Abstract
Background
The glutathione S-transferase (GST) enzyme GSTP1 utilizes byproducts of oxidative stress. We previously showed that alleles of GSTP1 that encode the Ile105→Val105 substitution are associated with the asthma phenotypes of atopy and bronchial hyperresponsiveness (BHR). However, a further polymorphic site (Ala114→Val114) has been identified that results in the following alleles: GSTP1*A (wild-type Ile105→Ala114), GSTP1*B (Val105→Ala114), GSTP1*C (Val105→Val114) and GSTP1*D (Ile105→Val114).
Methods
Because full identification of GSTP1 alleles may identify stronger links with asthma phenotypes, we describe an amplification refractory mutation system (ARMS) assay that allows identification of all genotypes. We explored whether the GSTP1 substitutions influence susceptibility to asthma, atopy and BHR.
Results
Among 191 atopic nonasthmatic, atopic asthmatic and nonatopic nonasthmatic individuals, none had the BD, CD, or DD genotypes. GSTP1 BC was significantly associated with reduced risk for atopy (P = 0.031). Compared with AA, trend test analysis identified a significant decrease in the frequency of GSTP1 BC with increasing severity of BHR (P = 0.031). Similarly, the frequency of GSTP1 AA increased with increasing BHR.
Conclusion
These data suggest that GSTP1*B and possibly GSTP1*C are protective against asthma and related phenotypes.
Keywords: amplification refractory mutation system, asthma, bronchial hyperresponsiveness, GSTP1
Introduction
Polymorphisms in members of the GST supergene family have been associated with individual susceptibility to lung diseases [1]. In the context of asthma GSTP1 — the predominant GST expressed in human lung [2] — is a candidate because this enzyme has a role in cellular protection against oxidative stress [3]. Thus, GSTP1 catalyzes the detoxification of byproducts of lipid and DNA oxidation [1]. Asthma is characterized by airway inflammation [4]. Indeed, BHR reflects the presence of inflammation, and is exhibited by virtually all asthmatic patients. Atopic individuals (as defined by serum IgE levels and skin prick tests) are very likely to have increased airway responsiveness [4]. Thus, studies designed to identify susceptibility genes for asthma must consider the possible interrelationship of BHR and atopy in the expression of the asthma phenotype.
We previously showed that the Ile105→Val105 substitution in GSTP1 is strongly associated with severity of BHR [5]. A further polymorphism is present at amino acid 114 (Ala114→Val114), however, indicating that unequivocal identification of GSTP1 alleles requires consideration of both substitutions. These polymorphisms give rise to wild-type GSTP1*A (Ile105→Ala114), GSTP1*B (Val105→Ala114), GSTP1*C (Val105→Val114) and GSTP1*D (Ile105→Val114) [6,7,8,9]. Although the Ile105 variant has a higher catalytic efficiency for 1-chloro-2,4-dinitrobenzene than does the Val105 variant [6], the Val105 variant appears to confer higher catalytic efficiency for polycyclic aromatic hydrocarbon diol epoxides [7,8,9,10,11]. The effect of the Ala114→Val114 substitution is unclear, although it may enhance the effect of the Ile105→Val105 substitution [11].
Because the substitution at amino acid 114 may modify the association of GSTP1 Ile105→Val105 with asthma phenotypes, we developed an ARMS assay in order to identify unambiguously those genotypes that result from the A, B, C and D alleles. This approach is necessary, because presently described assays do not differentiate AC and BD genotypes. We also determined the frequencies of these genotypes in atopic nonasthmatic, atopic asthmatic and nonatopic nonasthmatic healthy persons.
Materials and methods
Patients
Unrelated Northern European nonatopic nonasthmatic, atopic nonasthmatic and atopic asthmatic persons (n = 191) were recruited in North Staffordshire, UK [5]. They were stratified by degree of airway reactivity/obstruction as follows (Table 1): group 1, BHR negative (normal), with forced expiratory volume in 1 s (FEV1) greater than 80% predicted and PC20 greater than 16 mg/ml metacholine; group 2, borderline BHR (mild) with FEV1 greater than 80% predicted and PC20 positive with 8–16 mg/ml metacholine; group 3, BHR positive (moderate) with FEV1 greater than 80% predicted and provoking concentration of an inhaled substance that causes a 20% reduction in FEV1 (PC20) positive with 0.03–8 mg/ml metacholine; and group 4, severe airway dysfunction, with FEV1 of 80% or less than predicted, which was not challenged with metacholine for ethical reasons. A positive skin reaction (mean wheal diameter of at least 3 mm more than with saline control) in response to at least one of a panel of seven common aeroallergens (house dust mite, house dust, grass mix, tree pollen, cat fur, dog fur, feathers) and serum IgE levels greater than 100 IU/ml were used to define atopic status, together with personal history. The local Ethics Committee approved the study, and all participants provided written informed consent.
Table 1.
Clinical parameters of recruited subjects
| FEV1 >80% predicted* | ||||
| BHR negative | BHR positive | BHR positive | ||
| (PC20 >16 mg/ml; group 1) | (PC20 8–16 mg/ml; group 2) | (PC20 0.03–8 mg/ml; group3) | FEV1≤80% predicted* (group 4)† | |
| Parameters | ||||
| n (%) | 58 (33.9) | 15 (8.8) | 51 (29.8) | 47 (27.5) |
| Mean age ± SD (years) | 37.0 ± 11.9 | 29.3 ± 6.8 | 33.0 ± 11.1 | 41.4 ± 11.0 |
| Males (n [%]) | 18 (31.0) | 4 (26.7) | 13 (25.5) | 16 (34.0) |
| Females (n [%]) | 40 (69.0) | 11 (73.3) | 38 (74.5) | 31 (66.0) |
| Skin test negative (n [%]) | 32 (55.2) | 7 (46.7) | 12 (23.5) | 7 (14.9) |
| Skin test positive (n [%]) | 26 (44.8) | 8 (53.3) | 39 (79.5) | 40 (85.1) |
| IgE ≤100 IU/ml* (n [%]) | 38 (71.7) | 10 (83.3) | 21 (46.7) | 16 (40.0) |
| IgE >100 IU/ml* (n [%]) | 15 (28.3) | 2 (16.9) | 24 (53.3) | 24 (60.0) |
*In some patients we were unable to obtain a complete data set. Some individuals were unwilling to undertake a metacholine challenge test, whereas others did not wish to donate a blood sample (in this case DNA was isolated from a mouthwash sample). † Mean FEV1 = 67% predicted (min = 45%, max = 78%).
Determination of GSTP1 genotypes
Genotyping was performed using leucocyte DNA. The Ile105→Val105 substitution was identified using primers to exon 5 as previously described [7,12]. The Ala114→Val114 substitution was identified as described by Board et al [13], with an annealing temperature of 65°C. However, GSTP1 AC and BD gave heterozygous patterns using both of the above assays. An ARMS assay was therefore developed to differentiate between these genotypes. This included a forward primer upstream of the codon 105 substitution (5'-ACCCCAGGGCTC-TATGGGAA-3') and two reverse primers (primer A [Ala114 specific], 5'-TCACATAGTCATCCTTGCCGG-3'; and primer B [Val114 specific], 5'-TCACATAGTCATC-CTTGCCGA-3').
For each DNA sample two polymerase chain reactions (PCRs) were performed, amplifying a 998 base pair fragment. PCRs were carried out in 50 μl containing forward primer, reverse primer A or B (2 × 0.25 μmol/l), Taq polymerase (1 U), dNTP (4 × 200 μmol/l), 1 × polymerase buffer (10 mmol/l Tris-HCl pH9.0, 50 mmol/l KCl, 0.1% [vol/vol] Triton X-100, 1.5 mmol/l MgCl2), and target DNA (approximately 0.5 μg). Conditions were as follows: 94°C for 4 min, 30 cycles of denaturation (94°C, 1 min), primer annealing (62°C, 1 min) and elongation (72°C, 2 min).
The initial ARMS PCR was used to determine the Ala114→Val114 genotype. Thus, in persons with Ala114/Ala114 amplification occurred only with primer A, and only with primer B in persons with Val114/Val114; for Ala114/Val114 heterozygotes, amplification occurred with both primers. ARMS PCR products were then digested with Bsm AI to determine the cis/trans configuration (which variant at position 105 is paired with which allele at position 114) of the Ile105→Val105 encoding allele and resolved in 2% (vol/weight) agarose gels. Products gave fragments of 343, 322, 260 and 73 base pairs with Ile105/Ile105; 93, 250, 322, 260 and 73 base pairs with Val105/Val105; and all six fragments in heterozygotes (Ile105/Val105; Fig. 1).
Figure 1.
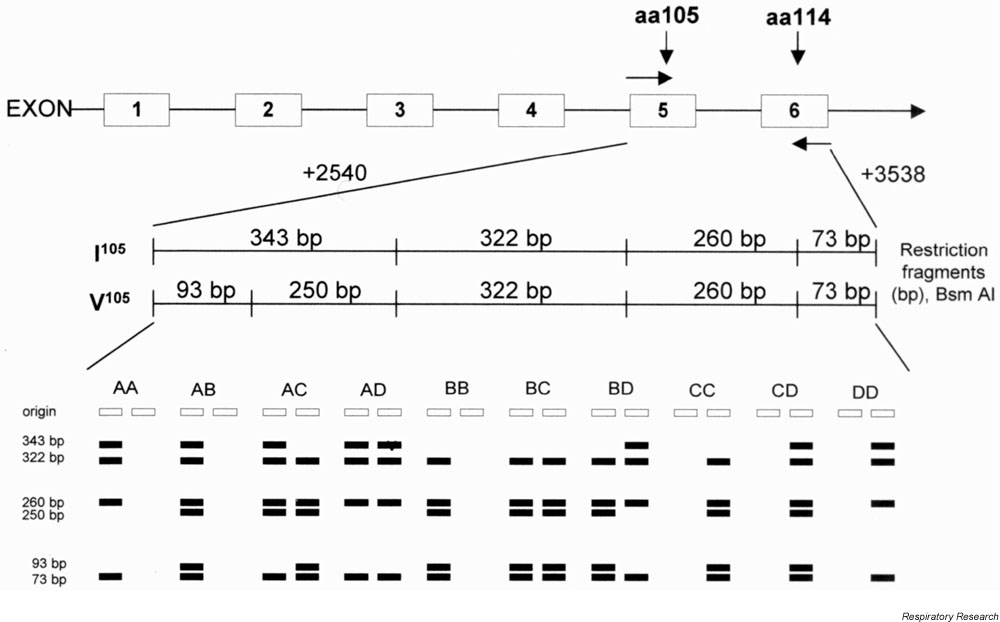
Illustration of the sites of primers and Bsm AI restriction endonuclease maps of the variant GSTP1, together with the banding patterns obtained from the ARMS assay for each genotype. Products gave fragments of 343, 322, 260 and 73 base pairs with Ile105/Ile105; 93, 250, 322, 260 and 73 base pairs with the Val105/Val105; and all six fragments in heterozygotes (Ile105/Val105). Base pair numbering according to Morrow et al [21].
Every run (including both restriction fragment length polymorphism and ARMS assays) included DNA samples of known genotype as positive controls and one negative control (no DNA). Samples with AA (Ala114/Ala114) and CC (Val114/Val114) were used to optimize the procedure, and as controls for the ARMS PCR. As an assessment of quality control, approximately 15% of DNA samples were reassayed at least once in order to confirm the assigned genotype. All results from the reassayed samples were consistent with the original genotype assignment.
Statistical analysis
χ2 tests were used to assess homogeneity between groups (eg skin test positive versus skin test negative). As some allele frequencies were small, the StatXact-Turbo (version 3; Cytel Software Corporation, Cambridge, MA, USA) statistical package was used when appropriate. All other statistical analyses were performed using the Stata statistical package (version 6; Stata Corporation, College Station, TX, USA). In order to correct for imbalances in age and sex between groups, logistic regression was used. The Armitage trend test was used to examine the relationship between genotypes and ordered categories (eg degree of BHR). In order to correct these trend test analyses for potential confounding factors such as age and sex, ordered logistic regression models were applied.
Results
Determination of GSTP1 genotypes
Using the two PCR assays it was possible to identify all genotypes unambiguously, except 24 heterozygotes for the Ile105→Val105 and Ala114→Val114 substitutions, which could have been GSTP1 AC or BD. Accordingly, ARMS PCR was used to identify these genotypes. Banding patterns are shown in Supplementary Figure 2.
Supplementary Figure 2.
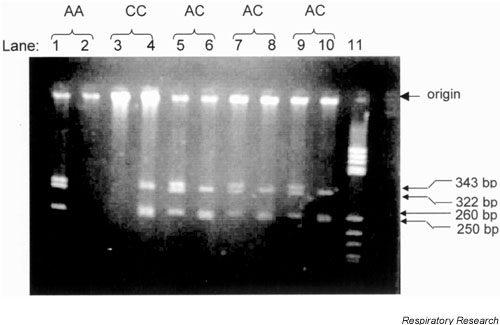
Lanes 1, 3, 5, 7 and 9 include the amplified product from Ala114 specific primer, whereas lanes 2, 4, 6, 8 and 10 include the product from the Val114 specific primer. PCR products from either primer give, after digestion with Bsm AI, fragments of either 343, 322, 260 and 73 base pairs for the Ile105 allele, or 322, 260, 250, 93 and 73 base pairs for the Val105 allele. Lane 11, molecular weight markers, pBR HaeIII. PCR products were visualized in 2% agarose gels.
The frequencies of GSTP1 genotypes are shown in Table 2. In 191 persons, none had BD, CD or DD, reflecting the rarity of GSTP1*D. The frequencies of GSTP1 genotypes achieved Hardy-Weinberg equilibrium. As reported previously [14], we found significant (P <0.0001, χ24 = 50.9) linkage disequilibrium between Ile105 and Ala114 and between Val105 and Val114.
Table 2.
The association ofGSTP1 genotype with atopic status and degree of airway reactivity/obstruction
| Total | Skin test | Skin test | IgE ≤ | IgE > | |||||
| Genotype | group | negative | positive | 100 IU/ml | 100 IU/ml | Group 1 | Group 2 | Group 3 | Group 4 |
| AA | 79 (42.4) | 29 (40.9) | 50 (41.7) | 42 (43.3) | 27 (38.6) | 17 (29.3) | 4 (26.7) | 21 (41.2) | 23 (48.9) |
| AB | 66 (34.6) | 21 (29.6) | 45 (37.5) | 30 (30.9) | 30 (42.9) | 24 (41.4) | 6 (40.0) | 21 (41.2) | 14 (29.8) |
| AC | 26 (13.6) | 10 (14.1) | 16 (13.3) | 11 (11.3) | 11 (15.7) | 7 (12.1) | 3 (20.0) | 6 (11.8) | 7 (14.9) |
| AD | 2 (1.1) | 0 (0) | 2 (1.7) | 1 (1.0) | 1 (1.4) | 1 (1.7) | 0 (0) | 0 (0) | 1 (2.1) |
| BB | 7 (3.7) | 4 (5.6) | 3 (2.5) | 5 (5.2) | 0 (0) | 3 (5.2) | 1 (6.7) | 1 (2.0) | 1 (2.1) |
| BC | 9 (4.7) | 7 (9.9) | 2 (1.7)* | 7 (7.2) | 0 (0) | 5 (8.6) | 1 (6.7) | 1 (2.0) | 1 (2.1) |
| BD | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| CC | 2 (1.1) | 0 (0) | 2 (1.7) | 1 (1.0) | 1 (1.4) | 1 (1.7) | 0 (0) | 1 (2.0) | 0 (0) |
| CD | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| DD | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Alleles | |||||||||
| GSTP1*A | 0.660 | 0.627 | 0.679 | 0.649 | 0.686 | 0.569 | 0.567 | 0.676 | 0.731 |
| GSTP1*B | 0.233 | 0.254 | 0.221 | 0.242 | 0.214 | 0.302 | 0.300 | 0.235 | 0.172 |
| GSTP1*C | 0.102 | 0.120 | 0.092 | 0.103 | 0.093 | 0.121 | 0.133 | 0.088 | 0.086 |
| GSTP1*D | 0.005 | 0 | 0.008 | 0.005 | 0.007 | 0.009 | 0 | 0 | 0.011 |
Atopic status was defined as positive skin test and IgE 100 IU/ml or less. Groups were defined as follows: group 1, percentage predicted >80%/PC20 >16 mg/ml; group 2, percentage predicted >80%/PC20 8-16 mg/ml; group 3, percentage predicted >80%/PC20 0-8 mg/ml; and group 4, percentage predicted ≤ 80%. Values are expressed as number (%) for the genotype frequencies, or as proportions for the allele frequencies.*Logistic regression analysis for skin test: uncorrected GSTP1 BC versus GSTP1 AA, OR 0.17, 95% CI 0.03-0.85 (P = 0.031); and corrected for age and sex, OR 0.18, 95% CI 0.04-0.96 (P = 0.045). Trend test analysis across groups 1-4: uncorrected GSTP1 BC versus GSTP1AA, P = 0.031; and corrected for age and sex, P = 0.02.
Association of GSTP1 genotype with atopic indices
Table 2 shows the association of GSTP1 genotypes with skin test positivity or IgE level. The frequencies of GSTP1 BB and BC were reduced in individuals with at least one positive skin test (2.5 and 1.7%, respectively) as compared with individuals who were skin test negative (5.6 and 9.9%, respectively). Although the frequency of GSTP1 BB was not significantly lower in the persons with positive skin tests (odds ratio [OR] 0.44; P = 0.297) or with IgE levels of 100 IU/ml or less, GSTP1 BC was significantly associated with skin test negativity (OR 0.17; P = 0.031; Table 2). There were no significant associations between GSTP1 genotypes and IgE level, although all individuals with GSTP1*B had IgE levels of 100 IU/ml or less.
Association of GSTP1 genotypes with airway obstruction/reactivity
Table 2 shows the frequencies of GSTP1 genotypes in relation to degree of airflow obstruction and BHR. The GSTP1 AA frequency increased with severity of airway reactivity/obstruction, whereas BB and BC frequencies displayed a reverse trend. Compared with GSTP1 AA, trend test analysis across the four groups revealed a significant decrease in frequency of GSTP1 BC (and GSTP1 BB, albeit not significant) with increasing airway reactivity/obstruction (P = 0.031), indicating a protective effect. This effect remained significant after correction for age and sex (P = 0.022) using ordered logistic regression analysis.
Association of GSTP1 genotype with presence of asthma
We further examined the association of GSTP1 genotypes with the clinical presence of asthma, as defined using the recognized cutoff of <8 mg/ml methacholine [15] on bronchial challenge (groups1 and 2 versus groups 3 and 4). The frequency of GSTP1 AA was increased in groups3 and 4 (44.9%) as compared with groups 1 and 2 (28.8%), whereas GSTP1 BB and BC frequencies were lower in groups 3 and 4 (2.0 and 2.0%, respectively) than in groups 1 and 2 (5.5 and 8.2%, respectively). Thus, GSTP1 BC was associated with a sixfold reduction in asthma risk compared with GSTP1 AA (OR 0.16, 95% confidence interval [CI] 0.03–0.86; P = 0.032). This association remained significant after correction for age and sex (OR 0.15, 95% CI 0.03–0.80; P = 0.027).
Discussion
We showed that the 105 substitution in GSTP1 is associated with atopy and BHR [5]. We have now extended these observations by examining whether the 114 substitution modulates this effect. We determined the GSTP1 genotype using two reported PCR assays for each polymorphic site. However, these assays do not differentiate GSTP1 AC and BD. Accordingly, we developed an ARMS assay to allow identification of all genotypes.
GSTP1 is of particular interest in asthma, because chromosome 11q13 is associated with its clinical phenotypes: atopy and BHR [16]. Several candidates (high affinity IgE receptor, clara cell secretory protein) have been identified, although the encoding genes do not account for the strength of the linkage to this region [16]. We proposed that GSTP1 is another candidate gene in this region [17], although it is of course possible that another gene may be the target, and that the observed associations with GSTP1 genotype reflect linkage disequilibrium with this gene. Thus, the efficiency of detoxification of reactive oxygen species products determined by polymorphism in GSTP1 may influence the development and/or severity of BHR and asthma.
We found that the frequency of GSTP1 AA was increased in patients with established asthma. Similar findings have been found in other proinflammatory conditions, such as chronic obstructive pulmonary disease [18] and multiple sclerosis [19]. However, studies in rheumatoid arthritis [20] and basal cell carcinoma [12] show that GSTP1 Ile105/Ile105 is protective, whereas Val105/Val105 is associated with a worse outcome. Thus, GSTP1-associated risk is likely to be disease-dependent, possibly reflecting differences in relevant substrates.
The data reported here also show that the frequencies of GSTP1 BC and BB were reduced in asthmatic persons, thus indicating a protective effect. This supports the view that there is little additional effect on disease risk from the Ala114→Val114 substitution, although the number of individuals with some genotypes was small and larger numbers would be required to confirm this observation. The advantage with using this ARMS assay is that it is possible to discriminate between genotypes AC and BD. It relies on a relatively large fragment (998 base pairs), however, and is therefore not suitable for genotyping of archival DNA. Thus, this technique is applicable for studies into population genetics.
In conclusion, we described an ARMS assay to identify genotypes resulting from the A, B, C and D alleles of GSTP1, and provided data on the effect of GSTP1 genotype on asthma risk. Our findings suggest that GSTP1*B and GSTP1*C confer similar protective effects. However, large patient groups are required to identify differential effects between GSTP1*B, GSTP1*C and GSTP1*D.
Abbreviations
ARMS = amplification refractory mutation system; BHR = bronchial hyperresponsiveness; CI = confidence interval; FEV1 = forced expiratory volume in 1 s; GST = glutathione S-transferase; OR = odds ratio; PC20 = provoking concentration of an inhaled substance that causes a 20% reduction in FEV1; PCR = polymerase chain reaction.
Acknowledgments
Acknowledgements
Supported by the British Lung Foundation (Project Grant P99/15) and the Danish Research Academy (Project Grant 0000120).
References
- Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological concequences. Pharmacology. 2000;61:151–166. doi: 10.1159/000028396. [DOI] [PubMed] [Google Scholar]
- Fryer AA, Hume R, Strange RC. The development of glutathione S-transferase and glutathione peroxidase activities in human lung. Biochim Biophys Acta. 1986;883:448–453. doi: 10.1016/0304-4165(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Berhane K, Widersten M, Engström Å, Kozarich JW, Mannervik B. Detoxification of base propenals and other α,β-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc Natl Acad Sci USA. 1994;91:1480–1484. doi: 10.1073/pnas.91.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ST. Issues in phenotype assessment. In the Genetics of Asthma Edited by Liggett SB, Meyers DA New York: Marcel Dekker, Inc, 1996. pp. 401–419.
- Fryer AA, Bianco A, Hepple M, Jones PW, Strange RC, Spiteri MA. Polymorphism at the glutathione S-transferase, GSTP1 locus: a new marker for bronchial hyperresponsiveness and asthma. Am J Respir Crit Care Med. 2000;161:1437–1442. doi: 10.1164/ajrccm.161.5.9903006. [DOI] [PubMed] [Google Scholar]
- Ali-Osman F, Akande O, Antoun G, Mao J-X, Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. J Biol Chem. 1997;272:10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- Harries LW, Stubbins MJ, Forman D, Howard GCW, Wolf CR. Identification of genetic polymorphism at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis. 1997;18:641–644. doi: 10.1093/carcin/18.4.641. [DOI] [PubMed] [Google Scholar]
- Sundberg K, Johansson A-S, Stenberg G, Wildersten M, Seidel A, Mannervik B, Jernstrom B. Differences in the catalytic efficiencies of allelic variants of glutathione transferase P1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocorbons. Carcinogenesis. 1998;19:433–436. doi: 10.1093/carcin/19.3.433. [DOI] [PubMed] [Google Scholar]
- Watson MA, Stewart RK, Smith GBJ, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275–280. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- Zimniak P, Pikula S, Bandorowicz Pikula J, Singhal SS, Srivastava SK, Awasthi S, Awasthi YC. Natural occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem. 1994;224:893–899. doi: 10.1111/j.1432-1033.1994.00893.x. [DOI] [PubMed] [Google Scholar]
- Hu X, O'Donnell R, Srivastava SK, Xia H, Zimniak P, Nanduri B, Bleicher RJ, Awasthi S, Awasthi YC, Ji X, Singh SV. Active site architecture of polymorphic forms of human glutathione S-transferase P1-1 accounts for their enantioselectivity and disparate activity in the glutathione conjugation of 7β,8α-dihydroxy-9α,10α-oxy-7,8,9,10-tetrahydrobenzo(a)pyrene. Biochem Biophys Res Commun. 1997;235:424–428. doi: 10.1006/bbrc.1997.6777. [DOI] [PubMed] [Google Scholar]
- Ramachamdran S, Hoban P, Ichii-Jones F, Pleasants L, Ali-Osman F, Lear J, Smith AD, Bowers PW, Fryer AA, Strange RC. Glutathione S-transferase GSTP1 and cyclin D1 genotypes: associations with numbers of basal cell carcinomas in a patient subgroup at high-risk of multiple tumours. Pharmacogenetics. 2000;10:545–556. doi: 10.1097/00008571-200008000-00008. [DOI] [PubMed] [Google Scholar]
- Board P, Harris M, Flanagan J, Langton L, Coggan M. Genetic heterogeneity of the structure and function of GSTT2 and GSTP1. Chem Biol Interact. 1998;111-112:83–89. doi: 10.1016/S0009-2797(97)00152-X. [DOI] [PubMed] [Google Scholar]
- Harris MJ, Coggan M, Langton L, Wilson SR, Board PG. Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients. Pharmacogenetics. 1998;8:27–31. doi: 10.1097/00008571-199802000-00004. [DOI] [PubMed] [Google Scholar]
- Corhay JL, Bury T, Louis R, Delavignette JP, Katembe JM, Weber G, Albert A, Radermecker MF. Bronchial responsiveness in active steelworkers. Eur Respir J. 1998;11:272–277. doi: 10.1183/09031936.98.11020272. [DOI] [PubMed] [Google Scholar]
- Thomas NS, Wilkinson J, Holgate ST. The candidate region approach to the genetics of asthma and allergy. Am J Respir Crit Care Med. 1997;156:S144–S151. doi: 10.1164/ajrccm.156.4.12-tac-13. [DOI] [PubMed] [Google Scholar]
- Spiteri MA, Bianco A, Strange RC, Fryer AA. Polymorphisms at the Glutathione S-transferase, GSTP1 locus: a novel mechanism for susceptibility and development of atopic airway inflammation. Allergy. 2000;55:15–20. doi: 10.1034/j.1398-9995.2000.00502.x. [DOI] [PubMed] [Google Scholar]
- Ishii T, Matsuse T, Teramoto S, Matsui H, Miyao M, Hosoi T, Takahashi H, Fukuchi Y, Ouchi Y. Glutathione S-transferase P1 (GSTP1) polymorphism in patients with chronic obstructive pulmonary disease. Thorax. 1999;54:693–696. doi: 10.1136/thx.54.8.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann CLA, Davies MB, Boggild MD, Aldersea J, Fryer AA, Jones PW, Ko Ko C, Young C, Strange RC, Hawkins CP. Glutathione S-transferase polymorphisms in multiple sclerosis. Their relationship to disability. Neurology. 2000;54:552–557. doi: 10.1212/wnl.54.3.552. [DOI] [PubMed] [Google Scholar]
- Mattey DL, Hassell AB, Plant M, Dawes PT, Ollier WR, Jones PW, Fryer AA, Alldersea JE, Strange RC. Association of polymorphism in glutathione S-transferase loci with susceptibility and outcome in rheumatoid arthritis: comparison with the shared epitope. Ann Rheum Dis. 1999;58:164–168. doi: 10.1136/ard.58.3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow CS, Cowan KM, Goldsmith ME. Structure of the human genomic glutathione S-transferase-pi gene. Gene. 1989;75:3–11. doi: 10.1016/0378-1119(89)90377-6. [DOI] [PubMed] [Google Scholar]


