA RGMB-AS1 Indicates a Poor Prognosis and Modulates Cell Proliferation, Migration and Invasion in Lung Adenocarcinoma. PLoS One. 2016;11:e0150790. doi: 10.1371/journal.pone.0150790. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Wang Z, Jin Y, Ren H, Ma X, Wang B, Wang Y. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am J Transl Res. 2016;8:680–687. [PMC free article] [PubMed] [Google Scholar]
- 45.He X, Liu Z, Su J, Yang J, Yin D, Han L, De W, Guo R. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumour Biol. 2016;37:9503–9510. doi: 10.1007/s13277-016-4787-6. [DOI] [PubMed] [Google Scholar]
- 46.Liang W, Lv T, Shi X, Liu H, Zhu Q, Zeng J, Yang W, Yin J, Song Y. Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine (Baltimore) 2016;95:e4608. doi: 10.1097/MD.0000000000004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin PC, Huang HD, Chang CC, Chang YS, Yen JC, Lee CC, Chang WH, Liu TC, Chang JG. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:583. doi: 10.1186/s12885-016-2569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu X, Li H, Liu C, Hu B, Li T, Wang Y. Long noncoding RNA AK126698 inhibits proliferation and migration of non-small cell lung cancer cells by targeting Frizzled-8 and suppressing Wnt/β-catenin signaling pathway. Onco Targets Ther. 2016;9:3815–3827. doi: 10.2147/OTT.S100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Lyu H, Liu H, Shi X, Song Y, Liu B. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep. 2016;6:31093. doi: 10.1038/srep31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 51.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jalal S, Earley JN, Turchi JJ. DNA repair: from genome maintenance to biomarker and therapeutic target. Clin Cancer Res. 2011;17:6973–6984. doi: 10.1158/1078-0432.CCR-11-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang J, Demaria S, Formenti S. Current clinical trials testing the combination of immunotherapy with radiotherapy. J Immunother Cancer. 2016;4:51. doi: 10.1186/s40425-016-0156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valdes M, Nicholas G, Goss GD, Wheatley-Price P. Chemotherapy in recurrent advanced non-small-cell lung cancer after adjuvant chemotherapy. Curr Oncol. 2016;23:386–390. doi: 10.3747/co.23.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 56.Fuertes MA, Alonso C, Pérez JM. Biochemical modulation of Cisplatin mechanisms of action: enhancement of antitumor activity and circumvention of drug resistance. Chem Rev. 2003;103:645–662. doi: 10.1021/cr020010d. [DOI] [PubMed] [Google Scholar]
- 57.Xiong Y, Huang BY, Yin JY. Pharmacogenomics of platinum-based chemotherapy in non-small cell lung cancer: focusing on DNA repair systems. Med Oncol. 2017;34:48. doi: 10.1007/s12032-017-0905-6. [DOI] [PubMed] [Google Scholar]
- 58.Gong WJ, Yin JY, Li XP, Fang C, Xiao D, Zhang W, Zhou HH, Li X, Liu ZQ. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37:8349–8358. doi: 10.1007/s13277-015-4497-5. [DOI] [PubMed] [Google Scholar]
- 59.Hu L, Chen SH, Lv QL, Sun B, Qu Q, Qin CZ, Fan L, Guo Y, Cheng L, Zhou HH. Clinical Significance of Long Non-Coding RNA CASC8 rs10505477 Polymorphism in Lung Cancer Susceptibility, Platinum-Based Chemotherapy Response, and Toxicity. Int J Environ Res Public Health. 2016;13:E545. doi: 10.3390/ijerph13060545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu MY, Li XQ, Gao TH, Cui Y, Ma N, Zhou Y, Zhang GJ. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J Thorac Dis. 2016;8:3314–3322. doi: 10.21037/jtd.2016.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishiguchi M, Kikuyama H, Kanazawa T, Tsutsumi A, Kaneko T, Uenishi H, Kawabata Y, Kawashige S, Koh J, Yoneda H. Increases in iPS Transcription Factor (Oct4, Sox2, c-Myc, and Klf4) Gene Expression after Modified Electroconvulsive Therapy. Psychiatry Investig. 2015;12:532–537. doi: 10.4306/pi.2015.12.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De Z, Wang W, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21 (WAF1/CIP1) expression. PLoS One. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Q, Cheng N, Li X, Pan H, Li C, Ren S, Su C, Cai W, Zhao C, Zhang L, Zhou C. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget. 2017;8:2558–2567. doi: 10.18632/oncotarget.13708. https://doi.org/10.18632/oncotarget.13708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J Transl Res. 2016;8:2385–2393. [PMC free article] [PubMed] [Google Scholar]
- 66.Chen H, Xin Y, Zhou L, Huang JM, Tao L, Cheng L, Tian J. Cisplatin and paclitaxel target significant long noncoding RNAs in laryngeal squamous cell carcinoma. Med Oncol. 2014;31:246. doi: 10.1007/s12032-014-0246-7. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. 2017 doi: 10.1002/jbt.21904. [DOI] [PubMed] [Google Scholar]
- 68.Yoon SM, Shaikh T, Hallman M. Therapeutic management options for stage III non-small cell lung cancer. World J Clin Oncol. 2017;8:1–20. doi: 10.5306/wjco.v8.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu AJ, Garay E, Foster A, Hsu M, Zhang Z, Chaft JE, Huang J, Rosenzweig KE, Rimner A. Definitive Radiotherapy for Local Recurrence of NSCLC After Surgery. Clin Lung Cancer. 2017 doi: 10.1016/j.cllc.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Youroukou A, Gkiozos I, Kalaitzi Z, Tsalafoutas I, Papalla K, Charpidou A, Kouloulias V. The potential role of brachytherapy in the irradiation of patients with lung cancer: a systematic review. Clin Transl Oncol. 2017 doi: 10.1007/s12094-017-1635-0. [DOI] [PubMed] [Google Scholar]
- 71.Li L, Zhu T, Gao YF, Zheng W, Wang CJ, Xiao L, Huang MS, Yin JY, Zhou HH, Liu ZQ. Targeting DNA Damage Response in the Radio (Chemo) therapy of Non-Small Cell Lung Cancer. Int J Mol Sci. 2016;17:E839. doi: 10.3390/ijms17060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahmad P, Sana J, Slavik M, Slampa P, Smilek P, Slaby O. MicroRNAs Involvement in Radioresistance of Head and Neck Cancer. Dis Markers. 2017:8245345. doi: 10.1155/2017/8245345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 74.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanan EJ, Eigenbrot C, Bryan MC, Burdick DJ, Chan BK, Chen Y, Dotson J, Heald RA, Jackson PS, La H, Lainchbury MD, Malek S, Purkey HE, et al. Discovery of selective and noncovalent diaminopyrimidine-based inhibitors of epidermal growth factor receptor containing the T790M resistance mutation. J Med Chem. 2014;57:10176–10191. doi: 10.1021/jm501578n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, Ladanyi M, Miller VA, Pao W. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 78.Yamaguchi F, Fukuchi K, Yamazaki Y, Takayasu H, Tazawa S, Tateno H, Kato E, Wakabayashi A, Fujimori M, Iwasaki T, Hayashi M, Tsuchiya Y, Yamashita J, et al. Acquired resistance L747S mutation in an epidermal growth factor receptor-tyrosine kinase inhibitor-naïve patient: A report of three cases. Oncol Lett. 2014;7:357–360. doi: 10.3892/ol.2013.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goyal S, Jamal S, Shanker A, Grover A. Structural investigations of T854A mutation in EGFR and identification of novel inhibitors using structure activity relationships. BMC Genomics. 2015;16:S8. doi: 10.1186/1471-2164-16-S5-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suda K, Mizuuchi H, Maehara Y, Mitsudomi T. Acquired resistance mechanisms to tyrosine kinase inhibitors in lung cancer with activating epidermal growth factor receptor mutation--diversity, ductility, and destiny. Cancer Metastasis Rev. 2012;31:807–814. doi: 10.1007/s10555-012-9391-7. [DOI] [PubMed] [Google Scholar]
- 81.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, Zhang Y, Xin DQ, Na YQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 82.Hong HH, Hou LK, Pan X, Wu CY, Huang H, Li B, Nie W. Long non-coding RNA UCA1 is a predictive biomarker of cancer. Oncotarget. 2016;7:44442–44447. doi: 10.18632/oncotarget.10142. https://doi.org/10.18632/oncotarget.10142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo S, Yang P, Jiang X, Li X, Wang Y, Zhang X, Sun B, Zhang Y, Jia Y. Genetic and epigenetic silencing of mircoRNA-506-3p enhances COTL1 oncogene expression to foster non-small lung cancer progression. Oncotarget. 2017;8:644–657. doi: 10.18632/oncotarget.13501. https://doi.org/10.18632/oncotarget.13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng N, Cai W, Ren S, Li X, Wang Q, Pan H, Zhao M, Li J, Zhang Y, Zhao C, Chen X, Fei K, Zhou C, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget. 2015;6:23582–23593. doi: 10.18632/oncotarget.4361. https://doi.org/10.18632/oncotarget.4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng N, Li X, Zhao C, Ren S, Chen X, Cai W, Zhao M, Zhang Y, Li J, Wang Q, Zhou C. Microarray expression profile of long non-coding RNAs in EGFR-TKIs resistance of human non-small cell lung cancer. Oncol Rep. 2015;33:833–839. doi: 10.3892/or.2014.3643. [DOI] [PubMed] [Google Scholar]
- 86.Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li X, Zhao C, Zhang L, Cai W, Zhou C. Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget. 2016;7:49948–49960. doi: 10.18632/oncotarget.10521. https://doi.org/10.18632/oncotarget.10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma P, Zhang M, Nie F, Huang Z, He J, Li W, Han L. Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed Pharmacother. 2017;87:20–26. doi: 10.1016/j.biopha.2016.12.079. [DOI] [PubMed] [Google Scholar]
- 88.Ni L, Dong C. New checkpoints in cancer immunotherapy. Immunol Rev. 2017;276:52–65. doi: 10.1111/imr.12524. [DOI] [PubMed] [Google Scholar]
- 89.Nurieva R, Thomas S, Nguyen T, Martin-Orozco N, Wang Y, Kaja MK, Yu XZ, Dong C. T-cell tolerance or function is determined by combinatorial costimulatory signals. EMBO J. 2006;25:2623–2633. doi: 10.1038/sj.emboj.7601146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 92.Soo RA, Kim HR, Asuncion BR, Fazreen Z, Omar MF, Herrera MC, Yun Lim JS, Sia G, Soong R, Cho BC. Significance of immune checkpoint proteins in EGFR-mutant non-small cell lung cancer. Lung Cancer. 2017;105:17–22. doi: 10.1016/j.lungcan.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 93.Daskivich TJ, Belldegrun A. Words of wisdom. Re: Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. Eur Urol. 2015;67:816–817. doi: 10.1016/j.eururo.2014.12.052. [DOI] [PubMed] [Google Scholar]
- 94.Tabchi S, Kourie HR, Kattan J. Adding checkpoint inhibitors to tyrosine kinase inhibitors targeting EGFR/ALK in non-small cell lung cancer: a new therapeutic strategy. Invest New Drugs. 2016;34:794–796. doi: 10.1007/s10637-016-0383-2. [DOI] [PubMed] [Google Scholar]
- 95.Ahn MJ, Sun JM, Lee SH, Ahn JS, Park K. EGFR TKI combination with immunotherapy in non-small cell lung cancer. Expert Opin Drug Saf. 2017;16:465–469. doi: 10.1080/14740338.2017.1300656. [DOI] [PubMed] [Google Scholar]
- 96.Yin L, Guo H, Zhao L, Wang J. The programmed death-1 gene polymorphism (PD-1.5C/T) is associated with non-small cell lung cancer risk in a Chinese Han population. Int J Clin Exp Med. 2014;7:5832–5836. [PMC free article] [PubMed] [Google Scholar]
- 97.Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y, Fan C, Zhang P, Guo C, Zhang S, Gong Z, Li X, Xiong F, et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001–39011. doi: 10.18632/oncotarget.16545. https://doi.org/10.18632/oncotarget.16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, Kelnar K, Martin D, Komaki R, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst. 2015:108. doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shin J, Xie D, Zhong XP. MicroRNA-34a enhances T cell activation by targeting diacylglycerol kinase ζ. PLoS One. 2013;8:e77983. doi: 10.1371/journal.pone.0077983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smolle MA, Calin HN, Pichler M, Calin GA. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J. 2017 doi: 10.1111/febs.14030. [DOI] [PubMed] [Google Scholar]
- 101.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, Wang X, Wang Y, Xu ZY, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 103.Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jenab-Wolcott J, Giantonio BJ. Bevacizumab: current indications and future development for management of solid tumors. Expert Opin Biol Ther. 2009;9:507–517. doi: 10.1517/14712590902817817. [DOI] [PubMed] [Google Scholar]
- 105.Eskens FA, Sleijfer S. The use of bevacizumab in colorectal, lung, breast, renal and ovarian cancer: where does it fit? Eur J Cancer. 2008;44:2350–2356. doi: 10.1016/j.ejca.2008.07.042. [DOI] [PubMed] [Google Scholar]
- 106.Roviello G, Bachelot T, Hudis CA, Curigliano G, Reynolds AR, Petrioli R, Generali D. The role of bevacizumab in solid tumours: A literature based meta-analysis of randomised trials. Eur J Cancer. 2017;75:245–258. doi: 10.1016/j.ejca.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 107.Li X, Song Y, Liu F, Liu D, Miao H, Ren J, Xu J, Ding L, Hu Y, Wang Z, Hou Y, Zhao G. Long Non-coding RNA MALAT1 Promotes Proliferation, Angiogenesis and Immunosuppressive Properties of Mesenchymal Stem Cells by Inducing VEGF and IDO. J Cell Biochem. 2017;118:2780–2791. doi: 10.1002/jcb.25927. [DOI] [PubMed] [Google Scholar]
- 108.Castellano JJ, Navarro A, Viñolas N, Marrades RM, Moises J, Cordeiro A, Saco A, Muñoz C, Fuster D, Molins L, Ramirez J, Monzo M. LincRNA-p21 Impacts Prognosis in Resected Non-Small Cell Lung Cancer Patients through Angiogenesis Regulation. J Thorac Oncol. 2016;11:2173–2182. doi: 10.1016/j.jtho.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 109.Peng H, Wang J, Li J, Zhao M, Huang SK, Gu YY, Li Y, Sun XJ, Yang L, Luo Q, Huang CZ. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–242. doi: 10.1016/j.lfs.2016.03.002. [DOI] [PubMed] [Google Scholar]