Abstract
Histone deacetylase 6 (HDAC6) function and dysregulation have been implicated in the etiology of certain cancers and more recently in central nervous system (CNS) disorders including Rett syndrome, Alzheimer’s and Parkinson’s diseases, and major depressive disorder. HDAC6-selective inhibitors have therapeutic potential, but in the CNS drug space the development of highly brain penetrant HDAC inhibitors has been a persistent challenge. Moreover, no tool exists to directly characterize HDAC6 and its related biology in the living human brain. Here, we report a highly brain penetrant HDAC6 inhibitor, Bavarostat, that exhibits excellent HDAC6 selectivity (>80-fold over all other Zn-containing HDAC paralogues), modulates tubulin acetylation selectively over histone acetylation, and has excellent brain penetrance. We further demonstrate that Bavarostat can be radiolabeled with 18F by deoxyfluorination through in situ formation of a ruthenium π-complex of the corresponding phenol precursor: the only method currently suitable for synthesis of [18F]Bavarostat. Finally, by using [18F]Bavarostat in a series of rodent and nonhuman primate imaging experiments, we demonstrate its utility for mapping HDAC6 in the living brain, which sets the stage for first-in-human neurochemical imaging of this important target.
Short abstract
Bavarostat, an HDAC6-selective inhibitor with high brain penetrance, was identified. Through innovative ruthenium-mediated radiofluorination PET imaging was enabled and shows promise for translation.
Introduction
Histone deacetylases have emerged as a pharmaceutical target with a broad range of potential indications. Several pan-HDAC inhibitors (not paralogue selective) of different chemotypes, which target several of the 11 paralogues of Zn-dependent HDACs and span different chemotypes, are approved by the FDA or are currently in clinical trials.1 However, these nonselective agents typically lead to undesired side effects, and considerable efforts are still spent toward the development of more selective chemical probes and drug candidates.2−4
HDAC6 has received recent interest because its structure is unique among the paralogues. Its cytosolic location reduces pharmaceutical interference with genetic material, and HDAC6-selective inhibitors have shown promise preclinically in terms of efficacy and safety.5 Paralogue selectivity has been generally difficult to engineer, but HDAC6 (along with HDAC3 and HDAC8 to a lesser extent2,6) has enough structural distinction from other paralogues to offer a starting point for rational design of selective inhibitors.7,8 Such paralogue-selective chemical probes are needed to understand the contribution of HDAC6 to disease: aberrant HDAC6 expression levels have been heavily implicated in the pathophysiology of glioblastoma multiforme,9−11 Rett syndrome,12,13 and Alzheimer’s14,15 and Parkinson’s diseases.16−18 Nonetheless, the design of brain-penetrant HDAC6-selective agents has proven challenging, and high doses are needed to achieve any functional effects of HDAC6 inhibition in the CNS.19
As a consequence, our understanding of the function and dysfunction of HDAC6 and the therapeutic potential of its inhibition in the living human brain remains limited. A positron emission tomography (PET) probe to study HDAC6 has great potential not only to gain insights into the molecular underpinnings of brain function and disease but also in the validation of therapeutic targets and therapeutic small molecules. Herein, we describe the de novo development of a brain-penetrant, selective HDAC6 inhibitor and its application in rodent and nonhuman primate PET imaging, enabled by a recently described deoxyfluorination method that leverages the reactivity of a phenol–ruthenium π-complex intermediate.
Results and Discussion
Inhibitor Design
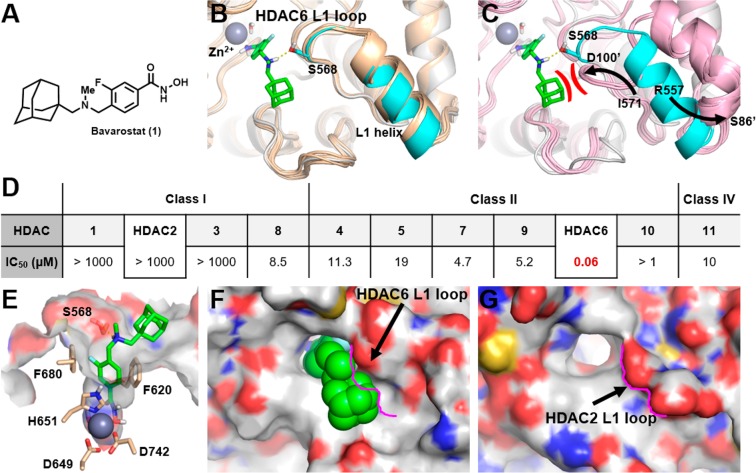
Several strategies exist to achieve selectivity for HDAC6 over other HDAC paralogues with hydroxamate inhibitors. These include the use of (1) large polycyclic capping groups,3,20,21 (2) a substituted sp2 hybridized α-carbon relative to the hydroxamate,22 and (3) sterically demanding substitution close to the chelating group.7,8 On the basis of these strategies, combined with our experience in PET radiotracer design for HDACs,23−26 we developed a small library of potential CNS-penetrant HDAC6 inhibitors. Of these, Bavarostat (Figure 1A) emerged as unique. In Figure 1B,C, the structure of Bavarostat is shown docked into the catalytic domain 2 (CD2) of human HDAC6, providing a model to the contribution of structural elements to its selectivity for HDAC6 over all other HDAC paralogues (Figure 1D) and exemplified by comparison to HDAC2.
Figure 1.
A: Structure of Bavarostat (1). B: Alignment of hHDAC6’s catalytic domain 2 (CD2) (white and cyan protein, PDB 5EDU) and the 15 zHDAC6 CD2 crystal structures available in the PDB database (wheat colored proteins), showing similar L1 loop and L1 helix conformations. HDAC6-selective inhibitor Bavarostat was docked into the hHDAC6 complex described in the Experimental Section. Bavarostat is predicted to form a hydrogen bond (yellow dashed line) with Ser568 on the hHDAC6 L1 loop segment represented in cyan. C: Alignment of hHDAC6 CD2 (white and cyan) and the five hHDAC2 catalytic site crystal structures (pink proteins) in PDB. Large movement of both the L1 loop segment and the L1 helix is represented by arrows (arrows depict movement of analogous amino acids from hHDAC6 to hHDAC2: left arrow depicts hHDAC6’s Ile571 going to corresponding hHDAC2’s Asp100′, and right arrow depicts hHDAC6’s Arg557 going to corresponding hHDAC2’s Ser86′). Prime indicates hHDAC2. The hypothesized steric and electrostatic clash between Bavarostat and the hHDAC2 L1 loop is represented in red. D: IC50 values for Bavarostat inhibition of HDAC1–11 were determined in a fluorescence assay (BPS Bioscience, San Diego, CA) measuring acetylation of a synthetic substrate. E: Bavarostat docked into a hHDAC6 CD2 complex incorporating a conserved water. Key amino acids chelating the catalytic zinc or lining the 10 Å channel are labeled. F/G: Significant topography differences between the surface of the catalytic domains of hHDAC6 (F) and hHDAC2 (G) are highlighted by the pink contouring of the L1 loop segment of hHDAC2.
Bavarostat was docked into an hHDAC6 complex to rationalize its selectivity (see Experimental Section for further details). Briefly, the hydroxamic acid of Bavarostat was modeled to chelate the catalytic zinc and form a hydrogen bond to an ordered water in a similar fashion as seen in the zHDAC6 structure (PDB 5EF7) recently published by Hai et al.27 Indeed, the rigidity introduced by the linker phenyl (sp2-hybridized hydroxamate α-carbon atom) in Bavarostat prevents an effective κ2-binding mode of the hydroxamate warhead. Since a high-resolution crystal structure of zHDAC6 bound to a phenyl-linked HDAC6-selective inhibitor, HPOB in PDB 5EF7, showed a unique mode of binding to the catalytic zinc ion via a conserved water molecule,27 we applied it to Bavarostat. The presence of this water molecule greatly improved the fit of Bavarostat to the protein, suggesting that the rigid phenyl linker moiety is partially conveying HDAC6 selectivity. Additional discriminating factors, such as sterically demanding moieties, were employed to convey paralogue selectivity and potency for HDAC6. First, the fluorophenyl of Bavarostat occupying the 10 Å channel leading to the protein surface forms a favorable noncovalent interaction with Phe620 via a parallel-displaced π-stacking configuration (Figure 1E). The fluorine substituent, installed with imaging applications in mind in a position not expected to interfere with HDAC binding,26 contributes to favorable van der Waals interactions by vectoring in a small divot in the 10 Å channel (Figure SI3).
Second, the proton of the tertiary amine off of the linker phenyl ring directly hydrogen bonds to Ser568 on the L1 loop (Figures 1B and 1E). Finally, the adamantyl group in Bavarostat is predicted to fill a hydrophobic surface groove adjacent to the L1 loop in hHDAC6 complex’s CD2 (Figure 1F). To understand if the conformation of the L1 loop and L1 helix found in the hHDAC6 CD2 structure (PDB 5EDU) was an anomaly due to the crystallization technique used or the result of the bound ligand that was crystallized with it, all zHDAC6 CD2 structures available were aligned to hHDAC6 CD227 (Figure 1B). The L1 loop and L1 helix in CD2 of hHDAC6 and CD2 of zHDAC6 structures consistently folded in the same conformation, suggesting that the L1 loop and L1 helix of hHDAC6 are in a stable low energy conformation. The selectivity profile of Bavarostat (Figure 1D) shows that it is highly selective for hHDAC6 over hHDAC2. To rationalize how Bavarostat may have gained such selectivity, the catalytic site of hHDAC2 was used as a comparison and a representative for class I HDACs. We aligned the CD2 structure of hHDAC6 (PDB 5EDU) to all available CD1 hHDAC2 structures to see if the L1 loop and L1 helix were in a similar conformation since the adamantyl of Bavarostat sits on a hydrophobic groove next to the L1 loop. The L1 loop and L1 helix of hHDAC2 were found to be in a consistent conformation across all available structures, suggesting that this folded state is also stable. This conformation, however, is significantly different relative to CD2 hHDAC6 as exemplified by the large movement of both the L1 loop segment and the L1 helix depicted by arrows in Figures 1B and 1C). The L1 loop/L1 helix conformation of CD1 of hHDAC2 would sterically clash with the bound Bavarostat compound (Figure 1C), preventing its positioning into the analogous hydrophobic groove (Figure 1G).
Our modeling suggests that bulky hydrophobic adamantyl group is best suited to mediate nonpolar surface interactions while affording HDAC6 selectivity and favorable physicochemical properties increasing Bavarostat’s likelihood to penetrate the blood–brain barrier (BBB). Further, the high protein density of HDAC6 in human brain (5–10 pmol per mg tissue)28 makes the IC50 of 60 nM promising for imaging applications.
Functional Selectivity for HDAC6 in Human iPSC-Derived Neuronal Cells
HDAC6 is a class IIb histone deacetylase, structurally unique among other HDACs, predominantly located in the cytosol where it displays distinct substrate selectivity.29−31 Most prominently α-tubulin and HSP90 are deacetylated by HDAC6, while nuclear proteins such as histones 3 and 4 (H3, H4) remain unaltered.5,31 Therefore, the inhibition of HDAC6 should increase the acetylation of α-tubulin (α-tubulin-ac) but not histone acetylation such as the acetylation of histone H3 lysine 9 (H3K9ac) and histone H4 lysine 12 (H4K12ac). To assess the selective inhibition of HDAC6 by Bavarostat in intact cells, we evaluated the cellular acetylation levels by Western blot analysis of a known substrate (α-tubulin-ac) along with two nonsubstrates of HDAC6 (H3K9ac and H4K12ac) in human induced pluripotent stem cell (iPSC) derived neural progenitor cells (NPCs).28 We treated NPCs with Bavarostat (10 μM) as well as several HDACi chemical probes including CI-99432 (a class I selective HDACi, 10 μM), ACY12155,33 (a mixed selectivity inhibitor, 5 μM), and Tubastatin A7 (a selective HDAC6 inhibitor, 10 μM) for 6 h (Figure 2).
Figure 2.
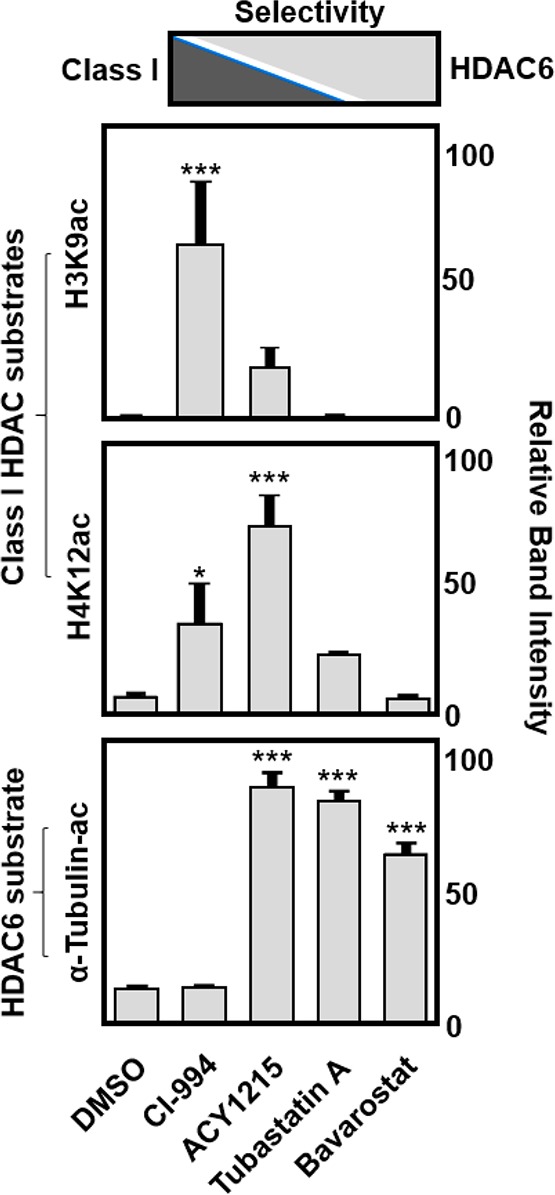
In vitro functional selectivity of Bavarostat for HDAC6. Histone 3 (lysine 9), histone 4 (lysine 12), and α-tubulin protein acetylation levels in IPS neuroprogenitor cells after treatment with Bavarostat (10 μM), a class I HDAC inhibitor (CI-994, 10 μM), an HDAC6-selective inhibitor (Tubastatin A, 10 μM), and a mixed selectivity inhibitor (ACY1215, 5 μM), in comparison to DMSO-treated cells, indicating functional HDAC6 selectivity for Bavarostat.
As expected, CI-994, the class I HDACi, showed a significant increase in H3K9ac and H4K12ac but not α-tubulin-ac. ACY-1215 also increased H3K9ac and H4K12ac, and additionally, it effectively increased α-tubulin-ac, consistent with its mixed selective nature for class I HDACs and HDAC6. Tubastatin A, the highly HDAC6 selective inhibitor, significantly increased α-tubulin-ac but not H3K9-ac or H4K12-ac. Bavarostat, the novel HDAC6-selective inhibitor, exhibited specific increased α-tubulin-ac only. Our data indicate that Bavarostat is able to access the cytoplasmic compartment of human neuronal cells, engage with endogenous HDAC6, and inhibit its deacetylase activity. In conclusion, the changes in cellular protein acetylation in response to HDACi treatment confirm the functional selectivity of Bavarostat for HDAC6 versus class I HDACs.
Radiochemical Synthesis
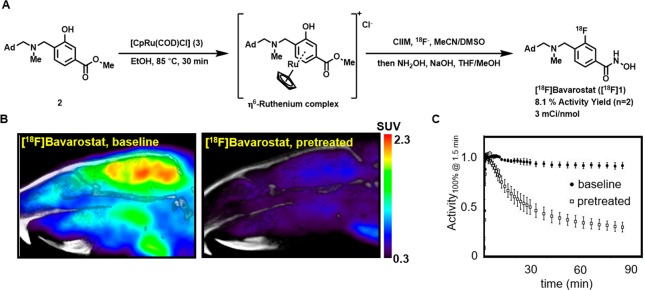
Radiolabeling of hydroxamic acids, particularly scaffolds like Bavarostat, has been difficult to achieve. Routes were lengthy, inefficient, low-yielding, and notoriously difficult to automate.26 Enabled by innovative ruthenium-mediated radiofluorination34 (Figure 3A), a highly efficient radiosynthesis afforded [18F]Bavarostat in high yield and high specific radioactivity. Using air-stable ruthenium complex 3, the labeling precursor 2 was converted into an η6-coordinated ruthenium–phenol complex solution and served without further purification to elute [18F]fluoride off an anion exchange cartridge with 83% elution efficiency. Within 30 min at 130 °C, the labeling proceeds with high conversion (>70% radiochemical yield estimated by TLC). Subsequent transacylation in the same pot afforded [18F]Bavarostat in 8.1% (n = 2) overall nondecay corrected radiochemical yield and high specific activity (4 mCi·nmol–1 at end of synthesis) after HPLC purification and reformulation.
Figure 3.
Rodent PET imaging with [18F]Bavarostat. A: Radiochemical synthesis of [18F]Bavarostat via ruthenium-mediated deoxyfluorination, ClIM = N,N-bis(2,6-diisopropylphenyl)-1-chloroimidazolium chloride. B: Representative sagittal slices summed from 30 to 90 min. C: Averaged (n = 3) time–activity curves of a whole-brain ROI of Sprague–Dawley rats injected with [18F]Bavarostat. In the blocked animals, 1.0 mg·kg–1 unlabeled Bavarostat was injected immediately prior to radiotracer administration, baseline animals treated with vehicle.
Rodent Imaging
To evaluate the potential of [18F]Bavarostat to serve as an HDAC6 radiotracer, PET imaging in rodents was performed. Sprague–Dawley rats were either pretreated with vehicle (1 mL·kg–1, saline:Tween 80:DMSO 8:1:1) or unlabeled Bavarostat (1 mg·kg–1, 1 mg·mL–1) immediately before radiotracer injection, and the dynamic PET data was acquired. As shown in the time–activity curves in Figure 3C, the compound exhibits excellent brain uptake and retention. Treatment of the animals with nonradioactive Bavarostat at 1 mg·kg–1 led to solid blocking of brain uptake, indicative of specific binding (Figure 3B). Normalized time–activity curves were highly reproducible, and signal magnitude was therefore set to 100% at t = 2 min to compare the outcome of several replicates. More work to determine a suitable input function will be needed to appropriately model the tracer kinetics for more in-depth analysis.
Autoradiography
Heterologous in vivo blocking studies were inconclusive due to the poor brain penetrance of known HDAC6-selective inhibitors. To circumvent the issue of blood–brain barrier penetrance, selectivity of the probe in brain tissue was demonstrated using autoradiography (Figure 4). Sagittal slices of rat brain tissue were exposed to [18F]Bavarostat in the presence of [19F]Bavarostat or Tubastatin A. At 10 μM [19F]Bavarostat, binding was reduced by almost 40% in the cerebellum, relative to corpus callosum, a region of high nonspecific binding. Tubastatin A, an HDAC6-selective compound, reduced the amount of bound radioactivity to a comparable level as Bavarostat. These data indicate that the binding to brain tissue occurs with selectivity for HDAC6.
Figure 4.
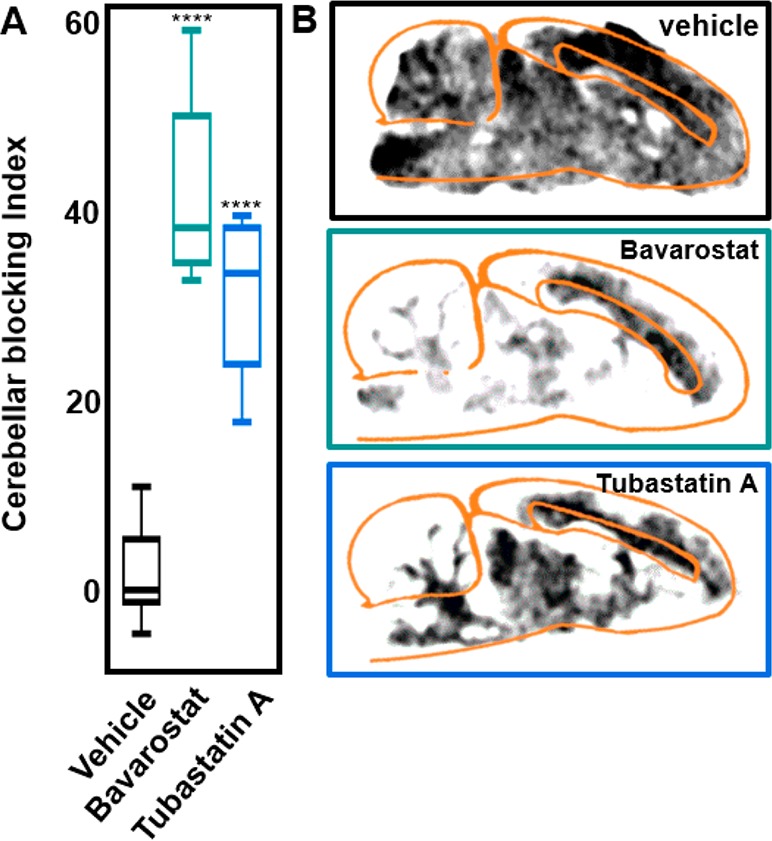
Autoradiography with [18F]Bavarostat. A: Signal intensity of cerebellum relative to corpus callosum at baseline and in sagittal slices of Sprague–Dawley rat brain pretreated with 10 μM Bavarostat or Tubastatin A respectively. Blocking index = 1 – [(PTCB·PTCC–1)/(BLCB·BLCC–1)], PT = pretreated, BL = vehicle treatment, CB = cerebellum, CC = corpus callosum. ****P < 0.0001. B: Representative sagittal slices of Sprague–Dawley rat brains pretreated with vehicle, 10 μM Bavarostat, or Tubastatin A respectively.
Nonhuman Primate (NHP) Imaging
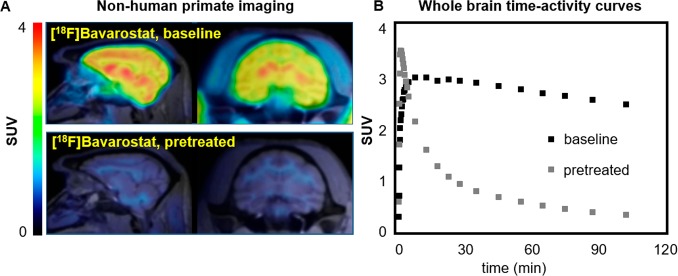
To assess the translational potential of [18F]Bavarostat for human imaging, the tracer properties were assessed in baboon (Papio anubis) through a series of preliminary experiments (Figure 5). The molecule exhibits excellent brain uptake, with a whole-brain standard uptake value of 3 around 30 min after tracer injection and high retention of signal over time. The uptake of [18F]Bavarostat is particularly high in subcortical areas and low in white matter, distinct from other HDAC imaging agents assessed in NHPs23,25,26,35−38 (see the Supporting Information). Particularly noteworthy is the low amount of nonspecific binding observed after pretreatment with 1 mg·kg–1 nonlabeled Bavarostat, and the localization of the residual signal mostly to white matter, which is also observed in autoradiography (Figure 4). An investigation of in vivo metabolism and kinetic modeling of [18F]Bavarostat binding is currently ongoing. In a preliminary analysis, a good correlation was obtained between standardized uptake value (SUV) and distribution volume (VT) derived by Logan plot from an arterial plasma input function (Supporting Information). In summary, the NHP imaging data are promising and warrant translation of [18F]Bavarostat for human HDAC6-imaging.
Figure 5.
Nonhuman primate imaging with [18F]Bavarostat. A: Sagittal and coronal slices of PET images acquired with [18F]Bavarostat in the baboon brain, baseline and pretreated with 1.0 mg·kg–1 unlabeled Bavarostat (averaged 60–120 min). B: Whole-brain SUV time–activity curves of [18F]Bavarostat in baboon brain, baseline and pretreated with 1.0 mg·kg–1 unlabeled Bavarostat.
Conceptually, the animal imaging experiments reveal two important pieces of information: (i) that the labeled small molecule [18F]Bavarostat at a microdose can easily access the brain compartment in living animals with high specific binding and (ii) that nonradioactive Bavarostat at a putative pharmacological dose of 1 mg·kg–1 is well tolerated by two species at least by acute iv treatment. The development of Bavarostat, a brain-penetrant and highly selective HDAC6 inhibitor, offers a powerful chemical probe for preclinical testing to reveal the impact of HDAC6 activity on brain chemistry and behavior.
Conclusion
We show that Bavarostat selectively inhibits HDAC6 in recombinant and cellular assays. Radiolabeling of [18F]Bavarostat was possible with innovative radiofluorination, and in vivo imaging in both rodent and nonhuman primate demonstrated the high brain uptake of the molecule. Its engagement of HDAC6 was confirmed by autoradiography. Bavarostat shows great promise as a radiotracer in rodent and nonhuman primate models and is expected to enable the study of HDAC6 in the living human brain.
Experimental Section
Chemical Synthesis
The synthesis of Bavarostat, the labeling precursor 2, and ruthenium complex 3 is described in the Supporting Information.
IC50
IC50 measurements were conducted by BPS Biosciences with an established fluorescence assay, using proprietary class-specific substrates and recombinant enzymes.
Molecular Modeling
Protein Preparation
The three HDAC structures studied in the modeling section of this work, human HDAC2 (hHDAC2, PDB 3MAX),39 zebrafish HDAC6 (zHDAC6, PDB 5EF7),27 and human HDAC6 (hHDAC6, PDB 5EDU),27 were aligned with the Protein Structure Alignment function of Maestro, Schrödinger Release 2017-1 (Maestro, Schrödinger, LLC, New York, NY, 2017) using the default settings.
The HDAC6 system used for the docking studies described in this work (referred to as the hHDAC6 complex from here on) was created from two structures, zHDAC6 (PDB 5EF7) and hHDAC6 (PDB 5EDU), to exploit key features from both. The zebrafish HDAC6 (zHDAC6, PDB 5EF7) system was selected since it is a holo structure bound to small molecule HPOB, which has the same warhead and linker moiety, a phenyl hydroxamic acid, as Bavarostat. The hydroxamic acid of HPOB in the zHDAC6 PDB 5EF7 structure makes two specific nonbonded contacts in the active site, one chelating the zinc and the other forming a hydrogen bond to an ordered water. In this regard, from the zHDAC6 structure (PDB 5EF7) the zinc, ligand (HPOB), and ordered water adjacent to the zinc were used to build the hHDAC6 complex. The protein and sequence from the unique hHDAC6 (PDB 5EDU) was also used to create the hHDAC6 complex. This new hHDAC6 complex system was then run through the Protein Preparation Wizard40 in Maestro, Schrödinger Release 2017-1 (Protein Preparation Wizard, Schrödinger, LLC, New York, NY, 2017), to protonate it and add any missing side chain residues using the OPLS3 force field.41,42 This preparation was then followed by an all atom minimization using the default settings to create a final chimeric hHDAC6 complex system.
Receptor Preparation for Docking
The final minimized hHDAC6 complex described above was prepared for docking using the Receptor Grid Generation program in Maestro. The HPOB ligand was removed from the hHDAC6 complex for docking. To enhance binding mode prediction a select number of binding pocket hydroxyls and thiols in the side chains were allowed to rotate during the docking simulation: these residues were Ser568, Cys618, and Tyr782. To address modeling into the tight receptor of the HDAC6 binding pocket channel, a GRID scaling factor of 0.6 with a partial charge cutoff of 0.25 was selected.
Ligand Preparation and Docking
The Bavarostat ligand studied in this work was prepared for docking using LigPrep in Maestro, Schrödinger Release 2017-1 (LigPrep, Schrödinger, LLC, New York, NY, 2017) and the OPLS3 force field.41,42 To enable ligand docking into the tight channel of the hHDAC6 complex a van der Waals radius of 0.7 and a partial charge cutoff of 0.15 were selected for the ligand. A targeted pH of 7.0 ±1.0 was selected. Glide, Schrödinger Release 2017-1 (Glide, Schrödinger, LLC, New York, NY, 2017), was used for docking of Bavarostat into the hHDAC6 complex described above.43 To gain accuracy in the docking simulation into the hHDAC6 complex system, the 3D coordinates of the four heavy atoms of the hydroxamic acid of Bavarostat were constrained to their equivalent atoms in HPOB in zHDAC6 (PDB 5EF7). The top scoring docking pose was selected for analysis. All modeling figures in this work were made with the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.
Cellular Protein Acetylation Levels
Human iPSC derived neural progenitor cells from a healthy control subject fibroblast cell line GM08330 (Coriell Institute for Medical Research) were generated as described previously28 and treated with DMSO or a solution of HDAC inhibitor (ACY1215, final concentration 5 μM; Bavarostat, CI-994, Tubastatin A, final concentration 10 μM) for 6 h at 37 °C. Lysis of cell pellets (n = 3 per condition) was performed in radioimmunoprecipitation assay (RIPA) buffer (Boston BioProducts #BP-115) with EDTA-free protease inhibitors (Sigma #4693159001). The lysates were centrifuged at 18,000 rpm at 4 °C for 15 min, and the supernatants were collected. The protein concentration was determined by BCA assay (Thermo Scientific #23227). Western blot analysis was conducted on samples adjusted to 6 μg of total protein/replicate.
Radiosynthesis of [18F]Bavarostat
Aqueous [18F]fluoride obtained from a cyclotron was passed through a SPE Chromafix 30-PS-HCO3 cartridge that had been previously conditioned with 5.0 mg·mL–1 aqueous potassium carbonate and then washed with 18 mL of Millipore Milli-Q water. The captured [18F]fluoride was washed by passing 1 mL of ethanol through the cartridge. At the beginning of the synthesis, 657 mCi was measured on the cartridge.
5 mg of methyl 4-((((adamantan-1-yl)methyl)(methyl)amino)methyl)-3-hydroxybenzoate, 10 mg of CpRu(cod)Cl, and 30 mg of N,N-bis(2,6-diisopropyl)phenyl-2-chloroimidazolium chloride were heated in 250 μL of ethanol at 85 °C for 30 min. The resulting solution was passed through the anion exchange cartridge and collected into a dram vial. The cartridge was flushed with 400 μL of acetonitrile and 400 μL of DMSO and collected into the same vial, which was subsequently sealed with a Teflon lined cap and heated to 130 °C for 30 min. Then, 1 mL of THF/MeOH (1:1), 0.4 mL of 50% aqueous NH2OH, and 0.1 mL of 5 M NaOH were added at room temperature and the reaction was stirred for 5 min.
The solution was diluted with water to 10 mL, loaded onto an OASIS MAX SPE cartridge (60 mg), washed with 5 mL of water, eluted with 2 mL of ethanol/0.1 M AcOH (1:1), and purified by semipreparative HPLC (Agilent Eclipse C-18, 9.4 × 250 mm, 5 μm; flow ramp 0.5 mL·min–1 to 5 mL·min–1 from 0 to 4 min, then 5 mL·min–1; 5% ACN in 0.01 N NaOH from 0 to 4 min, then ramp to 70% ACN in 0.01 NaOH at 45 min). The isolated fraction was reformulated on an OASIS MAX SPE cartridge (60 mg), washed with 5 mL of water, eluted with 2 mL of ethanol/0.1 M AcOH (1:1), diluted with 8 mL of 0.9% saline, and neutralized with 0.1 N NaOH to pH 5. Overall, 53.3 mCi was isolated (8.1% activity yield, 4 mCi·nmol–1 specific activity) within 94 min. For more analytical data, refer to the Supporting Information.
Autoradiography
Sagittal slices of Sprague–Dawley rat brains were sectioned (10 μm) with a cryostat (Thermo Scientific HM550), mounted onto ColorFrost Plus microscope slides (Fisher Scientific 12-550-18), and stored at −20 °C. The slides were submerged in baths of 50 mL of buffer (100 mM Tris, 50 mM NaCl, pH 7.5, 1% BSA) at room temperature and equilibrated for 15 min. The slides were then transferred into baths (50 mL of buffer) containing either 50 μL of DMSO or a solution of blocking compound in DMSO (Bavarostat at final concentrations of 100 nM, 1 μM, and 10 μM and Tubastatin A at a final concentration of 100 μM). After 15 min, 50 μCi of [18F]Bavarostat was added to each bath. After 15 min, all slides were washed by dipping 10× into buffer and subsequently submerged in 50 mL baths of buffer at 4 °C for 3 min. The slides were washed by dipping 10× into wash buffer (100 mM Tris, 50 mM NaCl, pH 7.5) and were dried at 35 °C in a vacuum chamber. A phosphorus screen (PerkinElmer 7001723) was exposed with the slides for 45 min and subsequently imaged with a Cyclone Plus Storage Phosphor (PerkinElmer) detector. ImageJ was used to apply a Gaussian blur (3.0 radius) smoothing, and a lookup table (Royal) with equivalent thresholds for brightness was applied. Raw intensity values from gray and white matter were quantified with the ImageJ measurement tool.
Animal Preparation
Rodent
Six male Sprague–Dawley rats (Charles River Laboratories) were used for PET imaging. According to institutional policy, animals were pair-housed until a weight of 500 g, and a 12 h/12 h light/dark cycle was applied. All treatment and imaging experiments were performed in accordance with procedures approved by the Institutional Animal Care and Use Committee at the Massachusetts General Hospital. Anesthesia was achieved with isoflurane in medical oxygen carrier (3% for induction, 2% for maintenance). For intravenous administration, a catheter with an extension line was placed in a lateral tail vein. Each animal received a bolus injection of either vehicle (1 mL·kg–1, 1:1:8 DMSO/Tween80/saline) or blocking agent in solution (1 mg·kg–1, 1 mg·mL–1 in 1:1:8 DMSO/Tween80/saline) immediately prior to injection of the radiotracer.
NHP
Two PET/MR studies (baseline/pretreatment) were carried out with one baboon (female, 16 kg, Papio Anubis) as approved by the Institutional Animal Care and Use Committee at the Massachusetts General Hospital. Nil per os was instructed 12 h prior to the study. Anesthesia was induced with intramuscular (im) ketamine (1.5 mL, 100 mg·mL-1) + xylazine (0.35 mL, 20 mg·mL-1), and refreshed 45 min later with ketamine (0.2 mL, 100 mg/mL) + xylazine (0.1 mL, 20 mg/mL). Anesthesia was continued during the study with 1–1.5% isoflurane in oxygen carrier. Pretreatment (1 mg·kg–1 Bavarostat, 5 mg·mL–1 in 10% DMSO, 10% Tween 80, 80% saline) was administered in one of the scans 5 min prior to radiotracer injection through a catheter in the saphenous vein. Radiotracer injection was performed through the same catheter. Vital signs (end-tidal CO2, oxygen saturation, heart rate, and respiration rate) were under continuous surveillance to maintain a normal physiological range, documented every 15 min.
PET/CT Image Acquisition
Rodent
After injection of a radiotracer bolus (ca. 700 μCi of [18F]Bavarostat), a 90 min dynamic PET scan was acquired in pairs for all animals. PET scans were performed on a GammaMedica Triumph PET/CT/SPECT scanner, corrected for attenuation with a μ-map derived from the corresponding CT image, which was acquired immediately following the PET scan. The dynamic PET data was binned into 38 time frames (8 × 15 s, 8 × 1 min, 10 × 2 min, 12 × 5 min) and reconstructed individually via an iterative MLEM (maximum likelihood expectation maximization) algorithm in 16 iterations.
NHP
PET/MRI acquisition was performed on a 3T Siemens TIM-Trio with a BrainPET insert (Siemens, Erlangen, Germany). A PET/MRI compatible eight-channel array coil customized for nonhuman primate brain imaging to increase image signal and quality was employed. After administration of the radiotracer (5.2 mCi baseline, 2.9 mCi block), dynamic PET image acquisition was initiated. Dynamic PET data were collected and stored in list mode for 120 min. Image reconstruction was performed using the 3D ordinary Poisson expectation maximization algorithm with detector efficiency, decay, dead time, attenuation, and scatter corrections. PET data were binned in 29 frames (6 × 10 s, 6 × 20 s, 2 × 30 s, 1 × 1 min, 5 × 5 min, 9 × 10 min). Image volumes were eventually reconstructed into 76 slices with 128 × 128 pixels and a 2.5 mm isotropic voxel size. Thirty minutes after scanner start, a high-resolution anatomical scan using multiecho MPRAGE sequence (TR = 2530 ms, TE1/TE2/TE3/TE4 = 1.64/3.5/5.36/7.22 ms, TI = 1200 ms, flip angle = 7°, and 1 mm isotropic) was acquired.
Image Analysis
Rodent
PET images were coregistered to the CT image acquired from the same animal using AMIDE. Data sets were cropped, and all further image analysis was conducted using PMOD 3.3 (PMOD Technologies, Zurich, Switzerland). For maximum consistency, the data were manually coregistered to the Schiffer Px Rat rat brain template44 and data was derived from a whole-brain VOI (volume of interest) for time–activity curves.
NHP
Using the Martinos jip analysis toolkit (www.nitrc.org/projects/jip), MPRAGE data was coregistered to the black baboon atlas45 and the transformation matrix applied to the dynamic PET data. Using PMOD 3.3, time–activity curves for VOIs were extracted, and time-averaged SUV values were obtained. Kinetic modeling was performed with a metabolite corrected arterial plasma input function in PMOD.
Acknowledgments
We thank Tonya Gilbert, Johanna Rokka, Yvonne Klingl, Hsiao-Ying Wey, and Nicole Zurcher for helpful discussions and assistance with experimental work. We thank Sofie Biosciences for assistance with the use of the Elixys platform. Research reported in this publication was supported by the National Institutes of Health (NIH) under Awards S10RR-017208 and R01NS099250. We acknowledge the U.S. Department of Energy (DE-SC0008430), the Phyllis and Jerome Lyle Rappaport MGH Research Scholar Award, the National Center for Research Resources (P41RR14075), and the Athinoula A. Martinos Center for Biomedical Imaging for resource support, including through NIH shared instrumentation Grants S10RR029495, 1S10RR017208-01A1, 1S10RR015728-01, S10RR022976, S10RR019933, and S10RR023401. We thank the NIH NIGMS (GM088237), the Phelps foundation, and the Max-Planck-Institut fuer Kohlenforschung for funding. The molecular docking was supported by a grant from the Stanley Medical Research Institute.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00274.
Chemical synthesis, radiochemical data, details on biochemical and imaging experiments, and docking studies (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Mottamal M.; Zheng S.; Huang T. L.; Wang G. Histone Deacetylase Inhibitors in Clinical Studies as Templates for New Anticancer Agents. Molecules 2015, 20 (3), 3898–3941. 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estiu G.; West N.; Mazitschek R.; Greenberg E.; Bradner J. E.; Wiest O. On the inhibition of histone deacetylase 8. Bioorg. Med. Chem. 2010, 18 (11), 4103–4110. 10.1016/j.bmc.2010.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estiu G.; Greenberg E.; Harrison C. B.; Kwiatkowski N. P.; Mazitschek R.; Bradner J. E.; Wiest O. Structural Origin of Selectivity in Class II-Selective Histone Deacetylase Inhibitors. J. Med. Chem. 2008, 51 (10), 2898–2906. 10.1021/jm7015254. [DOI] [PubMed] [Google Scholar]
- Wang D. Computational Studies on the Histone Deacetylases and the Design of Selective Histone Deacetylase Inhibitors. Curr. Top. Med. Chem. 2009, 9 (3), 241–256. 10.2174/156802609788085287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo L.; Hideshima T.; Kung A. L.; Tseng J.-C.; Tamang D.; Yang M.; Jarpe M.; van Duzer J. H.; Mazitschek R.; Ogier W. C.; Cirstea D.; Rodig S.; Eda H.; Scullen T.; Canavese M.; Bradner J.; Anderson K. C.; Jones S. S.; Raje N. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 2012, 119 (11), 2579–2589. 10.1182/blood-2011-10-387365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KrennHrubec K.; Marshall B. L.; Hedglin M.; Verdin E.; Ulrich S. M. Design and evaluation of ‘Linkerless’ hydroxamic acids as selective HDAC8 inhibitors. Bioorg. Med. Chem. Lett. 2007, 17 (10), 2874–2878. 10.1016/j.bmcl.2007.02.064. [DOI] [PubMed] [Google Scholar]
- Butler K. V.; Kalin J.; Brochier C.; Vistoli G.; Langley B.; Kozikowski A. P. Rational Design and Simple Chemistry Yield a Superior, Neuroprotective HDAC6 Inhibitor, Tubastatin A. J. Am. Chem. Soc. 2010, 132 (31), 10842–10846. 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Kozikowski A.; Pozzo-Miller L. A selective histone deacetylase-6 inhibitor improves BDNF trafficking in hippocampal neurons from Mecp2 knockout mice: implications for Rett syndrome. Front. Cell. Neurosci. 2014, 8, 68. 10.3389/fncel.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Liu X.; Chen X.; Zhang L.; Wang X. Histone deacetylase 6 promotes growth of glioblastoma through inhibition of SMAD2 signaling. Tumor Biol. 2015, 36 (12), 9661–9665. 10.1007/s13277-015-3747-x. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Hu P.; Tang F.; Lian H.; Chen X.; Zhang Y.; He X.; Liu W.; Xie C. HDAC6 promotes cell proliferation and confers resistance to Temozolomide in glioblastoma. Cancer Lett. 2016, 379 (1), 134–142. 10.1016/j.canlet.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Lucio-Eterovic A. K.; Cortez M. A.; Valera E. T.; Motta F. J.; Queiroz R. G.; Machado H. R.; Carlotti C. G.; Neder L.; Scrideli C. A.; Tone L. G. Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer 2008, 10.1186/1471-2407-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delépine C.; Meziane H.; Nectoux J.; Opitz M.; Smith A. B.; Ballatore C.; Saillour Y.; Bennaceur-Griscelli A.; Chang Q.; Williams E. C.; Dahan M.; Duboin A.; Billuart P.; Herault Y.; Bienvenu T. Altered microtubule dynamics and vesicular transport in mouse and human MeCP2-deficient astrocytes. Hum. Mol. Genet. 2016, 25 (1), 146–157. 10.1093/hmg/ddv464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold W. A.; Lacina T. A.; Cantrill L. C.; Christodoulou J. MeCP2 deficiency is associated with reduced levels of tubulin acetylation and can be restored using HDAC6 inhibitors. J. Mol. Med. 2015, 93 (1), 63–72. 10.1007/s00109-014-1202-x. [DOI] [PubMed] [Google Scholar]
- Anderson K. W.; Chen J.; Wang M.; Mast N.; Pikuleva I. A.; Turko I. V. Quantification of Histone Deacetylase Isoforms in Human Frontal Cortex, Human Retina, and Mouse Brain. PLoS One 2015, 10 (5), e0126592. 10.1371/journal.pone.0126592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado-Tejedor M.; Garcia-Barroso C.; Sanchez-Arias J. A.; Rabal O.; Perez-Gonzalez M.; Mederos S.; Ugarte A.; Franco R.; Segura V.; Perea G.; Oyarzabal J.; Garcia-Osta A. A First-in-Class Small-Molecule that Acts as a Dual Inhibitor of HDAC and PDE5 and that Rescues Hippocampal Synaptic Impairment in Alzheimer/’s Disease Mice. Neuropsychopharmacology 2017, 42 (2), 524–539. 10.1038/npp.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Ydewalle C.; Bogaert E.; Van Den Bosch L. HDAC6 at the Intersection of Neuroprotection and Neurodegeneration. Traffic 2012, 13 (6), 771–779. 10.1111/j.1600-0854.2012.01347.x. [DOI] [PubMed] [Google Scholar]
- Su M.; Shi J.-J.; Yang Y.-P.; Li J.; Zhang Y.-L.; Chen J.; Hu L.-F.; Liu C.-F. HDAC6 regulates aggresome-autophagy degradation pathway of α-synuclein in response to MPP+-induced stress. J. Neurochem. 2011, 117 (1), 112–120. 10.1111/j.1471-4159.2011.07180.x. [DOI] [PubMed] [Google Scholar]
- Du Y.; Wang F.; Zou J.; Le W.; Dong Q.; Wang Z.; Shen F.; Yu L.; Li Y. Histone deacetylase 6 regulates cytotoxic α-synuclein accumulation through induction of the heat shock response. Neurobiol. Aging 2014, 35 (10), 2316–2328. 10.1016/j.neurobiolaging.2014.04.029. [DOI] [PubMed] [Google Scholar]
- Jochems J.; Boulden J.; Lee B. G.; Blendy J. A.; Jarpe M.; Mazitschek R.; Van Duzer J. H.; Jones S.; Berton O. Antidepressant-Like Properties of Novel HDAC6-Selective Inhibitors with Improved Brain Bioavailability. Neuropsychopharmacology 2014, 39 (2), 389–400. 10.1038/npp.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty S. J.; Koeller K. M.; Wong J. C.; Grozinger C. M.; Schreiber S. L. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (8), 4389–4394. 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer S.; Saunders L.; Schlimme S.; Valkov V.; Wagner J. M.; Kratz F.; Sippl W.; Verdin E.; Jung M. Pyridylalanine-Containing Hydroxamic Acids as Selective HDAC6 Inhibitors. ChemMedChem 2009, 4 (2), 283–290. 10.1002/cmdc.200800196. [DOI] [PubMed] [Google Scholar]
- Wagner F. F.; Olson D. E.; Gale J. P.; Kaya T.; Weïwer M.; Aidoud N.; Thomas M.; Davoine E. L.; Lemercier B. C.; Zhang Y.-L.; Holson E. B. Potent and Selective Inhibition of Histone Deacetylase 6 (HDAC6) Does Not Require a Surface-Binding Motif. J. Med. Chem. 2013, 56 (4), 1772–1776. 10.1021/jm301355j. [DOI] [PubMed] [Google Scholar]
- Wang C.; Eessalu T. E.; Barth V. N.; Mitch C. H.; Wagner F. F.; Hong Y.; Neelamegam R.; Schroeder F. A.; Holson E. B.; Haggarty S. J.; Hooker J. M. Design, synthesis, and evaluation of hydroxamic acid-based molecular probes for in vivo imaging of histone deacetylase (HDAC) in brain. Am. J. Nucl. Med. Mol. Imaging 2014, 4 (1), 29–38. [PMC free article] [PubMed] [Google Scholar]
- Seo Y. J.; Kang Y.; Muench L.; Reid A.; Caesar S.; Jean L.; Wagner F.; Holson E.; Haggarty S. J.; Weiss P.; King P.; Carter P.; Volkow N. D.; Fowler J. S.; Hooker J. M.; Kim S. W. Image-Guided Synthesis Reveals Potent Blood-Brain Barrier Permeable Histone Deacetylase Inhibitors. ACS Chem. Neurosci. 2014, 5 (7), 588–596. 10.1021/cn500021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Schroeder F. A.; Wey H.-Y.; Borra R.; Wagner F. F.; Reis S.; Kim S. W.; Holson E. B.; Haggarty S. J.; Hooker J. M. In Vivo Imaging of Histone Deacetylases (HDACs) in the Central Nervous System and Major Peripheral Organs. J. Med. Chem. 2014, 57 (19), 7999–8009. 10.1021/jm500872p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebl M. G.; Wang C.; Schroeder F. A.; Placzek M. S.; Wey H.-Y.; Van de Bittner G. C.; Neelamegam R.; Hooker J. M. Development of a Fluorinated Class-I HDAC Radiotracer Reveals Key Chemical Determinants of Brain Penetrance. ACS Chem. Neurosci. 2016, 7 (5), 528–533. 10.1021/acschemneuro.5b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai Y.; Christianson D. W. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat. Chem. Biol. 2016, 12 (9), 741–747. 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey H.-Y.; Gilbert T. M.; Zürcher N. R.; She A.; Bhanot A.; Taillon B. D.; Schroeder F. A.; Wang C.; Haggarty S. J.; Hooker J. M. Insights into neuroepigenetics through human histone deacetylase PET imaging. Sci. Transl. Med. 2016, 8 (351), 351ra106. 10.1126/scitranslmed.aaf7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Yang X.-J. Tubulin acetylation: responsible enzymes, biological functions and human diseases. Cell. Mol. Life Sci. 2015, 72 (22), 4237–4255. 10.1007/s00018-015-2000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Fernández A.; Cabrero J. R.; Serrador J. M.; Sánchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell–cell interactions. Trends Cell Biol. 2008, 18 (6), 291–297. 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Spange S.; Wagner T.; Heinzel T.; Krämer O. H. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009, 41 (1), 185–198. 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Methot J. L.; Chakravarty P. K.; Chenard M.; Close J.; Cruz J. C.; Dahlberg W. K.; Fleming J.; Hamblett C. L.; Hamill J. E.; Harrington P.; Harsch A.; Heidebrecht R.; Hughes B.; Jung J.; Kenific C. M.; Kral A. M.; Meinke P. T.; Middleton R. E.; Ozerova N.; Sloman D. L.; Stanton M. G.; Szewczak A. A.; Tyagarajan S.; Witter D. J.; Paul Secrist J.; Miller T. A. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg. Med. Chem. Lett. 2008, 18 (3), 973–978. 10.1016/j.bmcl.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Santo L.; Hideshima T.; Kung A. L.; Tseng J.-C.; Tamang D.; Yang M.; Jarpe M.; van Duzer J. H.; Mazitschek R.; Ogier W. C.; Cirstea D. D.; Rodig S.; Eda H.; Scullen T. A.; Canavese M.; Bradner J. E.; Anderson K. C.; Jones S.; Raje N. S. Pharmacodynamic and Pharmacokinetic Properties of a Novel and Selective HDAC6 Inhibitor, ACY-1215, in Combination with Bortezomib in Multiple Myeloma. Blood 2011, 118 (21), 2912–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyzavi M. H.; Mandal D.; Strebl M.; Neumann C. N.; D’Amato E. M.; Chen J.; Hooker J. M.; Ritter T. 18F-Deoxyfluorination of Phenols via Ru π-Complexes. ACS Cent. Sci. 2017, 10.1021/acscentsci.7b00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey H.-Y.; Wang C.; Schroeder F. A.; Logan J.; Price J. C.; Hooker J. M. Kinetic Analysis and Quantification of [11C]Martinostat for in Vivo HDAC Imaging of the Brain. ACS Chem. Neurosci. 2015, 6 (5), 708–715. 10.1021/acschemneuro.5b00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A. E.; Hooker J.; Shumay E.; Logan J.; Shea C.; Kim S. W.; Collins S.; Xu Y.; Volkow N.; Fowler J. S. Evaluation of 6-([(18)F]fluoroacetamido)-1-hexanoicanilide for PET imaging of histone deacetylase in the baboon brain. Nucl. Med. Biol. 2009, 36 (3), 247–258. 10.1016/j.nucmedbio.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H.-H.; Tian M.; Hinz R.; Young D.; Shavrin A.; Mukhapadhyay U.; Flores L. G.; Balatoni J.; Soghomonyan S.; Jeong H. J.; Pal A.; Uthamanthil R.; Jackson J. N.; Nishii R.; Mizuma H.; Onoe H.; Kagawa S.; Higashi T.; Fukumitsu N.; Alauddin M.; Tong W.; Herholz K.; Gelovani J. G. Imaging Epigenetic Regulation by Histone Deacetylases in the Brain using PET/MRI with (18)F-FAHA. NeuroImage 2013, 64, 630–639. 10.1016/j.neuroimage.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y. J.; Muench L.; Reid A.; Chen J.; Kang Y.; Hooker J. M.; Volkow N. D.; Fowler J. S.; Kim S. W. Radionuclide Labeling and Evaluation of Candidate Radioligands for PET Imaging of Histone Deacetylase in the Brain. Bioorg. Med. Chem. Lett. 2013, 23 (24), 6700–6705. 10.1016/j.bmcl.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressi J. C.; Jennings A. J.; Skene R.; Wu Y.; Melkus R.; Jong R. D.; O’Connell S.; Grimshaw C. E.; Navre M.; Gangloff A. R. Exploration of the HDAC2 foot pocket: Synthesis and SAR of substituted N-(2-aminophenyl)benzamides. Bioorg. Med. Chem. Lett. 2010, 20 (10), 3142–3145. 10.1016/j.bmcl.2010.03.091. [DOI] [PubMed] [Google Scholar]
- Sastry G. M.; Adzhigirey M.; Day T.; Annabhimoju R.; Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27 (3), 221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Harder E.; Damm W.; Maple J.; Wu C.; Reboul M.; Xiang J. Y.; Wang L.; Lupyan D.; Dahlgren M. K.; Knight J. L.; Kaus J. W.; Cerutti D. S.; Krilov G.; Jorgensen W. L.; Abel R.; Friesner R. A. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12 (1), 281–296. 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- Shivakumar D.; Williams J.; Wu Y.; Damm W.; Shelley J.; Sherman W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput. 2010, 6 (5), 1509–1519. 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49 (21), 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Schiffer W. K.; Mirrione M. M.; Biegon A.; Alexoff D. L.; Patel V.; Dewey S. L. Serial microPET measures of the metabolic reaction to a microdialysis probe implant. J. Neurosci. Methods 2006, 155 (2), 272–284. 10.1016/j.jneumeth.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Black K. J.; Snyder A. Z.; Koller J. M.; Gado M. H.; Perlmutter J. S. Template images for nonhuman primate neuroimaging: 1. Baboon. NeuroImage 2001, 14 (3), 736–743. 10.1006/nimg.2001.0752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.