Abstract
B-lymphoid transcription factors (e.g. PAX5, IKZF1) are critical for early B-cell development1–2, yet genetic lesions occur in >80% of cases of B-cell acute lymphoblastic leukemia (ALL)3–4. The significance of these lesions in ALL remained unclear. Combining ChIP-seq and RNA-seq studies, we identified a novel B-lymphoid program for transcriptional repression of glucose and energy supply. Our metabolic analyses revealed that PAX5 and IKZF1 enforce a state of chronic energy deprivation, resulting in constitutive activation of the energy-stress sensor AMPK5–7. Dominant-negative mutants of PAX5 and IKZF1 relieved glucose and energy restriction. Studying a transgenic pre-B ALL mouse model, heterozygous deletion of Pax5 increased glucose uptake and ATP-levels by >25-fold. Reconstitution of PAX5 and IKZF1 in pre-B ALL patient samples restored a non-permissive state and induced energy crisis and cell death. A CRISPR/Cas9-based screen of PAX5- and IKZF1- transcriptional targets identified NR3C1 (glucocorticoid receptor)8, TXNIP (glucose feedback sensor)9 and CNR2 (cannabinoid receptor)10 as central effectors of B-lymphoid restriction of glucose and energy supply. Interestingly, transport-independent lipophilic methyl-conjugates of pyruvate and TCA cycle metabolites bypassed the gatekeeper function of PAX5 and IKZF1 and readily enabled leukemic transformation. Conversely, pharmacological TXNIP- and CNR2-agonists and a small molecule AMPK-inhibitor strongly synergized with glucocorticoids, identifying TXNIP, CNR2 and AMPK as potential therapy-targets. Furthermore, our results provide a mechanistic explanation for the empiric finding that glucocorticoids are effective in the treatment of B-lymphoid but not myeloid malignancies. We conclude that B-lymphoid transcription factors function as metabolic gatekeepers by limiting the amount of cellular ATP to levels that are insufficient for malignant transformation.
The transcription factors PAX5, IKZF1, EBF1 and TCF3 are critical for normal B-cell development11 and are opposed by CEBPA, a central driver of myeloid differentiation12. In adipocytes, EBF1 decreases glucose transport13, while CEBPA promotes glucose transport14. Transforming oncogenes (e.g. BCR-ABL1, RAS) impose significant metabolic requirements on glucose and energy supply. Given the high frequency of genetic lesions of B-lymphoid transcription factors in pre-B ALL, we tested the hypothesis that these transcription factors restrict glucose and energy supply and represent a metabolic barrier against malignant transformation.
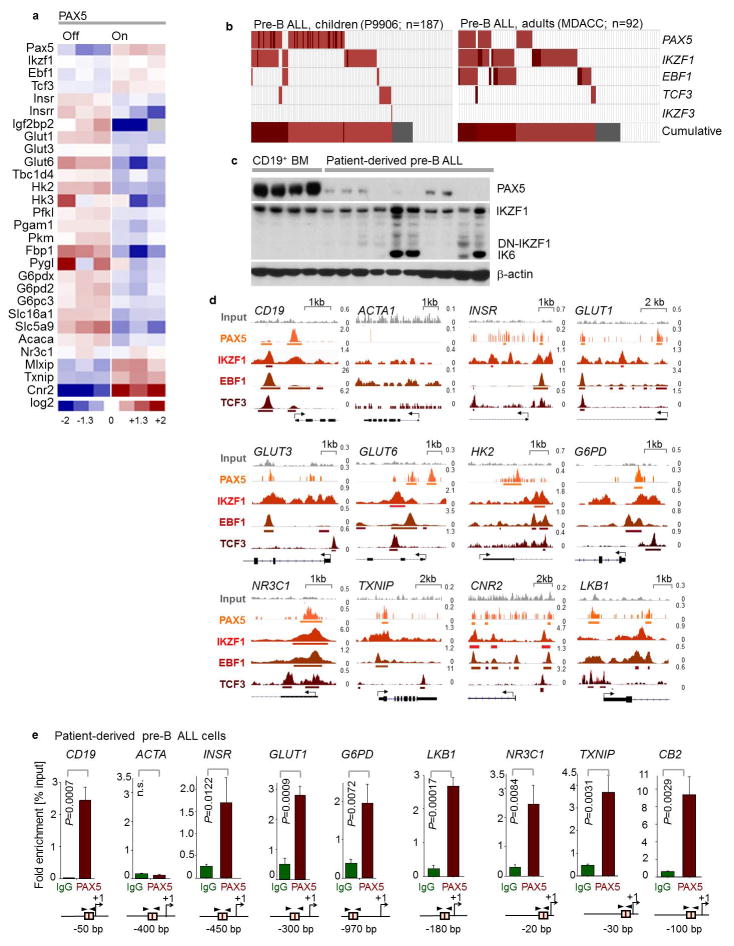
Studying inactivating lesions in PAX5, IKZF1, EBF1 and TCF3 in 279 patient samples from clinical trials for children and adults (P9906, MDACC), we found mutations or deletions in 209 cases. Patient-derived pre-B ALL xenografts studied here exhibited abnormal expression of PAX5 and IKZF1 proteins (Extended Data Fig. 1b–c). Analysis of ChIP-seq data of human B-cells revealed binding of PAX5, IKZF1, EBF1 and TCF3 to promoter regions of INSR, GLUT1, GLUT6, G6PD and HK2, which enable glucose uptake and utilization. We also observed binding peaks at promoter regions of genes that encode negative regulators of glucose uptake, including NR3C1, TXNIP and CNR2. Recruitment of PAX5 to these promoters was confirmed by single-locus quantitative ChIP (Extended Data Fig. 1d–e). Inducible reconstitution of Pax5 (GSE52870) in Pax5-deficient pre-B leukemia cells, downregulated multiple mediators of glucose uptake, while mRNA levels of glucose transport inhibitors increased (MLXIP, TXNIP, CNR2; Extended Data Fig. 1a).
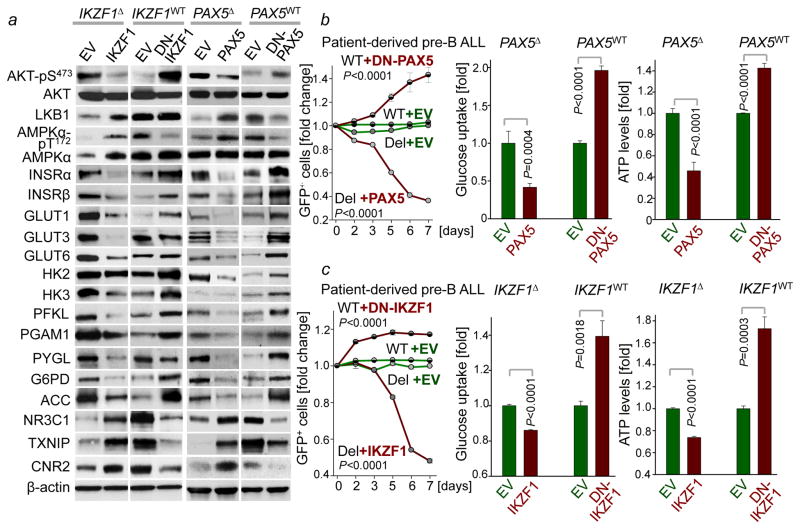
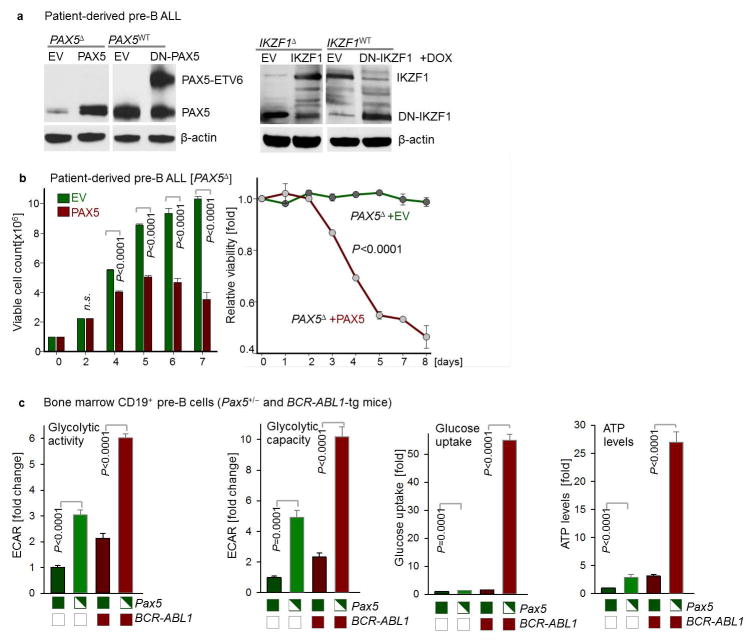
We verified these results in four patient-derived pre-B ALL samples. Inducible reconstitution of wildtype transcription factors in patient-derived pre-B ALL cells lacking PAX5- or IKZF1-function (Fig. 1a) induced activation of the energy-stress sensor LKB1-AMPK15. Consistent with glucose and energy depletion, reconstitution of IKZF1 and PAX5 diminished AKT-activation. Likewise, reconstitution of both IKZF1 and PAX5 decreased protein levels of the insulin receptor (INSR), glucose transporters (GLUT1, GLUT3, GLUT6) as well as glucose-metabolism effectors (HK2, HK3, PFKL, PGAM, PYGL and G6PD). On the contrary, PAX5 and IKZF1 strongly induced expression of glucose transport inhibitors (NR3C1, TXNIP and CNR2)8–10. For loss-of function studies, dominant-negative variants of IKZF1 (DN-IKZF1, lacking the zinc fingers 1–4) and PAX5 (DN-PAX5; PAX5-ETV6 fusion) were cloned from patient samples and inducibly expressed in two pre-B ALL xenografts carrying IKZF1 and PAX5 wildtype alleles (Extended Data Figure 2a). As expected, most of PAX5- and IKZF1-induced changes in protein expression were reversed by DN-IKZF1 and DN-PAX5 (Fig. 1a).
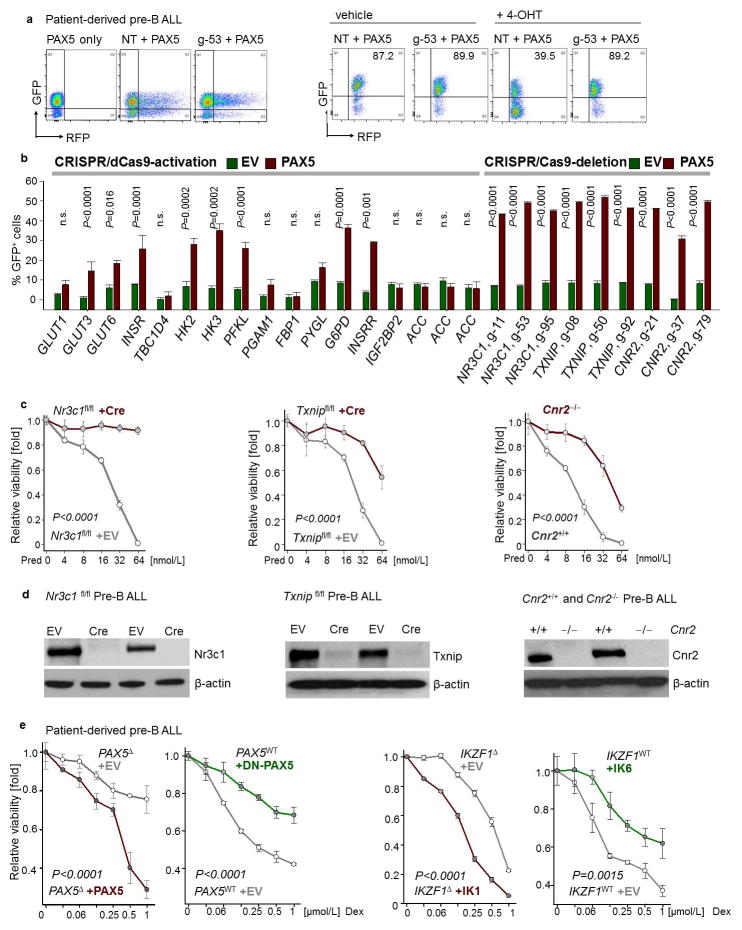
Figure 1. A B-lymphoid transcriptional program to regulate factors of glucose uptake and utilization.
a, Western blots of PAX5-, IKZF1-, DN-PAX5-, and DN-IKZF1-induced changes in patient-derived pre-B ALL cells. b, c, Enrichment or depletion (two-way ANOVA) of pre-B ALL cells carrying GFP-tagged PAX5 (b), IKZF1 (c), DN-PAX5 (b) or DN-IKZF1 (c). Glucose uptake and ATP levels were analyzed by two-tailed t-test. Data, mean ± s.d. (n = 3 independent experiments; b,c). For gel source data, see Supplementary Fig. 1.
Inducible reconstitution of PAX5 or IKZF1 in human pre-B ALL cells resulted in depletion of cells in competitive growth assays, in parallel with reduced glucose uptake and ATP-depletion. Conversely, DN-PAX5 and DN-IKZF1 conferred a net survival advantage as measured in competitive growth assays and increased glucose uptake and cellular ATP levels (Fig. 1b–c). Since oncogenic kinases (e.g. BCR-ABL1) require high amounts of ATP, B-lymphoid transcription factors may function as metabolic gatekeepers by limiting ATP-supply to levels that are insufficient for malignant transformation. To test this scenario, we studied glucose uptake, glycolysis and ATP levels in pre-B cells from Pax5 wildtype and Pax5 haploinsufficient mice16 in the presence and absence of a BCR-ABL1-transgene. In the presence of wildtype Pax5, the BCR-ABL1-transgene failed to increase glucose uptake and ATP levels and glycolytic activity increased only modestly. However, in Pax5-haploinsufficient pre-B cells, BCR-ABL1 massively increased glucose uptake (>50-fold), glycolytic capacity (>10-fold) and levels of cellular ATP (>25-fold). These findings provide genetic evidence for PAX5-mediated restraint of glucose uptake and energy supply in pre-B cells (Extended Data Figure 2).
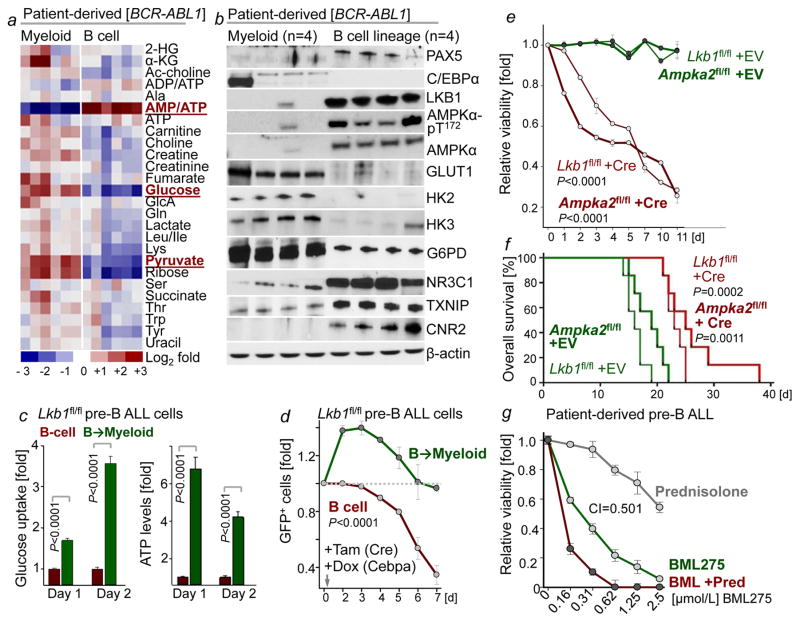
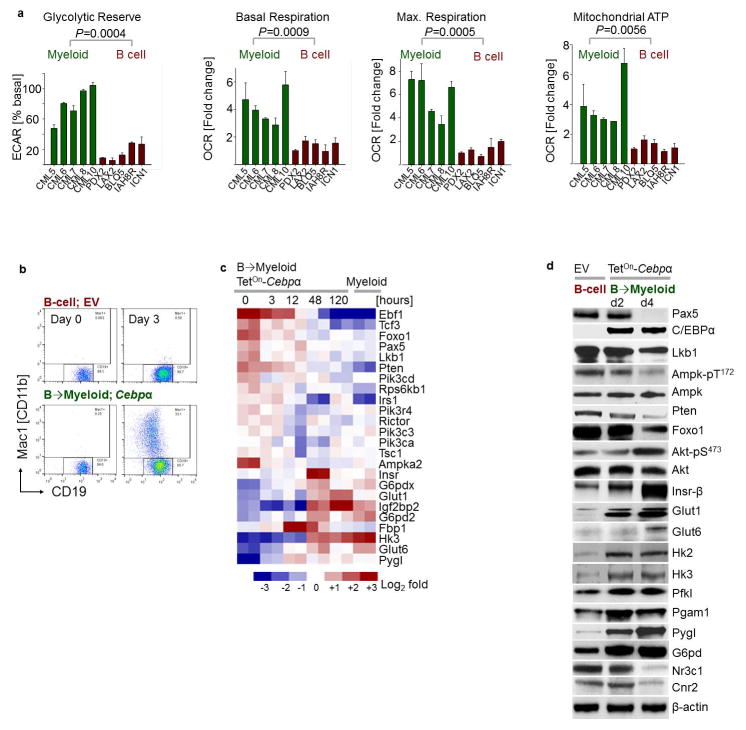
B-lymphoid and myeloid leukemia typically arise from a multipotent progenitor cell and often carry the same oncogenes. Despite their common origin, B-lymphoid and myeloid leukemia fundamentally differ in their biological and clinical characteristics. Given that myeloid (CEBPA) or B-lymphoid (IKZF1, PAX5) fates are instructed by transcription factors that regulate glucose and energy metabolism14–15, we tested the hypothesis that myeloid and B-cell identities are linked to distinct metabolic states. Comparing B-lineage Ph+ ALL and myeloid CML cells, our metabolomic studies revealed striking differences. Intracellular glucose (~10 to 13-fold) and pyruvate (~6 to 11-fold) were present at significantly higher levels in patient-derived myeloid compared to B-lymphoid leukemia cells. Myeloid leukemia cells had abundant ATP reserves, while ATP levels in B-lineage leukemia were strikingly low, indicating a state of chronic energy stress (Fig. 2a). Bioenergetic analyses revealed lower glycolytic reserves in B-lymphoid compared to myeloid leukemia cells (~5 to 14-fold). Basal and maximum respiration, as well as mitochondrial ATP levels were substantially lower in B-lineage leukemia cells (Extended Data Figure 3a), resulting in constuitutive activation of the energy stress sensor LKB1-AMPK. Ph+ ALL cells expressed effectors of glucose utilization (GLUT1, HK2, HK3, G6PD) at >10-fold lower levels, while expression levels of glucose transport inhibitors (NR3C1, TXNIP, CNR2; Fig. 2b) was significantly higher than in CML cells.
Figure 2. LKB1-AMPK is required to balance glucose and energy metabolism in pre-B ALL.
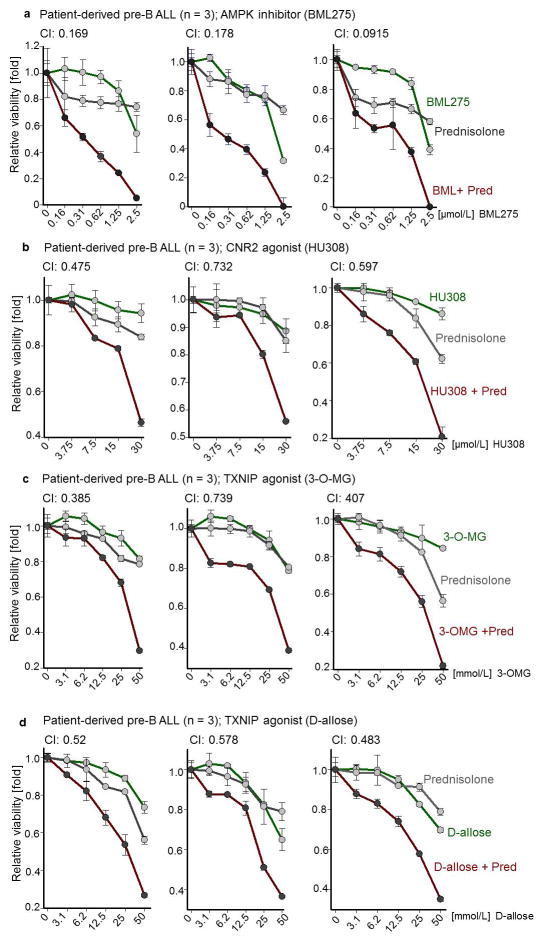
a, b, Heatmap of metabolomics (n = 2 per group, each in triplicate; a) and Western blots (n = 4; b) of patient-derived leukemia samples. c, Glucose uptake and ATP levels during B→ myeloid reprogramming (n = 4). d, Fold change of pre-B ALL cells carrying GFP-tagged Cre following Lkb1-deletion upon reprogramming (n = 6). e, Viability of pre-B ALL cells following deletion of Lkb1 or Ampkα2 (n = 3 independent experiments). f, Kaplan-Meier analysis (Mantel-Cox log-rank test) of recipient mice (n = 7 per group) injected with pre-B ALL cells following 4-OHT-induced deletion of Lkb1 or Ampkα2 (24 h). g, Patient-derived pre-B ALL cells treated with BML275 as indicated or in combination with prednisolone (n = 3), assessed by Combination Index (CI). Data, mean (± s.d), assessed by two-tailed t-test (c) or two-way ANOVA (d, e). For gel source data, see Supplementary Fig. 1.
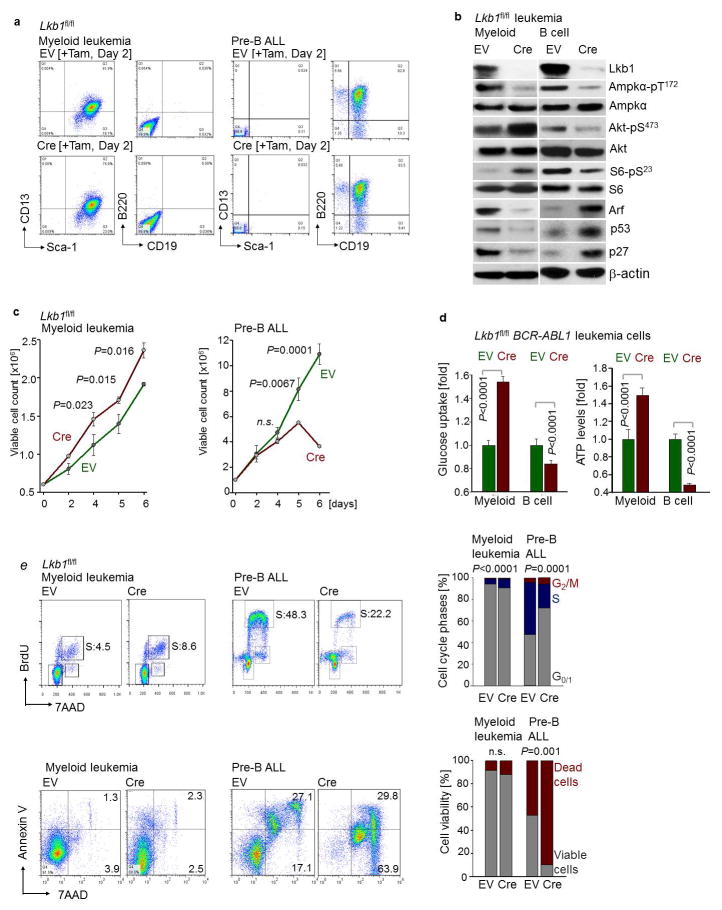
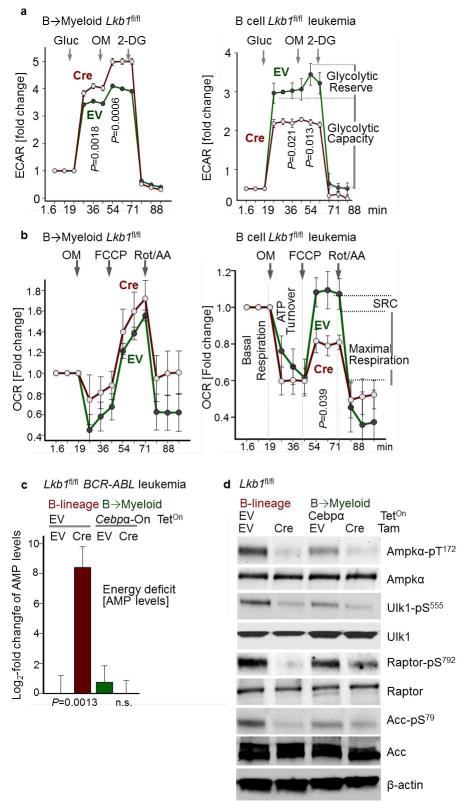
To examine the effects of B-cell- vs myeloid lineage identity in genetically identical murine leukemia cells, we studied reprogramming of B-cells based on inducible overexpression of the myeloid transcription factor CEBPα. Studying gene expression changes during B→myeloid reprogramming (GSE32330)12, we found upregulation of effector molecules of glucose transport and utilization (Insr, Glut1, Glut6, Hk2, Hk3, G6PD, Pygl). B→myeloid reprogramming markedly increased glucose uptake and cellular ATP levels (Fig. 2c), resulting in decreased activity of the energy stress sensor LKB1-AMPK (Extended Data Figure 3). Mirroring these differences, Cre-mediated deletion of Lkb1 induced cell death in B-lineage ALL cells, but accelerated proliferation in B → myeloid reprogrammed cells (Fig. 2d).
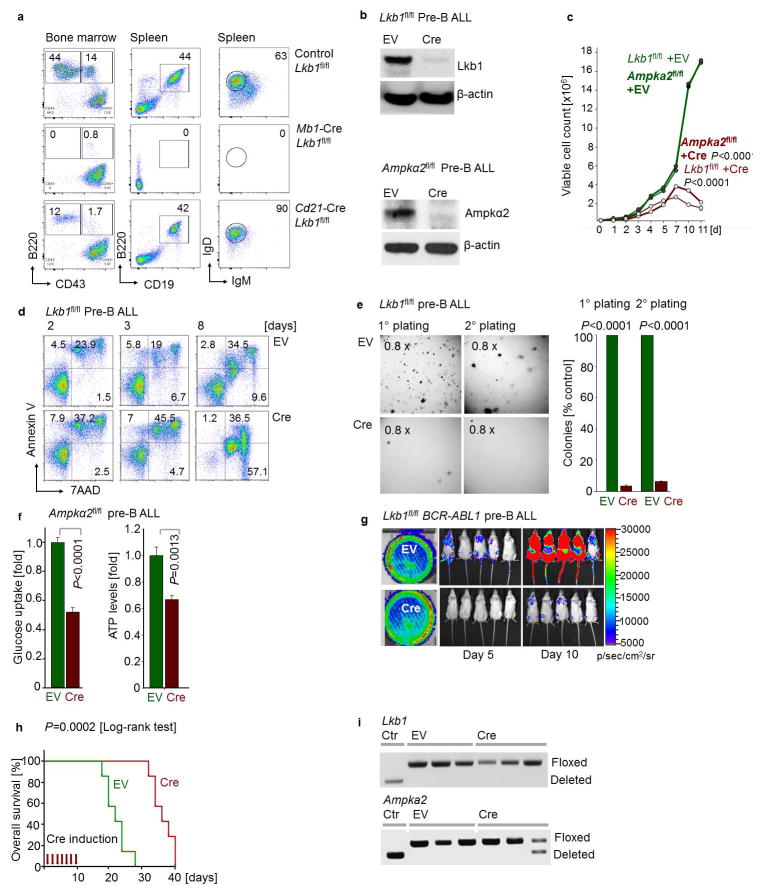
For this reason, we studied the consequences of inducible ablation of Lkb1 and Ampka2, of which expression levels were upregulated at the pre-B cell stage compared to later stages of B cell development (GSE38463). 4-hydroxytamoxifen (4-OHT)-inducible deletion of Lkb1 or Ampka2 induced rapid leukemia cell death, prevented malignant transformation of pre-B cells and affected development of leukemia in vivo. Deletion of Lkb1 or Ampka2 significantly prolonged overall survival of mouse recipients (Fig. 2e, f; Extended Data Figure 4). Genotyping of leukemias revealed that floxed alleles of Lkb1 and Ampka2 were retained in all cases (Extended Data Figure 4i), indicating strong positive selection of the few clones that escaped Cre-mediated deletion.
Seemingly at odds with our findings in pre-B ALL, a recent study showed that deletion of Ampka1 induced acceleration mature B-cell lymphoma17. Moreover, genetic lesions of PAX5, IKZF1, EBF1 and TCF3 are common in pre-B ALL but very rare in mature B-cell lymphomas (Extended Data Fig. 5). Hence, we tested the hypothesis that LKB1-AMPK function defines a stage-specific metabolic checkpoint during early B-cell development, when B-lymphoid transcription factors are most active. To this end, we crossed Lkb1fl/fl mice with Cre deleter-strains for deletion at early (Mb1) and late (Cd21) stages of B-cell development. While loss of Lkb1 at the pre-B cell stage resulted in a complete block of B-cell development, deletion of Lkb1 in mature CD21+ B-cells had no significant effect on survival and proliferation (Extended Data Figure 4a). These findings explain the apparent differences between pre-B ALL and mature B-cell lymphoma17, and also reveal a metabolic checkpoint function of Lkb1 at the pre-B cell stage.
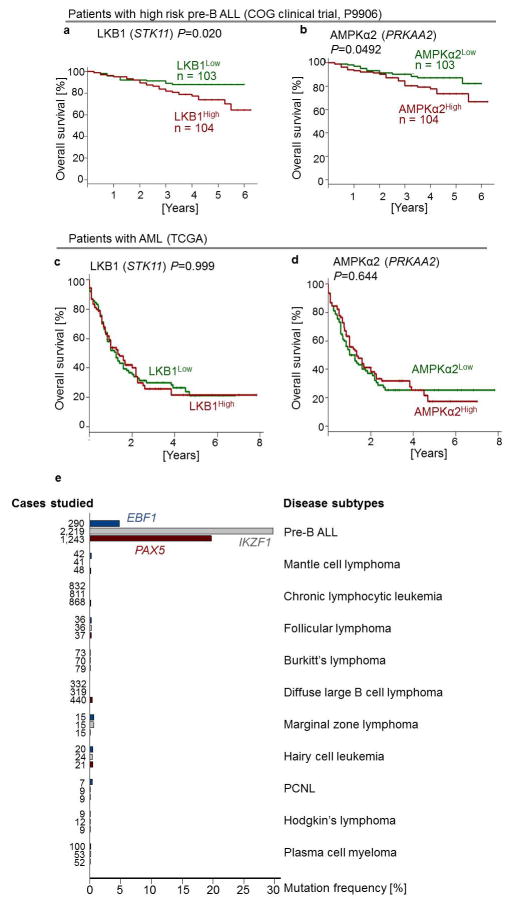
In both myeloid and B-lineage leukemia cells, acute deletion of Lkb1 resulted in loss of AMPK activity and inhibited phosphorylation of Ampk substrates (Extended Data Figures 6–7). Despite similar biochemical changes in response to Lkb1-deletion, functional outcomes, were strikingly different in myeloid and B-lymphoid cells: Lkb1 deletion in myeloid leukemia cells stimulated AKT signaling, increased ATP-, glucose uptake, glycolytic capacity and reserve and induced proliferation while cell cycle checkpoint molecules were downregulated. In contrast, deletion of Lkb1 or Ampka2 in B-lymphoid leukemia cells decreased cell viability and cell numbers, activated cell cycle checkpoint proteins in parallel with decreased glucose uptake, mitochondrial respiration and ATP levels (Extended Data Figures 6–7). Likewise, inducible deletion of Lkb1 in B-lymphoid ALL cells caused cell death, loss of colony formation capacity, and delayed leukemia-onset as well as prolonged survival of transplant recipient mice in vivo (Extended Data Figure 4). High expression levels of LKB1 and AMPK are associated with poor clinical outcome in patients with B-lymphoid but not myeloid leukemia (Extended Data Fig. 5).
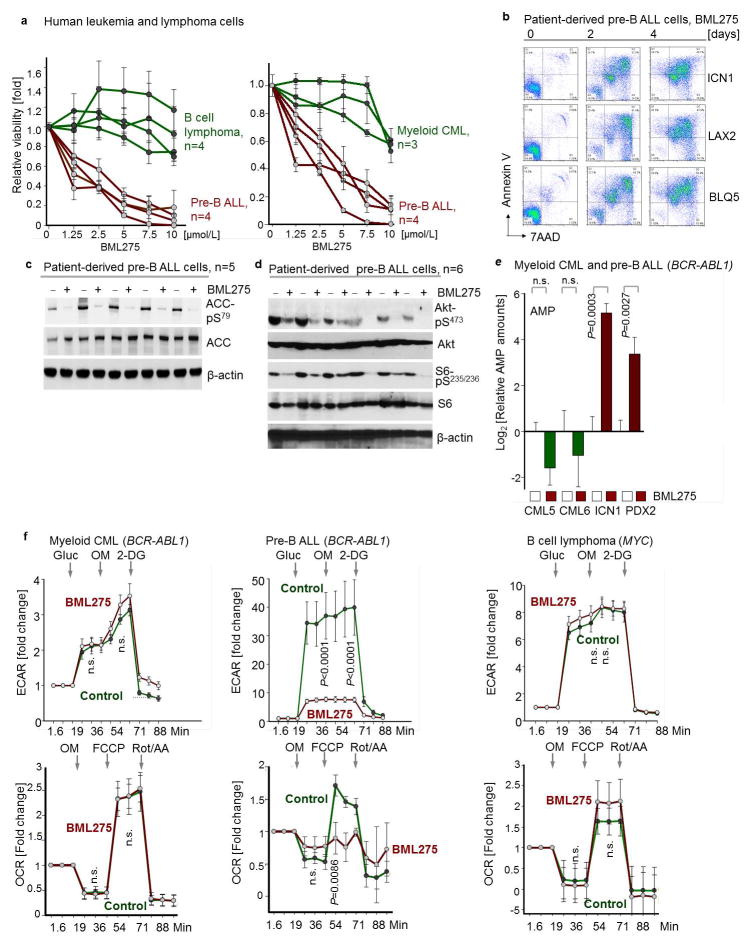
Unlike mature B-cells or myeloid cells, patient-derived pre-B ALL cells were highly sensitive to BML275, a small molecule inhibitor of AMPK18. BML275 abolished AMPK enzymatic activity and AKT-signaling and induced cell death associated with energy stress and AMP-accumulation. BML275 treatment of patient-derived pre-B ALL cells obliterated glycolytic flux and mitochondrial respiration, but had no significant effects on myeloid leukemia and mature B-cell lymphoma cells (Extended Data Figure 8). Given that both AMPK-inhibition and glucocorticoid-treatment reduced glucose uptake in pre-B ALL, we tested their pharmacological interactions. Combination studies of BML275 and prednisolone in patient-drived pre-B ALL cells demonstrated strong synergistic activity, suggesting that inhibition of AMPK represents a previously unrecognized vulnerability of pre-B ALL that can be leveraged by combination with glucocorticoids (Fig. 2g; Extended Data Figure 10a; Supplementary Table 1).
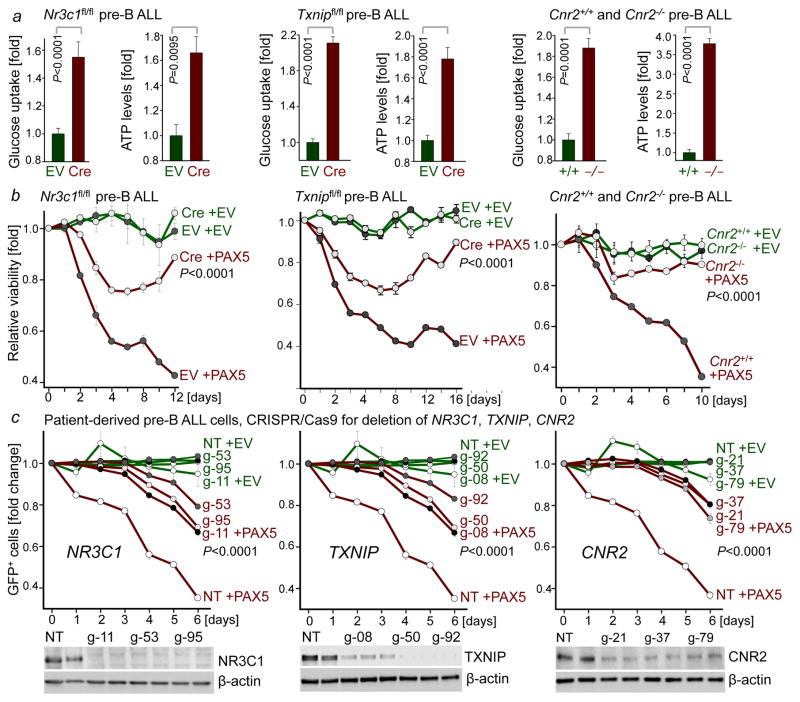
To measure the mechanistic contribution to PAX5-mediated tumor suppression, we performed a CRISPR-based genetic screen of PAX5-target genes. For this screen, we pursued two approaches: Transcriptional activation (CRISPR-a)19 to counteract repression by PAX5 resulted in a survival advantage for some PAX5-target genes, although the effects were relatively small. By comparison, CRISPR/Cas9-mediated deletion to counteract transcriptional activation of PAX5-targets NR3C1 (glucocorticoid receptor)8, TXNIP (glucose feedback sensor)9 and CNR2 (cannabinoid receptor)10 provided a strong survival advantage (Extended Figure 9). Genetic loss-of-function of Nr3c1, Txnip and Cnr2 (Extended Data Fig. 9d) significantly improved glucose uptake and increased ATP-levels (Fig. 3a). While inducible PAX5-expression caused cell death in murine pre-B ALL cells, the tumor-suppressive effect of Pax5 was mitigated upon genetic loss of Nr3c1, Txnip and Cnr2 (Fig. 3b). CRISPR/Cas9-mediated deletion of NR3C1, TXNIP and CNR2 in patient-derived pre-B ALL cells further corroborated the role of these negative regulators of glucose transport in human disease and PAX5-mediated tumor suppression (Fig. 3c).
Figure 3. Mechanistic contribution of PAX5 targets to regulation of glucose and energy metabolism in pre-B ALL.
a, Glucose uptake and ATP levels upon genetic loss of Nr3c1, Txnip or Cnr2 function, analyzed by two-tailed t-test. b, Viability upon inducible expression of Pax5 with or without loss of Nr3c1, Txnip or Cnr2 function. c, Fold change of GFP+ patient-derived pre-B ALL cells carrying Cas9 and gRNAs (NT: non-targeting) upon induction of GFP-tagged PAX5. Western blots of NR3C1, TXNIP and CNR2 to verify deletion (no induction). Data, mean ± s.d. (n = 3 independent experiments). Data assessed by two-way ANOVA (b, c). For gel source data, see Supplementary Fig. 1.
For the past five decades, glucocorticoids (GCs) have been a central component of therapy regimen for patients with pre-B ALL. GCs are highly active in B-lymphoid ALL, but ineffective in the treatment of myeloid leukemia20. However, the underlying reason for this difference remained elusive. Studying patient-derived leukemia samples, we found 6-to 20-fold higher NR3C1-protein levels in B-lymphoid compared to myeloid leukemia cells (Fig. 2b). While IKZF1 and PAX5 positively regulated NR3C1 levels, DN-IKZF1, DN-PAX5 as well as B→myeloid reprogramming erased NR3C1 expression (Fig. 1, Extended Data Figure 3). We tested whether B-lymphoid transcription factors set the threshold for glucocorticoid responses. Reconstitution of PAX5 and IKZF1 in pre-B ALL patient samples significantly shifted dose-response curves. Likewise, dexamethasone-responses were largely abolished upon induction of DN-IKZF1 and DN-PAX5 (Extended Data Figure 9e). Loss of Cnr2 and Txnip rescued prednisolone (Pred)-induced cell death in pre-B ALL cells (Extended Data Figure 9c), suggesting that glucocorticoid responses are influenced by factors that affect glucose uptake. On this basis, we tested drug-interactions between glucocorticoids and the CNR2-agonist H308 or the TXNIP-agonists 3-O-methylglucose (3-OMG) and D-allose21–22. Treating patient-derived pre-B ALL cells with HU308, 3-OMG or D-allose synergized with GC-treatment (Extended Data Figure 10; Supplementary Table 1), suggesting that Cnr2 and Txnip cooperate with Nr3c1 by exacerbating B-cell-intrinsic ATP-depletion.
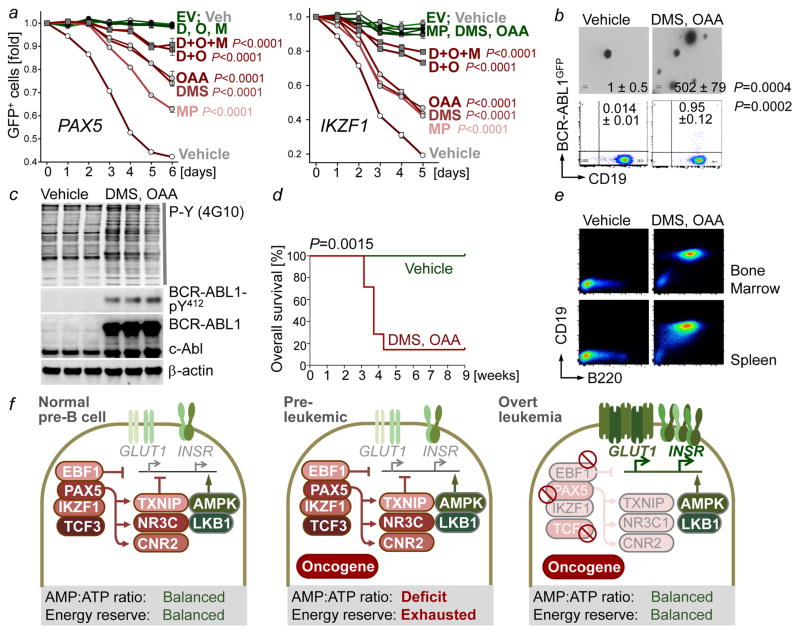
Our results indicate that B-lymphoid transcription factors, including PAX5 and IKZF1, exert their tumor suppressor function, at least in part, through transcriptional repression of glucose transport and restriction of metabolites that can fuel the TCA cycle. Restriction of glucose and energy supply imposed by B-lymphoid transcription factors is critical to prevent malignant transformation of pre-B cells. Based on this scenario, we predict that transport-independent glucose-metabolites can bypass the gatekeeper function of PAX5 and IKZF1. PAX5 and IKZF1 reconstitution was induced in haploinsufficient patient-derived pre-B ALL cells cultured either in the presence of vehicle or transport-independent lipophilic methyl-pyruvate (MP) or TCA cycle intermediates dimethyl-succinate (DMS) and oxaloacetate (OAA; Fig. 4a). Reconstitution of PAX5 and IKZF1 induced survival disadvantage in competitive growth assays in the presence of vehicle controls. However, dual (DMS and OAA) and triple (DMS, OAA and MP) combinations almost entirely rescued PAX5- and IKZF1-induced leukemia cell death (Fig. 4a).
Figure 4. Transcriptional restriction of glucose and energy supply prevents oncogenic signaling and pre-B cell transformation.
a, Fold change of GFP+ patient-derived pre-B ALL cells carrying GFP-tagged PAX5 (left) or IKZF1 (right) upon treatment with methyl pyruvate (MP; 5 mM), dimethyl succinate (DMS; 5 mM) and oxaloacetate (OAA; 5 mM; two-way ANOVA). b,c, Colony formation, GFP expression (b, two-tailed t-test), and Western blots of vehicle or OAA/DMS-treated murine BCR-ABL1+ pre-B cells (n = 3, c). d, Kaplan-Meier analysis (log-rank test) of recipient mice injected with treated BCR-ABL1+ pre-B cells (n = 7 per group). e, Representative FACS plots of bone marrow and spleens harvested from (d). f, Scenario for B-lymphoid transcription factors as metabolic gatekeepers. Data, mean from 3 independent experiments (± s.d; a, b). For gel source data, see Supplementary Fig. 1.
Conditional expression of GFP-tagged BCR-ABL1 from a recently developed conditional knockin allele23 was insufficient to drive malignant transformation of pre-B cells. BCR-ABL1+ pre-B cells of these mice remained in a pre-leukemic state and did not give rise to overt disease. When cultured under control conditions, BCR-ABL1+ pre-B cells formed few colonies and low BCR-ABL1 kinase activity (Fig. 4b–c). However, when BCR-ABL1+ pre-B cells were incubated for four weeks with dimethyl-succinate (DMS) and oxaloacetate (OAA), colony numbers increased (vehicle-treated: 1 ± 0.5; DMS/OAA-treated: 502 ± 79) and BCR-ABL1-expression and –activity were increased (Fig. 4b–c). Importantly, vehicle-treated BCR-ABL1+ pre-B cells failed to initiate leukemia when injected into sublethally irradiated NSG mouse recipients. However, 6 of 7 NSG recipients developed fatal disease within five weeks of injection when BCR-ABL1+ pre-B cells were cultured in the continuous presence of DMS and OAA (Fig. 4d–e). In the absence of DMS and OAA, only 0.014% of BCR-ABL1+ pre-B cells expressed the BCR-ABL1-GFP at detectable levels. Even when treated with DMS and OAA, the fraction of BCR-ABL1-GFP-expressing cells did not exceed 1% (Fig. 4b), suggesting that metabolite-induced transformation represents a rare event.
During early B-cell development, pre-B cells are under intense selective pressure. Collateral damage during V(D)J-recombination represents a main driver of malignant transformation24–25. We propose that B-lymphoid transcription factors function as metabolic gatekeepers by limiting glucose and energy supply to levels that are below minimum requirements for malignant transformation (Fig. 4f). Pre-leukemic B-cell clones carrying oncogenic lesions are frequently found in neonatal cord blood26–27. In addition, small fractions of normal B-cells in healthy adults carry silent oncogenes28–29. These findings indicate that pre-leukemic B-cell clones frequently occur in healthy individuals, both during childhood and later in life. We conclude that B-lymphoid repression of glucose uptake and energy supply represents a previously unrecognized metabolic barrier against malignant pre-B cell transformation and reveals TXNIP, CNR2 and AMPK as a novel targets for the treatment of pre-B ALL.
METHODS
Primary human samples and cell lines
Patient-derived pre-B acute lymphoblastic leukemia (ALL) cells were obtained with informed consent in compliance with the internal review board of the University of California San Francisco (Supplementary Table 2). Leukemia cells from bone marrow biopsy of patients with ALL were xenografted into sublethally irradiated NOD/SCID mice via tail vein injection. After passaging, leukemia cells were harvested. Cells were cultured on OP9 stroma cells in Alpha Minimum Essential Medium (MEMα; Invitrogen), supplemented with 20% fetal bovine serum (FBS), 2mM L-glutamine, 1mM sodium pyruvate, 100 IU/mL penicillin and 100 μg/mL streptomycin. Primary chronic myeloid leukemia (CML) cases were obtained with informed consent from the University Hospital Jena in compliance with institutional internal review boards (including the IRB of University of California San Francisco; Supplementary Table 3). Cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Invitrogen) supplemented with 20% BIT serum substitute (StemCell Technologies); 100 IU/mL penicillin and 100 μg/mL streptomycin; 25 μmol/L β-mercaptoethanol; 100 ng/mL SCF; 100 ng/mL G-CSF; 20 ng/mL FLT3; 20 ng/mL IL-3; and 20 ng/mL IL-6. Human cell lines (Supplementary Table 2) were obtained from DSMZ (Braunschweig, Germany) and were cultured in Roswell Park Memorial Institute medium (RPMI-1640; Invitrogen) supplemented with GlutaMAX containing 20% FBS, 100 IU/mL penicillin and 100 μg/mL streptomycin. Cell cultures were kept at 37°C in a humidified incubator under a 5% CO2 atmosphere. None of the cell lines used was found in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample. All cell lines were authenticated by STR profiles and were tested negative for mycoplasma.
Small molecule inhibitors and activators and metabolites
BML275 (water-soluble) and Imatinib were obtained from Santa Cruz Biotechnology and LC Laboratories, respectively. Stock solutions were prepared in DMSO or sterile water at 10 mmol/L and stored at −20°C. Prednisolone and dexamethasone (water-soluble) were purchased from Sigma-Aldrich and were resuspended in ethanol or sterile water at 10 mmol/L, respectively. Stock solutions were stored at −20°C. Fresh solutions (pH adjusted) of methyl pyruvate (MP), oxaloacetate (OAA), 3-O-methylglucose (3-OMG; an agonist of TXNIP), D-allose (an agonist of TXNIP) and recombinant insulin (Sigma-Aldrich) were prepared for each experiment. Dimethyl succinate (DMS) was obtained from Acros Organics, and fresh solutions (pH adjusted) were prepared prior to each experiment. For competitive growth assays, 5 mmol/L methyl pyruvate (MP), 5 mmol/L dimethyl succinate (DMS) and 5 mmol/L oxaloacetate (OAA) were used. CNR2 agonist HU308 was obtained from Cayman Chemical.
Murine BCR-ABL1-transformed pre-B ALL (Ph+ ALL) and myeloid leukemia (CML-like) models
To avoid inflammation-related effects in mice, bone marrow cells were extracted from mice (Supplementary Table 4) younger than 6 weeks of age without signs of inflammation. All mouse experiments were in compliance with institutional approval by the University of California, San Francisco Institutional Animal Care and Use Committee. Bone marrow cells were obtained by flushing cavities of femur and tibia with PBS. After filtration through a 70 μm filter and depletion of erythrocytes using a lysis buffer (BD PharmLyse, BD Biosciences), washed cells were either frozen for storage or subjected to further experiments.
Bone marrow cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Invitrogen) with GlutaMAX containing 20% fetal bovine serum, 100 IU/mL penicillin, 100 μg/mL streptomycin and 50 μM β-mercaptoethanol. To generate pre-B ALL (Ph+ ALL-like) cells, bone marrow cells were cultured in 10 ng/mL recombinant mouse IL-7 (PeproTech) and retrovirally transformed by BCR-ABL1. BCR–ABL1-transformed pre-B ALL cells were propagated only for short periods of time and usually not longer than for 2 months to avoid acquisition of additional genetic lesions during long-term cell culture. To generate myeloid leukemia (CML-like) cells, the myeloid-restricted protocol described previously30 was used. Bone marrow cells were cultured in 10 ng/mL recombinant mouse IL-3, 25 ng/mL recombinant mouse IL-6, and 50 ng/mL recombinant mouse SCF (PeproTech) and retrovirally transformed by BCR-ABL1. Immunophenotypic characterization was performed by flow cytometry. For conditional deletion, a 4-hydroxytamoxifen (4-OHT, Tam)-inducible, Cre-mediated deletion system was used. For retroviral constructs used, see Supplementary Table 5.
Retroviral and lentiviral constructs and transduction
Transfection of retroviral constructs (Supplementary Table S5) was performed using Lipofectamine 2000 (Invitrogen) with Opti-MEM media (Invitrogen). Retroviral supernatant was produced by co-transfecting HEK 293FT cells with the plasmids pHIT60 (gag-pol) and pHIT123 (ecotropic env). Lentiviral supernatant was produced by co-transfecting HEK 293FT cells with the plasmids pCDNL-BH and VSV-G or EM140. 293FT cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with GlutaMAX containing 10% fetal bovine serum, 100 IU/mL penicillin, 100 μg/mL streptomycin, 25 mmol/L HEPES, 1 mmol/L sodium pyruvate and 0.1 mmol/L non-essential amino acids. Regular medium was replaced after 16 h by growth medium containing 10 mmol/L sodium butyrate. After incubation for 8 h, the medium was changed back to regular growth medium. Twenty-four hours later, retroviral supernatant was collected, filtered through a 0.45 μm filter and loaded by centrifugation (2,000g, 90 min at 32 °C) onto 50 μg/mL RetroNectin- (Takara) coated non-tissue six-well plates. Lentiviral supernatant produced with VSV-G was concentrated using Lenti-X™ Concentrator (Clontech) and loaded onto RetroNectin-coated plates and incubated for 15 min at room temperature. Lentiviral supernatant produced with EM140 was collected and loaded onto RetroNectin-coated plates and incubated for 30 min at room temperature. Two to three million cells were transduced per well by centrifugation at 600g for 30 min and maintained for 48 h at 37 °C with 5% CO2 before transferring into culture flasks. For cells transduced with lentiviral supernatant produced with EM140, supernatant was removed the day after transduction and replaced with fresh culture medium. Cells transduced with estrogen receptor fusion proteins were induced with 4-hydroxytamoxifen (4-OHT, Tam; 1 μmol/L). Cells transduced with constructs carrying an antibiotic resistance marker were selected with its respective antibiotic.
Inducible gain- and loss-of-function of IKZF1 and PAX5
For loss-of-function studies, dominant-negative variants of IKZF1 (DN-IKZF1, lacking the IKZF1 zinc fingers 1–4) and PAX5 (DN-PAX5; PAX5-ETV6 fusion) were cloned from patient samples. Expression of DN-IKZF1 was induced by Dox (1 μg/mL), while activation of DN-PAX5 was induced by 4-OHT (1 μg/mL) in patient-derived pre-B ALL cells carrying IKZF1 and PAX5 wildtype alleles, respectively. Inducible reconstitution of wild-type IKZF1 and PAX5 in haploinsufficient pre-B ALL cells carrying deletions of either IKZF1 (IKZF1Δ) or PAX5 (PAX5Δ) were also studied. Lentiviral constructs used are listed in Supplementary Table 5. A doxycycline (Dox)-inducible TetOn vector system was used for inducible expression of PAX5 in murine BCR-ABL1 pre-B ALL. Retroviral constructs used are listed in Supplementary Table 5.
CEBPα-mediated reprogramming of pre-B cells into the myeloid lineage
To study the effects of B cell- vs. myeloid lineage identity in genetically identical murine leukemia cells, a doxycycline (Dox)-inducible TetOn-CEBPα vector system31 was used for reprogramming of B cells. Murine BCR-ABL1 pre-B ALL cells expressing Dox-inducible CEBPα or EV were induced with Dox (1 μg/mL). Conversion from the B cell lineage (CD19+ Mac1−) to the myeloid lineage (CD19− Mac1+) was monitored by flow cytometry. For Western blots, B-lineage cells (CD19+ Mac1−) and CEBPα-reprogrammed cells (CD19− Mac1+) were sorted from cells expressing EV or CEBPα, respectively, following Dox induction. For metabolic assays, sorted B-lineage cells and CEBPα-reprogrammed cells were cultured (+Dox) for 2 days following sorting, and were then seeded in fresh medium for measurements of glucose consumption (normalized to cell numbers) and total ATP levels (normalized to total protein).
To study Lkb1 deletion in the context of CEBPα-mediated reprogramming, BCR-ABL1-transformed Lkb1fl/fl pre-B ALL cells expressing Dox-inducible CEBPα were transduced with 4-hydroxytamoxifen- (4-OHT, Tam, 1 μg/mL) inducible Cre-GFP (Cre-ERT2-GFP). Without sorting for GFP+ cells, cells were induced with Dox and 4-OHT. Viability (expressed as relative change of GFP+ cells) was measured separately in B-lineage (gated on CD19+ Mac1−) and myeloid lineage (gated on CD19− Mac1+) populations.
To study whether Lkb1 deletion causes CEBPα-dependent effects on metabolism and signaling, Lkb1fl/fl BCR-ABL1 B-lineage ALL cells expressing Dox-inducible CEBPα or EV were transduced with 4-OHT-inducible Cre-GFP. Following sorting for GFP+ populations, cells were induced with Dox. B-lineage cells (CD19+ Mac1−) and CEBPα-reprogrammed cells (CD19− Mac1+) were sorted from cells expressing EV or CEBPα, respectively. Sorted cells were cultured (+Dox) and induced with 4-OHT. Protein lysates were harvested on day 2 following 4-OHT induction. For metabolomics, sorted cells were re-seeded in fresh medium on day 2 (4-OHT) and harvested for metabolite extraction.
CRISPR-mediated gene editing
For CRISPR/Cas9-mediated deletion of target genes, all constructs including lentiviral vectors expressing guide RNA (gRNA) and Cas9 nuclease were purchased from Transomic Technologies (Supplementary Table 5; see Supplementary Table 6 for gRNA sequences). Briefly, patient-derived pre-B ALL cells transduced with GFP-tagged, 4-OHT-inducible PAX5 or EV were transduced with pCLIP-hCMV-Cas9-Nuclease-Blast. Blasticidin-resistant cells were subsequently transduced with pCLIP-hCMV-gRNA-RFP. Non-targeting gRNA was used as control.
Constructs including lentiviral vectors expressing gRNA and dCas9-VPR used for CRISPR/dCas9-mediated activation of gene expression are listed in Supplementary Table 5. Nuclease-null Cas9 (dCas9) fused with VP64-p65-Rta (VPR) was cloned from SP-dCas9-VPR (gift from George Church; Addgene plasmid # 63798) and then subcloned into pCL6 vector with a Blasticidin-resistant marker. gRNA sequences (Supplementary Table 6) targeting the transcriptional start site of each specific gene were obtained from public databases (http://sam.genome-engineering.org/ and http://www.genscript.com/gRNA-database.html)32. gBlocks Gene Fragments were used to generate single-guide RNAs (sgRNAs) and were purchased from Integrated DNA Technologies, Inc. Each gRNA was subcloned into pCL6 vector with a dsRed reporter. Patient-derived pre-B ALL cells transduced with GFP-tagged inducible-PAX5 or EV were transduced with pCL6-hCMV-dCas9-VPR-Blast. Blasticidin-resistant cells were used for subsequent transduction of pCL6-hCMV-gRNA-dsRed, and dsRed+ cells were further analyzed by flow cytometry. For each target gene, two – three sgRNA clones were pooled together to generate lentiviruses. Non-targeting gRNA was used as control. To elucidate the mechanistic contribution of PAX5 targets, percentage of GFP+ cells carrying gRNA(s) for each target gene was monitored by flow cytometry upon inducible activation of GFP-tagged PAX5 or EV in patient-derived pre-B ALL cells in competitive growth assays.
Western blotting
Cells were lysed in CelLytic buffer (Sigma-Aldrich) supplemented with 1% protease inhibitor cocktail (Thermo Fisher Scientific). 20 μg of protein mixtures per sample were separated on NuPAGE (Invitrogen) 4–12% Bis-Tris gradient gels or 4–20% Mini-PROTEAN TGX™ precast gels, and transferred onto nitrocellulose membranes (Bio-Rad). The primary antibodies used are listed in Supplementary Table 7. For protein detection, the WesternBreeze Immunodetection System (Invitrogen) was used, and light emission was detected by either film exposure or the BioSpectrum Imaging system (UPV).
Flow cytometry
Approximately 106 cells per sample were resuspended in PBS blocked using Fc blocker for 10 minutes on ice, followed by staining with the appropriate dilution of the antibodies or their respective isotype controls for 15 minutes on ice. Cells were washed and resuspended in PBS with propidium iodide (PI, 0.2 μg/mL) or DAPI (0.75 μg/mL) as a dead cell marker. The antibodies used for flow cytometry are listed in Supplementary Table 7. For competitive growth assays, percentage of GFP+ cells was monitored by flow cytometry. For Annexin V staining, Annexin V binding buffer (BD Bioscience) was used instead of PBS and 7AAD (BD Bioscience) instead of PI. PE labeled-Annexin V was purchased from BD Bioscience. For BrdU staining, the BrdU Flow Kit was purchased from BD Bioscience and performed according to the manufacturer’s protocol.
Colony forming assay
The methylcellulose colony-forming assays were performed with 10,000 BCR-ABL1 pre-B ALL cells. Cells were resuspended in mouse MethoCult medium (StemCell Technologies) and cultured on 3-cm diameter dishes, with an extra water supply dish to prevent evaporation. Images were taken and colony numbers were counted after 14 days.
Cell viability assay and determination of viable cell counts
Cell viability upon genetic loss of function of target genes and/or inducible expression of Pax5 was monitored by flow cytometry using propidium iodide (PI, 0.2 μg/mL) as a dead cell marker. To study effects of an AMPK inhibitor (BML275), glucocorticoids (dexamethasone and prednisolone), CNR2 agonist (HU308), or TXNIP agonists (3-OMG and D-allose), forty thousand human or mouse leukemia cells were seeded in a volume of 80 μL in complete growth medium on opaque-walled, white 96-well plates (BD Biosciences). Compounds were added at the indicated concentrations in a total volume of 100 μL per well. After culturing for 3 days, cells were subjected to CellTiter-Glo® Luminescent Cell Viability Assay (Promega). Relative viability was calculated using baseline values of cells treated with vehicle control as a reference. Combination index (CI) was calculated using the CalcuSyn software to determine interaction (synergistic, CI < 1; additive, CI = 1; or antagonistic, CI > 1) between the two agents. Constant ratio combination design was used. Concentrations of BML275, D-allose, 3-OMG and HU308 used are indicated in the figures. Concentrations of Dex used were 10-fold lower than those of BML275. Concentrations of prednisolone used were 2-fold lower than those of BML275. To determine the number of viable cells, the trypan blue exclusion method was applied, using the Vi-CELL™ Cell Counter (Beckman Coulter).
Single-locus quantitative chromatin immunoprecipitation (ChIP) and ChIP-seq data
ChIP was performed as described previously33. Chromatin from fixed patient-derived Ph+ ALL cells (ICN1) was isolated and sonicated to 100–500 base pair DNA fragments. Chromatin fragments were immunoprecipitated with either IgG (as a control) or anti-Pax5 antibody (see Supplementary Table 7). Following reversal of crosslinking by formaldehyde, specific DNA sequences were analyzed by quantitative real-time PCR (see Supplementary Table 8 for primers). Primers were designed according to ChIP-seq tracks for PAX5 antibodies in B lymphocytes (ENCODE, Encyclopedia of DNA Elements, GM12878). ChIP-seq tracks for PAX5, IKZF1, EBF1 and TCF3 antibodies in a normal B cell sample (ENCODE GM12878, UCSC genome browser) on INSR, GLUT1, GLUT3, GLUT6, HK2, G6PD, NR3C1, TXNIP, CNR2 and LKB1 gene promoter regions are shown. CD19 and ACTA1 serve as a positive and a negative control gene, respectively. Y-axis represents the normalized number of reads per million reads for peak summit for each track. The ChIP-seq peaks were called by the MACS peak caller by comparing read density in the ChIP experiment relative to the input chromatin control reads, and are shown as bars under each wiggle track. Gene models are shown in UCSC genome browser hg19.
Glucose and ATP measurements
Extracellular glucose levels were measured using the Amplex Red Glucose/Glucose Oxidase Assay Kit (Invitrogen), according to the manufacturer’s protocol. Glucose concentrations were measured in fresh and spent medium. Total ATP levels were measured using the ATP Bioluminescence Assay Kit CLS II (Roche) according to the manufacturer’s protocol. One million cells per mL were seeded in fresh medium and treated as indicated in the figure legends. Relative levels of glucose consumed and total ATP are shown. All values were normalized to cell numbers (Fig. 1b, c; 2c, glucose uptake; Fig. 3a; Extended Data Fig. 2c; Extended Data Fig. 4f; Extended Data Fig. 6d) or total protein (Fig. 2c, ATP levels). Numbers of viable cells were determined by applying trypan blue dye exclusion, using the Vi-CELL™ Cell Counter (Beckman Coulter).
Metabolic assays using the XFe24 Flux Analyzer
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse XFe24 Flux Analyzer with the XF Cell Mito Stress Test Kit and XF Glycolysis Stress Test Kit (Seahorse Bioscience) according to the manufacturer’s instructions. All compounds and materials were obtained from Seahorse Bioscience. Briefly, 1.5 x105 cells per well were plated using Cell-Tak (BD Biosciences). Following incubation in XF-Base Medium supplemented with glucose and GlutaMAX for 1 h at 37°C (non-CO2 incubator) for pH stabilization, OCR was measured at the resting stage (basal respiration in XF Base Medium supplemented with GlutaMax and glucose) and in response to oligomycin (1 μmol/L; mitochondrial ATP production), mitochondrial uncoupler FCCP (5 μmol/L; maximal respiration), and respiratory chain inhibitor antimycin and rotenone (1 μmol/L). Spare respiratory capacity is the difference between maximal respiration and basal respiration. ECAR was measured under specific conditions to generate glycolytic profiles. Following incubation in glucose-free XF Base Medium supplemented with GlutaMAX™ for 1 h at 37°C (non-CO2 incubator) for pH stabilization, basal ECAR was measured. Following measurement of the glucose-deprived, basal ECAR, changes in ECAR upon the sequential addition of glucose (10 mmol/L; glycolysis), oligomycin (1 μmol/L; glycolytic capacity), and 2-deoxyglucose (2-DG; 0.1 mol/L) were measured. Glycolytic reserve was determined as the difference between oligomycin-stimulated glycolytic capacity and glucose-stimulated glycolysis. All values were normalized to cell numbers (Extended Data Fig. 2c) or total protein (Extended Data Fig. 3a; Extended Data Fig. 7a, b; Extended Data fig. 8f) and are shown as fold change relative to basal ECAR or OCR.
Metabolite extraction and mass spectrometry-based analysis
Metabolite extraction and mass spectrometry-based analysis was performed as described previously34. Metabolites were extracted from 2 × 105 cells per sample using the methanol/water/chloroform method. After incubation at 37°C for the indicated time, cells were rinsed with 150 mM ammonium acetate, pH 7.3, and 400 μL cold 100% methanol (Optima* LC/MS, Fisher) followed by 400 μL cold water (HPLC-Grade, Fisher) was added to cells. 10 nmol norvaline (Sigma) was added as internal control, followed by 400 μL cold chloroform (HPLC-Grade, Fisher). Samples were vortexed 3 times over 15 minutes and spun down at top speed for 5 minutes at 4°C. The top layer (aqueous phase) was transferred to a new Eppendorf tube, and samples were dried on Vacufuge Plus (Eppendorf) at 30°C. Extracted metabolites were stored at −80°C. For mass spectrometry-based analysis, the metabolites were resuspended in 70% ACN and 5 μl used for analysis with a mass spectrometer. The mass spectrometer (Q Exactive, Thermo Scientific) was coupled to an UltiMate3000 RSLCnano HPLC. The chromatography was performed with 5 mM NH4AcO (pH 9.9) (A) and ACN (B) at a flow rate of 300 μl/min starting at 85%B, going to 5% B at 18 min, followed by an isocratic step to 27 min and re-equilibration to 34 min. The separation was achieved on a Luna 3u NH2 100A (150 × 2 mm) (Phenomenex). The Q Exactive was run in polarity switching mode (+3 kV/−2.25 kV). Metabolites were detected based on retention time (tR) and on accurate mass (± 3 ppm). Metabolite quantification was performed as area-under-the-curve (AUC) with TraceFinder 3.1 (Thermo Scientific). Data analysis was in R, and data was normalized to the number of cells. Relative amounts were log2-transformed, median-centered and are shown as a heatmap.
Oncogenic priming of Mb1-Cre; Bcr+/LSL-BCR/ABL B-cell precursor cells
To generate a model for pre-leukemic B cell precursors expressing BCR-ABL1, BCR-ABL1 knockin mice were crossed with Mb1-Cre deleter strain (Mb1-Cre; Bcr+/LSL-BCR/ABL) for excision of a stop-cassette in early pre-B cells. Bone marrow cells harvested from Mb1-Cre; Bcr+/LSL-BCR/ABL mice cultured in the presence of IL-7 were primed with vehicle control or a combination of oxaloacetate (OAA; 8 mmol/L), dimethyl succinate (DMS; 8 mmol/L) and insulin (210 pmol/L). Following a week of priming, cells were maintained and expanded in the presence of IL-7 supplemented with vehicle control or a combination of OAA (0.8 mmol/L) and DMS (0.8 mmol/L) for 4 weeks. Pre-B cells of Mb1-Cre; Bcr+/LSL-BCR/ABL mice express low levels of BCR-ABL1 tagged to GFP, and were analyzed by flow cytometry for surface expression of GFP and CD19. The methylcellulose colony-forming assays were performed with 10, 000 cells treated with vehicle control or metabolites. Cells were resuspended in mouse MethoCult medium (StemCell Technologies) and cultured on 3-cm diameter dishes, with an extra water supply dish to prevent evaporation. Images were taken and colony numbers were counted after 14 days.
For in vivo transplantation experiment, cells were treated with vehicle control or metabolites (OAA/DMS) for 6 weeks. One million cells were intravenously injected into sublethally irradiated (250 cGy) 6–8 week old female NSG (NOD/SCID-IL2 receptor gamma chain knockout) mice (n = 7 per group). Mice were randomly allocated into each group, and the minimal number of mice in each group was calculated by using the ‘cpower’ function in R/Hmisc package. No blinding was used. Mouse was sacrificed when it became terminally sick and showed signs of leukemia burden (hunched back, weight loss and inability to move). Bone marrow and spleen were harvested for flow cytometry analyses for leukemia infiltration (CD19, B220). After 63 days, all remaining mice were sacrificed, and bone marrow and spleens from all mice were analyzed by flow cytometry. Statistical analysis was performed by Mantel-Cox log-rank test. All mouse experiments were in compliance with institutional approval by the University of California, San Francisco Institutional Animal Care and Use Committee.
In vivo experiments
Following cytokine-independent proliferation, BCR–ABL1-transformed Lkb1fl/fl or AMPKa2fl/fl pre-B ALL cells were transduced with Tam-inducible Cre or an empty vector control (EV). For ex vivo deletion, deletion was induced 24 h prior to injection. For in vivo deletion, deletion was induced by tamoxifen (0.4 mg/mouse; i.p. injection). 106 cells were injected into each sublethally irradiated (250 cGy) NOD/SCID mouse. Seven mice per group were injected via tail vein injection.
All mouse experiments were in compliance with institutional approval by the University of California, San Francisco Institutional Animal Care and Use Committee. 6–8 week old female NOD/SCID or NSG mice were randomly allocated into each group. The minimal number of mice in each group was calculated by using the ‘cpower’ function in R/Hmisc package. No blinding was used. When a mouse became terminally sick and showed signs of leukemia burden (hunched back, weight loss and inability to move), it was sacrificed, and bone marrow and/or spleen were collected for flow cytometry analyses for leukemia infiltration. Statistical analysis was performed by Mantel-Cox log-rank test. In vivo expansion and leukemia burden were monitored by luciferase bioimaging. Bioimaging of leukemia progression in mice was performed at the indicated time points using an in vivo IVIS 100 bioluminescence/optical imaging system (Xenogen). D-Luciferin (Promega) dissolved in PBS was injected intraperitoneally at a dose of 2.5 mg per mouse 15 min. before measuring the luminescence signal. General anaesthesia was induced with 5% isoflurane and continued during the procedure with 2% isoflurane introduced through a nose cone.
Statistical analysis and reproducibility
Data are generally shown as mean ± s.d. Statistical significance was analyzed by using Grahpad Prism software or R software (R Development Core Team 2009) by using two-tailed t-test, two-way ANOVA, or log-rank test as indicated in figure legends. Significance was considered at P < 0.05. For in vitro experiments, no statistical methods were used to predetermine the sample size. For in vivo transplantation experiments, the minimal number of mice in each group was calculated through use of the ‘cpower’ function in the R/Hmisc package. No animals were excluded. Overall survival (OS) and relapse-free survival (RFS) data were from GSE1187735,36 and TCGA. Kaplan-Meier survival analysis was used to estimate OS and RFS. Patients with high risk pre-B ALL (COG clinical trial, P9906, n = 207; Supplementary Table 10) were segregated into 2 groups based on high or low mRNA levels with respect to the median mRNA values of the probe-sets for the respective gene of interest. Log-rank test was used to compare survival differences between patient groups. R package “survival” Version 2.35–8 was used for the survival analysis and Cox proportional hazards regression model in R package for the multivariate analysis (R Development Core Team, 2009). The investigators were not blinded to allocation during experiments and outcome assessment. Experiments were repeated to ensure reproducibility of the observations.
Data availability
Gel scans are provided in Supplementary Fig. 1. All other data are available from the corresponding author upon reasonable request. Gene expression data were obtained from the GEO database accession numbers GSE3233012, GSE5287037, and GSE3846338. Patient outcome data were derived from the National Cancer Institute TARGET Data Matrix of the Children’s Oncology Group (COG) Clinical Trial P9906 (GSE11877)35, 36 and from TCGA (the Cancer Genome Atlas). GEO accession details are provided in Supplementary Tables 9 and 10. ChIP-seq tracks for PAX5, IKZF1, EBF1 and TCF3 antibodies in a normal B cell sample (ENCODE GM12878, UCSC genome browser) on INSR, GLUT1, GLUT3, GLUT6, HK2, G6PD, NR3C1, TXNIP, CNR2 and LKB1 gene promoter regions are shown UCSC genome browser hg19.
Extended Data
Extended Data Figure 1. Frequent genetic lesions of B-lymphoid transcription factors in B cell lineage leukemia.
a, Gene expression of B-lymphoid transcription factors (top) as well as positive (middle) and negative (bottom) regulators of glucose uptake and energy supply upon inducible restoration of Pax5 (GSE52870)38 in haploinsufficient pre-B leukemia cells. b, Lesions in PAX5, IKZF1, EBF1 and TCF3 were studied in clinical trials for B-lymphoid ALL in children (P9906; n = 187; top) and adults (MDACC; n = 92; bottom). Red and gray boxes denote patient samples with detected lesions. c, Protein expression of PAX5 and IKZF1 was examined by Western blot in 10 patient-derived pre-B ALL samples (control: CD19+ B cells from 4 healthy donors). d, ChIP-seq analysis for binding of B-lymphoid transcription factors in human B cells (ENCODE GM12878) to promoter regions of molecules implicated in positive (INSR, GLUT1, GLUT3, GLUT6, HK2, G6PD, LKB1) and negative (NR3C1, TXNIP, CNR2) regulation of glucose uptake and utilization. e, Recruitment of PAX5 was confirmed by qChIP in patient-derived pre-B ALL cells. Data shown as mean from 3 independent experiments (± s.d.) and assessed by two-tailed t-test. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2. B-lymphoid transcription factor PAX5 functions as a metabolic gatekeeper.
a, Protein levels of PAX5 and IKZF1 in haploinsufficient patient-derived pre-B ALL cells and pre-B ALL cells expressing functional PAX5 and IKZF1. b, Number of viable cells and cell viability upon inducible activation of PAX5 in haploinsufficient patient-derived pre-B ALL (PAX5Δ) cells. c, To test whether Pax5 functions as metabolic gatekeeper, BCR-ABL1-induced changes in glycolytic activity and capacity (ECAR), glucose uptake, and ATP levels (normalized to cell numbers) were studied in Pax5 wildtype and Pax5 haploinsufficient pre-B cells in the presence or absence of a BCR-ABL1-transgene. Data shown as mean from 3 independent experiments (± s.d.) and assessed by two-tailed t-test (b, left; c) or two-way ANOVA (b, right). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3. Divergent metabolic characteristics of myeloid and B-lineage leukemia.
a, Glycolytic reserve (ECAR) and mitochondrial functions (OCR) in patient-derived myeloid (CML) and B-lymphoid (Ph+ ALL) leukemia samples (n = 5, each in triplicate). Values were normalized to total protein and are shown as mean ± s.d., assessed by two-tailed t-test. b, Murine pre-B ALL cells were reprogrammed into myeloid differentiation using a Dox-inducible TetOn-Cebpa vector system, and characterized by flow cytometry (representative results from 3 independent experiments). c, Heatmap of gene expression of glucose uptake and metabolism regulators (GSE32330)12. d, Western blots of murine pre-B ALL cells upon B → myeloid reprogramming to verify gene expression changes. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 4. The energy stress sensor LKB1-AMPK plays a pro-survival role and modulates glucose uptake and energy supply in pre-B ALL.
a, Lkb1fl/fl mice were crossed with Cre deleter strains for deletion at early pre-B cell stages (Mb1) and in fully mature B cells (Cd21). B cell populations in bone marrow and spleen (n = 3 litter mates) were characterized by flow cytometry analysis. b, The catalytic subunit of AMPK has two isoforms – α1 and α2. Analysis of published gene expression data (GSE38463)39 revealed that expression of the α1-form peaks at later stages of B cell development, whereas expression levels of both Lkb1 and the α2-form of Ampk peak in pre-B cells. For this reason, we studied here the consequences of inducible ablation of Lkb1 and Ampkα2 in murine models for BCR-ABL1-transformed pre-B ALL cells. Protein levels of Lkb1 and Ampkα2 were verified by Western blots. c, Viable cell counts upon Cre-mediated deletion of Lkb1 or Ampkα2. d, Apoptosis following Lkb1 deletion was monitored by Annexin V/7AAD staining. e, Colony forming ability was assessed by serial replating upon deletion of Lkb1 in pre-B ALL cells. f, Glucose uptake and ATP levels (normalized to cell numbers) were measured following Cre-mediated deletion of Ampka2. g, Luciferase bioimaging of transplant recipient mice injected with Lkb1fl/fl pre-B ALL cells transduced with 4-OHT-inducible Cre or EV and treated with Tam (0.4 mg/mouse; n = 7 per group). h, Overall survival was assessed by a Kaplan-Meier analysis (P-value calculated by Mantel-Cox log-rank test). i, Leukemia samples developed in recipient mice (Fig. 2d) were genotyped for the presence of either floxed or deleted Lkb1 and Ampka2 alleles (n = 3 mice). Representative FACS plots and images from 3 independent experiments are shown (a, d, e). Data shown as mean from 3 independent experiments (± s.d.) and assessed by two-way ANOVA (c) or two-tailed t-test (e, f). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 5. LKB1 and AMPK are independent predictors of poor clinical outcome for patients with pre-B ALL.
a, b, Children with high risk pre-B ALL (COG clinical trial, P9906, n = 207) were divided into 2 groups based on higher or lower than the median mRNA levels of LKB1 (a) or AMPKα2 (b). c, d, Adults with acute myeloid leukemia (AML; the Cancer Genome Atlas, n = 184) were divided into 2 groups based on higher or lower than the median mRNA levels of LKB1 (c) or AMPKα2 (d). Overall survival of patients was assessed in the two groups by Kaplan-Meier analysis. Log rank test was used to assess statistical significance (a–d). e, Frequencies of somatic mutations in the coding regions of EBF1 (top, blue), IKZF1 (middle, gray) and PAX5 (bottom, red) from the Catalog of Somatic Mutations in Cancer (COSMIC) database are plotted for pre-B ALL and various subtypes of mature B cell lymphoma. Somatic mutations in 5′UTR regions of EBF1, IKZF1 and PAX5 frequently represent byproducts of somatic hypermutation during normal B cell development and are not included in this analysis. PCNL, primary central nervous system lymphoma.
Extended Data Figure 6. Divergent functions of Lkb1 in BCR-ABL1-driven pre-B ALL and myeloid leukemia.
a, Staining of Lkb1fl/fl BCR-ABL1 myeloid (CML-like) and B-lineage (Ph+ ALL-like) leukemia cells with (Cre) or without (EV) deletion of Lkb1 for surface markers CD19, B220 (B-lymphoid), Sca-1 and CD13 (myeloid). b, Phosphorylation of AMPKα-T172, AKT-S473 and S6-S235 and protein levels of cell cycle checkpoint molecules Arf, p53 and p27 following Lkb1-deletion. c, Viable cell counts upon deletion of Lkb1. d, Glucose uptake and ATP levels (normalized to cell numbers) in myeloid and B-lymphoid leukemia cells upon Lkb1-deletion (n = 6). Shown as mean ± s.d. e, Cell cycle analyses were performed by measuring BrdU incorporation in combination with 7AAD staining. Percentages of cells in the G0/1, S, and G2/M phases are shown. Viability following Lkb1 deletion was monitored by Annexin V/7AAD staining. Representative FACS plots from 3 independent experiments are shown (a, e). Data shown as mean (± s.d.) from 3 independent experiments (c, e). Significance assessed by two-tailed t-test (c, d, e). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 7. Activity of the energy-stress sensor LKB1 represents a specific vulnerability of B-lymphoid leukemia.
a, b, Glycolytic profiles (ECAR; a) and mitochondrial functions (OCR; b) upon Lkb1-deletion in B-lymphoid leukemia cells with (left) or without (right) B→myeloid reprogramming. Values were normalized to total protein (n = 6). c, AMP levels in sorted B-lymphoid and B→myeloid reprogrammed cells are shown as log2-transformed relative amounts (amount in Lkb1-deleted cells/average amount in control), and data was baseline-centered (baseline = average amount in control; n = 3). d, Phosphorylation of Ampkα-T172, Ulk-S555, Raptor-S792 and Acc-S79 was assessed by Western blots. Data shown as mean (± s.d.) and assessed by two-tailed t-test (a–c). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 8. Small molecule inhibition of AMPK in human pre-B ALL.
a, Human leukemia and lymphoma cells (n = 4 biological replicates for B cell lymphoma and pre-B ALL; n = 3 biological replicates for CML; each in triplicate) were treated with BML275 (72 hr), and relative viability was assessed. b, Apoptosis was examined by Annexin V/7AAD staining in patient-derived pre-B ALL samples (n = 3) upon treatment with BML275 (10 μmol/L). c, Phospho-ACC-S79 in patient-derived pre-B ALL samples (n = 5) following overnight treatment with control (−) or BML275 (+, 10 μmol/L) was assessed. d, Phosphorylation of S6-S235/236 and Akt-S473 in patient-derived pre-B ALL samples (n = 6) following overnight treatment with control (−) or BML275 (+, 10 μmol/L) was assessed. e, Levels of AMP (normalized to cell numbers) in patient-derived CML (CML5 and CML6) and Ph+ ALL (ICN1 and PDX2) cells following 12 h treatment with control or BML275 (10 μmol/L). Plotted is log2-transformed average relative amounts (amount in cells treated with BML275/average amount of in control; 2 cases per group; each in triplicate), and data were median-centered. Median-centering was performed separately for CML5, CML6, ICN1 and PDX2 samples. f, Patient-derived CML (left) and pre-B ALL (middle) cells as well as MYC-driven B cell lymphoma cell lines (right) were treated with BML275 (10 μmol/L) or vehicle control for 6 hr. Glycolytic profiles (ECAR) and mitochondrial functions (OCR) were measured (normalized to total protein; n = 6; f). Data shown as mean (± e.f.) and assessed by two-tailed t-test (e, f).
Extended Data Figure 9. Mechanistic contribution of PAX5 targets.
a, b, Representative FACS plots from CRISPR-based gene editing experiments (a). CRISPR complexes were delivered to patient-derived PAX5-haploinsufficient pre-B ALL cells along with RFP-tagged gRNAs to direct dCas9 (CRISPR-mediated gene activation) or Cas9 (CRISPR-mediated gene deletion) to specific PAX5 target genes (e.g. NR3C1; left). Upon inducible activation of GFP-tagged PAX5 or EV in patient-derived pre-B ALL cells, enrichment or depletion of GFP+ cells carrying RFP-tagged gRNAs (RFP+) was monitored by flow cytometry (right; NT: non-targeting; g-53: gRNA clone 53 for deletion of NR3C1). Changes in percentage of GFP+ cells carrying the indicated gRNAs following induction, as compared to cells carrying the NT gRNA (b). c, d, In murine pre-B ALL models for genetic loss of Nr3c1, Cnr2, and Txnip function, responses to prednisolone (Pred) were measured (c) and protein levels of Nr3c1, Txnip and Cnr2 were examined (d). (e) In a patient-derived PAX5-haploinsufficient pre-B ALL (PAX5Δ; left), Dex responses upon inducible activation of PAX5 or empty vector (EV) were measured. In a patient-derived PAX5-wildtype pre-B ALL (PAX5WT; right), effects of DN-PAX5 on Dex responses were measured. Likewise, dose-response curves for Dex were measured in two patient-derived pre-B ALL samples carrying either wildtype or deleted IKZF1 when DN-IKZF1 or IKZF1 were inducibly expressed, respectively. Data shown as mean from 3 independent experiments (± s.d.) and assessed by two-tailed t-test (b) or two-way ANOVA (c, e).
Extended Data Figure 10. Targeting AMPK, CNR2 and TXNIP in combination with glucocorticoids.
a, Three different patient-derived pre-B ALL samples were treated with AMPK inhibitor BML275 as indicated, prednisolone, or in combination for 72 hr. b, Three different patient-derived pre-B ALL cells were treated with CNR2 agonist HU308 as indicated, or prednisolone (Pred) alone, or in combination for 72 hr. c, d, Three different patient-derived pre-B ALL cells were treated with a TXNIP agonist 3-O-methylglucose (3-O-MG; c) or D-allose (d) as indicated, prednisolone alone, or in combination for 72 hr. Relative viability was assessed (a–d). Combination index (CI) values at ED50 are shown. Prednisolone concentrations used were 2-fold higher than BML275. All data shown as mean ± s.d. (n = 3 independent experiments).
Supplementary Material
Acknowledgments
This work is supported by NIH/NCI through Outstanding Investigator Award R35CA197628 (to M.M.), a Wellcome Trust Senior Investigator Award (to M.M.), the Howard Hughes Medical Institute (HHMI-55108547 to M.M.), the Melanoma Research Alliance Established Investigator Award (T.G.G.), the German Bundesministerium für Bildung und Forschung, BMBF (A.H.) and the German Carreras Foundation (DJCLS R13/26). T.G.G. is an American Cancer Society Research Scholar, M.M. is a Howard Hughes Medical Institute (HHMI) Faculty Scholar.
Footnotes
Author Contributions M.M. conceived the study. L.N.C. and M.M. wrote the paper, designed experiments. L.N.C., Z.C., D.B., J.-W.L., G.X., K.N.C., C.H., S.S., and V.C. performed experiments and analyzed data. S.K.M. performed functional proteomics. H.G. performed biostatistical analyses. T.A.M. T.E., A.H., M.K. and G.C. provided and characterized patient samples. I.S.G., A.B., T.S.R. G.J.L., and R.A.D. provided mouse models. M.A.P., K.R.Y., G.J.L., R.A.D., I.S.G., A.B., H.P.K. ans H.S. provided specific expertise in NR3C1, PAX5 and IKZF1 function. D.B. and T.G.G. provided experimentation, analysis and expertise in metabolomics.
The authors declare no competing financial interests.
References
- 1.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 2.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 3.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 4.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 5.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tara C, et al. Osteoblasts mediate the adverse effects of glucocorticoids on fuel metabolism. J Clin Invest. 2012;122:4172–4189. doi: 10.1172/JCI63377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu N, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49:1167–1175. doi: 10.1016/j.molcel.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nogueiras R, Diaz-Arteaga A, Lockie SH, Velásquez DA, Tschop J, López M, Cadwell CC, Diéguez C, Tschöp MH. The endocannabinoid system: role in glucose and energy metabolism. Pharmacol Res. 2009;60:93–98. doi: 10.1016/j.phrs.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B-cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Di Tullio A, Vu Manh TP, Schubert A, Castellano G, Mansson R, Graf T. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no onvert retrodiffentation. Proc Natl Acad Sci USA. 2011;109:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fretz JA, Nelson T, Xi Y, Adams DJ, Rosen CJ, Horowitz MC. Altered metabolism and lipodystrophy in the early B-cell factor 1-deficient mouse. Endocrinology. 2010;151:1611–1621. doi: 10.1210/en.2009-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 15.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martín-Lorenzo A, et al. Infection Exposure is a Causal Factor in B-cell Precursor Acute Lymphoblastic Leukemia as a Result of Pax5-Inherited Susceptibility. Cancer Discov. 2015;5:1328–1343. doi: 10.1158/2159-8290.CD-15-0892. [DOI] [PubMed] [Google Scholar]
- 17.Faubert B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 21.Hanus L, Breuer A, Tchilibon S, Shiloah S, Goldenberg D, Horowitz M, Pertwee RG, Ross RA, Mechoulam R, Fride E. HU-308: a specific agonist for CB2 (CNR2), a peripheral cannabinoid receptor. Proc Natl Acad Sci USA. 1999;96:14228–14233. doi: 10.1073/pnas.96.25.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoltzman CA, Kaadige MR, Peterson CW, Ayer DE. MondoA senses non-glucose sugars: regulation of thioredoxin-interacting protein (TXNIP) and the hexose transport curb. J Biol Chem. 2011;286:38027–38034. doi: 10.1074/jbc.M111.275503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foley SB, Hildenbrand ZL, Soyombo AA, Magee JA, Wu Y, Oravecz-Wilson KI, Ross TS. Expression of BCR/ABL p210 from a knockin allele enhances bone marrow engraftment without inducing neoplasia. Cell Rep. 2013;17:51–60. doi: 10.1016/j.celrep.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaminathan S, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16:766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cazzaniga G, et al. Developmental origins and impact of BCR-ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood. 2011;118:5559–5564. doi: 10.1182/blood-2011-07-366542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, Saha V, Biondi A, Greaves MF. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 28.Bose S, Deininger M, Gora-Tybor J, Goldman JM, Melo JV. The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood. 1998;92:3362–3367. [PubMed] [Google Scholar]
- 29.Damm F, et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014;4:1088–1101. doi: 10.1158/2159-8290.CD-14-0104. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 32.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:538–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immnol. 2012;13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thai M, Graham NA, Braas D, Nehil M, Komisopoulou E, Kurdistani SAK, McCormick F, Graeber TG, Christofk HR. Adenovirus E40RF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab. 2014;19:694–701. doi: 10.1016/j.cmet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang H, Chen IM, Wilson CS, Bedrick EJ, Harvey RC, Atlas SR, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu GJ, Cimmino L, Jude JG, Hu Y, Witkowski MT, McKenzie MD, et al. Pax5 loss imposes a reversible differentiation blck in B-progenitor acute lymphoblastic leukemia. Genes Dev. 2014;28:1337–1350. doi: 10.1101/gad.240416.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gel scans are provided in Supplementary Fig. 1. All other data are available from the corresponding author upon reasonable request. Gene expression data were obtained from the GEO database accession numbers GSE3233012, GSE5287037, and GSE3846338. Patient outcome data were derived from the National Cancer Institute TARGET Data Matrix of the Children’s Oncology Group (COG) Clinical Trial P9906 (GSE11877)35, 36 and from TCGA (the Cancer Genome Atlas). GEO accession details are provided in Supplementary Tables 9 and 10. ChIP-seq tracks for PAX5, IKZF1, EBF1 and TCF3 antibodies in a normal B cell sample (ENCODE GM12878, UCSC genome browser) on INSR, GLUT1, GLUT3, GLUT6, HK2, G6PD, NR3C1, TXNIP, CNR2 and LKB1 gene promoter regions are shown UCSC genome browser hg19.