Abstract
Periodontitis is a polymicrobial disease caused by complex interactions between distinct pathogens in a biofilm resulting in the destruction of periodontal tissues. It seems evident that unknown microorganisms might be involved in onset or progression of periodontitis. For many decades, research in the field of oral microbiology failed to identify certain subgingival microbiota due to technical limitations but, over a period of 12 years using molecular approaches and sequencing techniques, it became feasible to reveal the existence of new periodontal pathogens. Therefore, it is evident that in addition to conventional periodontal pathogens, other microbes might be involved in onset and progression of periodontitis. The novel pathogens enlisted under periodontal phylogeny include Cryptobacterium curtum, Dialister pneumosintes, Filifactor alocis, Mitsuokella dentalis, Slackia exigua, Selenomonas sputigena, Solobacterium moorei, Treponema lecithinolyticum, and Synergistes. The polymicrobial etiology of periodontitis has been elucidated by comprehensive techniques, and studies throwing light on the possible virulence mechanisms possessed by these novel periodontal pathogens are enlisted.
KEYWORDS: Novel microbiota, periodontitis, polymicrobial infection
INTRODUCTION
The environmental diversity of the oral cavity promotes colonization of distinct microbial communities. The breadth of this bacterial diversity was studied by many scientists through culture-independent studies. They implicated specific species or phylotypes in various oral diseases such as chronic periodontitis, necrotizing ulcerative diseases, and aggressive periodontitis.[1,2] Periodontitis is a polymicrobial disease caused by complex interactions between distinct pathogens in a biofilm resulting in the destruction of periodontal tissues.[3] The prime step in the treatment of periodontal disease is identification of the key pathogen. The establishment of an organism as a true pathogen is based on its high prevalence at disease sites and its reduction or absence with regression of disease. Since decades research in the field of oral microbiology failed to reveal complete taxonomic data to the species level by mere culture techniques. Due to these technical difficulties, the emergence of molecular and immunologic tests such as PCR, DNA probes, and immunoassays made research to progress toward the identification of uncultivable taxons also.[4] In 2001, Paster et al. used cloning and Sanger sequencing techniques to identify certain novel microbial species conforming their complex diversity in the etiology of periodontal disease.[1]
Periodontal pockets accommodate a multitude of bacterial phylotypes that make it difficult to differentiate between mere commensals and true pathogens. The profiles of these bacterial species differed on different oral surfaces, and this could be the reason why some Bacteria remain unidentified.[5,6] A few of these include, Filifactor alocis, Selenomonas, Synergistes, and Dialister pneumosintes that have been identified in a number of independent studies. Hence, the role of these novel pathogens in periodontal pathogenesis needs attention.[6,7]
REVIEW OF PHYLOGENY, MORPHOLOGY, AND PATHOGENESIS
Cryptobacterium curtum
The name Cryptobacterium curtum is derived from the Greek words “Kryptos” means hidden and “curtum” means shortened. It originally belonged to the genus Eubacterium but was reclassified into a new genus named Cryptobacterium.[8,9] They belongs to domain: Bacteria, phylum: Actinobacteria, class: Actinobacteria, order: Coriobacteriales, family: Coriobacteriaceae, genus: Cryptobacterium, and species: Curtum. These are short Gram-positive, obligatory anaerobic, nonmotile, nonsporing, mesophilic, asacharolytic rods varying in size from 0.8 μm to 1.0 μm. Ultrathin sections showed a single-layered cell wall of thickness 10 nm without Pili or flagella [Figure 1]. They form tiny translucent colonies of < 1 mm (0.3–0.5 mm) in diameter on brain heart infusion blood agar without hemolysis.[6,7,8] C. curtum growth in Peptone Yeast Glucose broth is enhanced by 0.5% arginine, derived by enzymatic degradation of peptides from argingine deaminase pathway. C. curtum exhibits poor growth due to limited utilization of substrates for metabolic activity.[7,8,9,10]
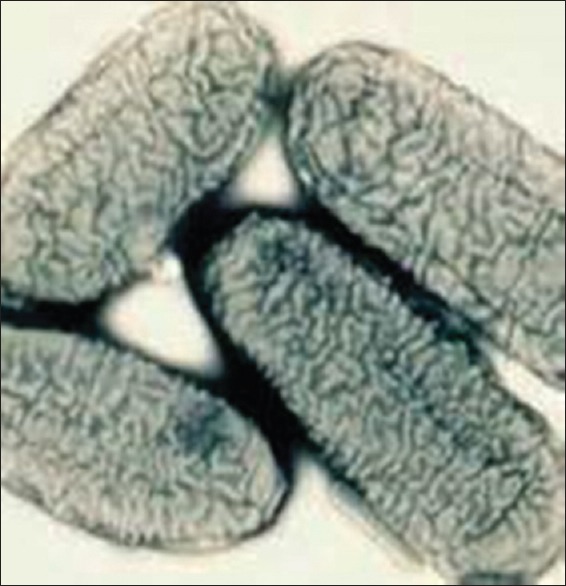
Figure 1.
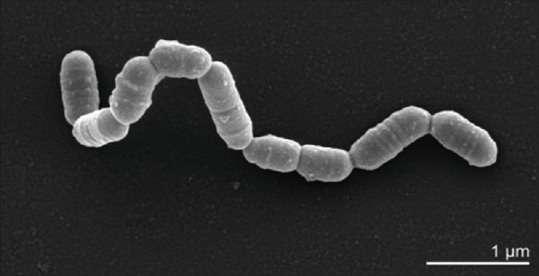
Cryptobacterium curtum scanning electron micrograph
C. curtum is nonreactive in most of the conventional biochemical tests. Glucose fermentation is negative, suggesting its asaccharolytic nature. Arginine stimulation is positive and nitrate reduction is negative for poor growth. C. curtum was isolated from patients with necrotic dental pulps, root canals, dental abscess, halitosis and was found associated with periodontal lesions.[8,9]
Filifactor alocis
The term filifactor is derived from Latin “filum”-thread, “factor”-maker that means a thread maker. F. alocis was isolated first in 1985 from gingival sulcus in gingivitis and periodontitis patients. F. alocis belongs to domain: Bacteria, phylum: Firmicutes, class: Clostridia, order: Clostridiales, family: Peptostreptococcaceae, genus: Filifacor, species: Alocis. F. alocis grows in brain heart infusion broth supplemented with yeast extract (0.5 mg/ml), L-cysteine (50 μg/ml), and 20% arginine anaerobically at 37°C in 10% H2, 10% CO2, and 80% N2. Particular amino acids such as arginine, lysine, cysteine stimulated the growth of F. alocis in the niche of periodontal pocket. The fastidious nature of F. alocis makes its isolation difficult by culture techniques.[9] It is the third most prevalent pathogen in generalized aggressive periodontitis (45%), second most prevalent in chronic periodontitis (90%) but shows the least prevalence in periodontitis resistant groups. F. alocis is detected more in the apical and middle thirds than in the cervical thirds of the pocket.[11,12]
It plays a key role in metabolic pathways and networks that influence the cell responses in periodontitis.[12] The high prevalence of F. alocis in periodontitis could be attributed to its unique virulence properties such as oxidative stress resistance, proinflammatory cytokine production, involvement in periodontal biofilms that triggers host response by secretion of battery of proteases. Enzymes such as the xaa pro-dipeptidase, o-sialoendopeptidase, oligoendopeptidase M3 families, are present only in the membrane fraction of F. alocis.[11,12,13] Protease (HMPREF 0389-00122) was identified in the extracellular fraction of F. alocis. It contains a collagenase peptidase function that could be implicated in tissue destruction in periodontal diseases. It was observed that F. alocis is more resistant than Porphyromonas gingivalis to hydrogen peroxide-induced oxidative stress. Therefore, this property of F. alocis favors its high existence in periodontal pockets.[14,15]
F. alocis contains several genes that have a well-developed mechanism for arginine metabolism which makes it conducive for the growth of several periodontal pathogens.[13] As they share common interacting proteins adherence and invasion of F. alocis is accentuated by P. gingivalis.[13,14] Coinfection between F. alocis and P. gingivalis exhibited filapodial projections on surface of host cells that mediated organisms internalization.[16] Proteomic analysis of F. alocis revealed increase in membrane adhesion proteins and microbial surface components recognizing adhesive matrix molecules that enhance virulence potential of F. alocis and P. gingivalis. The invasion of epithelial cells by F. alocis was examined by in situ hybridization [Figure 2]. F. alocis and P. gingivalis coexist forming a mixed species biofilm suggesting a symbiotic relationship between them due to their capacity to autoaggregate or express unique features such as tight colocalization and vesicle-mediated internalization.[15,16] The ability of F. alocis to interact with the variety of oral Bacteria enhances its participation in community development.[15,16] The investigations on community interactions of F. alocis revealed its relationship with microbes of varying pathogenicity such as the Streptococcus gordonii, Fusobacterium nucleatum, P. gingivalis, and Aggregatibacter actinomycetemcomitans as a major member of anaerobic niche.[15,16]
Figure 2.
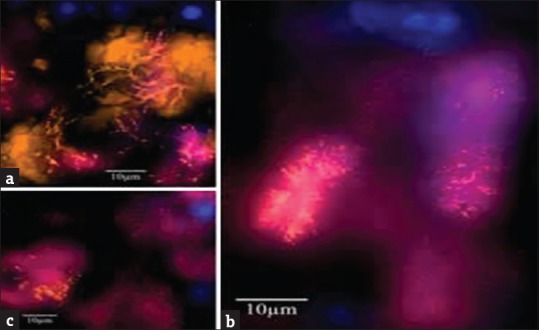
In situ hybridization image of Filifactor alocis (a) F. alocis forms tree like structures among coccoid and fusiform bacteria (b) F. alocis forms palisades with fusiform bacteria around large rod shaped eubacterial organisms (c) F. alocis being part of concentric bacterial aggregations
Virulence
F. alocis is resistant to oxidative stress better than P. gingivalis, attributing for its relative abundance in deeper portions of the soft tissue wall of periodontal pocket.[11] F. alocis exerts its effect on gingival epithelial cells and induces proinflammatory cytokine secretion leading to apoptotic cell death. F. alocis was frequently found in relation to subgingival plaque samples and saliva samples, supporting its increased prevalence.[13]
F. alocis induces apoptosis in gingival epithelial cells with a transient activation of MEK ½ and caspase-3 that has impact on both intrinsic and extrinsic pathways. F. alocis also possess genes encoding for bisulfur proteins and a ferrous iron transport system that facilitates efflux of reactive oxygen species.[14] F. alocis modulates host response by coinfection of gingival epithelial cells, host cell signaling, metabolic host response, cell–cell interaction, and activation of oncogenes.[16,17] F. alocis is susceptible to 100 g/nl of metronidazole, clindamycin (0.5 μg/ml), erythromycin (300 μg/ml), and carbenicillin (100 μg/ml).[18]
Mitsuokella dentalis
The members of genus Mitsuokella are named as Mitsuokella dentalis in honor of Mitsuoka, a Japanese bacteriologist and dentalis in Latin meaning “pertaining to teeth.” Mitsuoka isolated a large number of bacterial strains from humans, dogs, and pigs that appeared to be closely related to Bacteroides genus. However, they were excluded from genus bacteroides due to their high-DNA base composition analyzed through biochemical and ultrastructural methods.[19,20]
The genus Mitsuokella was created based on morphological, biochemical, and chemotaxonomic criteria to include the species Multacidus and Dentalis.[19] M. dentalis belongs to domain: Bacteria, phylum: Firmicutes, class: Clostridia, order: Clostridiales, family: Veillonellaceae, genus: Mitsuokella, species: Dentalis. It exhibits morpho; ogical similarities to Prevotella species except for its dissimilarities, such as absence of menaquinones and higher G + C contents (56–60 mol %) in M. dentalis. M. dentalis grows better on blood agar with hemolysed blood. On enriched horse blood agar after 3 days of incubation, it forms convex, irregular, translucent, wet, and mucoid colonies of 1–2 mm in diameter, with a water drop appearance[19,20] M. dentalis has a low virulence potential as a periodontal pathogen. It does not have the ability to activate latent human fibroblast type, neutrophil interstitial procollagenases that lead to degradation of Type I collagen that is an essential step for periodontal tissue invasion and disease progression. Low proportions of M. dentalis comprising 2% of organisms isolated from periodontal pockets implie its minimal role as a periodontopathogenic bacterium. M. dentalis being a strict anaerobe is susceptible to metronidazole.[21,22]
Slackia exigua
The name “slackia” is given to honor Geoffrey Slack a distinguished microbiologist and researcher. “Exigua” in Latin means scanty referring to the scanty growth of this organism.[23] Formerly, it was known as Eubacterium exigum (1996) which was later reclassified by Wade as Slackia exigua in 1999.[24] S. exigua belongs to domain: Bacteria, phylum: Actinobacteria, class: Actinobacteria, order: Coriobacteriales, family: Coriobacteriaceae, genus: Slackia, species: Exigua. It is Gram-positive, nonspore forming, nonmotile, asaccharolytic, and strictly anaerobic bacillus [Figure 3]. Colonies appear circular, convex, and translucent measuring < 1 mm in diameter. They are seen as single rods or in clumps with size varying from 0.5 μm × 1.0 μm that remain inert in most biochemical tests.[24]
Figure 3.
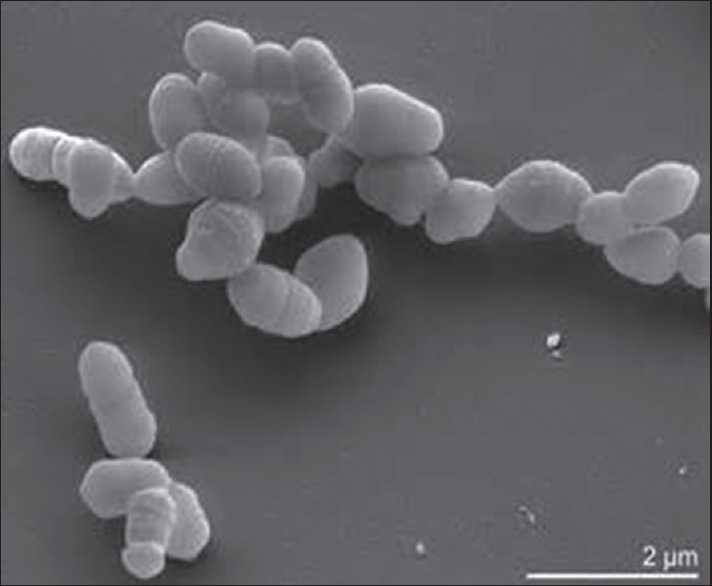
Slackia exigua scanning electron micrograph
They are asaccharolytic; hence, amino acids are important metabolic substrates for growth, particularly arginine and lysine which are produced by enzymatic degradation of peptides by trypsin-like proteinases. S. exigua does not produce butyrate from ornithine which could be one of the reasons for its poor growth. S. exigua have been significantly associated with periodontitis as observations revealed higher antibody titers in the serums of periodontitis patients.[25] Serum IgA levels against S. exigua were elevated in cases of refractory periodontitis, IgG levels in chronic periodontitis patients suggests that these species have breached the host defense to stimulate an immune response. S. exigua produced butyric acid from arginine that has a key role in promoting halitosis. Butyric acid also inhibits the proliferation of gingival fibroblasts and induces apoptosis in splenic T cells further leading to exacerbation of infectious lesions. Uematsu and Hoshino isolated strains from periodontal pockets of chronic periodontitis patients, of which 42% were Eubacteria species[25] and many studies have reported that S. exigua plays a pivotal role in moderately and severely affected periodontal disease as it is frequently isolated in periodontal lesions. S. exigua is susceptible to antimicrobials but resistant to sulfamethaxazole – trimethoprim.[25,26]
Dialister pneumosintes
Dialister pneumosintes is one among the newly cultivated organisms associated with periodontal disease. This novel pathogen was originally described by Olitsky and Gates. During 1921, its nomenclature was Bacterium pneumosintes. In 1923, it was placed in the genus Dialister but later, in 1970, it was regrouped by Holdeman and Moore in the genus Bacteroides. However, in 1980, Shah Collins excluded it from the genus Bacteroides. In 1994, it was reclassified into genus Dialister by Moore and Moore. Currently, this genus Dialister comprises four species D. pneumosintes, Dialister invisus, Dialister microaerophilus, Dialister propionifaciens[27,28] D. Pneumosintes belongs to domain: Bacteria, phylum: Firmicutes, class: Clostridia, order: Clostridiales, family: Veillonellaceae, genus: Dialister, species: Pneumosintes. D. pneumosintes is a nonmotile, nonsporing, nonfermenting, small asaccharolytic, obligatory anaerobic microaerophilic Gram-negative coccobacilli [Figure 4]. It exhibits small circular, tiny smooth, and transparent colonies on columbia blood agar.[27,28] Grows in 0.2% hemolyzed sheep erythrocytes, 0.0005% hemin, and 0.00005% menadione. Growth is most rapid in anaerobic environments at 37°C.
Figure 4.
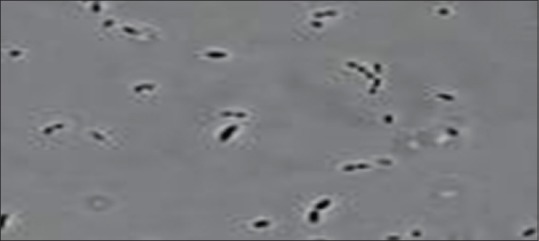
Transmission electron microscopic image of Dialister pneumosintes
Bacteroides or D. pneumosintes was first isolated during 1918–1921 from the nasopharyngeal secretions of patients with influenza and sinusitis. However, it could be isolated through molecular methods such as 16S rRNA gene sequencing, polymerase chain reaction methods in subgingival plaque from gingival crevice, and periodontal pockets in periodontitis patients.[27,28] It was seen in subgingival biofilm associated with periodontitis in smokers and patients with bacteremia. It was seen in < 2% of subgingival samples associated with gingivitis and periodontitis. D. pneumosintes are negative for all the biochemical reactions.[28,29] Lipopolysaccharides present in the cell wall of this microbe activate immune-mediated cells to release proinflammatory cytokines, prostaglandins, matrix metalloproteinases (MMP'S) that eventually lead to periodontal connective tissue destruction, and resorption of alveolar bone.[30,31] D. pneumosintes is reported to be significantly higher in prevalence among patients with refractory periodontitis, rapidly progressing periodontitis suggesting its role in disease pathogenesis.[29,30]
Selenomonas sputigena
The Selenomonas species are considered as not yet cultivated Gram-negative species of oral cavity.[32] Moore et al. isolated 5 new Selenomonas species S. artemidis, Selenomonas fluggei, Selenomonas diane, Selenomonas infelix, Selenomonas noxia. Although all Selenomonas species dominated disease sites, Selenomonas sputigena was most frequently detected.[33] It belongs to the Sporomusa, a subbranch of Clostridium cluster. S. sputigena belongs to domain: Bacteria, phylum: Firmicutes, class: Clostridia, order: Clostridiales, family: Veillonellaceae, genus: Selenomonas, species: Sputigena. Selenomonas sputigena was isolated from patients with progressive disease severity. S. sputigena was isolated using DNA probes, biochemical tests, SDS-PAGE, and CFA analysis. The Selenomonas species like other periodontal pathogens induced periodontal attachment loss. S. sputigena was detected in periodontal pockets of patients with chronic periodontitis, aggressive periodontitis suggesting its role as a potential pathogen and diagnostic marker for active periodontal disease.[33,34,35] S. sputigena can be segregated from other genera by its DNA-based composition such as Pectinatus, Quinello, Acidaminococcus, Megasphaera. S. sputigena grows on Mac conkey plates, chocolate agar, brain heat infusion agar supplemented with 5% sheep blood, hemin, menadione, anaerobically at 35°C after 4 days. It forms small (<0.5 mm) grey white opaque colonies.[34,36]
Today, S. sputigena has evolved as a chief periodontal pathogen due to its virulence factors and its key role in coaggregation and maturation of plaque. Lipopolysaccharides of S. sputigena could be one of the multitude of pathogenic factors involved in periodontal disease. It induces release of interleukin 6 (IL-6), IL-1α in macrophages thereby provoking inflammation. Its association with chronic periodontitis was confirmed by its high prevalence among periodontal pocket microbiota.[37,38]
Treponema lecithinolyticum
Numerous bacterial strains of Treponema lecithinolyticum were isolated from human oral cavities. Apart from Treponema denticola (Group II) recently cultivated Spirochaetes are T. lecithinolyticum, Treponema maltophilum and Treponema amylovorum. T. lecithinolyticum belongs to Group IV treponemes. T. lecithinolyticum belong to domain: Bacteria, phylim: Spirochaetes, class: Spirochaetes, order: Spirochetales, family: Spirochetaceae, genus: Treponema, species: Lecithinolyticum. The term lecithinolyticum in Greek means “lekithos-egg yolk,” “lytikos”-able to dissolve, for that reason lecithinolyticum produces effect similar to break down of egg yolk.[39] T. lecithinolyticum is a small, obligatory anaerobic, helically coiled, motile treponeme [Figure 5]. Its cells are 5 μm × 0.15 μm wide containing two endoflagella one per pole that overlap in the central region of the cell. Directional motility is not observed in liquid media but translational movement is detectable. It forms off white diffuse subsurface colonies up to 3 mm in diameter within 7 days of incubation at 37°C.[40]
Figure 5.
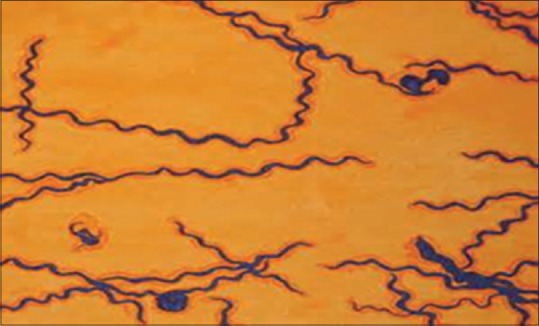
Morphology of Treponema lecithinolyticum (modified steiner silver stain)
T. lecithinolyticum was isolated from patients with gingivitis and was recently isolated from sites with aggressive periodontitis and endodontic infections. It was more frequently isolated from patients with rapidly progressive periodontitis than chronic periodontitis. T. Lecithinolyticum was detected more frequently than T. denticola in periodontal disease sites.[41] T. lecithinolyticum is dependent on N-acetyl glucosamine for growth and is stimulated by D-ribose, L-fucose, D-xylose, D-arabinose, D-fructose. T. lecithinolyticum shows prominent activities of acid phosphatase, alkaline phosphatase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, phospholipase A and C. T. lecithinolyticum can be distinguished from other treponemes by the 16sr RNA sequence, proteins, and antigen patterns.[42,43,44] T. lecithinolyticum may be inhibited by free fatty acids liberated by its own phospholipases.[43] T. lecithinolyticum synthesizes cellular fatty acids – C14, C15, C16, C17 (linear, iso, anterior forms) in diverse proportions. They show hemolytic activity when grown on solid media supplemented with 1% heat-inactivated human serum, and 2% washed human erythrocytes.[44,45,46]
T. lecithinolyticum is recently being considered as a powerful periodontal pathogen due to the virulence potential of T. lecithinolyticum major surface proteins in inducing periodontitis and acute necrotizing ulcerative gingivitis. Major surface protein is composed of β strands and loop regions. The N-terminus with surface exposed loops comes in contact with host cells.[47,48] These surface proteins play a pivotal role in cell adhesion and migration. They play a critical role in monocot adhesion and transendothelial migration responsible for initial infiltration of monocytes into periodontal tissues.
T. lecithinolyticum induced the activation of MMP-2 in gingival fibroblasts and periodontal ligament cells[49,50] It also promotes osteoclastogenesis by production of PGE2 and osteoclast differentiation factor[51,52] T. lecithinolyticum expressed operons for both genes protease complex-related proteins PrcA and PrtP.[49] T. lecithinolyticum has been strongly associated with human periodontal diseases.[50] T. lecithinolyticum can be considered a diagnostic marker due to its highest prevalence in generalized aggressive periodontitis, followed by chronic periodontitis, and least in periodontitis resistant group. T. lecithinolyticum is susceptible to metronidazole and nystatin.[51,53]
Solobacterium moorei
Solobacterium moorei is one of the dominant flora that coexists with other aerobes or anaerobes in oral cavity. It belongs to the domain: Bacteria, phylum: Firmicutes, class: Erysipelotrichi, order: Erysipelotrichales, family: Erysipelotrichaceae, genus: Solobacterium, species: Moorei. It was identified in the year 2000 and obtained its nomenclature from the Latin terms (so. lo.L.a solus, sole, n. bakterion a small rod). “Moorei” named in honour of W.E.C. Moore, a contemporary American microbiologist.[54] The cells are short, straight, or slightly curved and found single or in pairs varying in diameters of 0.2 and 0.4–0.7 μm.[54,55]
S. moorei is difficult to isolate due to differential exhibition of phenotypic characteristics. 16S rRNA gene sequences were used to detect the strains of S. moorei. S. moorei was identified from subgingival plaque of patients with refractory periodontitis, localized aggressive periodontitis, dentoalveolar abscess. It grows on brucella blood agar supplemented with hemin and vitamin K, anaerobic agar, a chocolate agar. S. moorei produces very few positive biochemical reactions. It is positive for enzymes arginine hydrolase, α- and β-galactosidase, α-glucosidase, arginine dihydrolase, leucine arylamidase, acid phosphatase, alkaline phosphatase, N-acetyl β and glucosaminidase. It reduces nitrates, ferments glucose, galactose, maltose but cannot ferment sucrose, mannitol.[54,55] S. moorei produced clinical features of fever as a symptom of infection due to bacteremia. Patients showed higher levels of C-reactive protein in infections or diseases associated with S. moorei. Biofilm formation is also a key step in production of halitosis by S. moorei. It adheres to oral epithelial cells through adhesins. It can also induce the secretion of IL-8 in gingival epithelial cells, promote osteoclast differentiation, and inhibit proliferation of osteoblasts.[53,55]
S. moorei has been reported to be associated with oral malodor. Vancauwenberghe et al.[56] reported significant correlation between volatile sulfur compound production and the presence of S. moorei in tongue coatings. S. moorei produces volatile sulfur compounds from mucin through a process involving the cell associated β galactosidase activity obtained through exogenous source of proteases. Higher levels of volatile sulfur compound production in the presence of cysteine are observed and cysteine was transformed into hydrogen sulfide, ammonia, pyruvate by cysteine desulfhydrase. Interestingly (Smoo-c-222-2), gene of this enzyme (cysteine desulfhydrase) was identified in genome of S. moorei. It modulates volatile sulfur compound production through efficient source of proteases produced by P. gingivalis. Hence, it requires an exogenous source of proteolytic enzymes to mediate hydrolysis of proteins and glycoproteins into peptides and amino acids.
Various mouth rinses containing essential oils such as eucalyptol, menthol, and thymol are found effective against halitosis-producing Bacteria particularly polyphenols from green tea extracts exert bactericidal action against halitosis producing Bacteria: Epigallocatechin 3-gallate (EGCG), a polyphenol present in green tea disrupts the biofilm formation of S. moorei. EGCG also reduced adherence to oral epithelial cells thereby disrupting biofilm formation. S. moorei are susceptible to antimicrobial agents. Any antimicrobial therapy regimen < 1 μg/ml that is suitable for nonresistant anaerobic Bacteria will be appropriate for Solobacteriummoorei.[57,58]
Synergistes
This bacterial division Synergistes comprises ubiquitous, diverse, and uncharacterized bacterial isolates. It belongs to domain: Bacteria, phylum: Synergistetes, class: Synergistia, order: Synergistales, family: Synergistaceae, genus: Synergistes, species: P4-G18p1. Synergistes plays a pivotal role as a pathogen as it is found in a variety of anaerobic ecosystems. Numerous bacterial strains of Synergistes were isolated from human oral cavities.[58,60] Hugenholtz et al.[61] thought Synergistes to be the sole representative of a novel division Synergistetes. It belongs to the cluster A of Synergistetes phylum. It was initially unnamed when isolated from sacral wounds but Horz et al.[62] later named it as Synergistes.
Synergistes are fastidious, slow growing, obligate anaerobic nonmotile, nonpigmenting, nonspore forming Gram-negative curved bacilli (0.7–0.8 μm wide, 0.8–2.2 μm long) arranged in pairs or short chains [Figure 6]. The colonies grown on Fastidious Anaerobic Agar were 0.7–1.1 mm in diameter, circular, convex to pyramidal, shiny with consistent opacity, and off white to watery steel grey in color.[59,60]
Figure 6.
Morphology of Synergistes
The Synergistes groups of organisms were retrieved by 16SrRNA sequences. Phylotypes have been isolated from sites with marginal periodontitis, endodontic infections, apical periodontitis, and dental caries. Fluorescent in situ hybridization was also used for Synergistes isolation from subgingival plaque.[59,60,61] Synergistes were grown on brucella agar and incubated at 37°C under anaerobic conditions. It grows in peptone yeast, glucose, brain heart infusion broth, and produced moderately turbid suspension. Growth was not stimulated by the addition of formate or fumarate or any of the carbohydrates to broth. Synergistes is positive for very few biochemical reactions. It is negative for catalase, indole, and nitrate reduction. It is positive for growth in bile. Synergistes does not produce urea, desulfoviridin. They can hydrolyse glycine-naphthylamide and thereby produce hydrogen sulfide.[59,60,61]
Synergistes is asacharolytic and does not produce acetic acid, isovaleric acid, propionic, isobutyric acid, succinic, phenylacetic acids as end products of carbohydrate metabolism.[60,62] Synergistes are most likely to be involved in periodontal pocket anaerobic environment. They utilize arginine and histidine as major energy-yielding substrates by participating in anaerobic metabolism of proteins. Godon et al. demonstrated Synergistes in periodontal pockets >6 mm. They are implicated mainly in anaerobic infections.[63] They have also been associated with dental plaque of necrotizing ulcerative gingivitis. They produced microecological changes such as increased pocket depth, inflammation, anaerobiosis, and gingival tissue destruction.[64] Synergistes were higher in proportion in severe stages of periodontitis than in early stages of disease Synergistes are susceptible to kanamycin, ampicillin-sulbactam, amoxicillin-clavulanate, metronidazole but were resistant to Colistin and Vancomycin.[63,64]
CONCLUSION
Research has focussed on the identification of novel hidden pathogens that might contribute to the pathogenesis of periodontal disease. Inspite of advanced therapeutic modalities aimed at complete elimination of periodontal pathogens, the prevalence of periodontal diseases is increasing. To develop effective therapeutic approaches, it requires identification of key and accessory pathogens and the role played by them in periodontal pathogenesis. These novel pathogens were grouped into high (Firmicutes), moderate (Synergistes, F. alocis, S. sputigena, T. lecithinolyticum), low (D. pneumosintes and M. dentalis) based on their association with periodontal disease. Thus revealing and understanding the role played by these pathogens in periodontal plethora is prudent for better prevention of periodontal disease.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 3.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–83. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park OJ, Yi H, Jeon JH, Kang SS, Koo KT, Kum KY, et al. Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. J Dent Res. 2015;94:921–7. doi: 10.1177/0022034515583531. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, Faveri M, Lobão E, Tamashiro N, et al. Newly identified pathogens associated with periodontitis: A systematic review. J Dent Res. 2014;93:846–58. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–44. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 7.Arora N, Mishra A, Chugh S. Microbial role in periodontitis: Have we reached the top? Some unsung Bacteria other than red comple. J Indian Soc Periodontol. 2014;18:9–13. doi: 10.4103/0972-124X.128192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uematsu H, Sato N, Djais A, Hoshino E. Degradation of arginine by Slackia exigua ATCC 700122 and Cryptobacterium curtum ATCC 700683. Oral Microbiol Immunol. 2006;21:381–4. doi: 10.1111/j.1399-302X.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- 9.Nakazawa F, Poco SE, Ikeda T, Sato M, Kalfas S, Sundqvist G, et al. Cryptobacterium curtum gen. nov. sp. nov. a new genus of gram-positive anaerobic rod isolated from human oral cavities. Int J Syst Bacteriol. 1999;49(Pt 3):1193–200. doi: 10.1099/00207713-49-3-1193. [DOI] [PubMed] [Google Scholar]
- 10.Mavrommatis K, Pukall R, Rohde C, Chen F, Sims D, Brettin T, et al. Complete genome sequence of Cryptobacterium curtum type strain (12-3) Stand Genomic Sci. 2009;1:93–100. doi: 10.4056/sigs.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aruni AW, Mishra A, Dou Y, Chioma O, Hamilton BN, Fletcher HM. Filifactor alocis – A new emerging periodontal pathogen. Microbes Infect. 2015;17:517–30. doi: 10.1016/j.micinf.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlafer S, Riep B, Griffen AL, Petrich A, Hübner J, Berning M, et al. Filifactor alocis – Involvement in periodontal biofilms. BMC Microbiol. 2010;10:66. doi: 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aruni AW, Roy F, Fletcher HM. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun. 2011;79:3872–86. doi: 10.1128/IAI.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Jotwani R, Le J, Krauss JL, Potempa J, Coventry SC, et al. Filifactor alocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infect Immun. 2014;82:1205–12. doi: 10.1128/IAI.01434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffatt CE, Whitmore SE, Griffen AL, Leys EJ, Lamont RJ. Filifactor alocis interactions with gingival epithelial cells. Mol Oral Microbiol. 2011;26:365–73. doi: 10.1111/j.2041-1014.2011.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aruni AW, Zhang K, Dou Y, Fletcher H. Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways. Infect Immun. 2014;82:3261–74. doi: 10.1128/IAI.01727-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng J, Fang B, Liao Y, Chresta CM, Smith PD, Roth JA. Apoptosis induction by MEK inhibition in human lung cancer cells is mediated by Bim. PLoS One. 2010;5:e13026. doi: 10.1371/journal.pone.0013026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Liu Y, Zhang M, Wang G, Qi Z, Bridgewater L, et al. A Filifactor alocis-centered co-occurrence group associates with periodontitis across different oral habitats. Sci Rep. 2015;5:9053. doi: 10.1038/srep09053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haapasalo M, Ranta H, Shah H, Ranta K, Lounatmaa K, Kroppenstedt RM, et al. Mitsuokella dentalis sp. nov. from dental root canals. Int J Syst Bacteriol. 1986;36:566–8. [Google Scholar]
- 20.Mitsuoka T, Tereda A, Watanabe K, Uchida K. Bacteroides multacidus, a new species from the faeces of humans and pigs. Int J Syst Bacteriol. 1974;24:35–41. [Google Scholar]
- 21.Haapasalo M, Ranta H, Shah H, Ranta K, Lounatmaa K, Kroppenstedt RM. Biochemical and structural characterization of an unusual group of gram-negative, anaerobic rods from human periapical osteitis. J Gen Microbiol. 1986;132:417–26. doi: 10.1099/00221287-132-2-417. [DOI] [PubMed] [Google Scholar]
- 22.Shah HN, Collins DM. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol. 1990;40:205–8. doi: 10.1099/00207713-40-2-205. [DOI] [PubMed] [Google Scholar]
- 23.Kim KS, Rowlinson MC, Bennion R, Liu C, Talan D, Summanen P, et al. Characterization of Slackia exigua isolated from human wound infections, including abscesses of intestinal origin. J Clin Microbiol. 2010;48:1070–5. doi: 10.1128/JCM.01576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poco SE, Jr, Nakazawa F, Ikeda T, Sato M, Sato T, Hoshino E. Eubacterium exiguum sp. nov. isolated from human oral lesions. Int J Syst Bacteriol. 1996;46:1120–4. doi: 10.1099/00207713-46-4-1120. [DOI] [PubMed] [Google Scholar]
- 25.Nakazawa F, Miyakawa H, Fujita M, Kamaguchi A. Significance of Asaccharolytic Eubacterium and closely related bacterial species in the human oral cavity. J Exp Clin Med. 2011;3:17–21. [Google Scholar]
- 26.Wade WG, Downes J, Dymock D, Hiom SJ, Weightman AJ, Dewhirst FE, et al. The family Coriobacteriaceae: Reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov. comb. nov. and Slackia heliotrinireducens gen. nov. comb. nov. and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov. comb. nov. Int J Syst Bacteriol. 1999;49(Pt 2):595–600. doi: 10.1099/00207713-49-2-595. [DOI] [PubMed] [Google Scholar]
- 27.Colombo AP, Martins do SR, Colombo AV. Molecular Detection of Human Bacterial Pathogens. 1st ed. Vol. 34. New York: CRC Press; 2011. Dialister dongyou liu; pp. 381–90. [Google Scholar]
- 28.Moore LV, Moore WE. Dialister pneumosintes. Int J Syst Evol Microbiol. 1994;44:187–92. doi: 10.1099/00207713-44-2-187. [DOI] [PubMed] [Google Scholar]
- 29.Doan N, Contreras A, Flynn J, Slots J, Chen C. Molecular identification of Dialister pneumosintes in subgingival plaque of humans. J Clin Microbiol. 2000;38:3043–7. doi: 10.1128/jcm.38.8.3043-3047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morio F, Jean-Pierre H, Dubreuil L, Jumas-Bilak E, Calvet L, Mercier G, et al. Antimicrobial susceptibilities and clinical sources of Dialister species. Antimicrob Agents Chemother. 2007;51:4498–501. doi: 10.1128/AAC.00538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumas-Bilak E, Jean-Pierre H, Carlier JP, Teyssier C, Bernard K, Gay B, et al. Dialister micraerophilus sp. nov. and Dialister propionicifaciens sp. nov. isolated from human clinical samples. Int J Syst Evol Microbiol. 2005;55(Pt 6):2471–8. doi: 10.1099/ijs.0.63715-0. [DOI] [PubMed] [Google Scholar]
- 32.Dighe AS, Shouche YS, Ranade DR. Selenomonas lipolytica sp. nov. an obligately anaerobic bacterium possessing lipolytic activity. Int J Syst Bacteriol. 1998;48(Pt 3):783–91. doi: 10.1099/00207713-48-3-783. [DOI] [PubMed] [Google Scholar]
- 33.Moore LV, Johnson JL, Moore WE. Selenomonas noxia sp. nov., Selenomonas fluggei sp. nov., Selenomonas infelix sp. nov., Selenomonas dianae sp. nov., Selenomonas artemedis sp. nov., from the human gingival crevice. Int J Syst Bacteriol. 1987;36:112–8. [Google Scholar]
- 34.Maiden MF, Tanner A, Moore WE. Identification of Selenomonas species by whole-genomic DNA probes, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, biochemical tests and cellular fatty acid analysis. Oral Microbiol Immunol. 1992;7:7–13. doi: 10.1111/j.1399-302x.1992.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 35.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, et al. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–26. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 36.Medikeri RS, Lele SV, Jain PM, Mali P, Medikeri MR. Quantification of Selenomonas sputigena in chronic periodontitis in smokers using 16S rDNA based PCR analysis. J Clin Diagn Res. 2015;9:ZC13–7. doi: 10.7860/JCDR/2015/12550.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofstad T, Naess V, Skaug N. Biological activity of lipopolysaccharides from oral Selenomonas. Scand J Dent Res. 1986;94:515–20. doi: 10.1111/j.1600-0722.1986.tb01794.x. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy LR, Carlson JR. Selenomonas sputigena septicemia. J Clin Microbiol. 1981;14:684–5. doi: 10.1128/jcm.14.6.684-685.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi BK, Paster BJ, Dewhirst FE, Göbel UB. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–95. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paster BJ, Dewhirst FE, Coleman BC, Lau CN, Ericson RL. Phylogenetic analysis of cultivable oral treponemes from the Smibert collection. Int J Syst Bacteriol. 1998;48(Pt 3):713–22. doi: 10.1099/00207713-48-3-713. [DOI] [PubMed] [Google Scholar]
- 41.Moter A, Hoenig C, Choi BK, Riep B, Göbel UB. Molecular epidemiology of oral treponemes associated with periodontal disease. J Clin Microbiol. 1998;36:1399–403. doi: 10.1128/jcm.36.5.1399-1403.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umemoto T, Nakazawa F, Hoshino E, Okada K, Fukunaga M, Namikawa I. Treponema medium sp. nov. isolated from human subgingival dental plaque. Int J Syst Bacteriol. 1997;47:67–72. doi: 10.1099/00207713-47-1-67. [DOI] [PubMed] [Google Scholar]
- 43.Wyss C, Choi BK, Schüpbach P, Guggenheim B, Göbel UB. Treponema maltophilum sp. nov. a small oral spirochete isolated from human periodontal lesions. Int J Syst Bacteriol. 1996;46:745–52. doi: 10.1099/00207713-46-3-745. [DOI] [PubMed] [Google Scholar]
- 44.Wyss C, Choi BK, Schüpbach P, Guggenheim B, Göbel UB. Treponema amylovorum sp. nov. a saccharolytic spirochete of medium size isolated from an advanced human periodontal lesion. Int J Syst Bacteriol. 1997;47:842–5. doi: 10.1099/00207713-47-3-842. [DOI] [PubMed] [Google Scholar]
- 45.Wyss C, Choi BK, Schüpbach P, Moter A, Guggenheim B, Göbel UB. Treponema lecithinolyticum sp. nov. a small saccharolytic spirochaete with phospholipase A and C activities associated with periodontal diseases. Int J Syst Bacteriol. 1999;49(Pt 4):1329–39. doi: 10.1099/00207713-49-4-1329. [DOI] [PubMed] [Google Scholar]
- 46.Wyss C. Fatty acids synthesized by oral treponemes in chemically defined media. FEMS Microbiol Lett. 2007;269:70–6. doi: 10.1111/j.1574-6968.2006.00609.x. [DOI] [PubMed] [Google Scholar]
- 47.Jun HK, Lee HR, Lee SH, Choi BK. Mapping of the proinflammatory domains of MspTL of Treponema lecithinolyticum. Microbiology. 2007;153(Pt 8):2386–92. doi: 10.1099/mic.0.2007/006650-0. [DOI] [PubMed] [Google Scholar]
- 48.Choi BK, Jung JH, Suh HY, Yoo YJ, Cho KS, Chai JK, et al. Activation of matrix metalloproteinase-2 by a novel oral spirochetal species Treponema lecithinolyticum. J Periodontol. 2001;72:1594–600. doi: 10.1902/jop.2001.72.11.1594. [DOI] [PubMed] [Google Scholar]
- 49.Lee SH, Kim KK, Choi BK. Upregulation of intercellular adhesion molecule1 and proinflammatory cytokines by the major surface proteins of Treponema lecithinolyticum, the phylogenetic group IV oral Spirochaetes associated with periodontitis and endodontic infections. Infect Immun. 2005;73:268–74. doi: 10.1128/IAI.73.1.268-276.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park KK, Heuner K, Göbel UB, Yoo YJ, Kim CK, Choi BK. Cloning and characterization of a major surface protein (MspTL) of Treponema lecithinolyticum associated with rapidly progressive periodontitis. FEMS Microbiol Lett. 2002;207:185–92. doi: 10.1111/j.1574-6968.2002.tb11049.x. [DOI] [PubMed] [Google Scholar]
- 51.Edwards AM, Jenkinson HF, Woodward MJ, Dymock D. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun. 2005;73:2891–8. doi: 10.1128/IAI.73.5.2891-2898.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SH, Kim KK, Choi BK. Upregulation of intercellular adhesion molecule 1 and proinflammatory cytokines by the major surface proteins of Treponema maltophilum and Treponema lecithinolyticum, the phylogenetic group IV oral spirochetes associated with periodontitis and endodontic infections. Infect Immun. 2005;73:268–76. doi: 10.1128/IAI.73.1.268-276.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fenno JC, Hannam PM, Leung WK, Tamura M, Uitto VJ, McBride BC. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect Immun. 1998;66:1869–77. doi: 10.1128/iai.66.5.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kageyama A, Benno Y. Phylogenic and phenotypic characterization of some Eubacterium-like isolates from human feces: Description of Solobacterium moorei Gen. Nov. Sp. Nov. Microbiol Immunol. 2000;44:223–7. doi: 10.1111/j.1348-0421.2000.tb02487.x. [DOI] [PubMed] [Google Scholar]
- 55.Haraszthy VI, Gerber D, Clark B, Moses P, Parker C, Sreenivasan PK, et al. Characterization and prevalence of Solobacterium moorei associated with oral halitosis. J Breath Res. 2008;2:017002. doi: 10.1088/1752-7155/2/1/017002. [DOI] [PubMed] [Google Scholar]
- 56.Vancauwenberghe F, Dadamio J, Laleman I, Van Tornout M, Teughels W, Coucke W, et al. The role of Solobacterium moorei in oral malodour. J Breath Res. 2013;7:046006. doi: 10.1088/1752-7155/7/4/046006. [DOI] [PubMed] [Google Scholar]
- 57.Tanabe S, Grenier D. Characterization of volatile sulfur compound production by Solobacterium moorei. Arch Oral Biol. 2012;57:1639–43. doi: 10.1016/j.archoralbio.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 58.Pedersen RM, Holt HM, Justesen US. Solobacterium moorei bacteremia: Identification, antimicrobial susceptibility, and clinical characteristics. J Clin Microbiol. 2011;49:2766–8. doi: 10.1128/JCM.02525-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vartoukian SR, Palmer RM, Wade WG. Diversity and morphology of members of the phylum “synergistetes” in periodontal health and disease. Appl Environ Microbiol. 2009;75:3777–86. doi: 10.1128/AEM.02763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siqueira JF, Jr, Rôças IN. Molecular detection and identification of Synergistes phylotypes in primary endodontic infections. Oral Dis. 2007;13:398–401. doi: 10.1111/j.1601-0825.2006.01301.x. [DOI] [PubMed] [Google Scholar]
- 61.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–74. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horz HP, Citron DM, Warren YA, Goldstein EJ, Conrads G. Synergistes group organisms of human origin. J Clin Microbiol. 2006;44:2914–20. doi: 10.1128/JCM.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belibasakis GN, Oztürk VÖ, Emingil G, Bostanci N. Synergistetes cluster A in saliva is associated with periodontitis. J Periodontal Res. 2013;48:727–32. doi: 10.1111/jre.12061. [DOI] [PubMed] [Google Scholar]
- 64.You M, Mo S, Watt RM, Leung WK. Prevalence and diversity of Synergistetes taxa in periodontal health and disease. J Periodontal Res. 2013;48:159–68. doi: 10.1111/j.1600-0765.2012.01516.x. [DOI] [PubMed] [Google Scholar]


