Abstract
The eukaryotic genome consists of DNA molecules far longer than the cells that contain them. They reach their greatest compaction during chromosome condensation in mitosis. This process is aided by condensin, a ‘Structural Maintenance of Chromosomes’ (SMC) family member1,2. The spatial organization of mitotic chromosomes and how condensin shapes chromatin architecture are not yet understood. Here, we use chromosome conformation capture (Hi-C)3,4 to study mitotic chromosome condensation in the fission yeast S. pombe5–7. This revealed that the interphase landscape characterized by small chromatin domains is replaced by fewer but larger domains in mitosis. Condensin achieves this by setting up longer-range, intra-chromosomal DNA interactions which compact and individualize chromosomes. At the same time, local chromatin contacts are constrained by condensin, with profound implications for local chromatin function during mitosis. Our results reveal condensin as a major determinant that changes the chromatin landscape as cells prepare their genomes for cell division.
Mitotic chromosomes are one of the most recognizable structures in eukaryotic cells. While their microscopic description goes back to the 19th century8, our knowledge of their molecular architecture remains scarce. Mitotic chromosome condensation is promoted by the chromosomal condensin complex, a ring-shaped multi-subunit protein assembly that topologically entraps DNA and that is thought to establish linkages between more than one DNA1,2. How condensin promotes chromosome condensation remains a topic of intense current interest. Chromosome conformation capture is a powerful tool to visualize patterns of chromosomal DNA contacts, yielding insight into how the DNA is arranged inside chromosomes3,4,9,10. This technique revealed large-scale homogenous folding of the chromatin chain within human mitotic chromosomes11. How local chromatin behavior changes as a result of mitotic chromosome compaction and how the condensin complex contributes to this process remain important open questions.
To understand the changes of mitotic chromatin organization at high resolution, we applied a high throughput chromosome conformation capture variant, Hi-C, to fission yeast. The small fission yeast genome size, distributed among three chromosomes, and its use of a fundamentally conserved chromosome condensation machinery, make this organism an attractive choice. We compared chromosomal DNA contact maps between asynchronously growing cells, >90% of which are in the G2 stage of interphase12, with cells arrested in mitosis by transcriptional repression of the Slp1Cdc20 activator of the anaphase promoting complex (Supplementary Fig. 1a)13. In our Hi-C analysis, mapped paired-end reads indicative of a DNA interaction were assigned to the nearest restriction site used for DNA fragmentation (DpnII), then these sites were grouped into 2 kb bins for further analysis (Supplementary Fig. 2a). This resulted in high resolution DNA interaction frequency maps of the fission yeast genome (Fig. 1a). High reproducibility of the interaction maps under these and each of the following biological conditions was confirmed by clustering of the Euclidean distance between the directionality plots of three independent repeats of each experiment (Supplementary Fig. 2b,c).
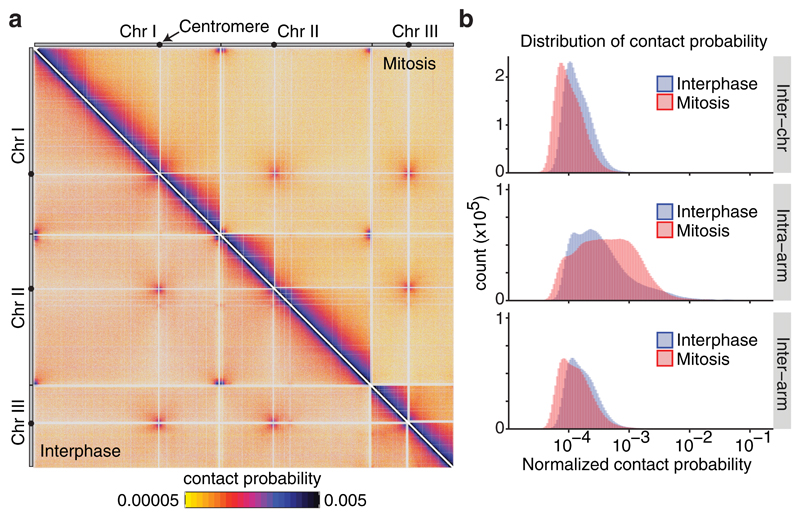
Figure 1. Hi-C reveals genome-wide contact changes during chromosome condensation.
(a) Normalized Hi-C contact probability maps in wild-type cells in interphase and mitosis (lower left and upper right triangles, respectively). The 3 fission yeast chromosomes are shown with centromere positions indicated (black dots). See Supplementary Figure 3 for an annotated schematic of the interactions that are recorded in this map. (b) Distribution of normalized contact probabilities between chromosomes (Inter-chr), within chromosome arms (Intra-arm) or between the two arms of the same chromosome (Inter-arm).
In addition to the expected richness of local DNA interactions along the three fission yeast chromosomes, seen in dark colors along the diagonal of the interphase contact map, prominent inter-centromeric and inter-telomeric interactions between chromosomes were apparent (Fig. 1a, lower left triangle; compare Supplementary Fig. 3 for an annotated map). This is expected from the Rabl orientation of fission yeast chromosomes in interphase9,12,14. In mitosis, interactions appeared to spread to greater distances from the diagonal, seen by the broadening of the dark region (Fig. 1a, upper right triangle). In contrast, interactions between chromosomes and between the two arms of each chromosome were reduced, indicated by the lighter orange color of these regions. Contact probability distribution plots confirmed a quantitative enrichment of interactions within chromosome arms in mitosis (Fig. 1b), at the expense of interactions between arms and between chromosomes. This provides a molecular correlate of the cytologically observed chromosome individualization and chromosome arm stiffening in mitosis (Supplementary Fig. 1a)7,8,15.
The changes in contact probabilities between interphase and mitosis are seen more clearly on Hi-C difference maps, obtained by simple subtraction of the normalized contact probabilities (Fig. 2a, upper right triangle). This confirmed an increased frequency of interactions within chromosome arms in mitosis, as well as the accompanying reduction in inter-arm and inter-chromosomal interactions. To elucidate the contribution of condensin to the altered contact probabilities in mitosis, we established a conditional shut-off allele for the condensin subunit Cut14Smc2 by combining promoter repression with an auxin-inducible degron (cut14SO; Supplementary Fig. 1a,b)5,16. The efficiency of Cut14 shut-off was confirmed by loss of mitotic chromosome compaction, measured by the distance between two fluorescent marks on chromosome I (Supplementary Fig. 1c-e)13. A Hi-C difference map between interphase and condensin-depleted mitotic cells revealed hardly any changes in contact probabilities (Fig. 2a, lower left triangle). Contact probability distribution plots confirmed that the previously seen changes in mitosis, namely increased intra-arm interactions, as well as reduced inter-chromosomal and inter-arm interactions, all depended on condensin (Fig. 2b, compare Fig. 1b). Similar results were obtained after depletion of the condensin subunit Cnd3CAP-G or following inhibition of Aurora B kinase, a positive regulator of mitotic condensation6,17–20, though Cnd3CAP-G was depleted less efficiently than Cut14Smc2 and Aurora B inhibition only partly blocked chromosome condensation (Supplementary Fig. 1b,f and 4a-c). A Hi-C difference map of mitotic cells containing or depleted of Cut14Smc2 further illustrated that condensin is responsible for most of the observed contact probability changes in mitosis (Fig. 2c).
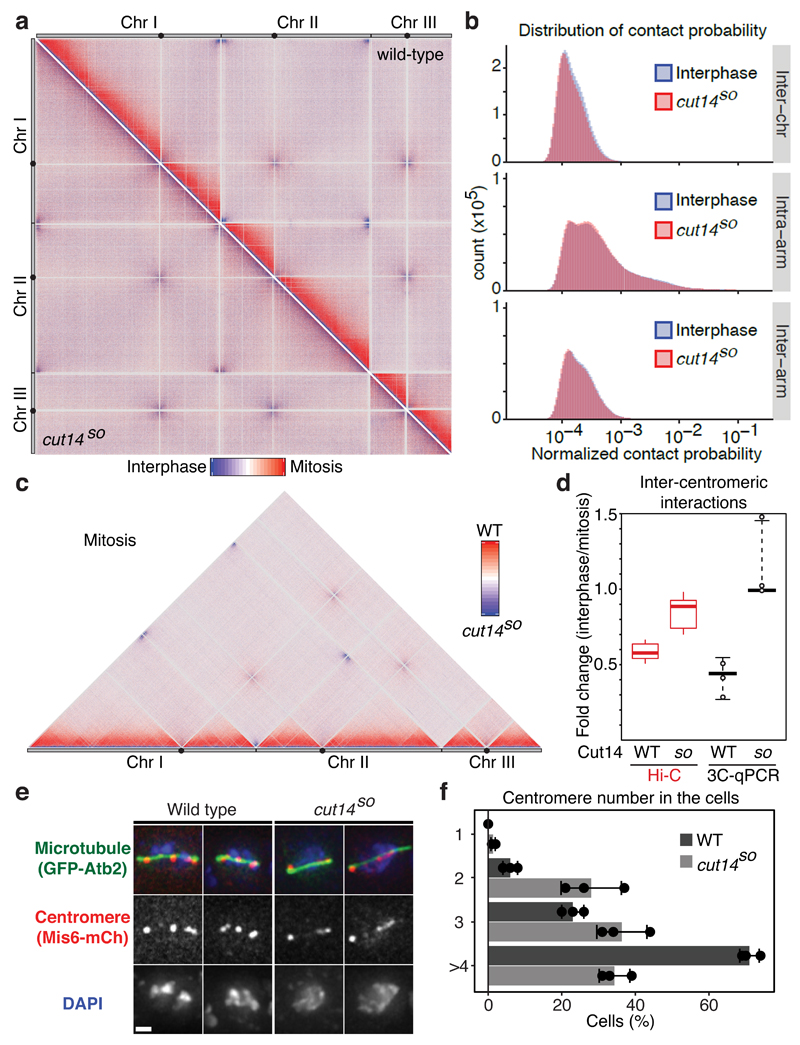
Figure 2. Mitotic conformational changes depend on the condensin complex.
(a) Hi-C difference maps between interphase and mitosis of wild-type and cut14SO cells (upper right and lower left triangles, respectively). (b) Distribution of normalized contact probabilities. As Fig. 1b, but comparing interphase and cut14SO mitosis. (c) Hi-C difference map comparing wild-type and cut14SO cells in mitosis. (d) Quantification of inter-centromeric interaction changes between interphase and mitosis. Box plots of the Hi-C contact probability changes between bins within central cores + 10 kb are compared to the medians and range of 3C-qPCR between centromere II and centromeres I and III of 2 technical replicates each of 3 independent experiments. (e) Localization of centromeres is shown in fixed mitotic cells. Mitotic spindles were also visualized and DNA was counterstained using 4',6-diamidino-2-phenylindole (DAPI). (f) The number of distinct centromere signals was counted. n = 100 cells were scored in three independent experiments, each. The mean ± s.d. is shown together with the individual results.
Hi-C differences reveal relative, but not necessarily absolute, changes of contact probabilities. We therefore used 3C followed by quantitative real time PCR (3C-qPCR) to calibrate our Hi-C difference maps. We chose inter-centromeric interactions as a benchmark, which are amongst the strongest inter-chromosomal contacts and are reliably detectable. 3C-qPCR showed that the interaction frequency between centromeres drops by just over 2-fold in mitosis, consistent with the almost 2-fold reduction of inter-centromeric interactions seen in the Hi-C difference maps (Fig. 2d). Both the Hi-C difference map and 3C-qPCR suggested that inter-centromeric contacts were retained in mitotic cells lacking condensin (Fig. 2d). To confirm this by an independent technique, we microscopically visualized centromere positioning in wild type and cut14SO cells arrested in mitosis. Centromeres were separated and distributed along the short metaphase spindle in wild type cells, as previously described (Fig. 2e,f)12. Centromeres remained clustered close to the spindle poles in Cut14-depleted cells, often displaying stretched signals. This pattern is consistent with persistent inter-centromere contacts and reminiscent of mitotic defects that have previously been reported in the absence of condensin19,21–23. Thus, the contact frequency changes reported in our Hi-C difference maps are a good reflection of absolute changes that occur during mitotic chromosome condensation.
We next focused our analysis on the most frequent DNA contacts, those within chromosome arms, as these are the most likely drivers of mitotic chromosome architecture changes. Figure 3a shows a magnified view of the Hi-C difference map along the chromosome II right arm, demonstrating increased longer-range interactions within the arm. Unexpectedly, we also noticed a marked reduction of short-range contacts, seen as a blue line along the base. Both changes depended on the condensin complex (Fig. 3b), as well as Aurora B kinase activity (Supplementary Fig. 4). Plotting the median contact probability as a function of genomic distance revealed fewer intra-arm interactions below 84 kb in mitosis, but increased interaction frequencies between 84 - 940 kb (Fig. 3c). Interactions beyond 940 kb were again depleted in mitosis (the transition points lie around 90 kb and 900 kb, respectively, on all chromosome arms). Again, these rearrangements of contact probabilities depended on condensin and Aurora B kinase activity (Fig. 3c and Supplementary Fig. 4). The reduction of megabase-scale contacts is reminiscent of reduced longest-range interactions as human chromosomes condense11, a probable reflection of chromosome arm stiffening as mitotic chromosomes gain shape.
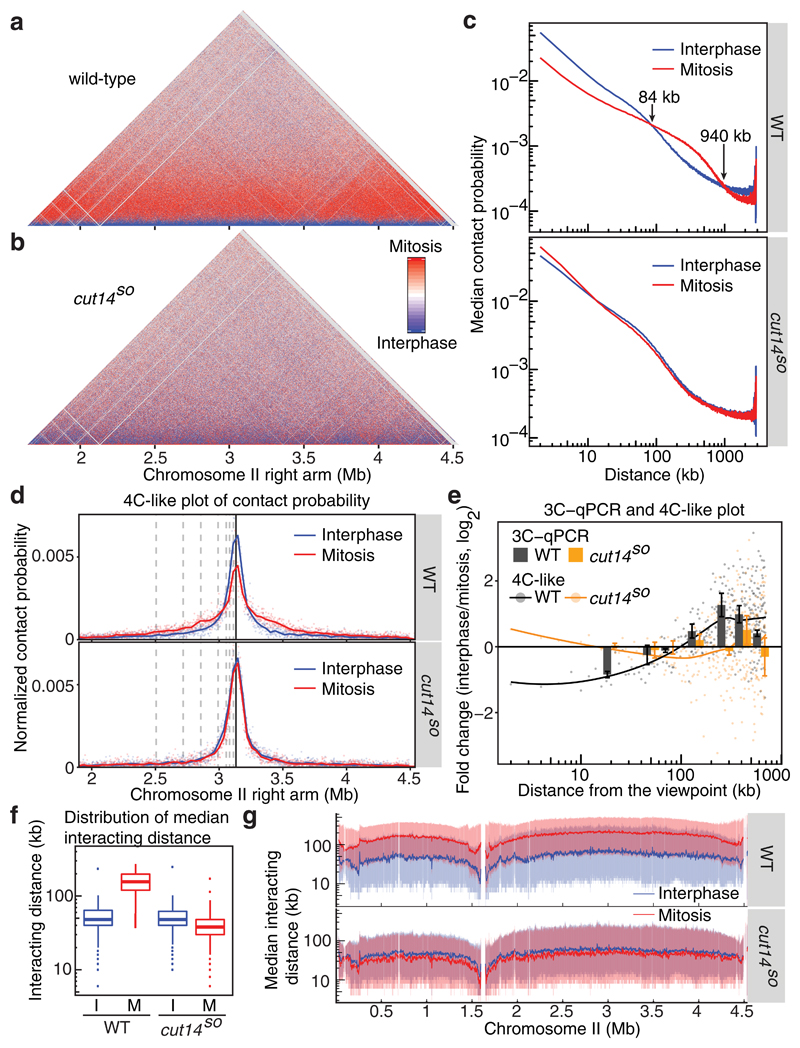
Figure 3. Condensin replaces local contacts with longer-range interactions in mitosis.
(a, b) Hi-C difference maps of the chromosome II right arm comparing interphase and mitosis in wild-type (a) and in cut14SO cells (b). (c) Median contact probabilities in interphase and mitosis as a function of distance along the chromosome II right arm are shown in wild-type (top) and cut14SO cells (bottom). (d) 4C-like plot of Hi-C contact probability from a viewpoint on the chromosome II right arm. The positions of 3C-qPCR primer-binding sites are shown (dotted and solid lines). (e) Interaction changes between interphase and mitosis determined by 3C-qPCR (mean ± s.e. of 3 biological repeats) are compared to 4C-like contact probability changes based on the Hi-C data (dots). Lines represent smoothed 4C-like contact probability changes. (f) Box plots of the distribution of contact frequencies within all chromosome arms under the indicated conditions. The box plot shows the medians and 25th and 75th percentiles, the whiskers indicate a 95% confidence interval. Outliers are also shown. (g) Moving intra-arm median interacting distances along chromosome II (solid lines) are shown together with shaded areas representing the 25th and 75th percentiles.
The chromosome volume decreases during mitotic condensation, resulting in chromatin compaction. A decrease of local chromatin contacts within mitotic chromosomes came therefore as a surprise. To further investigated this, we plotted interaction frequencies from one selected viewpoint in the Hi-C data to obtain a 4C-like plot4. Figure 3d shows such a plot from a viewpoint close to the middle of the chromosome II right arm. We selected 7 distances from this viewpoint and compared contact frequency in interphase and mitosis by 3C-qPCR (Fig. 3e). This provided quantitative confirmation of the 4C-like plot. Intra-arm contacts closer than 90 kb were reduced up to 2-fold in mitosis, while longer-range interactions increased to a similar extent, consistent with the changes seen in the Hi-C difference map. Both effects again depended on condensin. Thus, an unanticipated consequence of chromosome condensation is a reduction of local chromatin contacts.
4C-like analysis also allowed us to address whether chromosomes condense homogeneously, or whether compaction varies along chromosome arms. We used a sliding viewpoint and recorded the contact distributions at each position along chromosome II. The median of all interactions was around 50 kb in interphase, which increased to around 150 kb in mitosis (Fig. 3f). Plotting the sliding median along chromosome II revealed somewhat longer interactions in the middle of both the left and right chromosome arms and shorter interactions towards the centromeres and chromosome ends (Fig. 3g). This trend was not an artifact of the iteratively-corrected normalization used to generate our Hi-C maps and was similarly seen when using an alternative normalization strategy, as well as in the raw read counts (Supplementary Fig. 5). This implies that fission yeast chromosomes are slightly more compact in the middle of their arms compared to centromere proximal regions or chromosome ends.
Chromosomes are thought to consist of a series of topologically associated domains (TADs)9,24–26, within which chromatin interactions are enriched. We analyzed TADs along fission yeast chromosomes in interphase and mitosis by plotting the directionality of interactions along the chromosome (Supplementary Fig. 2b), from which we determined TAD boundary positions (Fig. 4a). These boundaries correlated with local minima of insulation scores, thus validating the boundary assignments (Supplementary Fig. 2d,e). This analysis confirmed the existence of numerous TADs in interphase with a median size of 267 kb. The median TAD size nearly doubled in mitosis to 481 kb, dependent on condensin as well as Aurora B kinase activity (Fig. 4a,b and Supplementary Fig. 6). The condensin-mediated mitotic TAD size expansion is compatible with the size range of condensin-mediated chromosome interactions previously determined by ChIA-PET27. Notably, 69% (25 out of 36) of the boundary positions in mitosis coincided with those observed in interphase, suggesting that interphase TADs fuse to give rise to larger chromosome domains in mitosis. We reached the same conclusions when we used an alternative algorithm, (TopDom)28, to assign domain boundaries (Supplementary Fig. 7).
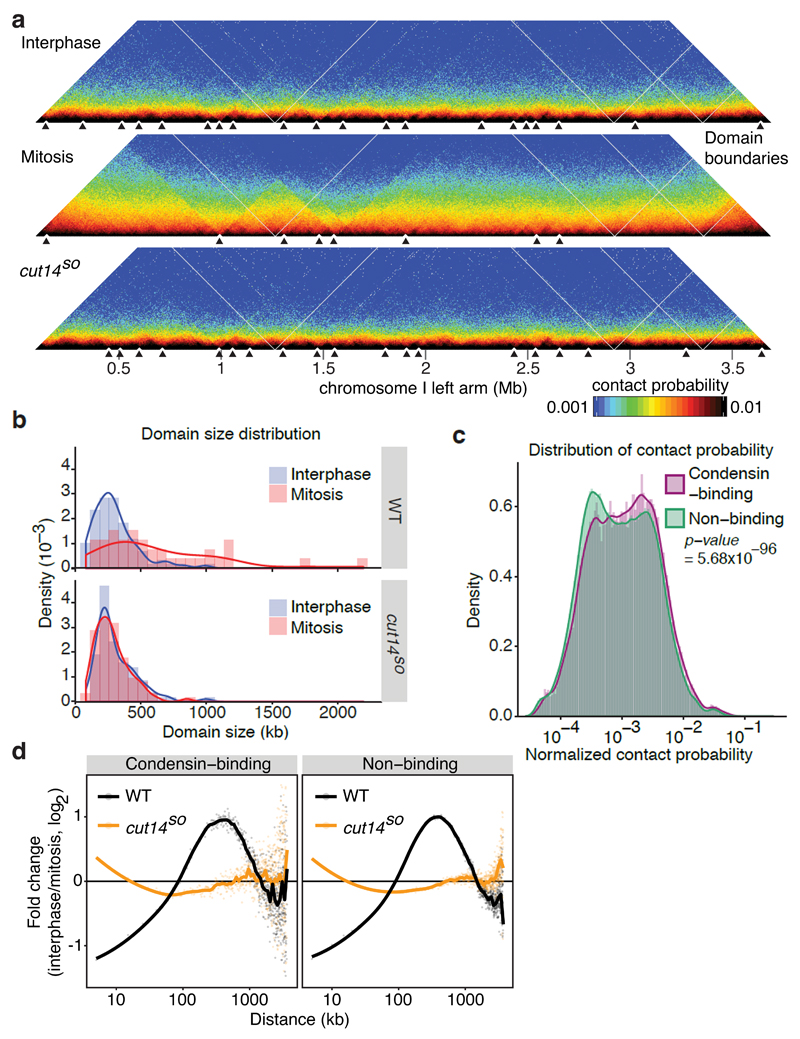
Figure 4. Condensin-dependent chromatin domain expansion in mitosis.
(a) Normalized Hi-C maps are shown with domain boundaries along a section of the chromosome I left arm under the indicated conditions. For domain visualization, a bin size of 5 kb was used. Black triangles indicate domain boundaries. (b) Density plots of domain size distributions. (c) Distribution of normalized contact probabilities of 5 kb bins containing or not containing a condensin binding site. A Wilcoxon Mann-Whitney test was used to test the null hypothesis that contact probabilities between condensin binding and non-binding sites are the same. (d) Contact probability changes between interphase and mitosis are plotted as a function of distance, separated into bins containing or not containing a condensin binding site.
We started to address how condensin promotes longer-range interactions by performing ChIP to determine condensin binding sites. We divided the chromosome into 5 kb bins that were categorized into those that did or did not contain a condensin-enriched site (Supplementary Table 1). The contact probability plot of these 5 kb bins revealed that condensin binding sites show a small but significantly greater probability of engaging in intra-arm interactions, compared to non-binding sites (Fig. 4c). This suggests that condensin acts by engaging in contacts between its binding sites. Next, we analyzed whether condensin binding sites show a preference for longer-range DNA interactions. The mitotic interaction frequency change of condensin binding sites as a function of genomic distance confirmed that these sites engage in mitotic longer-range interactions in a condensin-dependent manner. However, condensin non-binding sites showed a qualitatively similar behavior (Fig. 4d). Therefore, condensin binding sites are preferential targets for intra-arm interactions that facilitate contacts between neighboring DNA sequences. While site-specific looping between condensin binding sites has been seen by ChIA-PET27, our Hi-C approach finds that intra-arm interactions are widely spread. Condensin binding to chromosomes with limited sequence preference could be a reason for this.
To further clarify the role of condensin binding sites in chromatin organization, we analyzed their relationship with chromatin boundary positions. Most interphase and mitotic boundaries contained a condensin binding site (Supplementary Fig. 8), consistent with a role for condensin in domain formation and their fusion in mitosis.
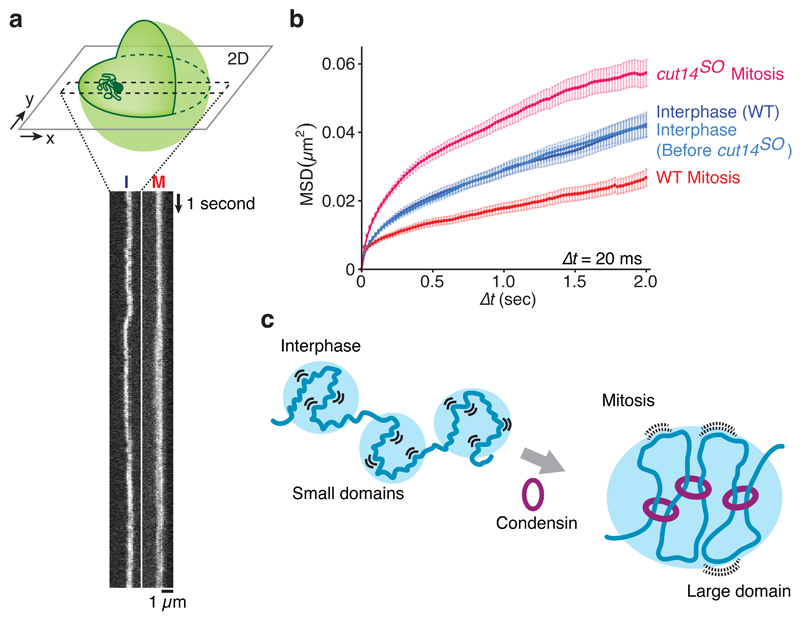
An unexpected consequence of mitotic chromosome condensation was the reduction of local chromatin contacts. To address whether this is due to restricted chromatin motility in mitosis, we monitored local movement of two loci on chromosomes I and II, visualized using LacOs bound by LacI-GFP and TetOs bound by TetR-tdTomato respectively, by high speed confocal microscopy. Kymographs of the LacI-GFP movements on chromosome I revealed greater motility in interphase compared to mitosis (Fig. 5a). To quantify this difference, we tracked the GFP locus at 20 ms intervals and prepared mean square displacement plots. This confirmed that chromatin motility is constrained in mitosis (Fig. 5b). The same was observed on chromosome II (Supplementary Fig. 9). Constrained mitotic chromatin motility depended on condensin. Motility markedly increased following condensin depletion, exceeding even that observed in interphase (Fig. 5b). We could not directly measure condensin’s impact on interphase motility, as condensin depletion that we achieve during a mitotic arrest requires longer than cells usually spend in interphase.
Figure 5. Condensin confines local chromatin motility.
(a) Kymographs of the LacO locus on chromosome 1, marked by LacI-GFP, in wild type cells in interphase (I) and mitosis (M). The schematic illustrates the experiment. (b) Mean square displacement (MSD) of the LacO locus under the indicated conditions. Mean ± S.E.M are plotted (n = 61-72). (c) A model for how chromosome compaction by condensin is achieved by longer-range interactions, accompanied by chromatin domain enlargement as well as reduced local chromatin motility.
Our study describes the conformational changes of eukaryotic chromatin during mitotic chromosome condensation. Condensin replaces predominantly short-range local contacts in interphase with longer-range interactions in mitosis. Our results do not distinguish whether condensin establishes longer-range interactions by extruding or expanding DNA loops, by stabilizing stochastic chromatin interactions, or both29,30. In either scenario, a slower dissociation rate of condensin from chromosomes, that has been observed in mitosis23, could promote longer-range interactions. It will be interesting to computationally study predictions from the two models and compare them to our high resolution experimental datasets. A notable consequence of mitotic longer-range interactions is restricted chromatin motility, accompanied by reduced local contacts (Fig. 5c). Establishment of gene silencing in budding yeast requires passage through mitosis, while transcriptional reprogramming in mammalian nuclei is promoted by mitotic factors in as yet unknown ways31–33. Future studies will explore whether reduced local chromatin motility facilitates either process. In line with the central role played by condensin during biochemically reconstituted chromosome assembly34, our findings illustrate condensin’s principal contribution to chromatin reorganization during mitotic chromosome condensation.
Online Methods
S. pombe strains and culture
All the strains used in this study are listed in Supplementary Table 2. PCR-based gene targeting35 and standard genetic manipulation36 were used during strain construction. To obtain the cut14 shut-off (cut14so) or cnd3 shut-off (cnd3so) strains, the C-terminus of the genes were fused to the auxin-inducible degron module IAA1737 and its endogenous promoter was replaced by the nmt81 promoter38 in cells harboring Skp1-Tir116. For the Z2-Cnd3-GFP strain, a plasmid containing GFP-tagged cnd3 gene under regulation of the endogenous cnd3 promoter was constructed by the module-based Golden Gate method for chromosomal integration plasmids in fission yeast39. The resultant plasmid was linearized by FseI and integrated at the Z2 locus. To replace the endogenous slp1 promoter, a kanR-nmt41 promoter-slp1 fragment was amplified from a previously described slp1 shut-off strain13 and integrated.
All strains were cultured in EMM medium supplemented with L-glutamic acid as nitrogen source. To shut off slp1 transcription and induce mitotic arrest, cells were cultured at 25 ºC for 3 hours after addition of 5 μg/ml thiamine. Following this, 60-75% of cells were arrested in mitosis, displaying a short mitotic spindle. The efficiency of this arrest was unaffected by condensin depletion or Aurora B kinase inactivation. To deplete Cut14 or Cnd3, cells were cultured at 25 ºC for 3 hours with 5 μg/ml thiamine as well as 0.5 mM of the auxin 1-naphthaleneacetic acid (NAA). To inhibit Aurora B kinase (Ark1), ark1-as3 cells18 were cultured with 5 μg/ml thiamine at 25 ºC for 1.5 hours, then 5 µM of 1NM-PP1, or the equivalent amount of DMSO as a control, was added to the medium for further 1.5 hours.
Fluorescent microscopy
Cells were fixed with 70 % ethanol and stained with 4’,6-diamidino-2-phenylindole (DAPI). Images were acquired as serial sections along the z-axis on a DeltaVision microscope system (Applied Precision) and combined using the quick projection algorithm in SoftWoRx. To measure the distance between genomic loci marked with LacO repeats bound by LacI-GFP and TetO repeats bound by TetR-tdTomato, z-stacks were merged and the distance was measured in two dimensions using Fiji40. To track chromatin motility, live-cell imaging was performed using an α Plan Apochromat 100×/1.46 NA oil objective (Carl Zeiss) in a temperature-controlled chamber. Images were captured on an Evolve 512 EMCCD camera (Photometrics), operated in streaming mode, mounted on an inverted microscope (AxioObserver.Z1; Carl Zeiss), constituents of a custom-built spinning-disc confocal system (Intelligent Imaging Innovations). All hardware was controlled with SlideBook software (Intelligent Imaging Innovations). Images were collected at 20 (TetR-tdTomato) or 50 Hz (LacI-GFP), following excitation with either a 488 or 568-nm laser. Fluorescent spot movement was automatically tracked and mean square displacement (MSD) was calculated using the ParticleTracker41 plug-in in Fiji (https://bitbucket.org/djpbarry/particletracker).
Western blotting
Cell extracts were prepared by glass bead cell breakage as detailed in the Hi-C/3C section. Protein levels were analyzed by SDS-polyacrylamide gel electrophoresis, followed by western blotting using α-AID (Cosmo Bio, CAC-APC004AM) or α-Tat1 (an in house generated mouse monoclonal antibody against α-tubulin) as primary antibodies.
Hi-C/3C sample preparation
The Hi-C experiment was performed as previously described, with modifications3. ~7.5x108 fission yeast cells were fixed with 0.5 % (w/v) formaldehyde at 25 ºC for 30 minutes. Excess formaldehyde was quenched with 0.125 M glycine at 25 ºC for 15 minutes. Fixed cells were washed 3 times with wash buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1 % (w/v) Triton X-100, 1 mg/ml NaN3) and resuspended in FA buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1 % (w/v) Triton X-100, 0.1 % (w/v) sodium-deoxycholate) containing cOmplete ULTRA protein inhibitor cocktail (Roche). Cells were broken by glass bead breakage in a Multi beads shocker (Yasui Kikai). The crude extract was centrifuged to retrieve the chromatin fraction at 15,000 rpm at 4 ºC for 15 minutes. Precipitated chromatin was washed once with DpnII restriction enzyme incubation buffer (New England Biolabs), then resuspended in DpnII buffer and incubated at 65 ºC for 10 minutes with 0.1 % (w/v) of SDS. After addition of 1 % (w/v) Triton X-100, chromatin was digested overnight with DpnII (New England Biolabs) and RNase A (Sigma) at 37 ºC. After heat inactivation of DpnII at 65 ºC for 20 minutes, digested chromatin was split into 3 parts, one for 3C library preparation and the other two for Hi-C library preparation. For Hi-C library preparation, DpnII overhangs were filled in with biotin-14-dATP, dCTP, dGTP, and dTTP using DNA polymerase I Klenow fragment (New England Biolabs) at 37 ºC for 45 minutes. To capture proximity, chromatin was diluted to 8 ml in ligase incubation buffer, followed by incubation at 16 ºC for 8 hours with T4 DNA ligase (New England Biolabs). For 3C library preparation, digested chromatin was similarly ligated, but without the fill-in step. DNA was in both cases recovered by Proteinase K digestion and de-crosslinking overnight at 65 ºC, followed by phenol-chloroform extraction and isopropanol precipitation. Hi-C library preparation progressed by removing Biotin-14-dATP at un-ligated ends by incubation at 20 ºC for 4 hours in the presence of 300 U of T4 DNA polymerase (New England Biolabs). After inactivating T4 DNA polymerase by addition of EDTA, DNA was fragmented using a focused ultrasonicator (Covaris S220) and purified on AMPure XP beads (Beckman Coulter). The ends of the fragmented DNA were repaired and sequencing adapters were ligated using the NEBNext Ultra DNA Library Prep kit reagents and NEBNext Multiplex Oligos for Illumina (Index Primers Set 1, New England Biolabs). Junction-containing biotinylated DNA fragments were adsorbed to MyOne Streptavidin C1 dynabeads (ThermoFisher) at room temperature for 15 minutes. Bead-bound DNA was amplified using the NEBNext High Fidelity 2x PCR Master Mix with index primer sets. DNA fragments in the 200 -700 bp range were purified by double selection on SPRI selection beads (BeckmanCoulter), followed by sequencing on the Illumina HiSeq platform (100 bp paired-end).
3C-qPCR
3C libraries were prepared as described above. The efficiency of proximal ligation was quantified using primer pairs corresponding to interactions of interest (listed in Supplementary Table 3). Real time quantitative PCR was performed with the PowerUp SYBR Green Master Mix using a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems). Ct values were normalized using internal controls (YK377-YK378 for inter-centromeric interactions, YK717-YK718 for intra-arm interactions), then ΔΔCt values comparing interphase and mitosis were calculated. For quantification of inter-centromeric interactions, ΔΔCt values were converted to a linear scale to derive fold change values. Two technical replicates were averaged in each of three biological repeats of the experiment. The median and range of values from the three repeat experiments are reported. To quantify intra-arm interactions, again two technical replicates were averaged in each of three biological repeats. The mean of ΔΔCt values between interphase and mitosis in the three biological repeats were plotted together with the standard error.
ChIP-seq sample preparation
Cells were fixed with 0.5 % (w/v) formaldehyde and then crude cell extracts were prepared by glass bead breakage as described above. Extracts were sonicated using a Bioruptor (Diagenode). After removing debris by centrifugation, extracts were split into input and ChIP sample. The ChIP sample was incubated with α-HA antibody (F-7, Santa Cruz sc-7392) overnight at 4 ºC. Precipitated DNA-Protein complexes were washed twice with FA buffer, twice with FA buffer containing 500 mM NaCl, twice with ChIP wash buffer (10 mM Tris-HCl pH 8, 250 mM LiCl, 0.5 % (w/v) NP-40, 0.5 % (w/v) sodium-deoxycholate, 1 mM EDTA), and once with TE. DNA ends were repaired by incubation with Klenow Enzyme (NEB) in the presence of dNTPs at 25 ºC for 30 minutes. End-repaired DNA-Protein complex was Eluted with TE containing 1 % (w/v) SDS by incubating at 25 ºC for 30 minutes. Eluted DNA-protein complexes were incubated with T4 DNA ligase, and then incubated at 65 ºC overnight, followed by incubation with RNase A at 37 ºC for 60 minutes. DNA was recovered by Proteinase K digestion and decrosslinking at 55 ºC for 2 hours, followed by phenol-chloroform extraction and ethanol precipitation. Sequencing adapters were ligated to the ends of purified DNA using the NEBNext Ultra DNA Library Prep kit and NEBNext Multiplex Oligos for Illumina. The adapter-ligated DNA was amplified using the NEBNext High Fidelity 2x PCR Master Mix with index primer sets. Amplified fragments were purified by double selection of SPRI selection beads (Beckman Coulter), followed by sequencing on the Illumina HiSeq platform.
Hi-C data analysis
Identification of ligation events
Reads containing DpnII recognition motifs were trimmed to immediately after the first occurrence of the motif. If either read of the resultant read pair was shorter than 20 bp then the pair was discarded. Each read of the read pair was aligned individually to the Schizosaccharomyces pombe genome (build ASM294v2.21) using the BWA-mem algorithm (version 0.7.12)42. Read pairs with unmapped reads, or read alignments with a mapping quality of <10, were discarded. To remove reads derived from un-ligated genomic fragments, concordantly mapped read pairs within 2 kb of each other on the genome, were also discarded. The sonication-fragmentation step within the library preparation protocol should generate unique read pairs from the ligation of any two DpnII sites within the genome. Identically mapped read pairs were therefore assumed to be PCR duplicates. A single read from these putative duplicate reads was randomly selected and the other reads were discarded. The now remaining reads were extended to the nearest DpnII recognition site downstream from the read start. Read pairs where either read start site was >1 kb upstream from a DpnII site were presumed to be derived from non-ligated fragments and discarded. Ligation between the remaining read pairs is then presumed to have occurred between the downstream DpnII sites. The number of read counts at each step for each of our Hi-C libraries is summarized in Supplementary Table 4.
Generation of interaction matrices
Count matrices were generated for chromosomes I, II and III. The genome was divided into equally sized bins (2 kb or 5 kb). Where the chromosome length is not an exact multiple of bin length, the maximum number of non-overlapping bins was generated and the most telomeric regions of the chromosome were not analyzed. Few reads, if any, are accurately assigned to these regions due to their non-unique sequences. Thus, no information is lost, while the equally sized bins ease subsequent statistical analyses. The count matrix is filled by ascribing individual DpnII ligation events to the bins containing their respective DpnII sites. Where the ligation event occurs at a bin boundary the event is ascribed to the bin containing the mapped read. To examine subsets of interactions, e.g. inter-chromosome, intra-arm or inter-arm ligations, the count matrices for these regions were simply extracted from the larger count matrix.
Normalization of interaction matrices
The observed interaction frequency between two genomic regions is affected by many factors other than their proximity. Normalization of the interaction matrices aims to remove these 'non-proximity' effects. Two techniques were used to normalize the matrices. The first technique, iterative correction43, does not attempt to model the source of these non-proximity factors. Instead, it operates on the principle that all genomic regions should be equally visible and partake in an equal number of interactions. The second technique, HiCNorm44, attempts to remove the impact of three non-proximity factors known to affect observed interactions, fragend frequency, GC content and mappability. The iteratively-corrected normalized matrices were used most of our analyses, except for Supplementary Figure 5a that utilized HiCNorm.
Prior to normalization the intra-bin ligations were removed from the matrix by setting the diagonal of the count matrix to zero. Additionally, bins involved in fewer inter-bin interactions than a specified threshold were removed from the matrix to ensure the optimal functioning of the normalization algorithm. A threshold of 500 interactions was used for the 2 kb bins (compare Supplementary Fig. 2a) and a threshold of 1000 for the 5 kb bins. 2 kb-binned matrices were used for our analysis, if not stated otherwise.
The iterative correction algorithm43 was used to normalize the altered count matrices such that all rows and columns were set to sum to 1. The resultant matrix can therefore be considered a probability matrix where the values at point 'i,j' is the probability that region 'i' is found to interact with region 'j' and vice-versa. HiCNorm (version 08.05.2012)44 was provided with the mappability, GC content and actie fragend size of all the 2 kb bins. Mappability scores and GC content were calculated from all regions within a bin that are within 200 bp of a DpnII cut site. The active fragend size is the cumulative length of these regions. Mappability scores were calculated by aligning tiled 50 bp reads (10 bp window shift), extracted from the regions of interest, and identifying the proportion of reads with a mapping quality of 10 or more as defined by BWA-mem (version 0.7.12)42. A floor value for mappability scores was set as 0.1.
Interaction directionality and Euclidean clustering
To calculate the interaction directionality for a given bin the interaction probability between the bin and its 50 upstream and 50 downstream bins were extracted from the intra-arm normalized matrix. The upstream and downstream probabilities are then paired by their absolute distance from bin of interest. The interaction directionality value is the mean value of the log2 ratio of each pair of upstream and downstream probabilities. A value greater than 0 (i.e. a positive log2 directionality value) indicates that the bin preferentially interacts with upstream regions, a value smaller than 0 (i.e. a negative log2 directionality value) indicates that downstream interactions are favored. Where one or more of the paired probability values are equal to zero, the pair is excluded from the calculation. Where less than 2 paired distance probabilities are available, due to zero values or the end of the chromosome, no interaction directionality is calculated. Samples were clustered by calculating the Euclidean distance between vectors of log2 interaction directionality values and performing complete-linkage agglomerative hierarchical clustering on the resulting distance matrix.
Histogram of contact probabilities
Contact probabilities in whole genome normalized matrices were categorized into inter-chromosomal, intra-arm and inter-arm interactions according to the position of the bins. Contact probabilities categorized above were used to plot histograms of count distribution in normalized contact probabilities using ggplot2 in R. Contact probabilities in intra-arm normalized matrices were further divided into condensin binding and non-binding according to the presence or absence of ChIP peaks, respectively (See the ChIP-seq data analysis section for detection of ChIP peak positions). Hi-C bins with ChIP peaks were identified using ChIPpeakAnno in Bioconductor45. Contact probabilities categorized as above were used to plot histograms.
Hi-C maps and Hi-C difference maps
Normalized matrices were plotted using ggplot2 in R. To prepare Hi-C difference maps, differential contact probability matrices were produced by subtraction of a normalized matrix in condition 1 (e.g. interphase) from a normalized matrix in condition 2 (e.g. mitosis). Then, matrix differences were plotted using ggplot2 in R. Normalized Hi-C maps with domain boundaries were prepared as below. 5 kb-binned normalized Hi-C matrices were plotted and then the positions of identified domain boundaries were plotted on top of that using ggplot2 in R.
Calculation of fold change of inter-centromeric interactions
Bins encompassing centromere sequences and the surrounding 10 kb were identified as centromeric bins. Fold change of inter-centromeric interaction probabilities were calculated after adding up the Hi-C contact probabilities within the centromeric bins from whole genome normalized matrices. Three biological replicates of Hi-C samples were used to generate boxplots for each condition.
Contact probability as a function of distance
All the contact probabilities in intra-arm normalized matrices were classified according to the distance between the two underlying bins. The median contact probability was calculated for each distance, then plotted against distance for each condition. To calculate probability-distance quartiles, all the interaction probabilities of a bin of interest were extracted from an intra-arm normalized matrix or from the raw count matrix in case of Supplementary Figure 5b. The bin probabilities were then ordered by their absolute distance from the bin of interest and the cumulative probability of the bins was calculated. The 25th, 50th and 75th percentiles are the distances at which the cumulative probability first equals or exceeds 0.25, 0.50 and 0.75, respectively. 25th, 50th (median) and 75th percentile distances for the bins along chromosome II were plotted for each condition using ggplot2 in R.
4C-like plot
A bin in which a primer for 3C-qPCR analysis anneals on the chromosome II right arm was selected as a viewpoint to generate 4C-like plots. Hi-C contact probabilities between the viewpoint and all other bins within the chromosome II right arm were then plotted as dots in each condition. Lines of smoothed 4C-like contact probabilities, calculated using the loess-smoothing method within ggplot2 in R, were also plotted.
Determination of domain boundaries
Domain boundaries were determined using either the log2 directionality or the TopDom software package (version 0.0.2)28 from the 2 kb binned intra-arm normalized matrices. The log2 directionality values were calculated as below. A 40 kb sliding window was used to traverse the chromosome arm. For each window position, the maximum log2 value (log2max) within the 20 kb upstream half of the window, and the minimum log2 value (log2min) within the 20 kb downstream half of the window, were identified. A log2shift value for the window is then calculated by subtracting log2min from log2max. A putative domain boundary was identified between the log2min and log2max values if log2shift is greater or equal to 0.5. The putative boundaries were then filtered by grouping overlapping boundary domains and discarding all domains except the one with the highest log2,shift value. The domain boundary is then assigned to the midpoint between the log2max and log2min values for the remaining domain boundaries. To improve reliability of domain boundary identification, only domain boundaries that were identified in the same location (±5 kb) in each of three biological replicates were classed as true boundary locations. To plot average log2 directionality at the boundaries, log2 directionality values within 40 kb upstream and downstream were normalized. The normalized log2 directionality values were plotted using the geom_smooth function provided by ggplot2 in R. Alternatively, TopDom was used as follows. A window size of 10 bins was used to identify the boundary positions. Common boundaries that were identified from three biological replicates were used for further analyses. Domain size was calculated as the distance between neighboring boundary positions.
Insulation Scores
Insulation scores were generated from the 2 kb binned intra-arm normalized matrices. Scores were calculated by taking the mean interaction probability across the 2 bins on either side of the bins of interest. To determine insulation score around the boundaries determined by log2 directionality, insulation scores within 50 kb upstream and downstream were normalized by log2 (insulation score/mean insulation score in the selected regions). Average normalized insulation scores were calculated from all the normalized insulation scores at every 10 kb insulation distances. The average normalized insulation scores as a function of insulation distance were plotted using ggplot2 in R.
ChIP-seq data analysis
Each read of the paired end ChIP-seq library was trimmed to a length of 50 bases. The reads were aligned to the Schizosaccharomyces pombe genome (build ASM294v2.21) using the BWA-mem algorithm (version 0.7.12)42. ChIP peaks were identified from the alignments using version 2.1.0 of the macs2 software with the input format set to BAMPE46. ChIP peaks are listed in Supplementary Table 1.
Supplementary Material
Acknowledgments
We would like to thank J. Abella and M. Way for their help with the high speed microscopy, A. Stewart for bioinformatics support, C. Haering and H. Masukata for reagents and P. Bates, E. Wershof, B. Khatri, Y. Murayama, T. Toda and our laboratory members for discussions and critical reading of the manuscript. This work was supported by the European Research Council and the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001198), the UK Medical Research Council (FC001198), and the Wellcome Trust (FC001198). Y.K. was supported by the Japanese Society for the Promotion of Science (JSPS).
Footnotes
Data availability. The Hi-C and ChIP-seq data reported in this manuscript have been deposited with the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE94478.
Code availability. The computer code used to generate the results reported in this manuscript has been deposited with GitHub, https://github.com/adam-rabinowitz/Condensin-mediated-remodeling-of-the-mitotic-chromatin-landscape.
Author Contributions
Y.K. and F.U. conceived the study, Y.K. performed the experiments, Y.K., A.R. and D.J.B. analyzed the data, Y.K. and F.U. wrote the manuscript with input from A.R.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Hirano T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell. 2016;164:847–57. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17:399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- 3.Belton JM, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–76. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saka Y, et al. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–52. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutani T, et al. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–83. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–58. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 8.Flemming W. F C W Vogel; 1882. Zellsubstanz, Kern und Zelltheilung. [Google Scholar]
- 9.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516:432–5. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–53. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–76. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrova B, et al. Quantitative analysis of chromosome condensation in fission yeast. Mol Cell Biol. 2013;33:984–98. doi: 10.1128/MCB.01400-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanizawa H, et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–77. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagasaka K, Hossain MJ, Roberti MJ, Ellenberg J, Hirota T. Sister chromatid resolution is an intrinsic part of chromosome organization in prophase. Nat Cell Biol. 2016;18:692–9. doi: 10.1038/ncb3353. [DOI] [PubMed] [Google Scholar]
- 16.Kanke M, et al. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 2011;12:8. doi: 10.1186/1471-2121-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen J, Hagan IM. S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr Biol. 2003;13:590–7. doi: 10.1016/s0960-9822(03)00205-7. [DOI] [PubMed] [Google Scholar]
- 18.Hauf S, et al. Aurora controls sister kinetochore mono-orientation and homolog bi-oritentation in meiosis-I. EMBO J. 2007;26:4475–4486. doi: 10.1038/sj.emboj.7601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- 20.Nakazawa N, Mehrotra R, Ebe M, Yanagida M. Condensin phosphorylated by the Aurora-B-like kinase Ark1 is continuously required until telophase in a mode distinct from Top2. J Cell Sci. 2011;124:1795–1807. doi: 10.1242/jcs.078733. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira RA, Coelho PA, Sunkel CE. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol Cell Biol. 2005;25:8971–84. doi: 10.1128/MCB.25.20.8971-8984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribeiro SA, et al. Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell. 2009;20:2371–80. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16:333–44. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 24.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crane E, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–4. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–29. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim K-D, Tanizawa H, Iwasaki O, Noma K. Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast. Nat Genet. 2016;48:1242–1252. doi: 10.1038/ng.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin H, et al. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res. 2016;44:e70. doi: 10.1093/nar/gkv1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng TM, et al. A simple biophysical model emulates budding yeast chromosome condensation. Elife. 2015;4:e05565. doi: 10.7554/eLife.05565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–49. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganier O, et al. Synergic reprogramming of mammalian cells by combined exposure to mitotic Xenopus egg extracts and transcription factors. Proc Natl Acad Sci USA. 2011;108:17331–6. doi: 10.1073/pnas.1100733108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li YC, Cheng TH, Gartenberg MR. Establishment of transcriptional silencing in the absence of DNA replication. Science. 2001;291:650–3. doi: 10.1126/science.291.5504.650. [DOI] [PubMed] [Google Scholar]
- 33.Martins-Taylor K, Dula ML, Holmes SG. Heterochromatin spreading at yeast telomeres occurs in M phase. Genetics. 2004;168:65–75. doi: 10.1534/genetics.103.020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shintomi K, Takahashi TS, Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol. 2015;17:1014–23. doi: 10.1038/ncb3187. [DOI] [PubMed] [Google Scholar]
- 35.Bahler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–51. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 36.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 38.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–6. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 39.Kakui Y, et al. Module-based construction of plasmids for chromosomal integration of the fission yeast Schizosaccharomyces pombe. Open Biol. 2015;5:150054. doi: 10.1098/rsob.150054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abella JV, et al. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2006;18:76–86. doi: 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu M, et al. HiCNorm: removing biases in Hi-C data via Poisson regression. Bioinformatics. 2012;28:3131–3133. doi: 10.1093/bioinformatics/bts570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.