Abstract
Formation of a totipotent blastocyst capable of implantation is one of the first major milestones in early mammalian embryogenesis, but less than half of in vitro fertilized embryos from most mammals will progress to this stage of development. Whole chromosomal abnormalities, or aneuploidy, are key determinants of whether human embryos will arrest or reach the blastocyst stage. Depending on the type of chromosomal abnormality, however, certain embryos still form blastocysts and may be morphologically indistinguishable from chromosomally normal embryos. Despite the implementation of pre-implantation genetic screening and other advanced in vitro fertilization (IVF) techniques, the identification of aneuploid embryos remains complicated by high rates of mosaicism, atypical cell division, cellular fragmentation, sub-chromosomal instability, and micro-/multi-nucleation. Moreover, several of these processes occur in vivo following natural human conception, suggesting that they are not simply a consequence of culture conditions. Recent technological achievements in genetic, epigenetic, chromosomal, and non-invasive imaging have provided additional embryo assessment approaches, particularly at the single-cell level, and clinical trials investigating their efficacy are continuing to emerge. In this review, we summarize the potential mechanisms by which aneuploidy may arise, the various detection methods, and the technical advances (such as time-lapse imaging, “-omic” profiling, and next-generation sequencing) that have assisted in obtaining this data. We also discuss the possibility of aneuploidy resolution in embryos via various corrective mechanisms, including multi-polar divisions, fragment resorption, endoreduplication, and blastomere exclusion, and conclude by examining the potential implications of these findings for IVF success and human fecundity.
Keywords: Aneuploidy, Chromothripsis, Fragmentation, Micronuclei, Pre-implantation embryo
Introduction
The formation of a blastocyst is one of the first major landmarks in early mammalian development and yet, only 30–50 % of pre-implantation embryos from the majority of mammals will typically reach this stage following in vitro fertilization (IVF; Fig. 1; Alper et al. 2001). In contrast, approximately 80 % of mouse embryos will form blastocysts when cultured in vitro since developmental arrest prior to this stage is far less frequent in this species. Although the cause(s) of embryo arrest may differ between species, an abnormal number of whole chromosomes, or aneuploidy, is thought to be a primary determinant of whether a human embryo will progress in development (Munne et al. 1994). Additional evidence from other mammals more closely related to humans suggests that the incidence of aneuploidy is not unique to human pre-implantation development (Dupont et al. 2010), although it may occur at a higher frequency in human embryos (Vanneste et al. 2009a, 2009b; Johnson et al. 2010b; Chavez et al. 2012; Chow et al. 2014). With the exception of trisomy 13, 18, and 21 and abnormalities in the X- or Y-chromosome, the vast majority of embryonic aneuploidies involving whole chromosomes are incompatible with live birth. Notably, high infant mortality rates are also associated with trisomy 13 (Patau syndrome) and trisomy 18 (Edwards syndrome), with only a small number of cases of survival past 1 year. For IVF-derived embryos, those with aneuploidy will probably arrest prior to transfer to the mother, fail to implant if transferred, or spontaneously abort early in gestation (for a review, see Delhanty 2005). In the case of in vivo human conceptions, an aneuploid embryo might undergo arrest even before a woman knows that she is pregnant. The introduction of pre-implantation genetic screening (PGS), first by DNA fluorescent in situ hybridization (DNA-FISH; Munne and Weier 1996), then via microarray-based approaches, and more recently using next-generation sequencing (NGS; Hou et al. 2013; Fiorentino et al. 2014; Wang et al. 2014), have provided a sufficient means for aneuploidy detection in IVF embryos. However, several challenges remain, including the potential detriment of the embryo following biopsy for PGS (Scott et al. 2013a, 2013b), chromosomal mosaicism between various cells or cell populations (Kuo et al. 1998; Daphnis et al. 2005; Baart et al. 2006; Mastenbroek et al. 2007), and the effects of sub-chromosomal aberrations (Vanneste et al. 2009a, 2009b; Johnson et al. 2010b; Chavez et al. 2012; Chow et al. 2014) on subsequent development. Recent identification of common genetic variants, or DNA sequence variation, near genes in patients that are at high-risk for producing aneuploid embryos further complicates the already complex process of pre-implantation chromosomal instability (McCoy et al. 2015). Moreover, despite the higher probability that aneuploid embryos will arrest at the cleavage stage, embryos with chromosomal abnormalities can still form blastocysts and are often morphologically indistinguishable from chromosomally normal (euploid) embryos, especially when evaluated by static observations (Baltaci et al. 2006). Thus, alternative approaches are needed for embryo selection, such as time-lapse imaging and the genetic, proteomic, or metabolomic analysis of culture media or other non-invasive methods following embryo culture for the potential detection of biomarkers that are associated with aneuploidy (Chavez et al. 2012; Seli et al. 2007; Wong et al. 2010). In this review, we summarize the methods used to distinguish euploid from aneuploid embryos, the complex mechanisms underlying chromosomal instability, and how aneuploidy is dealt with once it arises across diverse mammalian species. We also discuss the clinical consequences of aneuploidy generation, particularly in the context of IVF and other assisted reproduction technologies (ARTs), and the potential implications for early embryogenesis in natural human conceptions.
Fig. 1.
Chromosomal and genetic aspects of mammalian pre-implantation development. Recent advances in a mammalian pre-implantation embryology, including but not limited to, b time-lapse image analysis have greatly contributed to our knowledge of the key chromosomal and genetic characteristics impacting each major stage of pre-implantation development. c Approximate cell number of embryos at the morula and blastocyst stages. d Embryo chromosomal integrity may be influenced by the lack of and/or inheritance of aberrant parental mRNAs, proteins, or other factors such as paternal contribution of the centrosome, which mediates the first mitotic divisions. A select group of maternal mRNAs termed maternal-effect genes is recruited for translation following fertilization, whereas the remaining maternal mRNAs are degraded. In addition to individual maternal proteins, multi-protein complexes such as the subcortical maternal complex (SCMC) are important for embryonic progression beyond the 2-cell stage and for potential prevention of mitotic aneuploidy. e Additional mechanisms can contribute to the generation of chromosome instability, particularly in human embryos, including cellular fragmentation, anaphase lagging, chromosomal non-disjunction, and the lack of cell cycle checkpoints and overexpression of cell cycle drivers. f Unlike human embryos, which respond to chromosomal aberrations by fragmenting and continuing to divide, mouse embryos appear to deal with multi- and micronuclei formation by inducing blastomere lysis. g Chromosomes that are lagging during mitotic anaphase are encapsulated by the nuclear envelope to form a micronucleus, where they tend to undergo frequent double-strand DNA breaks, resembling events described during chromothripsis. This induces repair by non-homologous end joining, an error prone mechanism that can result in sub-chromosomal arrangements. As micronuclei might merge back with the primary nucleus upon nuclear envelope breakdown, rearranged chromosomes are able to incorporate into the genome and produce de novo mutations. h Despite the high incidence of whole and sub-chromosomal instability, there is evidence that potential corrective mechanisms exist, including the restoration of euploidy upon fragment resorption, multi-polar divisions in cases of trisomic rescue, endoreduplication followed by uniparental disomy for monosomic errors, and the exclusion of aneuploid blastomeres during the morula-blastocyst transition (see Fig. 3)
Methods for the detection of aneuploidy
DNA florescent in situ hybridization
In the past, the most frequently used method for diagnosing aneuploidy was PGS of day-3 biopsied blastomeres via DNA-FISH (Munne and Weier 1996). However, DNA-FISH can result in false positive or negative results due to procedural artifacts and is limited to the simultaneous analysis of only a select number of chromosomes, typically between five and twelve of the most frequently affected (Kuo et al. 1998; Daphnis et al. 2005; Baart et al. 2006; Mastenbroek et al. 2007). Thus, several studies initially underreported the incidence of human embryonic aneuploidy since all 24 chromosomes can potentially be affected, particularly in cases of mitotic errors (Chavez et al. 2012; Chow et al. 2014). In addition, cleavage-stage human embryos exhibit high rates of mosaicism between blastomeres (Kuo et al. 1998; Baart et al. 2006), which can complicate the interpretation of ploidy results if embryos contain a mixture of aneuploid and diploid cells. As the number of cells that can be biopsied from patient embryos is limited, supernumerary cleavage-stage embryos donated with informed consent for research have provided an alternative source for aneuploidy assessment. However, fresh embryos donated to research at the cleavage stage are usually of poorer quality than embryos that were selected for transfer and thus, not representative of the embryo population as a whole.
Microarray-based technologies
More recent studies with array comparative genomic hybridization (aCGH) or single nucleotide polymorphism (SNP) arrays to evaluate all 24 chromosomes in high quality, whole, human embryos have demonstrated that 50–80 % of human embryos at the cleavage stage have one or more blastomeres that are indeed aneuploid. In addition, the incidence of aneuploidy in cleaving human embryos appears to be irrespective of fertility status, maternal age, or whether a fresh versus frozen/thawed IVF cycle, since a similar frequency is observed in embryos from fertile couples, young women, and following cryopreservation (Vanneste et al. 2009a, 2009b; Johnson et al. 2010b; Chavez et al. 2012; Chow et al. 2014). Moreover, the incidence of aneuploidy in vitro is thought to reflect the situation in vivo considering that 20–30 % of natural human conceptions are estimated to result in a live birth (Zinaman et al. 1996; Macklon et al. 2002; Slama et al. 2002) and chromosomal abnormalities have been reported in the majority of spontaneous miscarriage cases (Fritz et al. 2001; Benkhalifa et al. 2005). This is in contrast to lower mammalian species, particularly the mouse, which is estimated to exhibit approximately 1 % embryonic aneuploidy rates, depending on the strain (Lightfoot et al. 2006). However, it should be noted that microarrays for the detection of chromosomal copy number are far more utilized for humans than for other mammalian species due, in large part, to PGS of human embryos in IVF clinics and genetic testing centers. Thus, the overall aneuploidy rates via “whole-genome” cytogenetic methods in pre-implantation embryos from non-human species, especially those more closely related to humans, remains to be determined. Aneuploidy analysis via DNA-FISH of rhesus macaque embryos produced in vitro (Dupont et al. 2010) and the frequent observations of abnormal nuclear structure in blastomeres from rhesus embryos as compared with other mammalian species (Chavez et al. 2012) does suggest that the incidence of chromosomal abnormalities in non-human primates is similar to that in humans.
Given the high incidence of mosaicism in cleavage-stage human embryos shown by aCGH (Vanneste et al. 2009a, 2009b; Chavez et al. 2012; Chow et al. 2014) and the potential detriment of polar body and blastomere biopsies on embryonic development, alternative approaches have recently been implemented to evaluate aneuploidy. These include extended culture of embryos to the blastocyst stage and analysis of chromosomal status via trophectoderm biopsy, wherein multiple extra-embryonic cells are removed from the outer layer of the embryo. Whereas human blastocysts are thought to exhibit less aneuploidy than cleavage-stage embryos (Magli et al. 2000), the presence of two or more cell populations with a different chromosomal make-up has been reported at the blastocyst stage and may be more prevalent in embryos from women of advanced maternal age (Fragouli et al. 2011; J. Liu et al. 2012). In addition, it is important to differentiate between the proportion of aneuploid cells in an embryo versus the percentage of embryos with mosaicism, as the latter increases at the blastocyst stage (van Echten-Arends et al. 2011). Besides mosaicism percentage, there are additional potential risks associated with prolonged embryo culture such as the introduction of epigenetic changes, monozygotic twinning, and other factors that may disrupt embryo integrity in humans and other mammalian species (Peramo et al. 1999; Khosla et al. 2001; Fernandez-Gonzalez et al. 2009; Katari et al. 2009; Kallen et al. 2010). Nevertheless, extended culture may prevent the selection of cleavage-stage embryos that are destined to arrest, which is why blastocyst transfer with or without the use of PGS has increased (Munne et al. 1994; Alper et al. 2001; Schoolcraft et al. 2010).
Quantitative polymerase chain reaction and DNA sequencing
In response to claims that DNA-FISH or whole-genome amplification in conjunction with SNP arrays (Northrop et al. 2010; Treff et al. 2011; Macaulay and Voet 2014) is inaccurate, time consuming, and not conducive to fresh embryo transfers, particularly at the blastocyst stage, fluorescent real-time quantitative polymerase chain reaction (qPCR) has been developed as an alternative to microarray-based approaches for PGS (Treff and Scott 2013). Whereas qPCR provides a more rapid method for ploidy assessment, the multiplex amplification and relative quantitation of only 96 presumably single copy loci (four on each chromosome) does not allow for the identification of copy number variants in specific chromosomal segments, termed segmental aneuploidy, in embryos. Given that certain sub-chromosomal aberrations, such as homozygous or hemizygous deletions, large duplications or amplifications, and regions of de novo uniparental isodisomy may induce embryonic/fetal lethality and are one of the major causes of developmental disorders in offspring that survive to birth (Hochstenbach et al. 2011; Vanneste et al. 2012), it is important to evaluate both whole and sub-chromosomal instability in embryos. Moreover, as this technology is still impacted by mosaicism and has only been applied to trophectoderm biopsies (Scott et al. 2013a, 2013b; Werner et al. 2014; Rienzi et al. 2015), which generally consist of between 5 and 10 cells, we do not yet know whether qPCR can reliably detect aneuploidy in single cells.
Next-generation whole-genome DNA sequencing (NGS or DNA-Seq) has also provided an additional platform besides qPCR and array-based methods to assess aneuploidy comprehensively in human embryos, including at the single-cell level (Hou et al. 2013; Fiorentino et al. 2014; Wang et al. 2014). Since aCGH covers approximately 30 % of the genome and can only detect gross chromosomal abnormalities at a lower threshold of approximately 1 MB (Bi et al. 2012), sub-chromosomal aberrations that are smaller than this resolution and missed by aCGH would be detectable by NGS in theory. In addition to examining copy number variation at a single nucleotide level, DNA-seq is also more cost-effective than aCGH since 48–96 samples or more can be simultaneously sequenced with high coverage by multiplexing with barcoded adapters. Furthermore, DNA-Seq does not require a priori knowledge of the genome being sequenced and thus, can be used for de novo genome assembly of multiple species (Hurd and Nelson 2009). Lastly, NGS also allows for the analysis of repetitive DNA sequences (Gisselsson et al. 2001), balanced chromosomal rearrangements, and an initial map of insertion and deletion (INDEL) variation in the genome (Talkowski et al. 2011). Until bioinformatics pipelines and NGS analysis software are developed and simplified for use in IVF clinics, however, microarray-based technologies are likely to continue to dominate the PGS field (Van der Aa et al. 2013).
Non-invasive methods for aneuploidy detection
Time-lapse imaging
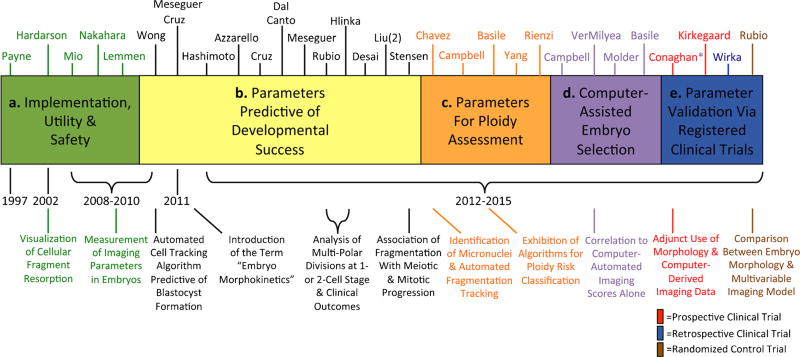
Despite increased accessibility of PGS for IVF patients, several randomized control trials (RCTs) using DNA-FISH to evaluate the ploidy status of a select number of chromosomes have demonstrated no benefit or a significant reduction in live birth rates from IVF cycles with PGS versus those without PGS (Mastenbroek et al. 2007; Hardarson et al. 2008; Staessen et al. 2008). More recent studies suggest that PGS via qPCR, microarrays, or NGS may improve IVF outcomes for certain patients, but this has not been evaluated by an RCT (Harper and Sengupta 2012). Nonetheless, embryo biopsy for PGS is invasive and potentially detrimental to subsequent development; this has led to the exploration of alternative non-invasive means of accessing embryo quality and ploidy. One such approach has been the implementation of time-lapse imaging to monitor embryos throughout development rather than at specific time points that may not reveal subtle differences in embryo behavior (Fig. 1). Whereas initial studies focused on the utility and safety of time-lapse monitoring (TLM; Payne et al. 1997; Hardarson et al. 2002; Lemmen et al. 2008; Mio and Maeda 2008; Nakahara et al. 2010), once TLM was determined not to be detrimental to embryo development, several groups began to investigate whether there were morphological, spatial, and/or temporal correlates between imaging behavior and embryo quality (Wong et al. 2010; Cruz et al. 2011; Meseguer et al. 2011; Fig. 2). In 2010, Wong and colleagues demonstrated that TLM could be used to predict blastocyst fate prior to embryonic genome activation (EGA) by measuring the duration of the first cytokinesis and the time between the second and third mitotic divisions (Wong et al. 2010). Since this initial report, other studies have confirmed the importance of early cleavage divisions and identified additional imaging parameters predictive of developmental success (Cruz et al. 2011; Meseguer et al. 2011; Azzarello et al. 2012; Cruz et al. 2012; Dal Canto et al. 2012; Hashimoto et al. 2012; Hlinka et al. 2012; Meseguer et al. 2012; Rubio et al. 2012; Desai et al. 2014; Y. Liu et al. 2014a, 2014b, 2015; Stensen et al. 2015; Fig. 2). Whether the first three mitotic divisions are similarly prognostic for mammalian species other than the human remains to be determined, but an examination of early mitotic timing in murine, bovine, and rhesus monkeys has suggested that this may be the case (Pribenszky et al. 2010; Sugimura et al. 2012; Burruel et al. 2014).
Fig. 2.
Evolutionary timeline of time-lapse imaging for embryo assessment. A timeline from 1997 to the present day showing the various “eras” of time-lapse image analysis with relevant references above and some of the major milestones below in the evaluation of embryo viability. The first time-lapse publications a examined the implementation, utility, and safety of this technology to monitor embryo development. Subsequently, studies began identifying cellular parameters predictive of success in the blastocyst stage b, followed by the identification of parameters predictive of embryo ploidy status c. More recent reports d have focused on the development of computer-assisted algorithms for automatic embryo assessment and classification. Other studies e have clinically applied and validated these parameters or newly identified parameters in either retrospective or prospective registered clinical trials. The first randomized control trial evaluating time-lapse image analysis versus traditional IVF assessment was recently published, with others likely to follow
In a follow-up study, Chavez et al. (2012) determined that the timing of the first three mitotic divisions, in conjunction with an assessment of a dynamic process called cellular fragmentation, might also be used largely to distinguish euploid from aneuploid human embryos at the cleavage stage. It is important to note, however, that approximately 12 % of aneuploid embryos from this study exhibited normal parameter timing and no cellular fragmentation, indicating that certain embryos can appear euploid at least up to the 4-cell stage. Upon further examination, these embryos all had incurred mitotic errors with 4 chromosomes or fewer affected, suggesting that if they were allowed to progress in development, perhaps additional later imaging parameters might be used to distinguish chromosomally normal and abnormal embryos. Indeed, other imaging parameters such as the time to 5th cell, initiation of cavitation, and the completion of blastulation may also differentiate between euploid and aneuploid cleavage-stage human embryos and blastocysts (Campbell et al. 2013a, 2013b; Basile et al. 2014; Yang et al. 2014; Fig. 2). Although it is known that aneuploidy often results in the absence of or delayed blastulation (Campbell et al. 2013a, 2013b; Kroener et al. 2012), no study has yet observed a direct correlation between the first cell division in aneuploid embryos and blastocyst formation, because this parameter was either not evaluated or not measured from a shared start point such as the time from insemination (Basile et al. 2014; Yang et al. 2014; Rienzi et al. 2015). This is imperative since meiotic errors are likely to cause a delay in the first mitotic division. In addition, cellular fragmentation, which frequently occurs at the 1- to 2-cell stage, appears to be associated with spindle formation and progression though meiosis and mitosis (Chavez et al. 2012; Stensen et al. 2015). Recently, Vera-Rodriguez and colleagues (2015) demonstrated that the duration of the first mitotic division is predictive of embryo chromosomal status up to approximately the 8-cell stage, but whether this imaging parameter is similarly predictive beyond 8-cells is unknown. Additional studies have also reported a significant difference in the duration of, or synchrony between, the second and subsequent mitotic divisions in euploid versus aneuploid embryos that reach the blastocyst stage, although these findings are not entirely clear given that others have observed no such correlation (Campbell et al. 2013a, 2013b; Basile et al. 2014; Yang et al. 2014; Rienzi et al. 2015). Taken together, these data suggest that aneuploidy can have differential effects on pre-implantation development and probably depends on the number of abnormal chromosomes and at which stage the error(s) occurred. Perhaps, future studies with high-resolution live-cell imaging of chromosome dynamics will more precisely determine how chromosomal mis-segregation impacts cell division and embryo behavior.
Of course, clinical validation of these findings is required, and recent retrospective or prospective registered clinical trials of patient embryos have confirmed the importance of certain cell cycle parameters in predicting blastocyst formation and/or aneuploidy risk (Conaghan et al. 2013; Kirkegaard et al. 2013; Athayde Wirka et al. 2014; Fig. 2). While there is some debate as to whether TLM is actually beneficial for embryo selection (Kaser and Racowsky 2014) and the assessment of ploidy status (Rienzi et al. 2015), the first RCT evaluating implantation rates, ongoing pregnancy, and early pregnancy loss suggests that TLM is more effective than conventional IVF techniques (Rubio et al. 2014). However, whether dynamic imaging analysis can also positively impact live birth rates, particularly in cases of single embryo transfers (SETs), remains to be determined. Furthermore, it is important to note that when comparing these time-lapse studies, any discrepancy in findings may be due to differences in the stage of pre-implantation development evaluated, whether only a portion of or whole embryos were analyzed, and how the imaging parameters were measured. Indeed, the use of a common procedural start point such as the time of intra-cytoplasmic sperm injection can have confounding effects on the measurement of other overlapping parameters, as recently shown by a study that implemented a biological start point instead (Y. Liu et al. 2014a). Lastly, which method(s) of ploidy assessment used is also a consideration, since array-based and sequencing approaches are far more comprehensive than qPCR and DNA-FISH of limited targets which might influence the interpretation of results. Nevertheless, despite evidence that the measurement of developmental kinetics can differentiate a large proportion of aneuploid embryos from those that are euploid, chromosomally normal and abnormal embryos that appear and behave similarly may be indistinguishable and still require PGS and/or additional screening (Chavez et al. 2012; Yang et al. 2014).
Based on observations that the measurement of imaging parameters can be diverse, subjective, and rather time-consuming, especially in cases of abnormal cell divisions, several studies have investigated the use of computer-assisted technologies for embryo selection (Campbell et al. 2013a, 2013b; Conaghan et al. 2013; Basile et al. 2014; VerMilyea et al. 2014; Molder et al. 2015; Fig. 2). The initial focus of these reports was on the implementation of algorithms for automated cell tracking and/or aneuploidy risk models for embryo classification. In addition, Conaghan and colleagues (2013; indicated by asterisk in registered clinical trials) also demonstrated that morphology plus automated cell tracking improved embryo selection particularly among high quality embryos and reduced inter-individual variability when used adjunctively. Additional studies have concentrated on the correlation between computer-derived embryo scoring and clinical pregnancy rate with promising results as well as on other algorithms for the classification of embryo potential or ploidy status (Campbell et al. 2013a, 2013b; Basile et al. 2014; VerMilyea et al. 2014; Molder et al. 2015). Given the inherent diversity in embryo kinetics and the vast differences in IVF routine between clinics, it will be important to determine whether clinic-specific or even patient-specific algorithms are necessary for embryo classification.
Embryo spent media
Alternatively, other non-invasive approaches, including the analysis of the media that was used for embryo culture, known as “spent media”, have also been explored in order to potentially diagnose the molecular health of developing embryos. The goal of these studies has been to identify biomarkers in spent media using advanced genetic, proteomic, or metabolomic technologies and to improve the assessment of embryo viability and/or ploidy status. If spent media analysis can support or replace embryo biopsies, which are technically challenging, invasive, and pose a risk for misdiagnosis because of chromosomal mosaicism, then perhaps the non-invasive screening of spent media could reduce implantation failures and increase IVF success. Whereas initial embryo metabolism studies evaluated specific molecules such as glucose, lactate, and pyruvate (Hardy et al. 1989; Gott et al. 1990; Conaghan et al. 1993; Turner et al. 1994; Gardner et al. 2001), more recent studies have performed metabolomics profiling of culture media (Seli et al. 2007, 2008, 2010b; Picton et al. 2010). However, since all of these studies used methods capable of identifying a limited number of metabolites involved in known cellular processes, the current focus has been on the discovery of important and yet to be characterized metabolomics pathways that allow for the simultaneous analysis of several hundred metabolites (Nadal-Desbarats et al. 2013). In a similar targeted-based approach, protein assessment of embryo spent media first concentrated on specific molecules such as HOXA10 (Sakkas et al. 2003) and leptin (Cervero et al. 2005), but more recent studies have been extended to the analysis of multiple known and unknown molecules by protein microarrays and mass spectrometry, respectively (Katz-Jaffe et al. 2006; Dominguez et al. 2010). By combining mass spectrometry with liquid chromatography for greater sensitivity in high-throughput samples, an additional study identified nine proteins (including lipocalin-1) that were differentially expressed between euploid and aneuploid blastocysts, but this finding has not yet been clinically validated (McReynolds et al. 2011). Thus, whether there is a clear association between a specific metabolomic or proteomic signature in spent media and ploidy status in embryos remains to be determined.
In addition to proteomic and metabolomic profiling, genetic analysis of spent media following embryo culture has also been evaluated as a non-invasive means for aneuploidy and mutation assessment. Notably, both nuclear and mitochondrial DNA have been detected in media from embryos that undergo blastomere or cellular fragmentation breakdown (Stigliani et al. 2013; Assou et al. 2014). In another study by Wu et al. (2015), the quantification of cell-free DNA (cfDNA) was measured in the spent media of embryos from couples at risk of the inherited disorder, α-thalassemia-SEA and shown to have a high efficiency for the detection of α-thalassemia-SEA in cfDNA at the blastocyst stage. Stigliani and colleagues (2013) also found that mitochondrial DNA quantity in culture media is significantly correlated with maternal age and the degree of cellular fragmentation and thus, may be an indication of poor embryo quality and aneuploidy. Lastly, “-omics” profiling of follicular fluid and genetic analysis of the oocyte-surrounding cumulus cells has been investigated for the purpose of pre-implantation screening and is reviewed extensively elsewhere (Assou et al. 2010; Seli et al. 2010a; Krisher et al. 2015). Taken together, we suggest that although these non-invasive aneuploidy detection methods may not serve as a substitute for PGS, especially when used alone, the analysis of a panel of biomarkers in conjunction with other approaches such as cfDNA or TLM analysis may assist in embryo diagnostics.
Potential mechanisms of aneuploidy generation
Introduction to meiosis and mitosis
To determine the possible causes of aneuploidy in pre-implantation embryos, research has primarily focused on the cellular and molecular players involved in the fidelity of correct chromosome segregation during meiosis and/or mitosis. As a brief summary, meiosis begins in spermatocytes and primary oocytes, wherein the DNA replicates to form pairs of homologous chromosomes (one paternal, one maternal), each containing sister chromatids. During the synapsis stage of prophase I, homologous chromosome pairs coalesce tightly through a zipper-like structure called the synaptonemal complex (Yuan et al. 2002; Page and Hawley 2004), and at the pachytene stage, non-sister homologous chromatids crossover, recombine, and exchange maternal and paternal chromosome regions at chiasmata loci. When all homologs have completed their crossover events, and once the meiotic recombination cell cycle checkpoints are fulfilled (Hochwagen and Amon 2006), the cell cycle progresses to the diplotene stage, during which the synaptonemal complex disassembles to leave bivalents tethered at chiasmata loci (Kleckner 2006). At this stage of meiosis, primary oocytes in neonatal ovaries undergo dictyate arrest and will not resume meiosis until just prior to ovulation after they are recruited during a menstrual cycle (Jones 2008). Upon exiting prophase I and progressing into metaphase I, homologous chromosome pairs still connected at chiasmata loci align on the metaphase plate. The protein structures called kinetochores at the centromeres of homologous pairs form amphitelic microtubule attachments, whereby microtubules emanating from centrioles on opposite spindle poles attach at sister kinetochores and create an evenly spaced bi-polar “tug” across the centromere of the chromosomes. Once tension is detected on both homologous kinetochores of all chromosome pairs, the microtubules shorten, the chiasmata are eliminated as the crossovers resolve, and the two homologs separate to opposite poles (Petronczki et al. 2003). For the oocyte, the first cytokinesis is highly asymmetrical, resulting in the extrusion of a small diploid polar body that is not able to participate in fertilization, whereas meiosis I spermatocytes divide symmetrically and have the potential to form functional gametes. The second meiotic division (MII) is similar to mitosis in that the now genetically unique sister chromatids become separated upon degradation of securin by the anaphase-promoting complex (APC) and dissolution of the glue-like cohesion protein complexes by the separase protease (Petronczki et al. 2003; Peters 2006). After clearing the spindle assembly checkpoint, the sister chromatids are pulled apart at the kinetochore by opposing amphitelically attached microtubules, and since the DNA was not replicated in MII, the resulting daughter cells are haploid (Petronczki et al. 2003). Whereas four identical round haploid spermatids will go on to differentiate into highly specialized spermatozoa, the extrusion of the haploid second polar body in MII oocytes does not occur until fertilization by a mature sperm (Saunders et al. 2002; Fan et al. 2003).
Maternal origins of aneuploidy
Given the complexity of meiosis during oogenesis as compared with spermatogenesis, it is not surprising that the majority of meiotic errors can be traced back to the maternal chromosomes (Hassold and Hunt 2001). Indeed, cytogenetic analysis of spontaneous miscarriages, stillbirths, and new-borns determined that up to 70 % of meiotic aneuploidies originated from the oocyte, depending on which chromosome(s) was affected (Lamb et al. 2005; Sherman et al. 2006; Handyside 2012). This percentage, of course, does not take into account pregnancies that spontaneously aborted during the first few weeks of gestation and therefore, were not detected. Nonetheless, IVF and other ART procedures have suggested that meiotic error rates in women are relatively high in comparison to other mammalian species, with approximately 20–25 % of human oocytes identified as aneuploid (Hassold and Hunt 2001). Estimates of aneuploidy in mouse oocytes, on the other hand, are typically between only 0.05 and 1 %. The incidence of human aneuploidy also increases with maternal age, since as many as 50 % of pregnancies are diagnosed as aneuploid in women over the age of 40 (Munne et al. 1995; Nagaoka et al. 2012). However, mosaic aneuploidies, which are mitotically derived, are generally not correlated with advanced maternal age (Silber et al. 2003; Munne 2006; Munne et al. 2007), except in cases of mitotic non-disjunction (Munne et al. 2002).
Although meiotic errors can occur during either fetal oogenesis or the resumption of meiosis several decades later, the considerable time between the initiation and the completion of meiosis is likely to contribute to the alteration or degradation of factors important for chromosome fidelity. Since the major wave of EGA does not occur until approximately the 4- to 8-cell stage in human embryos, the first cell divisions are highly dependent on the large pool of maternal mRNAs and proteins provided by the oocyte (Braude et al. 1988). This is supported by findings that the oocyte cytoplasm alone, which is devoid of nuclear content, is sufficient to reprogram a somatic cell capable of development to the blastocyst stage (Tachibana et al. 2013). Thus, defective maternal resources, together with the relaxation or absence of cell cycle checkpoints discussed below, may contribute to aneuploidy and allow embryos to proceed in mitosis (Harrison et al. 2000; Los et al. 2004; Mantikou et al. 2012). By definition, these factors are termed maternal-effect genes given that they were initially transcribed during oogenesis. In the mouse, several maternal-effect genes have been identified (for a review, see Li et al. 2010) and include factors associated with remodeling sperm chromatin, EGA, and de novo DNA methylation (Li et al. 2010; Fig. 1). As remodeling of the male genome is a complicated process that can lead to DNA breaks (Derijck et al. 2006), the maternal-derived ubiquitin-conjugating enzyme, UBE2A (Roest et al. 2004), repairs sperm chromatin in preparation for EGA. Other maternal-effect genes such as ZFP36L2 ring finger protein-like 2 (Ramos et al. 2004) and autophagy protein ATG5 (Tsukamoto et al. 2008) are involved in maternal mRNA and protein degradation, respectively.
Notably, the expression of certain maternal RNAs and proteins can persist beyond the oocyte-to-embryo transition. To complicate matters, particular maternal-effect genes may not be expressed until after EGA, making it difficult to differentiate between maternal and embryonic effects (Li et al. 2010). Multi-protein complexes of maternal origin, such as the sub-cortical maternal complex (SCMC), have also been shown to be important for embryonic progression beyond the 2-cell stage, at least in mouse embryos (Li et al. 2008; Fig. 1). The SCMC is a group of at least five proteins, including FLOPED/Ooep, MATER/Nlrp5, TLE6, Padi6, and FILIA/Khdc3, which localize to cytoplasmic lattices in mouse oocytes (Yurttas et al. 2008). Of these, Mater/Nlrp5 was one of the first proteins to be identified (Tong et al. 2000) and its depletion prevents SCMC formation, resulting in pre-implantation embryonic lethality in both mice (Li et al. 2008) and rhesus monkeys (Wu 2009). Similarly, embryonic lethality is also observed in embryos from mice lacking Floped/Ooep (Tashiro et al. 2010). Depletion of FILIA, on the other hand, in mouse oocytes does not lead to embryonic lethality, but rather to mitotic aneuploidies. This is most likely due to the role of FILIA in assembling key spindle components such as AURKA, PLK1, and γ-tubulin to the microtubule-organizing center as well as MAD2 to the kinetochores (Zheng and Dean 2009). In a study by Yu et al. (2014), the SCMC was also shown to control symmetrical divisions in mouse zygotes by influencing the central position of the mitotic spindle via Cofilin, a key regulator of F-actin assembly. These observations may be explained by evidence that Tle6-null mice exhibit unequal-sized blastomeres and cellular fragmentation, a phenotype more often observed in higher mammals such as cattle, rhesus monkeys, and human. To our knowledge, however, only a few maternal-effect genes have been studied in relation to mitotic aneuploidy in humans, even though SCMC members appear to be conserved across mammalian species (Li et al. 2010; Ohsugi et al. 2008; Akoury et al. 2015; Pierre et al. 2007). Since SCMC proteins play a role in maintaining both euploidy (Zheng and Dean 2009) and symmetrical divisions (Yu et al. 2014) in mouse pre-implantation embryos, it will be important to determine whether they have analogous functions in other mammalian species (Mantikou et al. 2012).
In addition to the potential inheritance of aberrant maternal RNA and proteins, there is also evidence that suboptimal maternal genetic variants or deleterious mutations within meiosis- or mitosis-associated genes and/or their regulators may also lead to abnormal chromosome segregation and cell division. Recently, McCoy and colleagues (2015) identified a genetic variant of polo-like kinase 4 (PLK4), a gene known to cause aneuploidy upon disruption of the centrosome cycle (Ko et al. 2005); this variant occurred at higher frequency in women who produced aneuploid embryos solely due to mitotic errors. Whether there are other genetic variants associated with mitotic errors remains to be determined, but this haplotype resides in a region of low recombination on chromosome 4 and includes additional genes such as INTU, SLC25A31, HSPA4L, MFSD8, LARP1B, and PGRMC2 (McCoy et al. 2015). Although these genes do not have known functions in maintaining proper chromosome segregation, it will be important to determine how their established roles in cell polarity, heat shock, and ATP translocation might relate to chromosomal instability in embryos (Chen et al. 2012). Perhaps, extensive research in the cancer field will lend insight into the key signaling pathways that are dysregulated to induce embryonic chromosomal instability and aneuploidy (Orr and Compton 2013).
Lastly, potential accumulation of maternal mitochondrial DNA (mtDNA) mutations over the several years between fetal oogenesis and meiotic resumption may also lead to defective mitochondria and possible aneuploidy generation. Whereas an association between maternal mtDNA and chromosomal instability is still speculative at the moment, oocytes from women of advanced maternal age have been shown to exhibit increased rates of mtDNA mutations (Keefe et al. 1995) and lower ATP production (Van Blerkom et al. 1995). Moreover, a significant decrease in mitochondrial activity has been documented in mosaic human embryos with chaotic profiles in comparison with euploid or mosaic embryos with non-chaotic profiles (Wilding et al. 2001) and in individual aneuploid versus euploid blastomeres from the same embryo (Picton et al. 2010). Since ATP production is required for the movement of microtubules during anaphase and cytoskeletal actin filament contraction during cytokinesis (Wilding et al. 2001; Eichenlaub-Ritter et al. 2004), maternal mtDNA mutations might indirectly impact chromosome segregation and cell division. Thus, the replacement of dysfunctional mitochondria with healthy mitochondria by nuclear transfer into an enucleated oocyte may provide an alternative means, especially for women of advanced maternal age, to successfully conceive as previously shown in both mice and cattle (Takeuchi et al. 2005; Chiaratti et al. 2011).
Paternal contribution to aneuploidy
Although maternal meiotic errors are far more frequent than those of paternal origin and increase with age, mitotically derived mosaic aneuploidies in embryos are generally not associated with advanced maternal age. This is supported by studies that determined that mitotic errors are just as frequent, if not more frequent, than meiotic errors in embryos from women of average maternal age (Vanneste et al. 2009a, 2009b; Chavez et al. 2012; Chow et al. 2014). Thus, when mitotic mosaicism is detected in embryos, it may be due to defects in the sperm used for fertilization (Silber et al. 2003; Munne 2006; Munne et al. 2007). Aside from providing the paternal genome and phospholipase C-zeta, which releases the oocyte from its MII arrest by activating calcium oscillations (Whitaker 2006; Yoon et al. 2008), the sperm also contributes its centrosome in the majority of mammalian species (Fig. 1). In contrast to all mammalian species studied to date, the centrosome is maternally inherited in the mouse embryo (Schatten et al. 1991), a finding that may help explain the relatively low aneuploidy rates observed in mouse embryos (Lightfoot et al. 2006). The sperm centrosome, consisting of two centrioles, duplicates and separates around the time of pronuclear syngamy in zygotes to generate spindle microtubules that pull the chromosomes apart during the first embryonic divisions (Sathananthan et al. 1991; Palermo et al. 1994). Palermo et al. (1994) demonstrated that the presence of two sperm in human zygotes led to mosaicism in embryos, whereas monospermic embryos and those that inherited an extra haploid set of chromosomes from the mother were euploid. Moreover, dispermic embryos also exhibit multi-polar mitotic spindles, and disruption of the sperm centrosome induces mosaicism in embryos (for a review, see Palermo et al. 1997). As further proof that the centrosome is paternally derived, the spindle complex in mature human oocytes has been shown to lack both centrioles and asters at the poles (Sathananthan et al. 2006). Studies have also found that certain forms of male infertility are due to centriole abnormalities (Rawe et al. 2002) and may contribute to mosaicism in embryos (Obasaju et al. 1999). Additional evidence is provided by observations of increased mitotic mosaicism in embryos fertilized by testicular sperm extraction from men with non-obstructive azoospermia and suggests that non-ejaculated spermatozoa may be immature and less effective in organizing the first mitotic spindle (Silber et al. 2003). Taken together, these studies suggest that further consideration should be given to paternal age and the potential inheritance of an age-related dysfunctional centrosome, similar to how an association between advanced maternal age and meiotic errors is regarded (Nagaoka et al. 2012).
Despite substantial evidence that the centrosome is paternally contributed in the majority of mammalian species, an apparent contradiction to this notion are findings of mitotic divisions following parthenogenetic activation. Indeed, parthenotes can develop to the blastocyst stage and are a source of highly proliferative parthenogenetic embryonic stem cells (pESCs; for a review, see Daughtry and Mitalipov 2014). Data from Brevini et al. (2009, 2012) have demonstrated that porcine parthenotes do indeed contain centrioles, but in an excessive number, suggesting that de novo centriole generation is dysregulated. Similar findings were observed in both porcine and human pESCs as well as observations of multipolar spindles and high rates of aneuploidy when evaluated by DNA-FISH. Furthermore, Q. Liu et al. (2014) showed that blastomeres from porcine parthenogenetically activated embryos were predominately aneuploid or haploid when tested by karyotyping. As further support for this, all biopsied blastomeres in spontaneous parthenogenetically activated embryos from an infertile patient were identified as aneuploid by DNA-FISH and SNP-microarray (Combelles et al. 2011). This suggests that, although the source of centrioles in parthenotes appears to be maternal, paternally inherited centrosomes are more proficient in early mitotic divisions.
In addition to the transfer of defective centrosomal components, the lack of and/or inheritance of aberrant paternal mRNAs, proteins, or other factors may also influence the chromosomal integrity of embryos (Xanthopoulou et al. 2012; Burruel et al. 2014; Vera-Rodriguez et al. 2015) (Fig. 1). Since sperm are generally considered transcriptionally silent, RNAs detected in the paternal gametes were initially assumed to be either products of transcript degradation or simply contaminants from surrounding cells in the testes or epididymis (Pessot et al. 1989). However, more recent studies have shown that sperm may actually contain up to thousands of different RNAs that retain both coding and specific non-coding RNAs, some of which have known roles in mitotic progression and the prevention of aneuploidy (Taylor et al. 1997; Dawlaty et al. 2008; Sendler et al. 2013). One of these microRNAs, miRNA-34c, is detected in zygotes, but not parthenotes, indicating it is inherited from the sperm. By blocking paternal miR-34c in the zygote, W.M. Liu and colleagues (2012) showed that the first cleavage division was significantly suppressed, at least in mouse embryos. Male mice deficient in all three members of the miR-34c family, however, are reported to be both viable and fertile, although the latter data was not shown (Concepcion et al. 2012). Nevertheless, additional studies also suggest that a small population of paternal transcripts inherited from human sperm is important for sustaining early embryogenesis beginning at the 3- to 4-cell stage in human pre-implantation development (Taylor et al. 1997; Sendler et al. 2013). The presence of functional sperm RNA indicates that, analogous to maternal-effect genes, males also assist in the oocyte-to-embryo transition. Determination as to which sperm RNAs persist and how their integrity might be influenced by ageing or environmental factors will be important.
Anaphase lagging and non-disjunction of chromosomes
Even with the presence of parental factors for the maintenance of euploidy, meiosis and mitosis are obviously complex cellular processes with several points during which errors could arise and impact subsequent development. As discussed above, if chromosomal mis-segregation occurs during meiosis, zygotes will inherit meiotic-based aneuploidies from the parental gamete(s) (Yuan et al. 2002), whereas chromosomal errors that take place in the first cell divisions lead to the propagation of mitotic-induced aneuploidies in the cleaving embryo. Regardless of whether meiotic or mitotic in origin, the majority of mis-segregation errors are thought to be the result of two different mechanisms: either lagging chromosomes during anaphase or non-disjunction (Fig. 1). Anaphase lagging occurs when a chromatid fails to connect or is dissociated from the spindle apparatus and is lost upon cytokinesis, resulting in a monosomy in one of the two daughter cells during meiosis or mitosis. For meiosis, this might occur as early as the MI stage if the spindle assembly checkpoint is bypassed, leading to precocious centromere division and abnormal chromatid allocation to the first polar body and MII oocyte (Angell 1997). Lagging chromosomes in anaphase may be due to the depletion and/or reduced stringency of the spindle assembly complex in oocytes and early preimplantation embryos, which rely on maternally supplied spindle assembly complex mRNAs and proteins until EGA. Evidence for this is provided by mouse embryo studies showing that the knockdown of the spindle assembly complex components, Bub3, BubR1, and Mad2, leads to improper chromosome segregation, aneuploidy generation, and the formation of micronuclei, or small nuclei that encapsulate chromosomes or pieces of chromosomes that failed to incorporate into the primary nucleus of one of the daughter cells during division (Wei et al. 2011). Furthermore, additional studies have demonstrated that chromosome lagging commonly arises from the formation of merotelic attachments in mitosis, whereby a single kinetochore on one side of the chromosome is attached to microtubules emanating from both spindle poles (for a review, see Gregan et al. 2011). Whereas lagging chromosomes arising from the formation of merotelic attachments have been show to occur in oocytes during meiosis (Kitajima et al. 2011; Lane et al. 2012), the exact mechanism(s) of chromosomal mis-segregaton during early mitotic divisions in mammalian embryos remains an enigma. This may be because, unlike other mitotic cell types, the cleavage-stage embryo undergoes mitosis to produce cells of decreasing cell size with each subsequent division; this can impact spindle length and symmetric versus asymmetric division, as discussed in a recent review (Howe and FitzHarris 2013).
Non-disjunction, on the other hand, is the failure of the sister chromatids to separate at the centromere during metaphase, resulting in the loss of a chromosome in one daughter cell and a reciprocal gain in the other. This event can be easily recognized as 3:1 chromosomal ratios between daughter cells and will give rise to an aneuploid zygote or mosaic aneuploid embryo depending on whether it occurs during meiosis or mitosis, respectively (Delhanty et al. 1993; Coonen et al. 2004; Daphnis et al. 2005, 2008; Katz-Jaffe et al. 2005). Although non-disjunction and anaphase lagging may have the same consequences, aneuploidy due to a single chromatid loss in MII oocytes far exceeds the prevalence of non-disjunction, at least during meiosis (Pellestor et al. 2002; Kuliev and Verlinsky 2004). Mitotic errors, however, appear to be equally distributed between chromosome lagging during anaphase and chromosomal non-disjunction (Chow et al. 2014).
Cell cycle drivers and checkpoints
To make matters worse, human pre-implantation embryos have also been shown to express both heightened cell cycle drivers and diminished cell cycle checkpoints, possibly making blastomeres more vulnerable to chromosome lagging during early mitotic divisions (Harrison et al. 2000; Kiessling et al. 2010) (Fig. 1). Only after EGA, from approximately the 4- to 8-cell stage onwards (Braude et al. 1988; Dobson et al. 2004), does cell cycle control become apparent in human embryos. Thus, permissive or lack of cycle checkpoints might allow aneuploid embryos to proceed through mitosis, even if chromosomes are not yet properly aligned on the mitotic spindle at the completion of metaphase (Harrison et al. 2000; Los et al. 2004). In turn, with robust expression of cell cycle drivers and shortened cell division intervals as compared with other cell types, the embryo enters anaphase and the lagging chromosomes become lost upon cytokinesis. Taken together, this suggests that embryos rely more on the circadian expression of genes, perhaps regulated by oscillators as previously described, rather than precise cell cycle control per se (Kiessling et al. 2009).
Aneuploidy tolerance and rescue
Cellular fragmentation and micronuclei formation
In light of the lack of cell cycle control, frequent findings of aneuploid diploid mosaicism (Kuo et al. 1998; Baart et al. 2006), and complex aneuploid mosaicism, whereby multiple chromosomes are affected in embryos without a single euploid blastomere, it is not surprising that the majority of chromosomal errors are unlikely to be corrected (Vanneste et al. 2009a, 2009b; Chavez et al. 2012). Nevertheless, some evidence suggests that “embryo self-corrective” mechanisms do indeed exist based on biopsy and PGS of an aneuploid blastomere(s) at the cleavage-stage followed by similar analysis of euploid trophectoderm cells from the same embryo a few days later (Munne et al. 2005; Barbash-Hazan et al. 2009). Although thought to occur only occasionally, the so-called embryo self-correction phenomenon might be explained by other processes, such as multi-polar divisions, blastomere exclusion, and cellular fragmentation, all of which are more frequently observed in embryos (Fig. 1). As discussed above, cellular fragmentation is relatively common in humans, with approximately 50 % or more of human embryos exhibiting fragmentation to some degree (Alikani et al. 1999; Antczak and Van Blerkom 1999). Pre-implantation embryos from other mammalian species, including rhesus monkeys, cattle, and even mice undergo cellular fragmentation, although to a lesser extent than humans and non-human primates (Enders et al. 1982; Sugimura et al. 2010; Fig. 3). Notably, cellular fragments are also detected in embryos conceived in vivo (Pereda and Croxatto 1978; Buster et al. 1985), indicating that fragmentation is not an artifact of in vitro culture. It is also important to note that cellular fragmentation is distinct from the cell-death-induced DNA fragmentation that can occur late in mammalian pre-implantation development, most significantly at the morula and blastocyst stages (Hardy 1999; Hardy et al. 2001; Xu et al. 2001). Cellular fragments can vary in size, in temporal and spatial distribution, and in organelle content (Jurisicova et al. 1996; Yang et al. 1998; Van Blerkom et al. 2001), making them sometimes difficult to differentiate from small blastomeres. High levels of fragmentation are associated with an increased incidence of aneuploidy (Pellestor et al. 1994; Munne 2006) and, depending on the timing of fragmentation, can be used to distinguish embryos with meiotic and/or mitotic errors (Chavez et al. 2012). Although extensive fragmentation may correlate with low implantation and clinical pregnancy potential (Giorgetti et al. 1995; Ziebe et al. 1997; Alikani et al. 1999; Hardy 1999; Ebner et al. 2001), by itself, it is not a clear indicator of poor embryo quality, as live births have resulted from the transfer of fragmented embryos (Edwards et al. 1984; Hoover et al. 1995; Alikani et al. 1999; Antczak and Van Blerkom 1999; Pelinck et al. 2010). Rather, it is the pattern and degree of cellular fragmentation, together with the stage at which it initially occurs, that are better predictors of embryo viability (Alikani et al. 1999; Antczak and Van Blerkom 1999).
Fig. 3.
Incidence of cellular fragmentation and multi-/micronuclei formation in cleavage-stage mammalian embryos. Top Representative micrographs of a human embryo taken by bright field imaging, b rhesus macaque embryo using Hoffmann modulation contrast, c bovine embryo using differential interference contrast (DIC), and d mouse embryo taken by dark field illumination time-lapse imaging. Note the appearance of several cellular fragments (black or white arrows) in cleavage-stage human and non-human primate embryos, but not in mouse embryos, with a lesser extent being observed in bovine embryos. Bottom Incidence of cellular fragmentation (arrows) is highly associated with multi- and micro-nuclei formation as indicated by confocal microscopy of LAMIN-B1 (green) expression in DAPI-stained (blue) cleavage-stage embryos from the same mammalian species (a’-d’); Scale bars 50 µm
Recent evidence presented by Chavez and colleagues (2012) suggests that cellular fragments can also encapsulate mis-segregated chromosomes following extrusion from the embryo, but exactly how this occurs remains unknown. Given that fragmentation was originally thought to represent membrane-bound extracellular cytoplasm, the idea that cellular fragments might contain nuclear DNA was intriguing, especially since microsurgical removal of fragments was once promoted (Eftekhari-Yazdi et al. 2006; Keltz et al. 2006). Nonetheless, the authors also reported frequent findings of micronuclei in cleavage-stage human embryos, suggesting that the formation of micronuclei provides a mechanism by which chromosomes are sequestered into fragments after mis-segregation (Chavez et al. 2012; Fig. 3). Since cellular fragmentation is a dynamic process, whereby chromosome-containing fragments can remain as either discrete units or be reabsorbed by the embryo (Hardarson et al. 2002; Lemmen et al. 2008; Chavez et al. 2012), it may also allow corrective means to overcome aneuploidy generation. More specifically, if a fragment fuses with the blastomere from which it originated or a neighboring blastomere that lacked the encapsulated chromosome(s), euploidy could potentially be restored in an embryo following the breakdown of the nuclear envelope (Fig. 4a). However, not all fragments necessarily contain DNA, and thus, fragment resorption may have no impact on ploidy status, or it may even increase genotypic complexity in embryos upon fusion with a euploid blastomere (Chavez et al. 2012). Regardless of the potential consequences, little is still known about what leads to cellular fragmentation and it will be important to determine the precise mechanism(s) by which it occurs and the way in which the timing, degree, and/or resorption of fragments contributes to the generation and potential correction of embryonic aneuploidy. As cancer cells also sequester mis-segregated chromosomes and chromatid fragments within micronuclei (Ford et al. 1988; Norppa and Falck 2003; Fenech et al. 2011; Hatch et al. 2013), findings during pre-implantation development may also be translatable to cancerous somatic tissues. In support of this, the blebbing of cytoplasmic membrane resembling cellular fragmentation in embryos has been shown to occur simultaneously with the extrusion of nuclear content in cancer cell lines during interphase, suggesting that micronuclei formation may be similarly regulated between somatic and embryonic cells (Shimizu et al. 2000; Utani et al. 2011).
Fig. 4.
Potential mechanisms of aneuploidy resolution during pre-implantation development. a Proposed model for the origins of complex embryonic aneuploidy based on fragmentation timing, fragment resorption, and underlying chromosomal abnormalities. Human embryos with meiotic errors (monosomies and trisomies) and those that appear to be triploid or tetraploid typically exhibit fragmentation at the 1-cell stage, suggesting that embryos respond to aneuploidy by fragmenting, perhaps as a survival mechanism. In contrast, fragmentation is most often detected at the 2-cell stage in embryos with mitotic errors. We also propose that the embryo probably divided before the lagging chromosomes were properly aligned on the mitotic spindle, resulting in the formation of micronuclei and the sequestering of the missing chromosomes into cellular fragments. As development proceeds, these fragments can either remain or be reabsorbed by the blastomere from which they originated to restore ploidy status or fuse with a neighboring blastomere to generate complex embryonic aneuploidies (adapted from Chavez et al. 2012). b Series of dark field time-lapse images, with the time in hours (hr) and minutes (mins) in the lower left corner showing of a rhesus macaque zygote with three cleavage furrows (white arrows) as it divides directly from 1-cell to 3-cells. c Similar bright field time-lapse image series demonstrating how one daughter cell divides twice (black arrows indicate cleavage furrows) before the other (star) to suggest a mechanism by which monosomic chromosomal errors may be resolved. d Single-channel (left) and multichannel (right) confocal analysis of histone modifications in human morulas that are undergoing cavitation to form a blastocyst reveals the presence of a large and likely aneuploid blastomere (arrows) excluded from each embryo (adapted from Chavez et al. 2014)
Unlike human embryos, murine pre-implantation embryos not only have a low incidence of aneuploidy, but also rarely exhibit micronucleation during development (Bond and Chandley 1983; Lightfoot et al. 2006; Chavez et al. 2012). However, a recent study indicates that the formation of micro- and multiple-nuclei (multi-nuclei) can be induced in mouse embryos by disrupting the function of particular cell cycle proteins critical for developmental progression. Chavez and colleagues (2014) determined that translational block of Rps6ka4/Msk2, a mitogenic factor known to be involved in the G1 phase of the cell cycle (Bettencourt-Dias et al. 2004), in mouse zygotes resulted in the appearance of multiple micro- or multi-nuclei at the cleavage stage. In addition, increased blastomere movement and lysis, resembling cellular events described during mitotic catastrophe (Vakifahmetoglu et al. 2008), was also observed in mouse embryos following mitotic arrest at the 3- to 8-cell stage. This suggests that, in contrast to human embryos, which appear to respond to aneuploidy generation by fragmenting, mouse embryos undergo blastomere lysis as a potential mechanism to avoid chromosomal instability (Fig. 1). It may also explain the low aneuploidy rates observed in mice, if micro- and multi-nuclei formation in blastomeres results in mitotic arrest rather than continued division as part of a selection process. Further support for this is provided by findings that deletion of BUB3, BUBR1, and MAD2 of the spindle assembly complex, which is essential for normal mitotic progression, leads to the formation of micronuclei, chromosome misalignment, aneuploidy, developmental delay, and decreased implantation rates. Overexpression of these spindle assembly complex components, on the other hand, prevents sister chromatid segregation (Wei et al. 2011). Disruption of α-thalassemia/mental retardation X-linked (ATRX), a protein that binds to pericentric heterochromatin in MII and mediates chromosome alignment along the metaphase plate, also causes micronuclei formation and aneuploidy in mouse oocytes and zygotes, suggesting that similar selection mechanisms also exist during meiosis (Baumann et al. 2010).
Chromothripsis and sub-chromosomal rearrangements
In comparison with primary nuclei, micronuclei display altered nuclear functions, including aberrant DNA replication, transcription, and DNA-damage repair as shown by in vitro experiments with cultured cells. The results of such studies also indicate a direct link between micronuclei formation and the incidence of “chromosome pulverization” or chromothripsis (Hoffelder et al. 2004; Terradas et al. 2009; Xu et al. 2011; Crasta et al. 2012). First described in cancer genomes (Stephens et al. 2011), chromothripsis is a phenomenon through which one or a few chromosomal segments are “pulverized” into many pieces and randomly reassembled in one unique cellular event during a single cell division (Pellestor et al. 2014). Chromothripsis has also been suggested to occur during pre-implantation development (Pellestor 2014; Pellestor et al. 2014) based on frequent observations of micronuclei formation, cellular fragmentation, segmental rearrangements, and abnormal mitotic divisions in cleavage-stage embryos. This pulverization and random reassembly can lead to chromosomal duplications, deletions, translocations, and inversions, which are often detrimental, resulting in subfertility or congenital abnormalities depending upon when it occurs (Loup et al. 2010; Kloosterman et al. 2011).
In incredibly rare cases, however, chromothripsis may lead to the spontaneous elimination of a deleterious gene and effectively “cure” a disease if the event occurs in an adult stem cell that can repopulate the affected niche (McDermott et al. 2015). Nevertheless, chromothripsis during gametogenesis or early embryogenesis is thought to be a likely explanation for the de novo occurrence of complex chromosomal rearrangements in offspring from parents with otherwise normal karyotypes (Pellestor 2014; Pellestor et al. 2014). Given that sub-chromosomal aberrations have also been reported in embryos from women of advanced maternal age or with more than one implantation failure (Voullaire et al. 2002; Wilton et al. 2003; Rius et al. 2010, 2011; Mantikou et al. 2012), chromothripsis may also be involved in recurrent pregnancy loss. The vast majority of chromothripsis events are thought to be catastrophic and result in embryonic lethality rather than being passed onto the next generation via the germ cells. Moreover, the de novo germline complex rearrangements that are detected in the offspring, but not inherited from the parents, also exhibit fewer chromosomal breaks in comparison with those detected in cancer genomes (Giardino et al. 2009; Kloosterman and Cuppen 2013). On the other hand, in cases of complex genomic rearrangements that are inherited, chromothripsis has been found to be one of the major culprits (Kloosterman et al. 2011, 2012; Chiang et al. 2012; Macera et al. 2015). However, how DNA damage in micronuclei can lead to chromothripsis and the role that this phenomenon plays, if any, in the high rates of aneuploidy observed in human embryos remains unclear.
As an explanation for the means by which DNA damage-induced chromothripsis might arise, a model correlating micronuclei formation with chromosome pulverization was proposed by Crasta et al. (2012) and confirmed by Zhang and colleagues (2015). Based on previous work with both cancer cells and solid tumors, Crasta et al. and others revealed that DNA damage via double-strand breaks appears irrespective of whether the micronucleus is intact (Crasta et al. 2012; Huang et al. 2012) or compromised by nuclear envelope collapse (Cimini et al. 2001; Hatch et al. 2013). Consequently, DNA repair, replication, and transcription become altered, leading to genomic instability as documented by the accumulation of gamma-H2A histone family, member X (γ-H2AX) foci. Recently, Zhang et al. (2015) used “LookSeq”, a combination of live-cell imaging and single-cell whole-genome sequencing, to confirm that micronuclei can partition into a daughter cell and undergo nuclear envelope rupture and defective DNA replication, leading to chromosomal copy number asymmetry between daughter cells. After only one cell division, micronuclei were reincorporated into the primary nucleus, and by sequencing the daughter cells with nuclear fusion, this process was determined to be restricted to mis-segregated chromosomes. Moreover, the authors observed that micronuclei formation resulted in diverse genomic rearrangements, some of which recapitulated all known hallmarks of chromothripsis (Zhang et al. 2015). In another study that involved time-lapse imaging to assess micronuclei formation in a cancer cell line, DNA-containing micronuclei extruded extracellularly were shown to fuse back with a daughter cell after cell division (Rao et al. 2008). Thus, analogous to pre-implantation embryo development, if extracellular micronuclei are resorbed and fuse with primary nuclei following attempted repair by non-homologous end joining and nuclear envelope breakdown, then the incorporation of compromised chromosomes might result in additional genetic mutations (Fig. 1). Chromothripsis is only one of the proposed mechanisms for generating the complex chromosomal rearrangements observed in congenital disorders and pre-implantation embryo loss; other possible mechanisms include DNA replication errors (P. Liu et al. 2012), telomeric dysfunction (Soler et al. 2009), and translocation events during meiosis (Pellestor et al. 2011). Although such events can be viewed as deleterious, similar mechanisms are thought to influence accelerated karyotypes in species evolution (Carbone et al. 2014), and it will be important to determine whether the sub-chromosomal rearrangements that emerge following chromothripsis are tolerated, or if they impact human fecundity.
Multi-polar cell divisions and trisomic rescue
Another relatively frequent event observed during pre-implantation development is the multi-polar division of zygotes or blastomeres, whereby embryos divide into three or more daughter cells rather than the typical two (Fig. 4b). Through the formation of multiple centrosomal spindle poles that pull anaphase chromosomes apart, embryos can undergo tri-polar or higher order divisions at the zygote stage or during early cleavage divisions (for a review, see Kalatova et al. 2015). Whereas the potential impact of multi-polar divisions at the 2-cell stage and beyond may not be as detrimental and is still being investigated, TLM studies estimate that approximately 12 % of 1-cell human embryos are characterized by multi-polar divisions (Chamayou et al. 2013) and, contrary to previous notions, are often associated with triploidy or tetraploidy. Moreover, embryos that exhibit 1:3 divisions at the zygote stage are also much less likely to reach the blastocyst stage than zygotes that undergo a bi-polar division and have been shown to correlate with poor implantation potential (Hlinka et al. 2012).
As a consequence of multi-polar divisions, diverse asymmetrical chromosome distribution can result, and one of the leading hypotheses is that centrosome duplication is to blame. In support of this, genetic variants of maternal PLK4, which is involved in centriole duplication and alters mitotic fidelity upon dysregulation, has been shown to correlate with mitotic aneuploidy and failed pregnancies in women undergoing IVF (McCoy et al. 2015). Notably, Ganem et al. (2009) determined that multi-polar divisions are rare in cancer cells and often become non-viable within the first two cell divisions. In contrast, human zygotes can undergo 1:3 cell divisions and are still able to progress further in pre-implantation development as discussed above. Ganem et al. (2009) also observed that in cancer cells, extra centrosomes spend the majority of mitosis in transient multi-polar intermediates before undergoing centrosome clustering. This type of configuration leads to an increase in merotelic kinetochore-microtubule attachment errors, whereby microtubules from both poles attach to a single kinetochore on one side of a chromosome. Consequently, even though cancer cells with extra centrosomes are able to proceed through bipolar anaphase, merotelic attachments result in increased anaphase lagging chromosomes and subsequent chromosomal instability. Whether similar mechanisms regulate pre-implantation development is unknown, but multi-polar divisions may not necessarily be detrimental and can constitute a means for trisomic embryos to undergo rescue via loss of the extra chromosome(s) upon division (Kalousek 2000; Fig. 1). If both of the retained chromosomes are inherited from the same parent, then uniparental disomy (UPD) will result, as is discussed in further detail below.
Endoreduplication and UPD
A process that is almost the opposite of multi-polar divisions is called endoreduplication, or the replication of a chromosome without cell division. This can result in several different outcomes depending on the original state of the chromosomes involved. For instance, if one chromosome undergoes endoreduplication in a diploid blastomere, one of the daughter cells in the subsequent cell division will be trisomic and the other disomic. Another well-known example of endoreduplication is the parthenogenetic activation of a haploid oocyte, resulting in the formation of a diploid parthenote capable of further development (Lin et al. 2007; Sritanaudomchai et al. 2010; Elling et al. 2011; Leeb et al. 2012; Paull et al. 2013; Daughtry and Mitalipov 2014).
Therefore, endoreduplication not only is a mechanism by which aneuploidy can be generated, but it can also be involved in the rescue of monosomic embryos (Fig. 4c). Analogous to embryos with trisomies, monosomic embryos could gain the missing chromosome(s) following endoreduplication to produce a diploid organism. However, unlike embryos that undergo trisomic rescue, monosomic rescue always results in UPD since the duplicated chromosome is either paternally or maternally inherited (Fig. 1). Thus, UPD can have clinical consequences such as the disruption of parent-specific genomic imprinting and other developmental disorders due to homozygosity. In a study by Conlin and colleagues (2010), two UPD patients exhibited homozygosity for all alleles genotyped on the afflicted chromosome, and the patient with UPD15 presented with Angelman syndrome, consistent with monosomic rescue by endoreduplication of paternal chromosome 15. The other patient with UPD14, however, displayed morphological and developmental abnormalities indicative of paternal UPD14 (Conlin et al. 2010). Given that, in both cases, the UPD was paternal, the most likely scenario was that a sperm fertilized a nullisomic oocyte produced from maternal meiotic non-disjunction. Subsequently, the haploid paternal chromatid in the fertilized oocyte underwent endoreduplication and led to the restoration of euploidy by UPD. While these patients exhibited obvious phenotypic manifestations due to the chromosome that was affected, certain UPD chromosomes can be tolerated, suggesting that endoreduplication and UPD can be an effective mechanism for monosomic rescue of embryos in some cases (Berend et al. 1999; Fig. 1).
Blastomere and fragment exclusion
Whereas the above-discussed corrective mechanisms might occur at the earliest stages of pre-implantation development, other means appear to exist for the resolution of aneuploidy later in embryogenesis during the transition from the cleavage to the blastocyst stage (Fig. 1). In particular, the complete exclusion of large fragments and blastomeres has been observed in both compact morulas and blastocysts from several mammalian species, including humans, rhesus macaques, cattle, and mice homozygous for the yellow allele (Ay) of the agouti locus (Calarco and Pedersen 1976; Lindner and Wright 1983; Alikani et al. 1999, 2002; Hardy 1999; Van Soom et al. 2003; Prados et al. 2012; Chavez et al. 2014). Although the frequency of blastomere exclusion is unknown since it is not a well-documented process, pre-implantation embryos are generally thought to tolerate the loss of one or more blastomeres based on blastomere biopsy for PGS. Moreover, given that lower aneuploidy rates are observed in blastocysts as compared with cleavage-stage embryos (Magli et al. 2000), one possible explanation for this may be the exclusion of aneuploid blastomeres from the rest of the embryo (Fig. 4d). To our knowledge, however, the chromosomal and/or genetic analysis of extruded blastomeres or fragments is still lacking; this may be due to the limitations of aCGH and other microarray-based methods for the detection of complex or polyploid genotypes. Nevertheless, the extrusion rather than the reincorporation of aneuploid or micronuclei-containing blastomeres may be beneficial for embryo genomic integrity based on the original chromosomal content and increased propensity for DNA damage in the micronuclear environment (Thompson and Compton 2011; Crasta et al. 2012; Huang et al. 2012; Hatch et al. 2013). Future work should focus on the precise frequency of this process and how blastomere exclusion might relate to the “embryo self-correction” phenomenon as a whole.
Confined placental mosaicism
Alternatively, another mechanism termed confined placental mosaicism has also been proposed to explain embryo correction and the reduced incidence of aneuploidy at the blastocyst stage (Magli et al. 2000). In this scenario, aneuploid cells are deliberately confined to the trophectoderm layer, whereas euploid cells are incorporated into the inner cell mass (ICM) and give rise to a chromosomally normal fetus (Kalousek and Dill 1983). Indeed, the presence of two or more cell populations with a different chromosomal make-up has been reported at the blastocyst stage, and this appears to be more prevalent in embryos from women of advanced maternal age (Fragouli et al. 2011; J. Liu et al. 2012). After careful separation of the ICM and trophectoderm cells, however, several studies demonstrated that chromosomally abnormal cells detected at the 8-cell stage are not preferentially allocated to the trophectoderm compartment but, instead evenly distributed throughout the blastocyst (Mottla et al. 1995; Evsikov and Verlinsky 1998; Magli et al. 2000; Derhaag et al. 2003; Fragouli et al. 2008; Johnson et al. 2010a; Capalbo et al. 2013). As further support, Johnson and colleagues (2010a) disaggregated over 50 blastocysts from mothers of average maternal age and observed discordance between trophectoderm and ICM samples in only 4 % of the embryos; this occurred in cases of structural chromosome aberrations (Johnson et al. 2010a). It is important to note, however, that this study evaluated ploidy status in multiple cells of the same cell type at once, rather than single cells, which may mask discordance between the trophectoderm and ICM. Nevertheless, other studies have reported the preferential allocation of aneuploid cells in chorionic villi samples as compared with fetal tissues, suggesting that confined placental mosaicism plays a role later in post-implantation development (Kalousek and Dill 1983; Ledbetter et al. 1992; Kalousek and Vekemans 1996; Phillips et al. 1996; Clouston et al. 1997; Goldberg and Wohlferd 1997; J. Liu et al. 2012). Taken together, this data indicates that the trophectoderm layer of blastocysts might still preferentially contain chromosomally abnormal cells and it will be necessary to determine whether this can be observed using more comprehensive methods than DNA-FISH and at the single cell level in whole embryos.
Clinical aspects of pre-implantation aneuploidy
Over 38 years have now passed since Louise Brown, the first “test tube” baby, was born (Steptoe and Edwards 1978). After many failed attempts involving several different methodologies and donors, Robert Edwards and Patrick Steptoe successfully produced, via IVF, an embryo that was able to implant, progress in development, and result in the birth of a healthy child. That such a relatively straightforward experimental procedure could produce a perfectly healthy baby demonstrates the extremely remarkable, and yet tightly controlled, cellular programs that a pre-implantation embryo must undergo in the first few days of development. Despite this extraordinary accomplishment, one alarming statistic is that even with the progressive increase in the number of IVF cycles performed each year, live birth rates from transferred embryos have hovered around 30–35 % for decades. Moreover, the implementation of more advanced IVF techniques such as intracytoplasmic sperm injection, PGS, and assisted hatching has not significantly changed the percentage of live births (Harper and Sengupta 2012; Kissin et al. 2014; Boulet et al. 2015). Thus, it will be essential to determine how more recently developed methods, including time-lapse imaging, DNA-Seq, and “-omic” profiling, either alone or in combination, impact IVF success rates over the next several years.
With over 5 million children born via IVF to date from hundreds of clinics worldwide, there are clear indications that the IVF procedure itself is not the only explanation for the plateau in live birth rates. Indeed, it is well recognized that the mammalian pre-implantation embryo does not develop cell-autonomously, and that the numerous extrinsic factors provided by the oviduct in vivo may not necessarily be emulated by culture media in vitro (Lee and Yeung 2006). Nevertheless, there is evidence that several of these developmental processes observed in IVF and described in this review do indeed occur naturally following in vivo conceptions (Pereda and Croxatto 1978; Buster et al. 1985). More specifically, a study from the 1970s, whereby a naturally conceived embryo was obtained from the oviduct following hysterectomy, demonstrated that in vivo pre-implantation embryos exhibit cellular fragmentation, blastomere asymmetry, and other morphological features, indicating that these events are not simply a consequence of in vitro embryo culture (Pereda and Croxatto 1978; Buster et al. 1985). Further evidence is provided by the high incidence of mosaicism detected in early spontaneous human abortions (Lebedev et al. 2004; Vorsanova et al. 2005) and the similar rates of mosaicism observed in chorionic villus samples (CVS) between IVF and natural pregnancies later in development (Huang et al. 2009; Mantikou et al. 2012).
Although much of the increase in the number of IVF cycles performed each year can be explained by couples continuing to delay child bearing for a variety of reasons, aneuploidy is not only associated with patients of an advanced maternal age. Mothers of all ages can produce a high percentage of aneuploid embryos as shown by a previous study, wherein 83 % and 70 % of embryos from both young and fertile women exhibited whole-chromosomal imbalances and sub-chromosomal abnormalities, respectively (Vanneste et al. 2009a, 2009b). Therefore, the argument that low IVF success rates are solely explained by advanced maternal age and infertility alone is slowly shifting since infertility affects both men and women equally and young fertile couples also produce aneuploid embryos (Mascarenhas et al. 2012; Taylor et al. 2014). Nevertheless, more healthy couples are using egg donors or surrogates for child bearing, and the cryogenic preservation of embryos for transfer later during a natural cycle is becoming increasingly popular (Fatemi et al. 2010; Evans et al. 2014). Lastly, some lesbian, gay, bisexual, and transgender couples are seeking artificial reproductive technologies for family planning, and yet, in all of these groups, no dramatic increase in live births has been observed when compared with more traditional IVF patients (Nordqvist et al. 2014). By subtracting factors such as advanced maternal age, infertility, and other clinical variables, healthy couples still produce embryos that arrest before the blastocyst stage and present abnormal karyotypes, indicating that aneuploidy is a universal phenomenon (Baart et al. 2006; Franasiak et al. 2014).
Summary
Infertility in humans is relatively common, and the use of IVF or other ARTs has more than doubled in the past decade and is expected to continue to increase as reproductive-age couples delay childbirth (Mascarenhas et al. 2012). Despite considerable advances in alleviating certain forms of this disorder, IVF success rates still hover around ~30–35 % (cdc.gov/art), and one of the main culprits is thought to be whole and sub-chromosomal abnormalities that arise during pre-implantation development (Munne et al. 1994). While it may occur at a higher frequency in human embryos, evidence from other closely related mammals indicates that aneuploidy is not unique to human pre-implantation development (Dupont et al. 2010). Moreover, given the complexity of early mammalian embryogenesis, it is likely that multiple factors such as parental contribution and inherent genomic vulnerability are associated with aneuploidy generation, multi/micronuclei formation, and cellular fragmentation during pre-implantation development. Although significant insight has been obtained from correlations with cancer cells and other mitotic cell types, certain aspects of genomic regulation and cell division are clearly unique to cleavage-stage embryos. As comprehensive chromosomal assessment techniques are further developed and optimized, especially in whole genomes and at the single cell level (Hou et al. 2013; Fiorentino et al. 2014; Wang et al. 2014), it is envisioned that the precise underlying cause(s) of embryonic chromosomal instability will be identified for potential therapeutic intervention. Ultimately, the overall goals of this research and future work are to select and transfer the embryo(s) with the highest probability of resulting in a normal term pregnancy and, preferably, single healthy baby. We also anticipate that the findings from these studies will enhance our understanding of natural human conception and the chromosomal requirements of early embryogenesis across diverse mammalian species.
Acknowledgments
We gratefully acknowledge the National Centers for Translational Research in Reproduction and Infertility (NCTRI)/NICHD (P50 HD071836), the Howard and Georgeanna Jones Foundation for Reproductive Medicine, the Medical Research Foundation of Oregon, and the Collins Medical Trust for funding to S.L.C. and the Reproductive Biology Training Grant (T32 HD007133) for funding to B.L.D.
References
- Akoury E, Zhang L, Ao A, Slim R. NLRP7 and KHDC3L, the two maternal-effect proteins responsible for recurrent hydatidiform moles, co-localize to the oocyte cytoskeleton. Hum Reprod. 2015;30:159–169. doi: 10.1093/humrep/deu291. [DOI] [PubMed] [Google Scholar]
- Alikani M, Cohen J, Tomkin G, Garrisi GJ, Mack C, Scott RT. Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil Steril. 1999;71:836–842. doi: 10.1016/s0015-0282(99)00092-8. [DOI] [PubMed] [Google Scholar]
- Alikani M, Sadowy S, Cohen J. Human embryo morphology and developmental capacity. In: van Soom A, Boerjan M, editors. Assessment of mammalian embryo quality. Springer Netherlands; Netherlands: 2002. pp. 1–31. [Google Scholar]
- Alper MM, Brinsden P, Fischer R, Wikland M. To blastocyst or not to blastocyst? That is the question. Hum Reprod. 2001;16:617–619. doi: 10.1093/humrep/16.4.617. [DOI] [PubMed] [Google Scholar]
- Angell R. First-meiotic-division nondisjunction in human oocytes. Am J Hum Genet. 1997;61:23–32. doi: 10.1086/513890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antczak M, Van Blerkom J. Temporal and spatial aspects of fragmentation in early human embryos: possible effects on developmental competence and association with the differential elimination of regulatory proteins from polarized domains. Hum Reprod. 1999;14:429–447. doi: 10.1093/humrep/14.2.429. [DOI] [PubMed] [Google Scholar]
- Assou S, Haouzi D, De Vos J, Hamamah S. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol Hum Reprod. 2010;16:531–538. doi: 10.1093/molehr/gaq032. [DOI] [PubMed] [Google Scholar]
- Assou S, Ait-Ahmed O, El Messaoudi S, Thierry AR, Hamamah S. Non-invasive pre-implantation genetic diagnosis of X-linked disorders. Med Hypotheses. 2014;83:506–508. doi: 10.1016/j.mehy.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Athayde Wirka K, Chen AA, Conaghan J, Ivani K, Gvakharia M, Behr B, Suraj V, Tan L, Shen S. Atypical embryo phenotypes identified by time-lapse microscopy: high prevalence and association with embryo development. Fertil Steril. 2014;101:1637–1648. e1631–e1635. doi: 10.1016/j.fertnstert.2014.02.050. [DOI] [PubMed] [Google Scholar]
- Azzarello A, Hoest T, Mikkelsen AL. The impact of pronuclei morphology and dynamicity on live birth outcome after time-lapse culture. Hum Reprod. 2012;27:2649–2657. doi: 10.1093/humrep/des210. [DOI] [PubMed] [Google Scholar]
- Baart EB, Martini E, van den Berg I, Macklon NS, Galjaard RJ, Fauser BC, Van Opstal D. Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum Reprod. 2006;21:223–233. doi: 10.1093/humrep/dei291. [DOI] [PubMed] [Google Scholar]
- Baltaci V, Satiroglu H, Kabukcu C, Unsal E, Aydinuraz B, Uner O, Aktas Y, Cetinkaya E, Turhan F, Aktan A. Relationship between embryo quality and aneuploidies. Reprod Biomed Online. 2006;12:77–82. doi: 10.1016/s1472-6483(10)60984-4. [DOI] [PubMed] [Google Scholar]
- Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, Ben-Yosef D. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92:890–896. doi: 10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, Garcia-Velasco J, Meseguer M. Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril. 2014;101:699–704. doi: 10.1016/j.fertnstert.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Baumann C, Viveiros MM, De La Fuente R. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet. 2010;6:e1001137. doi: 10.1371/journal.pgen.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkhalifa M, Kasakyan S, Clement P, Baldi M, Tachdjian G, Demirol A, Gurgan T, Fiorentino F, Mohammed M, Qumsiyeh MB. Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat Diagn. 2005;25:894–900. doi: 10.1002/pd.1230. [DOI] [PubMed] [Google Scholar]
- Berend SA, Feldman GL, McCaskill C, Czarnecki P, Van Dyke DL, Shaffer LG. Investigation of two cases of paternal disomy 13 suggests timing of isochromosome formation and mechanisms leading to uniparental disomy. Am J Med Genet. 1999;82:275–281. doi: 10.1002/(sici)1096-8628(19990129)82:3<275::aid-ajmg15>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Bi W, Breman A, Shaw CA, Stankiewicz P, Gambin T, Lu X, Cheung SW, Jackson LG, Lupski JR, Van den Veyver IB, et al. Detection of >/=1Mb microdeletions and microduplications in a single cell using custom oligonucleotide arrays. Prenat Diagn. 2012;32:10–20. doi: 10.1002/pd.2855. [DOI] [PubMed] [Google Scholar]
- Bond DCA. Aneuploidy: the origins and causes of aneuploidy in experimental organisms. Oxford University Press; Oxford: 1983. pp. 86–91. [Google Scholar]
- Boulet SL, Mehta A, Kissin DM, Warner L, Kawwass JF, Jamieson DJ. Trends in use of and reproductive outcomes associated with intracytoplasmic sperm injection. JAMA. 2015;313:255–263. doi: 10.1001/jama.2014.17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332:459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Brevini TA, Pennarossa G, Antonini S, Paffoni A, Tettamanti G, Montemurro T, Radaelli E, Lazzari L, Rebulla P, Scanziani E, et al. Cell lines derived from human parthenogenetic embryos can display aberrant centriole distribution and altered expression levels of mitotic spindle check-point transcripts. Stem Cell Rev. 2009;5:340–352. doi: 10.1007/s12015-009-9086-9. [DOI] [PubMed] [Google Scholar]
- Brevini TA, Pennarossa G, Maffei S, Tettamanti G, Vanelli A, Isaac S, Eden A, Ledda S, de Eguileor M, Gandolfi F. Centrosome amplification and chromosomal instability in human and animal parthenogenetic cell lines. Stem Cell Rev. 2012;8:1076–1087. doi: 10.1007/s12015-012-9379-2. [DOI] [PubMed] [Google Scholar]
- Burruel V, Klooster K, Barker CM, Pera RR, Meyers S. Abnormal early cleavage events predict early embryo demise: sperm oxidative stress and early abnormal cleavage. Sci Rep. 2014;4:6598. doi: 10.1038/srep06598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster JE, Bustillo M, Rodi IA, Cohen SW, Hamilton M, Simon JA, Thorneycroft IH, Marshall JR. Biologic and morphologic development of donated human ova recovered by nonsurgical uterine lavage. Am J Obstet Gynecol. 1985;153:211–217. doi: 10.1016/0002-9378(85)90116-4. [DOI] [PubMed] [Google Scholar]
- Calarco PG, Pedersen RA. Ultrastructural observations of lethal yellow (Ay/Ay) mouse embryos. J Embryol Exp Morphol. 1976;35:73–80. [PubMed] [Google Scholar]
- Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod Biomed Online. 2013a;26:477–485. doi: 10.1016/j.rbmo.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod Biomed Online. 2013b;27:140–146. doi: 10.1016/j.rbmo.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L, Nagy ZP. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Hum Reprod. 2013 doi: 10.1093/humrep/det245. [DOI] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, Meyer TJ, Herrero J, Roos C, Aken B, et al. Gibbon genome and the fast karyotype evolution of small apes. Nature. 2014;513:195–201. doi: 10.1038/nature13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervero A, Horcajadas JA, Dominguez F, Pellicer A, Simon C. Leptin system in embryo development and implantation: a protein in search of a function. Reprod Biomed Online. 2005;10:217–223. doi: 10.1016/s1472-6483(10)60943-1. [DOI] [PubMed] [Google Scholar]
- Chamayou S, Patrizio P, Storaci G, Tomaselli V, Alecci C, Ragolia C, Crescenzo C, Guglielmino A. The use of morphokinetic parameters to select all embryos with full capacity to implant. J Assist Reprod Genet. 2013;30:703–710. doi: 10.1007/s10815-013-9992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, Behr B, Reijo Pera RA. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun. 2012;3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez SL, McElroy SL, Bossert NL, De Jonge CJ, Rodriguez MV, Leong DE, Behr B, Westphal LM, Reijo Pera RA. Comparison of epigenetic mediator expression and function in mouse and human embryonic blastomeres. Hum Mol Genet. 2014;23:4970–4984. doi: 10.1093/hmg/ddu212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SR, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012;44:390–397. S391. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaratti MR, Ferreira CR, Perecin F, Meo SC, Sangalli JR, Mesquita LG, de Carvalho Balieiro JC, Smith LC, Garcia JM, Meirelles FV. Ooplast-mediated developmental rescue of bovine oocytes exposed to ethidium bromide. Reprod Biomed Online. 2011;22:172–183. doi: 10.1016/j.rbmo.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Chow JF, Yeung WS, Lau EY, Lee VC, Ng EH, Ho PC. Array comparative genomic hybridization analyses of all blastomeres of a cohort of embryos from young IVF patients revealed significant contribution of mitotic errors to embryo mosaicism at the cleavage stage. Reprod Biol Endocrinol. 2014;12:105. doi: 10.1186/1477-7827-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouston HJ, Fenwick J, Webb AL, Herbert M, Murdoch A, Wolstenholme J. Detection of mosaic and non-mosaic chromosome abnormalities in 6- to 8-day old human blastocysts. Hum Genet. 1997;101:30–36. doi: 10.1007/s004390050581. [DOI] [PubMed] [Google Scholar]
- Combelles CM, Kearns WG, Fox JH, Racowsky C. Cellular and genetic analysis of oocytes and embryos in a human case of spontaneous oocyte activation. Hum Reprod. 2011;26:545–552. doi: 10.1093/humrep/deq363. [DOI] [PubMed] [Google Scholar]
- Conaghan J, Hardy K, Handyside AH, Winston RM, Leese HJ. Selection criteria for human embryo transfer: a comparison of pyruvate uptake and morphology. J Assist Reprod Genet. 1993;10:21–30. doi: 10.1007/BF01204436. [DOI] [PubMed] [Google Scholar]
- Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, Baker VL, Adamson GD, Abusief ME, Gvakharia M, et al. Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril. 2013;100:412–419. e415. doi: 10.1016/j.fertnstert.2013.04.021. [DOI] [PubMed] [Google Scholar]
- Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D’Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, Deardorff MA, Krantz ID, Hakonarson H, Spinner NB. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19:1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coonen E, Derhaag JG, Dumoulin JC, van Wissen LC, Bras M, Janssen M, Evers JL, Geraedts JP. Anaphase lagging mainly explains chromosomal mosaicism in human preimplantation embryos. Hum Reprod. 2004;19:316–324. doi: 10.1093/humrep/deh077. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M, Gadea B, Garrido N, Pedersen KS, Martinez M, Perez-Cano I, Munoz M, Meseguer M. Embryo quality, blastocyst and ongoing pregnancy rates in oocyte donation patients whose embryos were monitored by time-lapse imaging. J Assist Reprod Genet. 2011;28:569–573. doi: 10.1007/s10815-011-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz M, Garrido N, Herrero J, Perez-Cano I, Munoz M, Meseguer M. Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod Biomed Online. 2012;25:371–381. doi: 10.1016/j.rbmo.2012.06.017. [DOI] [PubMed] [Google Scholar]
- Dal Canto M, Coticchio G, Mignini Renzini M, De Ponti E, Novara PV, Brambillasca F, Comi R, Fadini R. Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod Biomed Online. 2012;25:474–480. doi: 10.1016/j.rbmo.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Daphnis DD, Delhanty JD, Jerkovic S, Geyer J, Craft I, Harper JC. Detailed FISH analysis of day 5 human embryos reveals the mechanisms leading to mosaic aneuploidy. Hum Reprod. 2005;20:129–137. doi: 10.1093/humrep/deh554. [DOI] [PubMed] [Google Scholar]
- Daphnis DD, Fragouli E, Economou K, Jerkovic S, Craft IL, Delhanty JD, Harper JC. Analysis of the evolution of chromosome abnormalities in human embryos from Day 3 to 5 using CGH and FISH. Mol Hum Reprod. 2008;14:117–125. doi: 10.1093/molehr/gam087. [DOI] [PubMed] [Google Scholar]
- Daughtry B, Mitalipov S. Concise review: parthenote stem cells for regenerative medicine: genetic, epigenetic, and developmental features. Stem Cells Transl Med. 2014;3:290–298. doi: 10.5966/sctm.2013-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhanty JD. Mechanisms of aneuploidy induction in human oogenesis and early embryogenesis. Cytogenet Genome Res. 2005;111:237–244. doi: 10.1159/000086894. [DOI] [PubMed] [Google Scholar]
- Delhanty JD, Griffin DK, Handyside AH, Harper J, Atkinson GH, Pieters MH, Winston RM. Detection of aneuploidy and chromosomal mosaicism in human embryos during preimplantation sex determination by fluorescent in situ hybridisation, (FISH) Hum Mol Genet. 1993;2:1183–1185. doi: 10.1093/hmg/2.8.1183. [DOI] [PubMed] [Google Scholar]
- Derhaag JG, Coonen E, Bras M, Bergers Janssen JM, Ignoul-Vanvuchelen R, Geraedts JP, Evers JL, Dumoulin JC. Chromosomally abnormal cells are not selected for the extra-embryonic compartment of the human preimplantation embryo at the blastocyst stage. Hum Reprod. 2003;18:2565–2574. doi: 10.1093/humrep/deg485. [DOI] [PubMed] [Google Scholar]
- Derijck AA, van der Heijden GW, Giele M, Philippens ME, van Bavel CC, de Boer P. gammaH2AX signalling during sperm chromatin remodelling in the mouse zygote. DNA Repair. 2006;5:959–971. doi: 10.1016/j.dnarep.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RA. The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- Desai N, Ploskonka S, Goodman LR, Austin C, Goldberg J, Falcone T. Analysis of embryo morphokinetics, multinucleation and cleavage anomalies using continuous time-lapse monitoring in blastocyst transfer cycles. Reprod Biol Endocrinol. 2014;12:54. doi: 10.1186/1477-7827-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez F, Gadea B, Mercader A, Esteban FJ, Pellicer A, Simon C. Embryologic outcome and secretome profile of implanted blastocysts obtained after coculture in human endometrial epithelial cells versus the sequential system. Fertil Steril. 2010;93:774–782. e771. doi: 10.1016/j.fertnstert.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Dupont C, Segars J, DeCherney A, Bavister BD, Armant DR, Brenner CA. Incidence of chromosomal mosaicism in morphologically normal nonhuman primate preimplantation embryos. Fertil Steril. 2010;93:2545–2550. doi: 10.1016/j.fertnstert.2009.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner T, Yaman C, Moser M, Sommergruber M, Polz W, Tews G. Embryo fragmentation in vitro and its impact on treatment and pregnancy outcome. Fertil Steril. 2001;76:281–285. doi: 10.1016/s0015-0282(01)01904-5. [DOI] [PubMed] [Google Scholar]
- van Echten-Arends J, Mastenbroek S, et al. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum Reprod Update. 2011;17:620–627. doi: 10.1093/humupd/dmr014. [DOI] [PubMed] [Google Scholar]
- Edwards RG, Fishel SB, Cohen J, Fehilly CB, Purdy JM, Slater JM, Steptoe PC, Webster JM. Factors influencing the success of in vitro fertilization for alleviating human infertility. J In Vitro Fert Embryo Transf. 1984;1:3–23. doi: 10.1007/BF01129615. [DOI] [PubMed] [Google Scholar]
- Eftekhari-Yazdi P, Valojerdi MR, Ashtiani SK, Eslaminejad MB, Karimian L. Effect of fragment removal on blastocyst formation and quality of human embryos. Reprod Biomed Online. 2006;13:823–832. doi: 10.1016/s1472-6483(10)61031-0. [DOI] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Vogt E, Yin H, Gosden R. Spindles, mitochondria and redox potential in ageing oocytes. Reprod Biomed Online. 2004;8:45–58. doi: 10.1016/s1472-6483(10)60497-x. [DOI] [PubMed] [Google Scholar]
- Elling U, Taubenschmid J, Wirnsberger G, O’Malley R, Demers SP, Vanhaelen Q, Shukalyuk AI, Schmauss G, Schramek D, Schnuetgen F, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders AC, Hendrickx AG, Binkerd PE. Abnormal development of blastocysts and blastomeres in the rhesus monkey. Biol Reprod. 1982;26:353–366. doi: 10.1095/biolreprod26.2.353. [DOI] [PubMed] [Google Scholar]
- Evans J, Hannan NJ, Edgell TA, Vollenhoven BJ, Lutjen PJ, Osianlis T, Salamonsen LA, Rombauts LJ. Fresh versus frozen embryo transfer: backing clinical decisions with scientific and clinical evidence. Hum Reprod Update. 2014;20:808–821. doi: 10.1093/humupd/dmu027. [DOI] [PubMed] [Google Scholar]
- Evsikov S, Verlinsky Y. Mosaicism in the inner cell mass of human blastocysts. Hum Reprod. 1998;13:3151–3155. doi: 10.1093/humrep/13.11.3151. [DOI] [PubMed] [Google Scholar]
- Fan HY, Huo LJ, Meng XQ, Zhong ZS, Hou Y, Chen DY, Sun QY. Involvement of calcium/calmodulin-dependent protein kinase II (CaMKII) in meiotic maturation and activation of pig oocytes. Biol Reprod. 2003;69:1552–1564. doi: 10.1095/biolreprod.103.015685. [DOI] [PubMed] [Google Scholar]
- Fatemi HM, Kyrou D, Bourgain C, Van den Abbeel E, Griesinger G, Devroey P. Cryopreserved-thawed human embryo transfer: spontaneous natural cycle is superior to human chorionic gonadotropin-induced natural cycle. Fertil Steril. 2010;94:2054–2058. doi: 10.1016/j.fertnstert.2009.11.036. [DOI] [PubMed] [Google Scholar]
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, de Dios Hourcade J, Lopez-Vidriero I, Benguria A, De Fonseca FR, Gutierrez-Adan A. Analysis of gene transcription alterations at the blastocyst stage related to the long-term consequences of in vitro culture in mice. Reproduction. 2009;137:271–283. doi: 10.1530/REP-08-0265. [DOI] [PubMed] [Google Scholar]
- Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, Kokocinski F, Michel CE, Minasi MG, Greco E. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod. 2014;29:2802–2813. doi: 10.1093/humrep/deu277. [DOI] [PubMed] [Google Scholar]
- Ford JH, Schultz CJ, Correll AT. Chromosome elimination in micronuclei: a common cause of hypoploidy. Am J Hum Genet. 1988;43:733–740. [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, Gordon A, Wells D. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod. 2011;26:480–490. doi: 10.1093/humrep/deq344. [DOI] [PubMed] [Google Scholar]
- Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT., Jr The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101:656–663. e651. doi: 10.1016/j.fertnstert.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Fritz B, Hallermann C, Olert J, Fuchs B, Bruns M, Aslan M, Schmidt S, Coerdt W, Muntefering H, Rehder H. Cytogenetic analyses of culture failures by comparative genomic hybridisation (CGH)-Re-evaluation of chromosome aberration rates in early spontaneous abortions. Eur J Hum Genet. 2001;9:539–547. doi: 10.1038/sj.ejhg.5200669. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schoolcraft WB. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil Steril. 2001;76:1175–1180. doi: 10.1016/s0015-0282(01)02888-6. [DOI] [PubMed] [Google Scholar]
- Giardino D, Corti C, Ballarati L, Colombo D, Sala E, Villa N, Piombo G, Pierluigi M, Faravelli F, Guerneri S, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat Diagn. 2009;29:257–265. doi: 10.1002/pd.2215. [DOI] [PubMed] [Google Scholar]
- Giorgetti C, Terriou P, Auquier P, Hans E, Spach JL, Salzmann J, Roulier R. Embryo score to predict implantation after in-vitro fertilization: based on 957 single embryo transfers. Hum Reprod. 1995;10:2427–2431. doi: 10.1093/oxfordjournals.humrep.a136312. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci U S A. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JD, Wohlferd MM. Incidence and outcome of chromosomal mosaicism found at the time of chorionic villus sampling. Am J Obstet Gynecol. 1997;176:1349–1352. doi: 10.1016/s0002-9378(97)70356-9. discussion 1352–1343. [DOI] [PubMed] [Google Scholar]
- Gott AL, Hardy K, Winston RM, Leese HJ. Non-invasive measurement of pyruvate and glucose uptake and lactate production by single human preimplantation embryos. Hum Reprod. 1990;5:104–108. doi: 10.1093/oxfordjournals.humrep.a137028. [DOI] [PubMed] [Google Scholar]
- Gregan J, Polakova S, Zhang L, Tolic-Norrelykke IM, Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handyside AH. Molecular origin of female meiotic aneuploidies. Biochim Biophys Acta. 2012;1822:1913–1920. doi: 10.1016/j.bbadis.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Hardarson T, Lofman C, Coull G, Sjogren A, Hamberger L, Edwards RG. Internalization of cellular fragments in a human embryo: time-lapse recordings. Reprod Biomed Online. 2002;5:36–38. doi: 10.1016/s1472-6483(10)61594-5. [DOI] [PubMed] [Google Scholar]
- Hardarson T, Hanson C, Lundin K, Hillensjo T, Nilsson L, Stevic J, Reismer E, Borg K, Wikland M, Bergh C. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate: a randomized controlled trial. Hum Reprod. 2008;23:2806–2812. doi: 10.1093/humrep/den217. [DOI] [PubMed] [Google Scholar]
- Hardy K. Apoptosis in the human embryo. Rev Reprod. 1999;4:125–134. doi: 10.1530/ror.0.0040125. [DOI] [PubMed] [Google Scholar]
- Hardy K, Hooper MA, Handyside AH, Rutherford AJ, Winston RM, Leese HJ. Non-invasive measurement of glucose and pyruvate uptake by individual human oocytes and preimplantation embryos. Hum Reprod. 1989;4:188–191. doi: 10.1093/oxfordjournals.humrep.a136869. [DOI] [PubMed] [Google Scholar]
- Hardy K, Spanos S, Becker D, Iannelli P, Winston RM, Stark J. From cell death to embryo arrest: mathematical models of human preimplantation embryo development. Proc Natl Acad Sci U S A. 2001;98:1655–1660. doi: 10.1073/pnas.98.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JC, Sengupta SB. Preimplantation genetic diagnosis: state of the art 2011. Hum Genet. 2012;131:175–186. doi: 10.1007/s00439-011-1056-z. [DOI] [PubMed] [Google Scholar]
- Harrison RH, Kuo HC, Scriven PN, Handyside AH, Ogilvie CM. Lack of cell cycle checkpoints in human cleavage stage embryos revealed by a clonal pattern of chromosomal mosaicism analysed by sequential multicolour FISH. Zygote. 2000;8:217–224. doi: 10.1017/s0967199400001015. [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Kato N, Saeki K, Morimoto Y. Selection of high-potential embryos by culture in poly(dimethylsiloxane) microwells and time-lapse imaging. Fertil Steril. 2012;97:332–337. doi: 10.1016/j.fertnstert.2011.11.042. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlinka D, Kalatova B, Uhrinova I, Dolinska S, Rutarova J, Rezacova J, Lazarovska S, Dudas M. Time-lapse cleavage rating predicts human embryo viability. Physiol Res. 2012;61:513–525. doi: 10.33549/physiolres.932287. [DOI] [PubMed] [Google Scholar]
- Hochstenbach R, Buizer-Voskamp JE, Vorstman JA, Ophoff RA. Genome arrays for the detection of copy number variations in idiopathic mental retardation, idiopathic generalized epilepsy and neuropsychiatric disorders: lessons for diagnostic workflow and research. Cytogenet Genome Res. 2011;135:174–202. doi: 10.1159/000332928. [DOI] [PubMed] [Google Scholar]
- Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM, Saunders WS. Resolution of anaphase bridges in cancer cells. Chromosoma. 2004;112:389–397. doi: 10.1007/s00412-004-0284-6. [DOI] [PubMed] [Google Scholar]
- Hoover L, Baker A, Check JH, Lurie D, O’Shaughnessy A. Evaluation of a new embryo-grading system to predict pregnancy rates following in vitro fertilization. Gynecol Obstet Investig. 1995;40:151–157. doi: 10.1159/000292326. [DOI] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- Howe K, FitzHarris G. Recent insights into spindle function in mammalian oocytes and early embryos. Biol Reprod. 2013;89:71. doi: 10.1095/biolreprod.113.112151. [DOI] [PubMed] [Google Scholar]
- Huang A, Adusumalli J, Patel S, Liem J, Williams J, 3rd, Pisarska MD. Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertil Steril. 2009;91:2355–2360. doi: 10.1016/j.fertnstert.2008.03.044. [DOI] [PubMed] [Google Scholar]
- Huang Y, Jiang L, Yi Q, Lv L, Wang Z, Zhao X, Zhong L, Jiang H, Rasool S, Hao Q, et al. Lagging chromosomes entrapped in micronuclei are not ‘lost’ by cells. Cell Res. 2012;22:932–935. doi: 10.1038/cr.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd PJ, Nelson CJ. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief Funct Genomic Proteomic. 2009;8:174–183. doi: 10.1093/bfgp/elp013. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010a;25:1066–1075. doi: 10.1093/humrep/dep452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M, Ryan A, Smotrich D, Rabinowitz M, Murray MJ. Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod. 2010b;16:944–949. doi: 10.1093/molehr/gaq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KT. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum Reprod Update. 2008;14:143–158. doi: 10.1093/humupd/dmm043. [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Varmuza S, Casper RF. Programmed cell death and human embryo fragmentation. Mol Hum Reprod. 1996;2:93–98. doi: 10.1093/molehr/2.2.93. [DOI] [PubMed] [Google Scholar]
- Kalatova B, Jesenska R, Hlinka D, Dudas M. Tripolar mitosis in human cells and embryos: occurrence, pathophysiology and medical implications. Acta Histochem. 2015;117:111–125. doi: 10.1016/j.acthis.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Kallen B, Finnstrom O, Lindam A, Nilsson E, Nygren KG, Olausson PO. Blastocyst versus cleavage stage transfer in in vitro fertilization: differences in neonatal outcome? Fertil Steril. 2010;94:1680–1683. doi: 10.1016/j.fertnstert.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Kalousek DK. Pathogenesis of chromosomal mosaicism and its effect on early human development. Am J Med Genet. 2000;91:39–45. doi: 10.1002/(sici)1096-8628(20000306)91:1<39::aid-ajmg7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Kalousek DK, Dill FJ. Chromosomal mosaicism confined to the placenta in human conceptions. Science. 1983;221:665–667. doi: 10.1126/science.6867735. [DOI] [PubMed] [Google Scholar]
- Kalousek DK, Vekemans M. Confined placental mosaicism. J Med Genet. 1996;33:529–533. doi: 10.1136/jmg.33.7.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser DJ, Racowsky C. Clinical outcomes following selection of human preimplantation embryos with time-lapse monitoring: a systematic review. Hum Reprod Update. 2014;20:617–631. doi: 10.1093/humupd/dmu023. [DOI] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Trounson AO, Cram DS. Chromosome 21 mosaic human preimplantation embryos predominantly arise from diploid conceptions. Fertil Steril. 2005;84:634–643. doi: 10.1016/j.fertnstert.2005.03.045. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Gardner DK, Schoolcraft WB. Proteomic analysis of individual human embryos to identify novel biomarkers of development and viability. Fertil Steril. 2006;85:101–107. doi: 10.1016/j.fertnstert.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Keefe DL, Niven-Fairchild T, Powell S, Buradagunta S. Mitochondrial deoxyribonucleic acid deletions in oocytes and reproductive aging in women. Fertil Steril. 1995;64:577–583. [PubMed] [Google Scholar]
- Keltz MD, Skorupski JC, Bradley K, Stein D. Predictors of embryo fragmentation and outcome after fragment removal in in vitro fertilization. Fertil Steril. 2006;86:321–324. doi: 10.1016/j.fertnstert.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Reik W, Feil R. Culture of preimplantation embryos and its long-term effects on gene expression and phenotype. Hum Reprod Update. 2001;7:419–427. doi: 10.1093/humupd/7.4.419. [DOI] [PubMed] [Google Scholar]
- Kiessling AA, Bletsa R, Desmarais B, Mara C, Kallianidis K, Loutradis D. Evidence that human blastomere cleavage is under unique cell cycle control. J Assist Reprod Genet. 2009;26:187–195. doi: 10.1007/s10815-009-9306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling AA, Bletsa R, Desmarais B, Mara C, Kallianidis K, Loutradis D. Genome-wide microarray evidence that 8-cell human blastomeres over-express cell cycle drivers and under-express checkpoints. J Assist Reprod Genet. 2010;27:265–276. doi: 10.1007/s10815-010-9407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K, Kesmodel US, Hindkjaer JJ, Ingerslev HJ. Time-lapse parameters as predictors of blastocyst development and pregnancy outcome in embryos from good prognosis patients: a prospective cohort study. Hum Reprod. 2013;28:2643–2651. doi: 10.1093/humrep/det300. [DOI] [PubMed] [Google Scholar]
- Kissin DM, Kawwass JF, Monsour M, Boulet SL, Session DR, Jamieson DJ. Assisted hatching: trends and pregnancy outcomes, United States, 2000–2010. Fertil Steril. 2014;102:795–801. doi: 10.1016/j.fertnstert.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Cuppen E. Chromothripsis in congenital disorders and cancer: similarities and differences. Curr Opin Cell Biol. 2013;25:341–348. doi: 10.1016/j.ceb.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011a;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011b;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- Krisher RL, Schoolcraft WB, Katz-Jaffe MG. Omics as a window to view embryo viability. Fertil Steril. 2015;103:333–341. doi: 10.1016/j.fertnstert.2014.12.116. [DOI] [PubMed] [Google Scholar]
- Kroener L, Ambartsumyan G, Briton-Jones C, Dumesic D, Surrey M, Munne S, Hill D. The effect of timing of embryonic progression on chromosomal abnormality. Fertil Steril. 2012;98:876–880. doi: 10.1016/j.fertnstert.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Kuliev A, Verlinsky Y. Meiotic and mitotic nondisjunction: lessons from preimplantation genetic diagnosis. Hum Reprod Update. 2004;10:401–407. doi: 10.1093/humupd/dmh036. [DOI] [PubMed] [Google Scholar]
- Kuo HC, Ogilvie CM, Handyside AH. Chromosomal mosaicism in cleavage-stage human embryos and the accuracy of single-cell genetic analysis. J Assist Reprod Genet. 1998;15:276–280. doi: 10.1023/A:1022588326219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb NE, Sherman SL, Hassold TJ. Effect of meiotic recombination on the production of aneuploid gametes in humans. Cytogenet Genome Res. 2005;111:250–255. doi: 10.1159/000086896. [DOI] [PubMed] [Google Scholar]
- Lane SI, Yun Y, Jones KT. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development. 2012;139:1947–1955. doi: 10.1242/dev.077040. [DOI] [PubMed] [Google Scholar]
- Lebedev IN, Ostroverkhova NV, Nikitina TV, Sukhanova NN, Nazarenko SA. Features of chromosomal abnormalities in spontaneous abortion cell culture failures detected by interphase FISH analysis. Eur J Hum Genet. 2004;12:513–520. doi: 10.1038/sj.ejhg.5201178. [DOI] [PubMed] [Google Scholar]
- Ledbetter DH, Zachary JM, Simpson JL, Golbus MS, Pergament E, Jackson L, Mahoney MJ, Desnick RJ, Schulman J, Copeland KL, et al. Cytogenetic results from the U.S. Collaborative Study on CVS. Prenat Diagn. 1992;12:317–345. doi: 10.1002/pd.1970120503. [DOI] [PubMed] [Google Scholar]
- Lee KF, Yeung WS. Gamete/embryo - oviduct interactions: implications on in vitro culture. Hum Fertil. 2006;9:137–143. doi: 10.1080/14647270600636467. [DOI] [PubMed] [Google Scholar]
- Leeb M, Walker R, Mansfield B, Nichols J, Smith A, Wutz A. Germline potential of parthenogenetic haploid mouse embryonic stem cells. Development. 2012;139:3301–3305. doi: 10.1242/dev.083675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmen JG, Agerholm I, Ziebe S. Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod Biomed Online. 2008;17:385–391. doi: 10.1016/s1472-6483(10)60222-2. [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot DA, Kouznetsova A, Mahdy E, Wilbertz J, Hoog C. The fate of mosaic aneuploid embryos during mouse development. Dev Biol. 2006;289:384–394. doi: 10.1016/j.ydbio.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Lin G, OuYang Q, Zhou X, Gu Y, Yuan D, Li W, Liu G, Liu T, Lu G. A highly homozygous and parthenogenetic human embryonic stem cell line derived from a one-pronuclear oocyte following in vitro fertilization procedure. Cell Res. 2007;17:999–1007. doi: 10.1038/cr.2007.97. [DOI] [PubMed] [Google Scholar]
- Lindner GM, Wright RW., Jr Bovine embryo morphology and evaluation. Theriogenology. 1983;20:407–416. doi: 10.1016/0093-691x(83)90201-7. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang W, Sun X, Liu L, Jin H, Li M, Witz C, Williams D, Griffith J, Skorupski J, et al. DNA microarray reveals that high proportions of human blastocysts from women of advanced maternal age are aneuploid and mosaic. Biol Reprod. 2012a;87:148. doi: 10.1095/biolreprod.112.103192. [DOI] [PubMed] [Google Scholar]
- Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A. 2012b;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012c;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chapple V, Roberts P, Ali J, Matson P. Time-lapse videography of human oocytes following intracytoplasmic sperm injection: events up to the first cleavage division. Reprod Biol. 2014a;14:249–256. doi: 10.1016/j.repbio.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang M, Hou D, Han X, Jin Y, Zhao L, Nie X, Zhou X, Yun T, Zhao Y, et al. Karyotype characterization of in vivo- and in vitro-derived porcine parthenogenetic cell lines. PLoS One. 2014b;9:e97974. doi: 10.1371/journal.pone.0097974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chapple V, Feenan K, Roberts P, Matson P. Time-lapse videography of human embryos: Using pronuclear fading rather than insemination in IVF and ICSI cycles removes inconsistencies in time to reach early cleavage milestones. Reprod Biol. 2015;15:122–125. doi: 10.1016/j.repbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Los FJ, Van Opstal D, van den Berg C. The development of cytogenetically normal, abnormal and mosaic embryos: a theoretical model. Hum Reprod Update. 2004;10:79–94. doi: 10.1093/humupd/dmh005. [DOI] [PubMed] [Google Scholar]
- Loup V, Bernicot I, Janssens P, Hedon B, Hamamah S, Pellestor F, Anahory T. Combined FISH and PRINS sperm analysis of complex chromosome rearrangement t(1;19;13): an approach facilitating PGD. Mol Hum Reprod. 2010;16:111–116. doi: 10.1093/molehr/gap105. [DOI] [PubMed] [Google Scholar]
- Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macera MJ, Sobrino A, Levy B, Jobanputra V, Aggarwal V, Mills A, Esteves C, Hanscom C, Pereira S, Pillalamarri V, et al. Prenatal diagnosis of chromothripsis, with nine breaks characterized by karyotyping, FISH, microarray and whole-genome sequencing. Prenat Diagn. 2015;35:299–301. doi: 10.1002/pd.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklon NS, Geraedts JP, Fauser BC. Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Hum Reprod Update. 2002;8:333–343. doi: 10.1093/humupd/8.4.333. [DOI] [PubMed] [Google Scholar]
- Magli MC, Jones GM, Gras L, Gianaroli L, Korman I, Trounson AO. Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod. 2000;15:1781–1786. doi: 10.1093/humrep/15.8.1781. [DOI] [PubMed] [Google Scholar]
- Mantikou E, Wong KM, Repping S, Mastenbroek S. Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta. 2012;1822:1921–1930. doi: 10.1016/j.bbadis.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 2012;9:e1001356. doi: 10.1371/journal.pmed.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, Vogel NE, Arts EG, de Vries JW, Bossuyt PM, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science. 2015;348:235–238. doi: 10.1126/science.aaa3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott DH, Gao JL, Liu Q, Siwicki M, Martens C, Jacobs P, Velez D, Yim E, Bryke CR, Hsu N, et al. Chromothriptic cure of WHIM syndrome. Cell. 2015;160:686–699. doi: 10.1016/j.cell.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McReynolds S, Vanderlinden L, Stevens J, Hansen K, Schoolcraft WB, Katz-Jaffe MG. Lipocalin-1: a potential marker for noninvasive aneuploidy screening. Fertil Steril. 2011;95:2631–2633. doi: 10.1016/j.fertnstert.2011.01.141. [DOI] [PubMed] [Google Scholar]
- Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- Meseguer M, Rubio I, Cruz M, Basile N, Marcos J, Requena A. Embryo incubation and selection in a time-lapse monitoring system improves pregnancy outcome compared with a standard incubator: a retrospective cohort study. Fertil Steril. 2012;98:1481–1489. e1410. doi: 10.1016/j.fertnstert.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Mio Y, Maeda K. Time-lapse cinematography of dynamic changes occurring during in vitro development of human embryos. Am J Obstet Gynecol. 2008;199:660, e661–e665. doi: 10.1016/j.ajog.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Molder A, Drury S, Costen N, Hartshorne GM, Czanner S. Semiautomated analysis of embryoscope images: using localized variance of image intensity to detect embryo developmental stages. Cytometry A. 2015;87:119–128. doi: 10.1002/cyto.a.22611. [DOI] [PubMed] [Google Scholar]
- Mottla GL, Adelman MR, Hall JL, Gindoff PR, Stillman RJ, Johnson KE. Lineage tracing demonstrates that blastomeres of early cleavage-stage human pre-embryos contribute to both trophectoderm and inner cell mass. Hum Reprod. 1995;10:384–391. doi: 10.1093/oxfordjournals.humrep.a135949. [DOI] [PubMed] [Google Scholar]
- Munne S. Chromosome abnormalities and their relationship to morphology and development of human embryos. Reprod Biomed Online. 2006;12:234–253. doi: 10.1016/s1472-6483(10)60866-8. [DOI] [PubMed] [Google Scholar]
- Munne S, Weier HU. Simultaneous enumeration of chromosomes 13, 18, 21, X, and Y in interphase cells for preimplantation genetic diagnosis of aneuploidy. Cytogenet Cell Genet. 1996;75:263–270. doi: 10.1159/000134497. [DOI] [PubMed] [Google Scholar]
- Munne S, Grifo J, Cohen J, Weier HU. Chromosome abnormalities in human arrested preimplantation embryos: a multiple-probe FISH study. Am J Hum Genet. 1994;55:150–159. [PMC free article] [PubMed] [Google Scholar]
- Munne S, Alikani M, Tomkin G, Grifo J, Cohen J. Embryo morphology, developmental rates, and maternal age are correlated with chromosome abnormalities. Fertil Steril. 1995;64:382–391. [PubMed] [Google Scholar]
- Munne S, Sandalinas M, Escudero T, Marquez C, Cohen J. Chromosome mosaicism in cleavage-stage human embryos: evidence of a maternal age effect. Reprod Biomed Online. 2002;4:223–232. doi: 10.1016/s1472-6483(10)61810-x. [DOI] [PubMed] [Google Scholar]
- Munne S, Velilla E, Colls P, Garcia Bermudez M, Vemuri MC, Steuerwald N, Garrisi J, Cohen J. Self-correction of chromosomally abnormal embryos in culture and implications for stem cell production. Fertil Steril. 2005;84:1328–1334. doi: 10.1016/j.fertnstert.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Munne S, Chen S, Colls P, Garrisi J, Zheng X, Cekleniak N, Lenzi M, Hughes P, Fischer J, Garrisi M, et al. Maternal age, morphology, development and chromosome abnormalities in over 6000 cleavage-stage embryos. Reprod Biomed Online. 2007;14:628–634. doi: 10.1016/s1472-6483(10)61057-7. [DOI] [PubMed] [Google Scholar]
- Nadal-Desbarats L, Veau S, Blasco H, Emond P, Royere D, Andres CR, Guerif F. Is NMR metabolic profiling of spent embryo culture media useful to assist in vitro human embryo selection? MAGMA. 2013;26:193–202. doi: 10.1007/s10334-012-0331-x. [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara T, Iwase A, Goto M, Harata T, Suzuki M, Ienaga M, Kobayashi H, Takikawa S, Manabe S, Kikkawa F, et al. Evaluation of the safety of time-lapse observations for human embryos. J Assist Reprod Genet. 2010;27:93–96. doi: 10.1007/s10815-010-9385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordqvist S, Sydsjo G, Lampic C, Akerud H, Elenis E, Skoog Svanberg A. Sexual orientation of women does not affect outcome of fertility treatment with donated sperm. Hum Reprod. 2014;29:704–711. doi: 10.1093/humrep/det445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norppa H, Falck GC. What do human micronuclei contain? Mutagenesis. 2003;18:221–233. doi: 10.1093/mutage/18.3.221. [DOI] [PubMed] [Google Scholar]
- Northrop LE, Treff NR, Levy B, Scott RT., Jr SNP microarray-based 24 chromosome aneuploidy screening demonstrates that cleavage-stage FISH poorly predicts aneuploidy in embryos that develop to morphologically normal blastocysts. Mol Hum Reprod. 2010;16:590–600. doi: 10.1093/molehr/gaq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obasaju M, Kadam A, Sultan K, Fateh M, Munne S. Sperm quality may adversely affect the chromosome constitution of embryos that result from intracytoplasmic sperm injection. Fertil Steril. 1999;72:1113–1115. doi: 10.1016/s0015-0282(99)00391-x. [DOI] [PubMed] [Google Scholar]
- Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development. 2008;135:259–269. doi: 10.1242/dev.011445. [DOI] [PubMed] [Google Scholar]
- Orr B, Compton DA. A double-edged sword: how oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front Oncol. 2013;3:164. doi: 10.3389/fonc.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Palermo G, Munne S, Cohen J. The human zygote inherits its mitotic potential from the male gamete. Hum Reprod. 1994;9:1220–1225. doi: 10.1093/oxfordjournals.humrep.a138682. [DOI] [PubMed] [Google Scholar]
- Palermo GD, Colombero LT, Rosenwaks Z. The human sperm centrosome is responsible for normal syngamy and early embryonic development. Rev Reprod. 1997;2:19–27. doi: 10.1530/ror.0.0020019. [DOI] [PubMed] [Google Scholar]
- Paull D, Emmanuele V, Weiss KA, Treff N, Stewart L, Hua H, Zimmer M, Kahler DJ, Goland RS, Noggle SA, et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature. 2013;493:632–637. doi: 10.1038/nature11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D, Flaherty SP, Barry MF, Matthews CD. Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum Reprod. 1997;12:532–541. doi: 10.1093/humrep/12.3.532. [DOI] [PubMed] [Google Scholar]
- Pelinck MJ, Hoek A, Simons AH, Heineman MJ, van Echten-Arends J, Arts EG. Embryo quality and impact of specific embryo characteristics on ongoing implantation in unselected embryos derived from modified natural cycle in vitro fertilization. Fertil Steril. 2010;94:527–534. doi: 10.1016/j.fertnstert.2009.03.076. [DOI] [PubMed] [Google Scholar]
- Pellestor F. Chromothripsis: how does such a catastrophic event impact human reproduction? Hum Reprod. 2014;29:388–393. doi: 10.1093/humrep/deu003. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Girardet A, Andreo B, Arnal F, Humeau C. Relationship between morphology and chromosomal constitution in human preimplantation embryo. Mol Reprod Dev. 1994;39:141–146. doi: 10.1002/mrd.1080390204. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Andreo B, Arnal F, Humeau C, Demaille J. Mechanisms of non-disjunction in human female meiosis: the coexistence of two modes of malsegregation evidenced by the karyotyping of 1397 in-vitro unfertilized oocytes. Hum Reprod. 2002;17:2134–2145. doi: 10.1093/humrep/17.8.2134. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Anahory T, Lefort G, Puechberty J, Liehr T, Hedon B, Sarda P. Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update. 2011;17:476–494. doi: 10.1093/humupd/dmr010. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Gatinois V, Puechberty J, Genevieve D, Lefort G. Chromothripsis: potential origin in gametogenesis and preimplantation cell divisions. A review. Fertil Steril. 2014;102:1785–1796. doi: 10.1016/j.fertnstert.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Peramo B, Ricciarelli E, Cuadros-Fernandez JM, Huguet E, Hernandez ER. Blastocyst transfer and monozygotic twinning. Fertil Steril. 1999;72:1116–1117. doi: 10.1016/s0015-0282(99)00412-4. [DOI] [PubMed] [Google Scholar]
- Pereda J, Croxatto HB. Ultrastructure of a seven-cell human embryo. Biol Reprod. 1978;18:481–489. doi: 10.1095/biolreprod18.3.481. [DOI] [PubMed] [Google Scholar]
- Pessot CA, Brito M, Figueroa J, Concha II, Yanez A, Burzio LO. Presence of RNA in the sperm nucleus. Biochem Biophys Res Commun. 1989;158:272–278. doi: 10.1016/s0006-291x(89)80208-6. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Phillips OP, Tharapel AT, Lerner JL, Park VM, Wachtel SS, Shulman LP. Risk of fetal mosaicism when placental mosaicism is diagnosed by chorionic villus sampling. Am J Obstet Gynecol. 1996;174:850–855. doi: 10.1016/s0002-9378(96)70312-5. [DOI] [PubMed] [Google Scholar]
- Picton HM, Elder K, Houghton FD, Hawkhead JA, Rutherford AJ, Hogg JE, Leese HJ, Harris SE. Association between amino acid turnover and chromosome aneuploidy during human preimplantation embryo development in vitro. Mol Hum Reprod. 2010;16:557–569. doi: 10.1093/molehr/gaq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre A, Gautier M, Callebaut I, Bontoux M, Jeanpierre E, Pontarotti P, Monget P. Atypical structure and phylogenomic evolution of the new eutherian oocyte- and embryo-expressed KHDC1/DPPA5/ECAT1/OOEP gene family. Genomics. 2007;90:583–594. doi: 10.1016/j.ygeno.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Prados FJ, Debrock S, Lemmen JG, Agerholm I. The cleavage stage embryo. Hum Reprod. 2012;27(Suppl 1):i50–i71. doi: 10.1093/humrep/des224. [DOI] [PubMed] [Google Scholar]
- Pribenszky C, Losonczi E, Molnar M, Lang Z, Matyas S, Rajczy K, Molnar K, Kovacs P, Nagy P, Conceicao J, et al. Prediction of in-vitro developmental competence of early cleavage-stage mouse embryos with compact time-lapse equipment. Reprod Biomed Online. 2010;20:371–379. doi: 10.1016/j.rbmo.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, Blackshear PJ. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- Rao X, Zhang Y, Yi Q, Hou H, Xu B, Chu L, Huang Y, Zhang W, Fenech M, Shi Q. Multiple origins of spontaneously arising micronuclei in HeLa cells: direct evidence from long-term live cell imaging. Mutat Res. 2008;646:41–49. doi: 10.1016/j.mrfmmm.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Rawe VY, Terada Y, Nakamura S, Chillik CF, Olmedo SB, Chemes HE. A pathology of the sperm centriole responsible for defective sperm aster formation, syngamy and cleavage. Hum Reprod. 2002;17:2344–2349. doi: 10.1093/humrep/17.9.2344. [DOI] [PubMed] [Google Scholar]
- Rienzi L, Capalbo A, Stoppa M, Romano S, Maggiulli R, Albricci L, Scarica C, Farcomeni A, Vajta G, Ubaldi FM. No evidence of association between blastocyst aneuploidy and morphokinetic assessment in a selected population of poor-prognosis patients: a longitudinal cohort study. Reprod Biomed Online. 2015;30:57–66. doi: 10.1016/j.rbmo.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Rius M, Obradors A, Daina G, Cuzzi J, Marques L, Calderon G, Velilla E, Martinez-Passarell O, Oliver-Bonet M, Benet J, et al. Reliability of short comparative genomic hybridization in fibroblasts and blastomeres for a comprehensive aneuploidy screening: first clinical application. Hum Reprod. 2010;25:1824–1835. doi: 10.1093/humrep/deq118. [DOI] [PubMed] [Google Scholar]
- Rius M, Daina G, Obradors A, Ramos L, Velilla E, Fernandez S, Martinez-Passarell O, Benet J, Navarro J. Comprehensive embryo analysis of advanced maternal age-related aneuploidies and mosaicism by short comparative genomic hybridization. Fertil Steril. 2011;95:413–416. doi: 10.1016/j.fertnstert.2010.07.1051. [DOI] [PubMed] [Google Scholar]
- Roest HP, Baarends WM, de Wit J, van Klaveren JW, Wassenaar E, Hoogerbrugge JW, van Cappellen WA, Hoeijmakers JH, Grootegoed JA. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol Cell Biol. 2004;24:5485–5495. doi: 10.1128/MCB.24.12.5485-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio I, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escriba MJ, Bellver J, Meseguer M. Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil Steril. 2012;98:1458–1463. doi: 10.1016/j.fertnstert.2012.07.1135. [DOI] [PubMed] [Google Scholar]
- Rubio I, Galan A, Larreategui Z, Ayerdi F, Bellver J, Herrero J, Meseguer M. Clinical validation of embryo culture and selection by morphokinetic analysis: a randomized, controlled trial of the EmbryoScope. Fertil Steril. 2014 doi: 10.1016/j.fertnstert.2014.07.738. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Lu C, Zulfikaroglu E, Neuber E, Taylor HS. A soluble molecule secreted by human blastocysts modulates regulation of HOXA10 expression in an epithelial endometrial cell line. Fertil Steril. 2003;80:1169–1174. doi: 10.1016/s0015-0282(03)02163-0. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Kola I, Osborne J, Trounson A, Ng SC, Bongso A, Ratnam SS. Centrioles in the beginning of human development. Proc Natl Acad Sci U S A. 1991;88:4806–4810. doi: 10.1073/pnas.88.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathananthan AH, Selvaraj K, Girijashankar ML, Ganesh V, Selvaraj P, Trounson AO. From oogonia to mature oocytes: inactivation of the maternal centrosome in humans. Microsc Res Tech. 2006;69:396–407. doi: 10.1002/jemt.20299. [DOI] [PubMed] [Google Scholar]
- Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K, Lai FA. PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development. 2002;129:3533–3544. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- Schatten G, Simerly C, Schatten H. Maternal inheritance of centrosomes in mammals? Studies on parthenogenesis and polyspermy in mice. Proc Natl Acad Sci U S A. 1991;88:6785–6789. doi: 10.1073/pnas.88.15.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoolcraft WB, Fragouli E, Stevens J, Munne S, Katz-Jaffe MG, Wells D. Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil Steril. 2010;94:1700–1706. doi: 10.1016/j.fertnstert.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Scott RT, Jr, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril. 2013a;100:624–630. doi: 10.1016/j.fertnstert.2013.04.039. [DOI] [PubMed] [Google Scholar]
- Scott RT, Jr, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, Tao X, Treff NR. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril. 2013b;100:697–703. doi: 10.1016/j.fertnstert.2013.04.035. [DOI] [PubMed] [Google Scholar]
- Seli E, Sakkas D, Scott R, Kwok SC, Rosendahl SM, Burns DH. Noninvasive metabolomic profiling of embryo culture media using Raman and near-infrared spectroscopy correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2007;88:1350–1357. doi: 10.1016/j.fertnstert.2007.07.1390. [DOI] [PubMed] [Google Scholar]
- Seli E, Botros L, Sakkas D, Burns DH. Noninvasive metabolomic profiling of embryo culture media using proton nuclear magnetic resonance correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2008;90:2183–2189. doi: 10.1016/j.fertnstert.2008.07.1739. [DOI] [PubMed] [Google Scholar]
- Seli E, Vergouw CG, Morita H, Botros L, Roos P, Lambalk CB, Yamashita N, Kato O, Sakkas D. Noninvasive metabolomic profiling as an adjunct to morphology for noninvasive embryo assessment in women undergoing single embryo transfer. Fertil Steril. 2010a;94:535–542. doi: 10.1016/j.fertnstert.2009.03.078. [DOI] [PubMed] [Google Scholar]
- Seli E, Robert C, Sirard MA. OMICS in assisted reproduction: possibilities and pitfalls. Mol Hum Reprod. 2010b;16:513–530. doi: 10.1093/molehr/gaq041. [DOI] [PubMed] [Google Scholar]
- Sendler E, Johnson GD, Mao SH, Goodrich RJ, Diamond MP, Hauser R, Krawetz SA. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013;41:4104–4117. doi: 10.1093/nar/gkt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman SL, Lamb NE, Feingold E. Relationship of recombination patterns and maternal age among non-disjoined chromosomes 21. Biochem Soc Trans. 2006;34:578–580. doi: 10.1042/BST0340578. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Shimura T, Tanaka T. Selective elimination of acentric double minutes from cancer cells through the extrusion of micronuclei. Mutat Res. 2000;448:81–90. doi: 10.1016/s0027-5107(00)00003-8. [DOI] [PubMed] [Google Scholar]
- Silber S, Escudero T, Lenahan K, Abdelhadi I, Kilani Z, Munne S. Chromosomal abnormalities in embryos derived from testicular sperm extraction. Fertil Steril. 2003;79:30–38. doi: 10.1016/s0015-0282(02)04407-2. [DOI] [PubMed] [Google Scholar]
- Slama R, Eustache F, Ducot B, Jensen TK, Jorgensen N, Horte A, Irvine S, Suominen J, Andersen AG, Auger J, et al. Time to pregnancy and semen parameters: a cross-sectional study among fertile couples from four European cities. Hum Reprod. 2002;17:503–515. doi: 10.1093/humrep/17.2.503. [DOI] [PubMed] [Google Scholar]
- Soler D, Pampalona J, Tusell L, Genesca A. Radiation sensitivity increases with proliferation-associated telomere dysfunction in nontransformed human epithelial cells. Aging Cell. 2009;8:414–425. doi: 10.1111/j.1474-9726.2009.00488.x. [DOI] [PubMed] [Google Scholar]
- Sritanaudomchai H, Ma H, Clepper L, Gokhale S, Bogan R, Hennebold J, Wolf D, Mitalipov S. Discovery of a novel imprinted gene by transcriptional analysis of parthenogenetic embryonic stem cells. Hum Reprod. 2010;25:1927–1941. doi: 10.1093/humrep/deq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staessen C, Verpoest W, Donoso P, Haentjens P, Van der Elst J, Liebaers I, Devroey P. Preimplantation genetic screening does not improve delivery rate in women under the age of 36 following single-embryo transfer. Hum Reprod. 2008;23:2818–2825. doi: 10.1093/humrep/den367. [DOI] [PubMed] [Google Scholar]
- Stensen MH, Tanbo TG, Storeng R, Abyholm T, Fedorcsak P. Fragmentation of human cleavage-stage embryos is related to the progression through meiotic and mitotic cell cycles. Fertil Steril. 2015;103:374–381. e374. doi: 10.1016/j.fertnstert.2014.10.031. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet. 1978;2:366. doi: 10.1016/s0140-6736(78)92957-4. [DOI] [PubMed] [Google Scholar]
- Stigliani S, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA content in embryo culture medium is significantly associated with human embryo fragmentation. Hum Reprod. 2013;28:2652–2660. doi: 10.1093/humrep/det314. [DOI] [PubMed] [Google Scholar]
- Sugimura S, Akai T, Somfai T, Hirayama M, Aikawa Y, Ohtake M, Hattori H, Kobayashi S, Hashiyada Y, Konishi K, et al. Time-lapse cinematography-compatible polystyrene-based microwell culture system: a novel tool for tracking the development of individual bovine embryos. Biol Reprod. 2010;83:970–978. doi: 10.1095/biolreprod.110.085522. [DOI] [PubMed] [Google Scholar]
- Sugimura S, Akai T, Hashiyada Y, Somfai T, Inaba Y, Hirayama M, Yamanouchi T, Matsuda H, Kobayashi S, Aikawa Y, et al. Promising system for selecting healthy in vitro-fertilized embryos in cattle. PLoS One. 2012;7:e36627. doi: 10.1371/journal.pone.0036627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Neri QV, Katagiri Y, Rosenwaks Z, Palermo GD. Effect of treating induced mitochondrial damage on embryonic development and epigenesis. Biol Reprod. 2005;72:584–592. doi: 10.1095/biolreprod.104.032391. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Ernst C, Heilbut A, Chiang C, Hanscom C, Lindgren A, Kirby A, Liu S, Muddukrishna B, Ohsumi TK, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro F, Kanai-Azuma M, Miyazaki S, Kato M, Tanaka T, Toyoda S, Yamato E, Kawakami H, Miyazaki T, Miyazaki J. Maternal-effect gene Ces5/Ooep/Moep19/Floped is essential for oocyte cytoplasmic lattice formation and embryonic development at the maternal-zygotic stage transition. Genes Cells. 2010;15:813–828. doi: 10.1111/j.1365-2443.2010.01420.x. [DOI] [PubMed] [Google Scholar]
- Taylor DM, Ray PF, Ao A, Winston RM, Handyside AH. Paternal transcripts for glucose-6-phosphate dehydrogenase and adenosine deaminase are first detectable in the human preimplantation embryo at the three- to four-cell stage. Mol Reprod Dev. 1997;48:442–448. doi: 10.1002/(SICI)1098-2795(199712)48:4<442::AID-MRD4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Terradas M, Martin M, Tusell L, Genesca A. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair. 2009;8:1225–1234. doi: 10.1016/j.dnarep.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci U S A. 2011;108:17974–17978. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- Treff NR, Scott RT., Jr Four-hour quantitative real-time polymerase chain reaction-based comprehensive chromosome screening and accumulating evidence of accuracy, safety, predictive value, and clinical efficacy. Fertil Steril. 2013;99:1049–1053. doi: 10.1016/j.fertnstert.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Treff NR, Su J, Tao X, Northrop LE, Scott RT., Jr Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Mol Hum Reprod. 2011;17:335–343. doi: 10.1093/molehr/gaq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Turner K, Martin KL, Woodward BJ, Lenton EA, Leese HJ. Comparison of pyruvate uptake by embryos derived from conception and non-conception natural cycles. Hum Reprod. 1994;9:2362–2366. doi: 10.1093/oxfordjournals.humrep.a138453. [DOI] [PubMed] [Google Scholar]
- Utani K, Okamoto A, Shimizu N. Generation of micronuclei during interphase by coupling between cytoplasmic membrane blebbing and nuclear budding. PLoS One. 2011;6:e27233. doi: 10.1371/journal.pone.0027233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15:1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Davis PW, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod. 1995;10:415–424. doi: 10.1093/oxfordjournals.humrep.a135954. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Davis P, Alexander S. A microscopic and biochemical study of fragmentation phenotypes in stage-appropriate human embryos. Hum Reprod. 2001;16:719–729. doi: 10.1093/humrep/16.4.719. [DOI] [PubMed] [Google Scholar]
- Van der Aa N, Zamani Esteki M, Vermeesch JR, Voet T. Preimplantation genetic diagnosis guided by single-cell genomics. Genome Med. 2013;5:71. doi: 10.1186/gm475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, van der Veen F, Repping S. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum Reprod Update. 2011;17:620–627. doi: 10.1093/humupd/dmr014. [DOI] [PubMed] [Google Scholar]
- Van Soom A, Mateusen B, Leroy J, De Kruif A. Assessment of mammalian embryo quality: what can we learn from embryo morphology? Reprod Biomed Online. 2003;7:664–670. doi: 10.1016/s1472-6483(10)62089-5. [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009a;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Melotte C, Debrock S, Sermon K, Staessen C, Liebaers I, Fryns JP, D’Hooghe T, Vermeesch JR. What next for preimplantation genetic screening? High mitotic chromosome instability rate provides the biological basis for the low success rate. Hum Reprod. 2009b;24:2679–2682. doi: 10.1093/humrep/dep266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste E, Van der Aa N, Voet T, Vermeesch JR. Aneuploidy and copy number variation in early human development. Semin Reprod Med. 2012;30:302–308. doi: 10.1055/s-0032-1313909. [DOI] [PubMed] [Google Scholar]
- Vera-Rodriguez M, Chavez SL, Rubio C, Pera RA, Simon C. Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis. Nat Commun. 2015a;6:7601. doi: 10.1038/ncomms8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Rodriguez M, Chavez SL, Rubio C, Reijo Pera RA, Simon C. Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis. Nat Commun. 2015b;6:7601. doi: 10.1038/ncomms8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerMilyea MD, Tan L, Anthony JT, Conaghan J, Ivani K, Gvakharia M, Boostanfar R, Baker VL, Suraj V, Chen AA, et al. Computer-automated time-lapse analysis results correlate with embryo implantation and clinical pregnancy: a blinded, multi-centre study. Reprod Biomed Online. 2014;29:729–736. doi: 10.1016/j.rbmo.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorsanova SG, Kolotii AD, Iourov IY, Monakhov VV, Kirillova EA, Soloviev IV, Yurov YB. Evidence for high frequency of chromosomal mosaicism in spontaneous abortions revealed by inter-phase FISH analysis. J Histochem Cytochem. 2005;53:375–380. doi: 10.1369/jhc.4A6424.2005. [DOI] [PubMed] [Google Scholar]
- Voullaire L, Wilton L, McBain J, Callaghan T, Williamson R. Chromosome abnormalities identified by comparative genomic hybridization in embryos from women with repeated implantation failure. Mol Hum Reprod. 2002;8:1035–1041. doi: 10.1093/molehr/8.11.1035. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang X, Zhang J, Song Z, Wang S, Gao Y, Wang J, Luo Y, Niu Z, Yue X, et al. Detection of chromosomal aneuploidy in human preimplantation embryos by next-generation sequencing. Biol Reprod. 2014;90:95. doi: 10.1095/biolreprod.113.116459. [DOI] [PubMed] [Google Scholar]
- Wei Y, Multi S, Yang CR, Ma J, Zhang QH, Wang ZB, Li M, Wei L, Ge ZJ, Zhang CH, et al. Spindle assembly checkpoint regulates mitotic cell cycle progression during preimplantation embryo development. PLoS One. 2011;6:e21557. doi: 10.1371/journal.pone.0021557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MD, Leondires MP, Schoolcraft WB, Miller BT, Copperman AB, Robins ED, Arredondo F, Hickman TN, Gutmann J, Schillings WJ, et al. Clinically recognizable error rate after the transfer of comprehensive chromosomal screened euploid embryos is low. Fertil Steril. 2014;102:1613–1618. doi: 10.1016/j.fertnstert.2014.09.011. [DOI] [PubMed] [Google Scholar]
- Whitaker M. Calcium at fertilization and in early development. Physiol Rev. 2006;86:25–88. doi: 10.1152/physrev.00023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding M, Dale B, Marino M, di Matteo L, Alviggi C, Pisaturo ML, Lombardi L, De Placido G. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum Reprod. 2001;16:909–917. doi: 10.1093/humrep/16.5.909. [DOI] [PubMed] [Google Scholar]
- Wilton L, Voullaire L, Sargeant P, Williamson R, McBain J. Preimplantation aneuploidy screening using comparative genomic hybridization or fluorescence in situ hybridization of embryos from patients with recurrent implantation failure. Fertil Steril. 2003;80:860–868. doi: 10.1016/s0015-0282(03)01162-2. [DOI] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, Reijo Pera RA. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- Wu X. Maternal depletion of NLRP5 blocks early embryogenesis in rhesus macaque monkeys (Macaca mulatta) Hum Reprod. 2009;24:415–424. doi: 10.1093/humrep/den403. [DOI] [PubMed] [Google Scholar]
- Wu H, Ding C, Shen X, Wang J, Li R, Cai B, Xu Y, Zhong Y, Zhou C. Medium-based noninvasive preimplantation genetic diagnosis for human alpha-thalassemias-SEA. Medicine. 2015;94:e669. doi: 10.1097/MD.0000000000000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthopoulou L, Ghevaria H, Mantzouratou A, Serhal P, Doshi A, Delhanty JD. Chromosome breakage in human preimplantation embryos from carriers of structural chromosomal abnormalities in relation to fragile sites, maternal age, and poor sperm factors. Cytogenet Genome Res. 2012;136:21–29. doi: 10.1159/000334836. [DOI] [PubMed] [Google Scholar]
- Xu J, Cheung T, Chan ST, Ho P, Yeung WS. The incidence of cytoplasmic fragmentation in mouse embryos in vitro is not affected by inhibition of caspase activity. Fertil Steril. 2001;75:986–991. doi: 10.1016/s0015-0282(01)01687-9. [DOI] [PubMed] [Google Scholar]
- Xu B, Sun Z, Liu Z, Guo H, Liu Q, Jiang H, Zou Y, Gong Y, Tischfield JA, Shao C. Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS One. 2011;6:e18618. doi: 10.1371/journal.pone.0018618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Hwang KJ, Kwon HC, Kim HS, Choi KW, Oh KS. Detection of reactive oxygen species (ROS) and apoptosis in human fragmented embryos. Hum Reprod. 1998;13:998–1002. doi: 10.1093/humrep/13.4.998. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, Liu J. Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: a prospective study with sibling oocytes. BMC Med Genomics. 2014;7:38. doi: 10.1186/1755-8794-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Jellerette T, Salicioni AM, Lee HC, Yoo MS, Coward K, Parrington J, Grow D, Cibelli JB, Visconti PE, et al. Human sperm devoid of PLC, zeta 1 fail to induce Ca(2+) release and are unable to initiate the first step of embryo development. J Clin Invest. 2008;118:3671–3681. doi: 10.1172/JCI36942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, Ou-Yang Y, Wang ZB, Zheng P, Zhu MS, et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun. 2014;5:4887. doi: 10.1038/ncomms5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Hoja MR, Wilbertz J, Nordqvist K, Hoog C. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science. 2002;296:1115–1118. doi: 10.1126/science.1070594. [DOI] [PubMed] [Google Scholar]
- Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A. 2009;106:7473–7478. doi: 10.1073/pnas.0900519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebe S, Petersen K, Lindenberg S, Andersen AG, Gabrielsen A, Andersen AN. Embryo morphology or cleavage stage: how to select the best embryos for transfer after in-vitro fertilization. Hum Reprod. 1997;12:1545–1549. doi: 10.1093/humrep/12.7.1545. [DOI] [PubMed] [Google Scholar]
- Zinaman MJ, Clegg ED, Brown CC, O’Connor J, Selevan SG. Estimates of human fertility and pregnancy loss. Fertil Steril. 1996;65:503–509. [PubMed] [Google Scholar]