The metabolic enzyme IMPDH assembles into octamers that can polymerize and form micron-scale structures in cells. Octamers can adopt active, expanded or inactive, compressed conformations driven by allosteric nucleotide and substrate binding. Both forms are accommodated within polymers, and polymerization alone does not alter catalytic activity.
Abstract
Several metabolic enzymes undergo reversible polymerization into macromolecular assemblies. The function of these assemblies is often unclear, but in some cases they regulate enzyme activity and metabolic homeostasis. The guanine nucleotide biosynthetic enzyme inosine monophosphate dehydrogenase (IMPDH) forms octamers that polymerize into helical chains. In mammalian cells, IMPDH filaments can associate into micron-length assemblies. Polymerization and enzyme activity are regulated in part by binding of purine nucleotides to an allosteric regulatory domain. ATP promotes octamer polymerization, whereas guanosine triphosphate (GTP) promotes a compact, inactive conformation whose ability to polymerize is unknown. Also unclear is whether polymerization directly alters IMPDH catalytic activity. To address this, we identified point mutants of human IMPDH2 that either prevent or promote polymerization. Unexpectedly, we found that polymerized and nonassembled forms of recombinant IMPDH have comparable catalytic activity, substrate affinity, and GTP sensitivity and validated this finding in cells. Electron microscopy revealed that substrates and allosteric nucleotides shift the equilibrium between active and inactive conformations in both the octamer and the filament. Unlike other metabolic filaments, which selectively stabilize active or inactive conformations, recombinant IMPDH filaments accommodate multiple states. These conformational states are finely tuned by substrate availability and purine balance, while polymerization may allow cooperative transitions between states.
INTRODUCTION
An increasing number of noncytoskeletal proteins undergo reversible polymerization. Screens have identified hundreds of proteins that dynamically form assemblies in response to nutrient stress (Alberti et al., 2009; Narayanaswamy et al., 2009; Noree et al., 2010; O’Connell et al., 2012, 2014; Aughey and Liu, 2015; Liu, 2016; Shen et al., 2016). For unknown reasons, this phenotype appears disproportionately associated with metabolic enzymes (O’Connell et al., 2012). In many cases the role of polymerization has not yet been determined, but for the few metabolic filaments that have been characterized, polymerization regulates enzyme activity and is important for maintaining homeostasis (Beaty and Lane, 1983; Barry et al., 2014; Petrovska et al., 2014; Lynch et al., 2017).
Inosine monophosphate dehydrogenase (IMPDH) reversibly assembles into helical polymers of stacked octamers in an ATP-dependent manner in vitro (Labesse et al., 2013) and into larger filamentous bundles in cells (Ji et al., 2006). This enzyme catalyzes the rate-limiting step in de novo guanine nucleotide biosynthesis, nicotinamide adenine dinucleotide (NAD+)-dependent oxidation of IMP into xanthosine monophosphate (Hedstrom, 2009). Humans possess two differentially expressed IMPDH genes (IMPDH1 and 2) sharing 84% amino acid sequence identity (Carr et al., 1993) and both can assemble into filaments (Gunter et al., 2008; Thomas et al., 2012). Although substrate channeling has been implicated in nucleotide biosynthesis, IMPDH filaments have not been shown to colocalize with other purine biosynthesis enzymes and their purpose is unclear. However, the pyrimidine biosynthetic enzyme cytidine triphosphate (CTP) synthase (CTPS) coassembles with IMPDH in some circumstances (Keppeke et al., 2015), suggesting the potential for coordination of purine and pyrimidine biosynthesis. Filaments of bacterial CTPS are inhibited while eukaryotic filaments are active, in both cases due to stabilization of specific conformational states in the polymer, and this has been proposed as a general mechanism by which metabolic filaments may regulate enzyme activity (Barry et al., 2014; Strochlic et al., 2014; Lynch et al., 2017).
A challenge in assessing the impact of assembly on IMPDH catalytic activity is the fact that IMPDH is generally not polymerized in cell lines grown in rich media (Carcamo et al., 2014). Polymerization can be triggered, however, with inhibitors of purine biosynthesis, including IMPDH inhibitors mycophenolic acid or ribavirin, inhibitors of enzymes acting either upstream or downstream of IMPDH (Carcamo et al., 2014) or by depleting the culture media of essential purine precursors (Calise et al., 2014). This responsiveness of IMPDH to reduced flux through the guanine biosynthetic pathway suggests that assembly could be associated with a homeostatic mechanism to restore guanine nucleotide levels. Alternatively, IMPDH filaments could serve as a storage depot for inactive enzyme. These experimental manipulations, however, dramatically affect IMPDH activity independently of filament assembly, confounding their use to assess filament catalytic activity. We sought to develop approaches to address the effect of IMPDH polymerization on catalytic activity. Here, using novel IMPDH2 point mutants, electron microscopy, biochemical assays and isotopic tracer studies in live cells, we demonstrate that IMPDH polymerization alone does not alter its catalytic activity and that allosteric regulation of enzyme activity occurs in both the free and polymer forms.
RESULTS AND DISCUSSION
Identification of polymerization altering mutants of IMPDH2
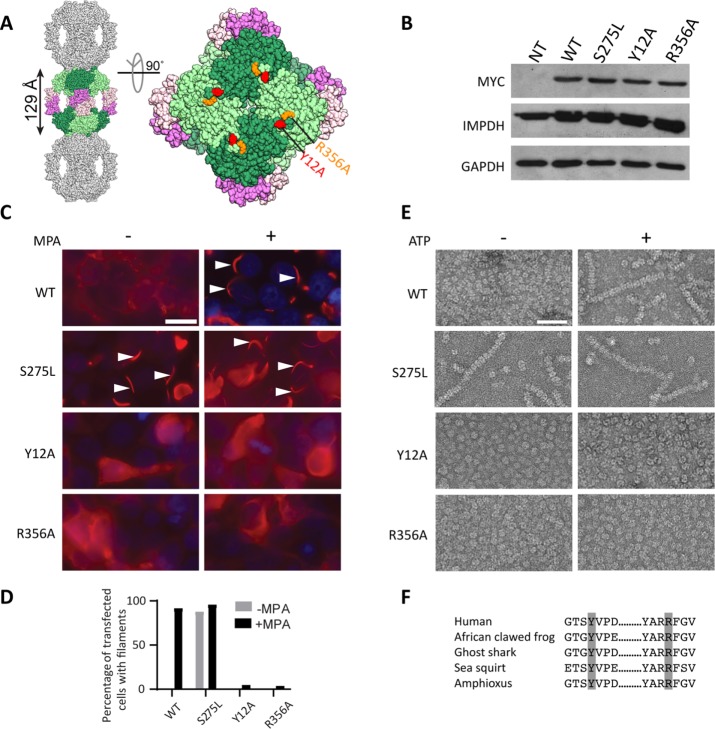
We sought to identify residues at the putative octamer polymerization interface that when mutated to alanine would disrupt IMPDH2 polymerization. The crystal structure of human IMPDH2 (Protein Data Bank [PDB] code:1NF7) contains stacked octamers that resemble IMPDH filaments (Labesse et al., 2013). On the assumption that these crystal contacts are the same as polymerization interfaces in filaments, we designed mutants that we predicted might block polymerization (Figure 1A). Individual myc-tagged alanine mutants were transiently transfected in HEK293 cells (Figure 1B), and filament assembly was induced by treatment with the IMPDH inhibitor mycophenolic acid (MPA) and visualized with anti-myc antibodies. Among other amino acids examined (threonine 10 and aspartate 15), only alanine substitution of tyrosine 12 (Y12A) or arginine 356 (R356A) disrupted filament induction by MPA while wild-type assembled as expected (Figure 1, C and D). These residues are evolutionarily conserved among chordates, supporting their functional importance (Figure 1F).
FIGURE 1:
Identification of filament disrupting and promoting IMPDH2 mutants. (A) The human IMPDH2 octamer (PDB file 1NF7). One octamer (greens, catalytic domains; pinks, Bateman domains) is shown with a subset of crystal packing neighbors (gray). Tyrosine 12 (red) and arginine 356 (orange) are indicated. (B) Immunoblot of HEK293 cell lysates with antibodies against myc-tagged IMPDH2, total IMPDH, or loading control glyceraldehyde 3-phosphate dehydrogenase (GAPDH). NT, not transfected. (C) Anti-myc immunofluorescence of HEK293 cells transfected with the indicated IMPDH2-myc constructs (red). 4’6-diamidino-2-phenylindole (DAPI) staining in blue. Cells were either treated with 10 µM mycophenolic acid (MPA) to induce IMPDH2 filaments (arrowheads) or not, as indicated. Images are representative of three experiments. Scale bar, 20 µm. (D) Quantification of transfected cells with filaments; n = 100 cells per sample. (E) Negative-stain electron microscopy of purified IMPDH2 proteins incubated with 5 mM NAD+, with and without 1 mM ATP. Images represent two separate experiments. Scale bar, 50 nm. (F) Evolutionary conservation of tyrosine 12 and arginine 356. IMPDH amino acid sequences from Homo sapiens, Xenopus laevis, Callorhinchus milii, Ciona savignyi, and Branchiostoma belcheri were retrieved and aligned using Consurf (Ashkenazy et al., 2016).
To confirm that these mutations directly prevent IMPDH2 self-assembly (as opposed to disrupting protein folding or interactions with assembly partners), we used electron microscopy to visualize recombinant IMPDH2 (see Coomassie-stained gel in Supplemental Figure 1). Wild-type IMPDH2 appears as a mixture of octamers and tetramers with occasional short filaments in the absence of ligands and formed long polymers in the presence of ATP (Figure 1E). In the absence of ligands, Y12A and R356A appear as a mixture of octamers and tetramers without polymers, and no polymers were observed in the presence of ATP, indicating that the mutations directly interfere with polymerization. While we observed only single filaments of wild-type IMPDH2, lateral bundles of filaments have been reported in cells (Thomas et al., 2012; Juda et al., 2014), suggesting that other cellular factors may mediate bundling.
A study of human IMPDH sequence polymorphisms in healthy individuals identified a rare, nonsynonymous polymorphism altering serine 275 to leucine (S275L) in IMPDH1 (Wu et al., 2010) that constitutively assembled into filaments in cells (Wu, 2011). We engineered this mutation into IMPDH2 and confirmed the propensity of S275L to assemble in cells, even in the absence of MPA (Figure 1, C and D). Unlike wild-type protein, purified S275L robustly assembles polymers in both the presence and absence of ATP, indicating that the propensity of S275L to polymerize is an intrinsic consequence of the mutation (Figure 1E). S275L is localized adjacent to the NAD+ binding site and the basis for its constitutive filamentation is unclear. Nevertheless, this construct served as a useful experimental counterpoint to the nonassembling mutants.
Catalytic activity of IMPDH2 mutants
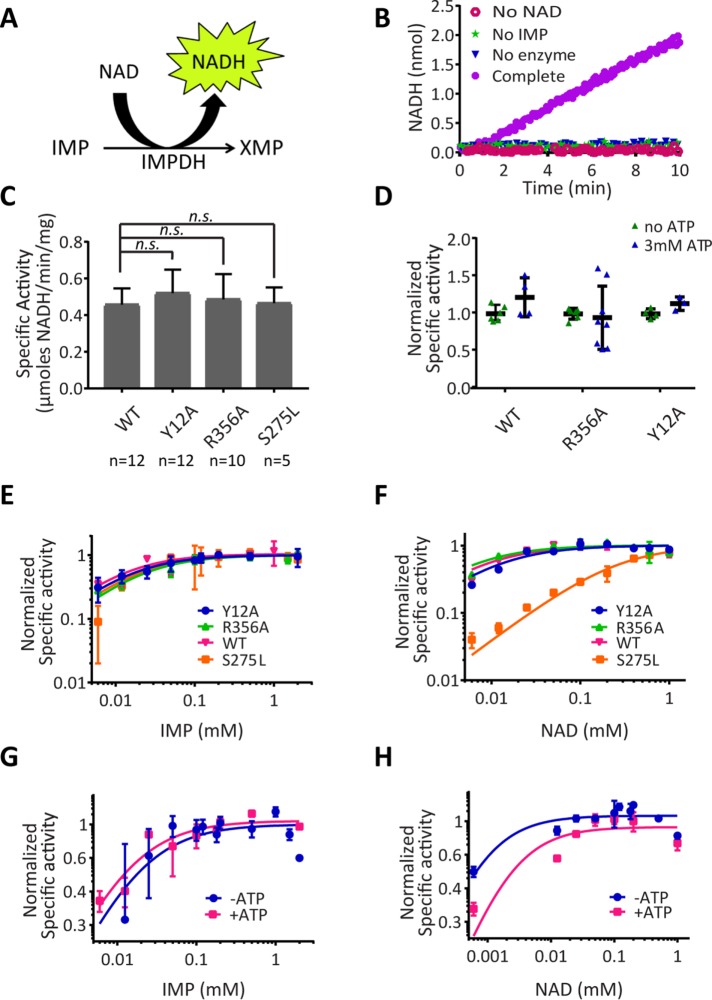
Next we assessed the catalytic activity of recombinant IMPDH2 by monitoring NADH production in real time (Figure 2, A and B). Initial reaction rates were measured for unpolymerized IMPDH under conditions of saturating substrates NAD+ and IMP in the absence of ATP. We observed no significant difference in the specific activity of wild type and any of the IMPDH2 mutants (Figure 2C). We next measured their activity in the absence or presence of ATP, which promotes polymerization of wild-type IMPDH2 but not Y12A or R356A (Figure 1E). We observed no evidence for increased activity of wild-type IMPDH2 with ATP, consistent with prior reports (Mortimer and Hedstrom, 2005; Pimkin and Markham, 2008; Labesse et al., 2013; Buey et al., 2015) (Figure 2D). Nonpolymerizing IMPDH2 mutants were also unaffected by ATP. Taken together, the comparable activity of IMPDH2 under polymerizing and nonpolymerizing conditions indicates that assembly of purified IMPDH2 does not affect its activity.
FIGURE 2:
IMPDH2 polymerization does not alter its catalytic activity in vitro. (A) Schematic of the IMPDH reaction. (B) NADH fluorescence was monitored in reactions lacking NAD+, IMP, IMPDH2 (no enzyme), or a reaction containing all components (Complete). (C) Wild-type and mutant IMPDH2 activity under nonassembling conditions. All mutants showed no significant difference from wild type (two-sided Student’s t test; p > 0.05). Bars denote mean and SD. (D) IMPDH activity assays as in C were conducted in the absence or presence of ATP to induce assembly. ATP did not significantly alter activity in any case (two-sided Student’s t test). (E, F) Initial reaction rates are plotted as a function of substrate concentration for IMP or NAD+ comparing wild-type and mutant IMPDH2 under polymerization conditions (1 mM ATP) or for wild type in the presence or absence of ATP (G, H). Michaelis constants determined from these titrations are given in Supplemental Table 1. Two to five replicates per substrate concentration.
Measuring catalytic activity under saturating substrate concentrations might mask subtle differences in IMPDH2 substrate affinity in the polymer compared with free enzyme. We therefore determined apparent Michaelis constants (Km) of mutant and wild-type IMPDH2 for substrates IMP and NAD+ by measuring initial reaction velocity at a range of substrate concentrations under polymerizing and nonpolymerizing conditions. Constitutively assembling S275L exhibited similar affinity for IMP (Figure 2E) but an ∼30-fold decrease in NAD affinity compared with wild type (Figure 2F) (see also Supplemental Table 1), consistent with this residue directly contacting NAD+. By contrast, nonpolymerizing and wild-type IMPDH2 had indistinguishable affinity for both IMP (Figure 2E) and NAD+ (Figure 2F), implying that wild-type polymers and unassembled mutants have comparable substrate affinities. Consistent with this, we found that triggering wild-type IMPDH2 polymerization with ATP also did not affect IMP affinity (Figure 2G). A 10-fold increase in the Km(app.) for NAD+ is likely due to competitive binding of ATP to the NAD+ site rather than an effect of polymerization since no difference in NAD+ substrate affinity was observed comparing wild type and nonpolymerizing mutants (Figure 2F). We conclude that substrate affinity is not substantially altered by IMPDH2 polymerization. More broadly, these results demonstrate that Y12A and R356A are separation-of-function mutations, abolishing polymerization while maintaining catalytic activity.
IMPDH assembly does not alter guanine nucleotide biosynthetic flux in vivo
Next we asked whether IMPDH2 polymerization alters its catalytic activity in the cellular context in a way not detectable for the purified protein in vitro, for example, by regulating IMPDH2 interaction with other proteins. We measured IMPDH activity in live cells by monitoring incorporation of isotopically labeled glycine, provided in the culture media, into GMP, a downstream metabolic product of IMPDH, or AMP as a control. Because IMPDH is rate limiting for guanine nucleotide synthesis, we reasoned that alterations in IMPDH activity would impact labeled glycine incorporation into guanosine monophosphate (GMP).
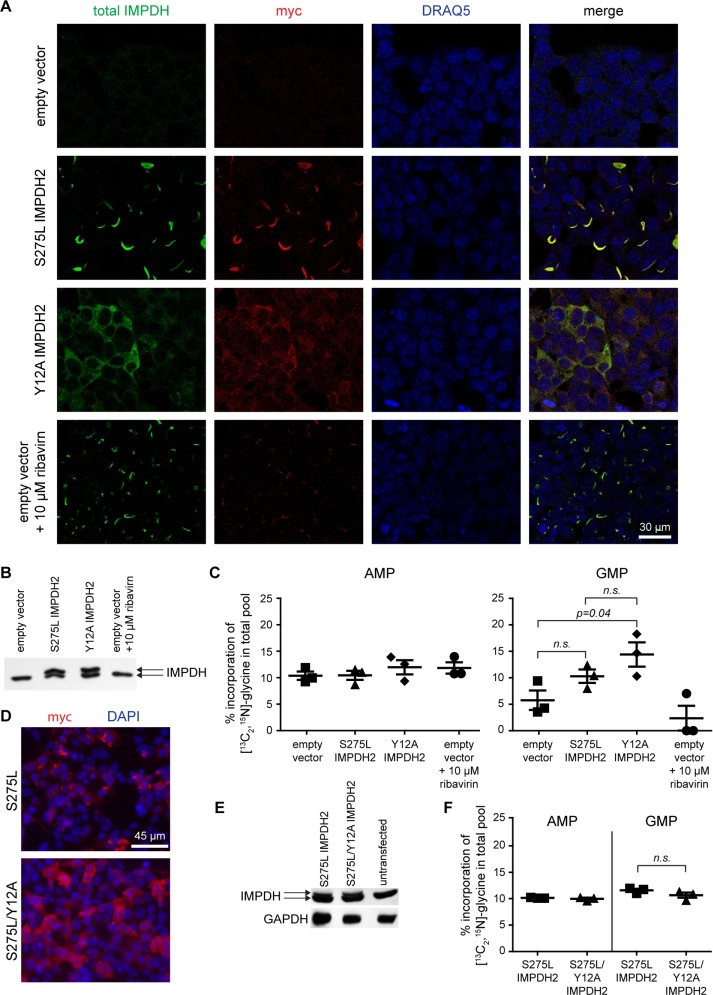
We validated this assay by transfecting cells with S275L or Y12A IMPDH2 (Figure 3, A and B). Parallel cultures were incubated with [13C2, 15N]glycine, resulting in a three-unit mass increase in glycine-derived metabolites and allowing a robust quantitation of pulse-labeled AMP/GMP by liquid chromatography-tandem mass spectrometry. Unlike the percentage of isotopically labeled AMP, the isotopically labeled GMP fraction was modestly increased in both IMPDH2-transfected cells compared with empty vector control (reaching statistical significance only for Y12A), consistent with an increase in total IMPDH activity (Figure 3C). Conversely, treatment of cells with the IMPDH inhibitor ribavirin potently suppressed incorporation of the glycine label (Figure 3C) and triggered assembly of endogenous IMPDH (Figure 3A). Thus this assay effectively reports cellular IMPDH catalytic activity.
FIGURE 3:
IMPDH filament assembly does not alter guanine nucleotide biosynthesis. (A) HEK293 cells transfected with the indicated constructs were visualized with antibodies against IMPDH2-myc, total IMPDH, or a DNA stain (DRAQ5). S275L assembles into filaments while Y12A remains diffusely cytoplasmic. One sample was treated with the IMPDH inhibitor ribavirin. (B) IMPDH immunoblot showing endogenous protein expression (lower band) and myc-tagged IMPDH2 (upper band). (C) Incorporation of [13C2 15N]glycine into AMP and GMP. Bars denote mean and standard error for three biological replicates conducted on different days. No statistical differences (two-sided Student’s t test; p > 0.05) were found between samples for incorporation of [13C2, 15N]glycine into AMP pools. (D) Immunofluorescence localization of myc-IMPDH2 (red) and DAPI (blue). (E) Representative IMPDH or GAPDH immunoblot. (F) [13C2 15N]glycine incorporation into AMP and GMP as in C. No statistically significant differences were found (two-sided Student’s t test; p > 0.05).
Since S275L and Y12A exhibit substantially different NAD+ affinities (Figure 2F), we next sought to compare the catalytic activity of polymeric and diffuse IMPDH forms with identical catalytic properties. We transfected cells with either the constitutively polymerizing S275L mutant or a double mutant of S275L/Y12A (Figure 3, D and E). As expected, the Y12A mutation prevented polymerization triggered by S275L. Importantly, Y12A does not affect IMPDH2 catalytic parameters (Figure 2, E and F), allowing us to directly compare the activity of filamentous versus diffuse IMPDH2. Incorporation of the glycine label into both AMP and GMP was indistinguishable between S275L and S275L/Y12A (Figure 3F), demonstrating comparable flux through IMPDH regardless of its polymerization state. While this result is based on the comparison of cells modestly overexpressing mutant IMPDH2 forms, it supports our in vitro results and suggests that diffuse and filamentous forms of IMPDH2 may also share comparable catalytic activity in vivo.
The unexpected similarity in the activity of diffuse and filamentous IMPDH2 begs the question of why IMPDH assembles in cells under conditions of limited nucleotide biosynthetic flux. Our methods may not detect subtle activity differences due to the background activity of endogenous wild-type IMPDH. Alternatively, filament assembly could fulfill a different function such as sequestering IMPDH from noncatalytic functions (Kozhevnikova et al., 2012) or scaffolding interactions with other proteins. The present work introduces separation-of-function mutants of IMPDH that will be useful for addressing the role of assembly in vivo.
GTP effects on IMPDH octamer conformation and catalytic activity
Conformational changes in the IMPDH octamer have been associated with altered catalytic activity. For example, eukaryotic IMPDHs are subject to feedback inhibition by low-millimolar guanosine diphosphate (GDP) or GTP (Buey et al., 2015). The crystal structure of Ashbya gossypii IMPDH bound to GDP (PDB: 4Z87) revealed a collapsed conformation of the IMPDH octamer mediated by the binding of three GDP molecules to sites within the Bateman domain (Buey et al., 2015). Two sites overlap with the canonical Bateman domain ATP-binding sites, and competition between adenine and guanine nucleotides for these allosteric regulatory sites and/or binding of a guanine nucleotide to the third site can trigger a dramatic change in the IMPDH octamer from an expanded, catalytically active form to a collapsed, inactive form (Buey et al., 2017). Residues that mediate GTP binding are conserved in eukaryotic but not prokaryotic IMPDHs, though the ability of human IMPDH to adopt the collapsed state has not been reported. To examine this, we carried out a comprehensive investigation of IMPDH2 conformational states using negative stain and cryoelectron microscopy. We sought to determine which conformations the human octamer can adopt in the free and filament contexts and what role ligands play in determining these conformations.
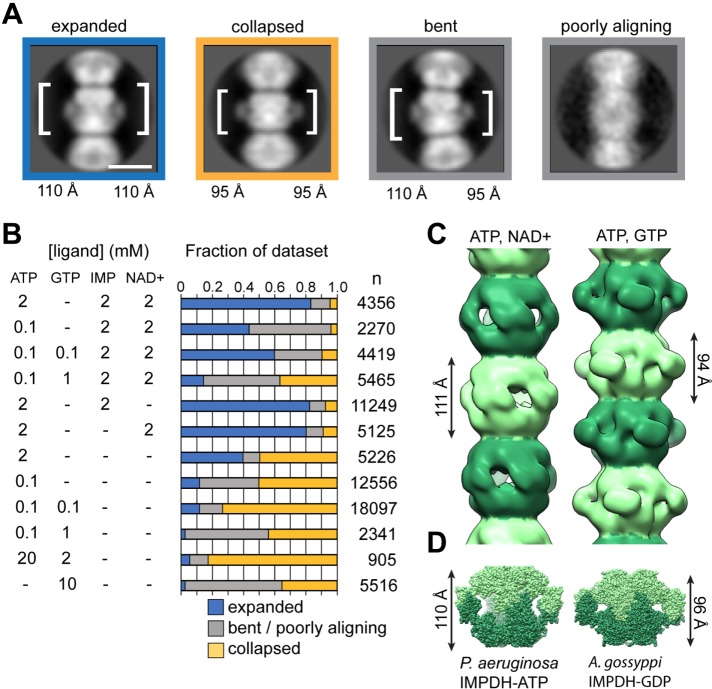
We explored a range of combinations and concentrations of ATP, GTP, and substrates and assessed the structural consequences using negative stain images and two-dimensional, reference-free classification of IMDPH2 filaments. We identified four major classes of filament segments as follows: expanded, collapsed, bent, and “poorly aligning” (Figure 4A). The spacing between octamer centers in the expanded conformation is ∼110 Å compared with ∼95 Å in the collapsed conformation. The bent conformation is well resolved with one side matching the spacing of the collapsed conformation and the other side matching the expanded conformation. We defined as “poorly aligning” segments that were assigned to classes without detailed features. This finding demonstrates the surprising plasticity of the filaments to accommodate both expanded and collapsed octamer conformations, and individual filaments frequently contain octamers in different conformational states. We quantified the frequency of these classes in the presence or absence of ATP, GTP, and substrates (Figure 4B). This analysis shows that binding of either substrate shifts the conformational equilibrium toward the expanded octamer and that GTP promotes the collapsed conformation, suggesting that the balance of GTP and substrates tunes the conformational state of the IMPDH filament. A number of studies have reported highly curved IMPDH filaments in vivo, which is consistent with the flexibility we observe in vitro.
FIGURE 4:
Ligands influence IMPDH filament architecture by altering the protomer conformation. (A) Representative class averages of IMPDH filaments in four different conformational states. The height of one octamer is indicated. (B) Quantification of the fraction of IMPDH helical segments in each conformational state as a function of ligand concentration. Number of particles counted for each condition is shown. (C) Negative-stain EM reconstruction of IMPDH2 filaments in the expanded, substrate-bound (0.1 mM ATP, 2 mM NAD+) and collapsed, GTP-bound (0.1 mM ATP, 0.1 mM GTP) conformations. The refined helical rotation and rise for the expanded filament are 30° and 111 Å and for the collapsed filament 35.5° and 94 Å. (D) The heights of crystal structures of Pseudomonas aeruginosa IMPDH-ATP and Ashbya gossypii IMPDH-GDP closely match the refined helical rise of the human IMPDH2 filaments in the expanded and collapsed states, respectively.
We performed three-dimensional reconstructions of IMPDH2 filaments under conditions where the equilibrium was shifted most strongly toward the expanded conformation (0.1 mM ATP, 3 mM IMP, 5 mM NAD+) or the collapsed conformation (0.1 mM ATP, 0.1 mM GTP) (Figure 4C), yielding structures at ∼20 Å resolution. Both structures are helical polymers formed from stacked octamers. The structures differ significantly from each other in the rise between octamers (111 Å for expanded and 95 Å for the collapsed state), consistent with the two-dimensional class averages. In the collapsed conformation there is a 30.5° rotation between octamers, and the collapsed filament has a 35.5° rotation between octamers. It appears that the filament assembly contacts between octamers are relatively fixed, and the differences in helical symmetry arise from conformational changes within the octamer that alter the relative positioning of the Bateman and catalytic domains.
Strikingly, neither filament conformation matches the ∼130 Å spacing of the human IMPDH2 octamer in its crystal lattice (Figure 1A). Rather, the expanded conformation most closely resembles the ATP-bound active octamer conformation first reported for Pseudomonas IMPDH (Labesse et al., 2013), while the collapsed conformation is most similar to the Ashbya GDP-bound inhibited form (Buey et al., 2015) (Figure 4D). The identification of conditions that shift the conformation of IMPDH between collapsed and expanded conformations suggests the potential for cooperativity in the transition of polymerized octamers between states. This model would imply that polymers may function to coordinate octamer conformational changes en masse.
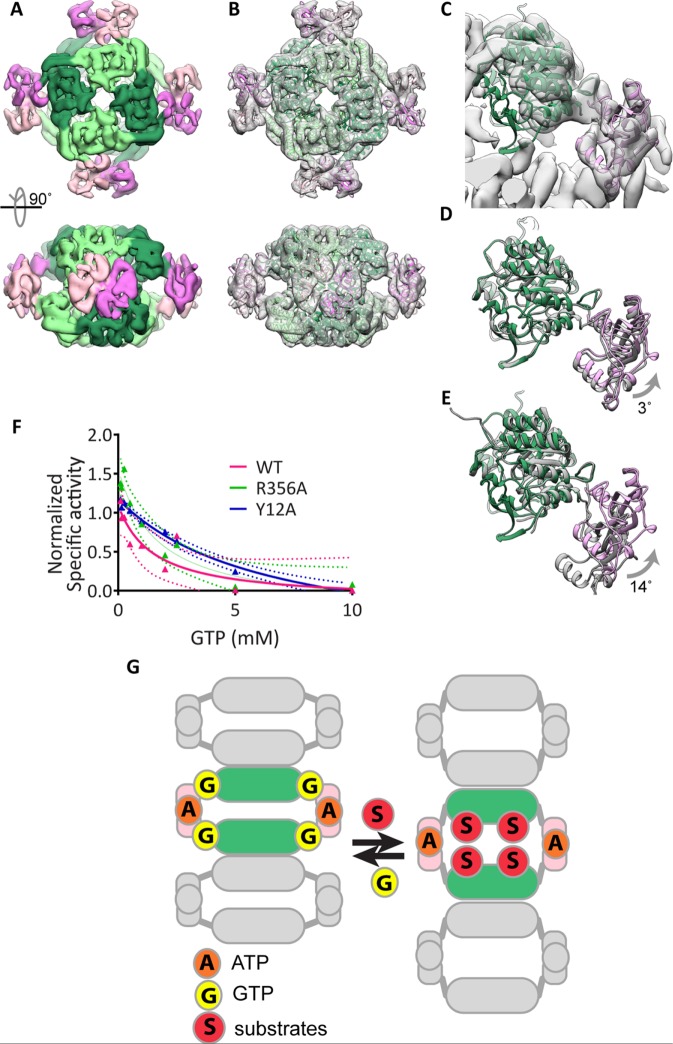
We sought to determine whether human IMPDH2 can adopt a collapsed conformation similarly to Ashbya IMPDH using high-resolution cryoelectron microscopy. We determined the 8.7-Å structure of GTP-bound Y12A IMPDH2 (Figure 5A). At this resolution, alpha-helices are well defined, allowing us to model the overall conformation of the protein by rigid-body fitting of the human IMPDH catalytic and Bateman domains into the electron microscopy (EM) density (Figure 5, B and C). Comparison of the atomic model of GTP-bound Y12A IMPDH2 to the Ashyba GDP-bound IMPDH2 crystal structure (PDB ID 4Z87) reveals that, in the presence of GTP, human IMPDH2 also assumes the collapsed conformation (Figure 5D) and not the extended form (Figure 5E).
FIGURE 5:
GTP-bound human IMPDH2 adopts the collapsed, inhibited conformation. (A) Cryo-EM reconstruction of Y12A in the presence of 0.1 mM ATP and GTP at 8.7 Å resolution. Catalytic (greens) and Bateman (pinks) domains are well resolved. (B) Atomic model of IMPDH2-ATP-GTP, generated by fitting the catalytic domain and Bateman domains into the cryo-EM structure as two separate rigid bodies. (C) Close-up view of a single IMPDH monomer fitted in the cryo-EM structure. (D) A monomer of the A. gossypii IMPDH-GDP crystal structure (gray) (PDB ID 4Z87) aligned to the human IMPDH2-ATP-GTP model (color) or to the P. aueriginosa IMPDH-ATP monomer (E). (F) Dose-dependent inhibition of wild-type and mutant IMPDH2 by GTP. IMPDH2 was incubated with 1 mM ATP and 3 mM IMP to promote polymerization and then GTP was added for 10 min prior to reaction initiation by with 5 mM NAD+. Mean values are plotted and fitted to a three-parameter dose–response curve. Each data point represents three to six replicate reactions. Ninety-five-percent confidence intervals are plotted as dotted lines of the corresponding color. (G) Model: human IMPDH exists in a conformational equilibrium between an expanded, active conformation and a collapsed, inactive conformation that can be shifted by binding to substrates or GTP. Unlike other metabolic filaments, the conformational equilibrium of IMPDH is unaffected by polymerization because the filament form can accommodate both active and inactive conformations.
Since both polymerized and octomeric IMPDH2 can adopt the expanded and collapsed conformations, we hypothesized that polymerization might stabilize the expanded conformation, reducing negative feedback inhibition and allowing higher steady-state GTP accumulation. To test this, we measured the effect of GTP on the activity of wild-type and mutant IMPDH2 under polymerization conditions. Contrary to our hypothesis, we observed comparable low-millimolar GTP-mediated inhibition of all three forms (Figure 5F), although we cannot rule out subtle differences in the kinetics or cooperativity of inhibition.
The ability of IMPDH2 filaments to accommodate multiple functional states of the enzyme in different conformations is unexpected. Recent structures of the metabolic filament CTP synthase have shown that, while different ligands influence the conformation of the enzyme, a single conformation is stabilized in the filament, creating a direct link between assembly and enzyme activity (Barry et al., 2014; Lynch et al., 2017). Thus the ability of IMPDH filaments to accommodate different functionally important conformational states while remaining assembled in the filament is unusual. One possibility is that IMPDH assembly provides a means for effectors to rapidly modulate the activity of the entire IMPDH population, for example, through cooperative transitions between expanded and collapsed states along the polymer, to provide a more switch-like transition of IMPDH activity. Alternatively, assembly may provide a platform for recruitment of other enzymes or regulatory proteins, with the conformational state of IMPDH providing a physical signal for the level of guanine biosynthetic activity. For example, colocalization of IMPDH and CTPS filaments may provide a mechanism for coordinately regulating purine and pyrimidine biosynthesis. The nonassembling IMPDH mutants introduced here should facilitate investigations of the physiological role of polymerization.
MATERIALS and METHODS
Plasmids and mutagenesis
For expression in mammalian cells, human IMPDH2 was cloned into pcDNA3.1/myc-HIS B. The resulting IMPDH2 protein retains the native N-terminus and has an appended C-terminal myc and 6-histidine tag. Expression is driven by the CMV promoter. Mutagenesis was conducted using QuickChange site-directed mutagenesis (Agilent) according to manufacturer’s instructions. Mutagenesis was conducted with the following forward primers and their corresponding reverse complements:
R356A 5′-CAGAGTATGCACGGGCCTTTGGTGTTCCG-3′
Y12A 5′-GATTAGTGGGGGCACGTCCGCAGTGCCAGACGACGGACTC-3′
S275L 5′-GGATGTAGTGGTTTTGGACTTATCCCAGGGAAATTCCATCTTC-3′
For recombinant IMPDH2 expression in Escherichia coli, wild-type and mutant IMPDH2 was cloned in pSMT3-Kan (Mossessova and Lima, 2000). This vector appends an N-terminal SMT3/SUMO tag, which can be cleaved by ULP1 protease following purification. As a cautionary note, we found that some N-terminal epitope tags disrupt polymerization of wild-type IMPDH2. Consequently, our studies were conducted with IMPDH2 generated with this cleavable tag, which leaves only five residual, nonnative amino acids. The coding region of all mutated constructs were fully sequenced.
Cell culture and transfection
HEK293 cells (kindly provided by Marc Kirschner, Harvard University) were cultured in DMEM (Cellgro) supplemented with 10% fetal bovine serum (FBS) (HyClone), 2 mM l-glutamine (Life Technologies), 1% penicillin, and 1% streptomycin. Cells were transfected using Lipofectamine 2000 in Opti-MEM Reduced Serum Medium (ThermoFisher Scientific) as per manufacturer’s instructions.
Immunoblotting
SDS–PAGE gels were transferred by semidry transfer apparatus (Amersham) to nitrocellulose membranes. Membranes were blocked using 5% milk in Tris-buffered saline (TBS). Anti-myc (Santa Cruz), anti-IMPDH (Abcam), and anti-GAPDH (Santa Cruz) antibodies were used and detected with peroxidase-coupled secondary antibodies and enhanced chemiluminescence detection (Amersham).
Immunofluoresence
Cells were grown on coverslips and fixed for 20 min at room temperature with 4% formaldehyde followed by permeabilitation in 0.5% Triton X-100 for 10 min. Blocking and antibody dilution was in phosphate-buffered saline (PBS) with 0.1% Triton X-100, 2% bovine serum albumin (BSA), and 0.1% sodium azide. Anti-myc antibodies (Santa Cruz) were used and DAPI was used to counterstain nuclei. Images were collected using MetaVue software (Molecular Devices) controlling a CoolSnap ES camera (Photometrics) on a Nikon TE2000-U inverted microscope using a 60× Plan Apo 1.40 NA Nikon objective.
Recombinant IMPDH expression and purification
pSMT3-Kan IMPDH2 plasmids were transformed into BL21 (DE3) competent E. coli cells using standard procedures. Transformants were inoculated into 25-ml overnight precultures, which were then used to inoculate 1-l cultures of Luria broth grown at 37°C until they reached an OD600 of 0.8. Flasks were then cooled on ice for ∼5 min and then induced with 1 mM IPTG (GoldBio) for 4 h at 30°C. Cells were then pelleted for 5 min at 8000 × g in a Beckmann JA-10 rotor. Cell pellets were stored at –80°C.
Thawed cell pellets were resuspended with stirring in 20 ml (per liter of original culture volume) of lysis buffer (50 mM potassium phosphate (KPO4), 300 mM potassium chloride (KCl), 20 mM imidazole, 0.8 M urea, pH 8.0) containing Benzonase nuclease (Sigma). Cells were homogenized using the Emulsiflex-C3 (Avestin) with three cycles at 10,000–15,000 psi. Lysates were then cleared at 18,000 rpm for 20 min at 4°C in a Beckman Coulter JA 25.5 rotor. SUMO-tagged IMPDH was recovered from the lysates using 1 ml (per liter of original culture volume) of nickel-nitrilotriacetic acid agarose (Qiagen) and incubation for 2 h at 4°C. Beads were washed three times with 10 bead-volumes each of lysis buffer before elution (with 50 mM KPO4, 300 mM KCl, 500 mM imidazole, pH 8.0). Peak elution fractions were pooled and incubated with ULP1 protease (purified as described in Mossessova and Lima, 2000) (1 mg ULP1 per 100 mg IMPDH2) overnight at 4°C. Next 1 mM dithiothreitol (DTT) and 0.8-M urea were added, and the protein was concentrated to using a 30,000 molecular weight cutoff (MWCO) Amicon filter and loaded onto either a Superdex 200 or a Superose 6 gel filtration column preequilibrated in gel filtration buffer (50 mM Tris, 100 mM KCl, 1 mM DTT, pH 7.4). Peak fractions from the included volume were pooled, aliquotted, flash frozen in liquid nitrogen, and stored at –80°C.
IMPDH activity assay
NADH production was measured in real time in a 100-µl cuvette (1-cm path length) in a Cary Eclipse fluorometer (excitation: 340 nm; emission: 440 nm) equipped with a temperature-controlled multicuvette holder maintained at 37°C. Fluorescence was converted to µmoles NADH produced by comparison with an NADH standard curve. In the standard reaction, 50 mM Tris pH, 7.4, 100 mM KCl, 1 mM DTT, 5 mM NAD+, and 3 mM IMP. Reactions were initiated by the addition of 13.8 µg of IMPDH2 per 100-µl reaction. Preliminary studies showed these substrate concentrations to be saturating. IMPDH concentration and buffer conditions were chosen to precisely mimic those used for negative-stain electron microscopy analysis below. In reactions containing ATP, IMPDH2 was preincubated for 10 min at room temperature with the indicated concentration of ATP prior to reaction initiation with substrate. In substrate titration experiments, the constant substrate was included in the preincubation and the reaction was initiated by the addition of the variable substrate. In GTP-containing reactions, IMPDH2 was first preincubated with 1 mM ATP and 3 mM IMP to induce assembly, and then GTP was added to the indicated concentration for an additional 10 min before the reaction was initiated by the addition of 5 mM NAD+. Specific activity was calculated by linear interpolation of the reaction slope within the initial 3 min (approximately nine measurements per minute).
[13C2, 15N]glycine incorporation into AMP and GMP
HEK293 cells were maintained in DMEM (Cellgro) containing 10% FBS (HyClone) and 2 mM l-glutamine (Life Technologies). Twenty-four hours prior to the glycine addition, two 10-cm dishes of 40% confluent HEK293 cells were transfected with the empty vector pcDNA 3.1/myc-His B, S275L IMPDH2 or Y12A IMPDH2 constructs. The next day, cells were washed once in Dulbecco’s PBS (Life Technologies), and the media of the cells was replaced with DMEM containing 10% dialyzed FBS, 2 mM l-glutamine, and 1.2 mM [13C2, 15N]glycine (Cambridge Isotope Laboratories). After 6 h, cells were washed once with Dulbecco’s PBS, released from plates with 0.25% trypsin-EDTA, harvested, washed twice more with Dulbecco’s PBS, and counted. Cells from paired tissue culture dishes were pooled, resulting in (1–10) × 106 cells per sample.
Samples were then processed and analyzed as described previously (Laourdakis et al., 2014). Briefly, 70 μl of a solution containing 0.5 M perchloric acid and 200 μM of the internal standard [13C9, 15N3]CTP was added to each cell pellet. The cells were vortexed for 10 s and incubated on ice for 20 min. Lysates were then neutralized with 7 μl of 5 M potassium hydroxide, vortexed for 10 s, and incubated on ice for an additional 20 min. Cell debris was harvested by centrifugation at 11,000 × g for 10 min at 4°C. Supernatants were further clarified using Amicon Ultra 0.5-ml centrifugal filters (3 kDa MWCO) as per manufacturer’s instructions.
Collected extracts were analyzed by an ultra-performance liquid chromatography (UPLC, Waters Acquity I-class) coupled to a triple quadrupole mass spectrometer (Waters Xevo TQ-S micro). A Waters Acquity UPLC HSS T3 column (1.8 mm, 2.1 mm × 50 mm) and a column in-line filter were used. Quantitative analysis of nucleotides was performed using the multiple reaction monitoring mode, and the peaks of nucleotides were analyzed using MassLynx V4.1. In addition, calibration standards were freshly prepared for every sample set analysis, and linear calibration curves were established. All reported data represent the mean and standard error of three independent biological replicates. P values were calculated using two-tailed, unpaired Student’s t tests in Graphpad Prism.
For each replicate, samples for immunofluorescence and immunoblotting were prepared in parallel to samples for nucleotide extraction. Cells for immunofluorescence were stained as described above except that DRAQ5 (Biolegend) was used as the nuclear stain. Images were obtained using a Leica DM 5000 microscope coupled to a Leica TCS SP5 confocal laser scanner. For immunoblot analysis, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with Complete protease inhibitor cocktail (Roche). Proteins were separated by SDS–PAGE and transferred to nitrocellulose. Blots were stained with anti-IMPDH2 antibody EPR8365(B) (Abcam).
Negatively stained electron microscopy and particle averaging/reconstruction
Aliquots of purified protein, including WT IMPDH and the mutants Y12A, R356A, and S275L, were diluted to 2.5 µM in assembly buffer (50 mM Tris, 100 mM KCl, 1 mM DTT, pH 7.4) and incubated for 15 min at room temperature in the presence of various concentrations of ATP, GTP, IMP, and NAD+, as outlined in Figures 1E and 4C. Following incubation, samples were applied to glow-discharged continuous carbon film EM grids and negatively stained using 1% uranyl formate. Grids were imaged by transmission electron microscopy using an FEI Tecnai G2 Spirit operating at 120 kV and a Gatan Ultrascan 4000 CCD via the Leginon software package (Suloway et al., 2005). Image processing, including CTF estimation, particle picking, and two-dimensional reference-free classification, was performed using the Appion and RELION software packages (Lander et al., 2009; Scheres, 2012). The image data collected from all ATP/GTP/IMP/NAD+ combinations were combined and processed en masse (77,525 helical repeats in total). Following two-dimensional classification, the resulting class averages were manually grouped according to the observed rise into one of three categories: “collapsed” (rise ∼94 Å), “expanded” (rise ∼114 Å), or “undetermined” (filament ends, distorted and bent octamers, and poorly aligned particles). The proportion of helical repeats from each experimental data set belonging to these three categories was then back-calculated.
For three-dimensional reconstructions of IMPDH filaments, negative-stain images were acquired at a pixel size of 2.07 Å, and individual filaments were identified manually in Appion (Lander et al., 2009). Reconstructions were performed by iterative helical real space refinement (Egelman, 2010) in SPIDER (Shaikh et al., 2008), imposing D4 symmetry on the helical asymmetric unit throughout. The structures of the collapsed and expanded IMPDH filament have been deposited in the EMDB (Lawson et al., 2011) (EMD–8691, expanded; EMD-8690, collapsed).
Electron cryomicroscopy and particle averaging/reconstruction
Purified Y12A protein was diluted in assembly buffer to 4 µM and incubated at room temperature in the presence of 0.1 mM ATP and 0.1 mM GTP for 15 min. Following incubation, the sample was applied to a glow-discharged C-Flat holey carbon EM grid (Protochips) and blotted/plunged into liquid ethane using a Vitrobot plunging apparatus (FEI). Imaging was performed using an FEI Tecnai G2 F20 operating at 300 kV and a Gatan K-2 direct electron detector via the Leginon software package (CITE Suloway 2005, JSB). Image processing, including CTF estimation, particle picking, two-dimensional reference-free classification, three-dimensional classification and three-dimensional refinement, was performed using the Appion and RELION software packages (CITE Lander 2009, JSB & Scheres 2012 JSB). The map resolution, as determined by the relion_postprocess script using the Fourier shell correlation (FSC) = 0.143 criterion was 8.7 Å. The final electron density map was further sharpened using a b-factor of -987. The structure has been deposited with the EMDB (EMD-8692).
Statistics
Statistical tests used are indicated in the figure legends and p values <0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) R01 GM083025 and a grant from the Pennsylvania Department of Health to J.R.P. and NIH grants GM118396 to J.M.K. and GM102503 to A.A. K.C.D. and J.C.S. were supported, in part, by NIH T32 CA009035. We thank Alana O’Reilly for the use of her confocal microscope and insightful discussions and Liz Hedstrom for IMPDH expression constructs.
Abbreviations used:
- CTPS
CTP synthase
- IMPDH
inosine monophosphate dehydrogenase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-04-0263) on August 9, 2017.
REFERENCES
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey GN, Liu JL. Metabolic regulation via enzyme filamentation. Crit Rev Biochem Mol Biol. 2015;51:282–293. doi: 10.3109/10409238.2016.1172555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry RM, Bitbol AF, Lorestani A, Charles EJ, Habrian CH, Hansen JM, Li HJ, Baldwin EP, Wingreen NS, Kollman JM, Gitai Z. Large-scale filament formation inhibits the activity of CTP synthetase. Elife. 2014;3:e03638. doi: 10.7554/eLife.03638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty NB, Lane MD. The polymerization of acetyl-CoA carboxylase. J Biol Chem. 1983;258:13051–13055. [PubMed] [Google Scholar]
- Buey RM, Fernandez-Justel D, Marcos-Alcalde I, Winter G, Gomez-Puertas P, de Pereda JM, Luis Revuelta J. A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci Rep. 2017;7:2648. doi: 10.1038/s41598-017-02805-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Ledesma-Amaro R, Velazquez-Campoy A, Balsera M, Chagoyen M, de Pereda JM, Revuelta JL. Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat Commun. 2015;6:8923. doi: 10.1038/ncomms9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calise SJ, Carcamo WC, Krueger C, Yin JD, Purich DL, Chan EK. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell Mol Life Sci. 2014;71:2963–2973. doi: 10.1007/s00018-014-1567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calise SJ, Purich DL, Nguyen T, Saleem DA, Krueger C, Yin JD, Chan EK. ‘Rod and ring’ formation from IMP dehydrogenase is regulated through the one-carbon metabolic pathway. J Cell Sci. 2016;129:3042–3052. doi: 10.1242/jcs.183400. [DOI] [PubMed] [Google Scholar]
- Carcamo WC, Calise SJ, von Muhlen CA, Satoh M, Chan EK. Molecular cell biology and immunobiology of mammalian rod/ring structures. Int Rev Cell Mol Biol. 2014;308:35–74. doi: 10.1016/B978-0-12-800097-7.00002-6. [DOI] [PubMed] [Google Scholar]
- Carr SF, Papp E, Wu JC, Natsumeda Y. Characterization of human type I and type II IMP dehydrogenases. J Biol Chem. 1993;268:27286–27290. [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter JH, Thomas EC, Lengefeld N, Kruger SJ, Worton L, Gardiner EM, Jones A, Barnett NL, Whitehead JP. Characterisation of inosine monophosphate dehydrogenase expression during retinal development: differences between variants and isoforms. Int J Biochem Cell Biol. 2008;40:1716–1728. doi: 10.1016/j.biocel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Hedstrom L. IMP dehydrogenase: structure, mechanism, and inhibition. Chem Rev. 2009;109:2903–2928. doi: 10.1021/cr900021w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Gu J, Makhov AM, Griffith JD, Mitchell BS. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic Acid by GTP. J Biol Chem. 2006;281:206–212. doi: 10.1074/jbc.M507056200. [DOI] [PubMed] [Google Scholar]
- Juda P, Smigova J, Kovacik L, Bartova E, Raska I. Ultrastructure of cytoplasmic and nuclear inosine-5’-monophosphate dehydrogenase 2 “rods and rings” inclusions. J Histochem Cytochem. 2014;62:739–750. doi: 10.1369/0022155414543853. [DOI] [PubMed] [Google Scholar]
- Keppeke GD, Calise SJ, Chan EK, Andrade LE. Assembly of IMPDH2-based, CTPS-based, and mixed rod/ring structures is dependent on cell type and conditions of induction. J Genet Genomics. 2015;42:287–299. doi: 10.1016/j.jgg.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Kozhevnikova EN, van der Knaap JA, Pindyurin AV, Ozgur Z, van Ijcken WF, Moshkin YM, Verrijzer CP. Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol Cell. 2012;47:133–139. doi: 10.1016/j.molcel.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Labesse G, Alexandre T, Vaupre L, Salard-Arnaud I, Him JL, Raynal B, Bron P, Munier-Lehmann H. MgATP regulates allostery and fiber formation in IMPDHs. Structure. 2013;21:975–985. doi: 10.1016/j.str.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laourdakis CD, Merino EF, Neilson AP, Cassera MB. Comprehensive quantitative analysis of purines and pyrimidines in the human malaria parasite using ion-pairing ultra-performance liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;967:127–133. doi: 10.1016/j.jchromb.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL. The Cytoophidium and Its Kind: Filamentation and Compartmentation of Metabolic Enzymes. Annu Rev Cell Dev Biol. 2016;32:349–372. doi: 10.1146/annurev-cellbio-111315-124907. [DOI] [PubMed] [Google Scholar]
- Lynch EM, Hicks DR, Shepherd M, Endrizzi JA, Maker A, Barry RM, Gitai Z, Baldwin EP, Kollman JM. Human CTP synthase filament structure reveals the active enzyme conformation. Nat Struct Mol Biol. 2017;24:507–514. doi: 10.1038/nsmb.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SE, Hedstrom L. Autosomal dominant retinitis pigmentosa mutations in inosine 5’-monophosphate dehydrogenase type I disrupt nucleic acid binding. Biochem J. 2005;390:41–47. doi: 10.1042/BJ20042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Nat Acad Sci USA. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JD, Tsechansky M, Royall A, Boutz DR, Ellington AD, Marcotte EM. A proteomic survey of widespread protein aggregation in yeast. Mol Biosyst. 2014;10:851–861. doi: 10.1039/c3mb70508k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JD, Zhao A, Ellington AD, Marcotte EM. Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu Rev Cell Dev Biol. 2012;28:89–111. doi: 10.1146/annurev-cellbio-101011-155841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovska I, Nuske E, Munder MC, Kulasegaran G, Malinovska L, Kroschwald S, Richter D, Fahmy K, Gibson K, Verbavatz JM, Alberti S. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. Elife. 2014;3:e02409. doi: 10.7554/eLife.02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkin M, Markham GD. The CBS subdomain of inosine 5’-monophosphate dehydrogenase regulates purine nucleotide turnover. Mol Microbiol. 2008;68:342–359. doi: 10.1111/j.1365-2958.2008.06153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QJ, Kassim H, Huang Y, Li H, Zhang J, Li G, Wang PY, Yan J, Ye F, Liu JL. Filamentation of Metabolic Enzymes in Saccharomyces cerevisiae. J Genet Genomics. 2016;43:393–404. doi: 10.1016/j.jgg.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Stavrides KP, Thomas SV, Nicolas E, O’Reilly AM, Peterson JR. Ack kinase regulates CTP synthase filaments during Drosophila oogenesis. EMBO Rep. 2014;15:1184–1191. doi: 10.15252/embr.201438688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Thomas EC, Gunter JH, Webster JA, Schieber NL, Oorschot V, Parton RG, Whitehead JP. Different characteristics and nucleotide binding properties of inosine monophosphate dehydrogenase (IMPDH) isoforms. PloS one. 2012;7:e51096. doi: 10.1371/journal.pone.0051096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY. Pharmacogenetics and Pharmacogenomics of Mycophenolic Acid. Doctoral Thesis. Mayo Graduate School College of Medicine, UMI Dissertation Publishing. 2011.
- Wu TY, Peng Y, Pelleymounter LL, Moon I, Eckloff BW, Wieben ED, Yee VC, Weinshilboum RM. Pharmacogenetics of the mycophenolic acid targets inosine monophosphate dehydrogenases IMPDH1 and IMPDH2: gene sequence variation and functional genomics. Br J Pharmacol. 2010;161:1584–1598. doi: 10.1111/j.1476-5381.2010.00987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.