Abstract
Osteoporosis is a common disease diagnosed primarily by measurement of bone mineral density (BMD). We undertook a genome-wide association study in 142,487 individuals from the UK Biobank to identify loci associated with BMD estimated by quantitative ultrasound of the heel (“eBMD”). We identified 307 conditionally independent SNPs attaining genome-wide significance at 203 loci, explaining approximately 12% of the phenotypic variance. These included 153 novel loci, and several rare variants with large effect sizes. To investigate underlying mechanisms we undertook: 1) bioinformatic, functional genomic annotation and human osteoblast expression studies; 2) gene function prediction; 3) skeletal phenotyping of 120 knockout mice with deletions of genes adjacent to lead independent SNPs; and 4) analysis of gene expression in mouse osteoblasts, osteocytes and osteoclasts. These studies strongly implicate GPC6 as a novel determinant of BMD and also identify abnormal skeletal phenotypes in knockout mice for a further 100 prioritized genes.
Keywords: bone mineral density, genome-wide association, GPC6, osteoporosis, UK Biobank, ultrasound
Introduction
Osteoporosis is a common age-related disorder characterised by low bone mass and deterioration in bone micro-architecture leading to an increase in skeletal fragility and fracture risk. Low bone mineral density (BMD) is a strong risk factor for osteoporosis and is a key indicator for its diagnosis and treatment 1. BMD is highly heritable 2, and genome-wide association studies (GWAS) have identified common variants at 73 loci associated with the trait, including many significantly associated with fracture risk 3,4. Recently, deep imputation based on whole-genome sequencing, has also identified low frequency variants of large effect associated with BMD and fracture risk 4. Despite these advances, common and rare variants only explain 5.8% of the total phenotypic variance in BMD 3,4.
Most previous genetic studies of BMD have analysed data derived from dual energy X-ray absorptiometry (DXA). However, DXA is expensive and consequently the largest GWAS to date of DXA-derived BMD includes only 32,965 individuals4, compromising the ability to detect genetic loci. An alternative method of estimating BMD that is quick, safe, relatively inexpensive and can therefore be assessed in very large samples of individuals, is derived from ultrasound, typically at the heel calcaneus (referred to as estimated BMD “eBMD” in this manuscript). Measures of eBMD derived from ultrasound are highly heritable (in the order of 50% to 80%) 5–8, are independently associated with fracture risk 9,10, and are moderately correlated with DXA derived BMD at the hip and spine (r = 0.4 to 0.6) 11. A previous GWAS that used heel ultrasound parameters (N = 15,514) identified variants at nine loci, including seven previously associated with lumbar spine/hip BMD 12.
Since genetic loci associated with BMD are strongly enriched for the targets of clinically relevant osteoporosis therapies13,14, the discovery of new genetic loci and the biological pathways they implicate may help identify drug targets for the prevention and treatment of fragility fracture. In order to identify novel genetic determinants of BMD, we performed genome-wide association in the UK Biobank Study, which has measured eBMD and genome-wide genotypes on 142,487 individuals. We subsequently performed three systematic and complementary approaches to prioritize genes for functional validation (Supplementary Fig. 1).
Results
Genome-wide Association Study of eBMD
BMD was measured by quantitative ultrasound of the heel, a non-invasive estimate of BMD (eBMD) that predicts fracture 9,10. After stringent quality control of both eBMD and genome-wide genotypes (see Online Methods and Supplementary Fig. 2), data were available from 142,487 individuals (53% women) (Supplementary Table 1). We tested the additive effect of 17,166,351 single nucleotide polymorphisms (SNPs) with minor allele frequency (MAF) > 0.1% and imputation quality (info) score >0.4 on eBMD, controlling for age, sex and genotyping array. In total, 307 conditionally independent SNPs at 203 loci surpassed our revised genome-wide significant threshold (P ≤ 6.6 x 10-9, which accounts for the larger number of independent SNPs deeply imputed in the UK Biobank, see Online Methods) and jointly explained ~12% of the variance in eBMD (Supplementary Fig. 3, Supplementary Table 2). Together the 307 SNPs explained about a third of eBMD SNP heritability estimated by BOLT-REML (h2SNP = 0.36). Although there was substantial inflation of the test statistics relative to the null (λGC = 1.37), linkage disequilibrium (LD) score regression15 indicated that the majority of inflation was due to polygenicity rather than population stratification (LD Score regression intercept = 1.05). Of the 203 loci identified, 153 (75%) regions had not been implicated in previous GWAS of BMD (Supplementary Table 2, Supplementary Fig. 3).3,4,16–22 Interestingly, the list of novel associations included multiple variants (e.g. SNPs at TBX1, ZNRF3) for which there was extremely strong evidence of association with heel eBMD (i.e. P < 10-30), but little evidence of association (p > 0.05 any trait) in the previous GEFOS-Seq GWAS of DXA derived BMD4 (Supplementary Table 3).
Our study also replicated SNPs in 55 out of 73 regions (>75%) that had been reported as genome-wide significant in previous GWAS of BMD at other body sites (i.e. P < 0.05 and consistent direction of effect), and we replicated all genome-wide significant loci identified in a previous GWAS of ultrasound heel eBMD12 (Supplementary Table 4). Our list of known BMD associated SNPs is deliberately broad and comprehensive in terms of previous GWAS. This comprehensive inclusion policy, however, means that we have also incorporated the results of some smaller GWAS studies that may include false positives. When we restrict our attention to the 64 SNPs reported in the large Estrada et al (2012) GEFOS meta-analysis3 (which are unlikely to represent type 1 errors), we replicate 54 of 64 (84%) SNPs. Possible reasons for non-replicating loci include site specificity, differences in phenotype (ultrasound derived versus DXA derived BMD), differences in ancestral population between studies, and type 1 error in the smaller previous study.
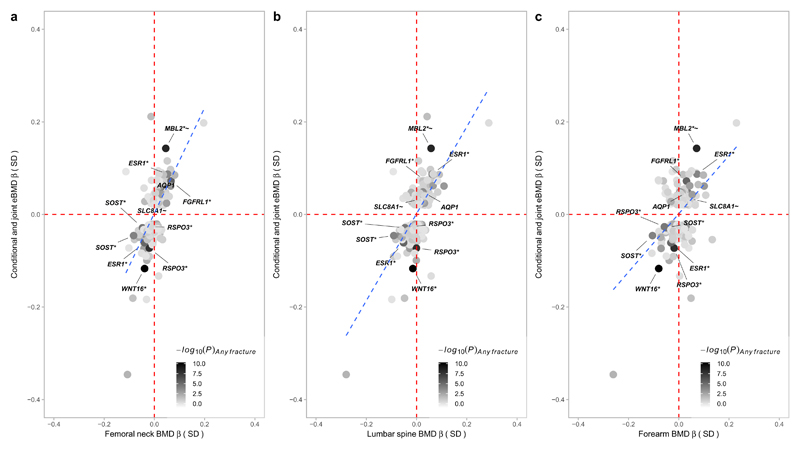
Notably, across six loci (RSPO3, LINC00326, CPED1, MPP7, KCNMA1, TMEM263), there were SNPs that exhibited different directions of effect in the current eBMD study compared to previous BMD studies. In the case of the SNPs at CPED1, these also showed an association with fracture in UK Biobank (see below), but in the direction predicted by eBMD rather than the direction predicted by BMD in previous studies (i.e. alleles that predispose to low eBMD are associated with increased risk of fracture). Although these opposite directions of association are harder to explain, differences in the phenotype measured by DXA and ultrasound technologies are likely to be responsible. For example, whereas heel ultrasound primarily measures trabecular bone, DXA-based BMD measurements reflect a combination of trabecular and cortical bone. In addition, ultrasound-based measurements are independent of bone size, while areal BMD as measured by DXA is not fully size corrected. In fact, of the six loci showing opposite associations between DXA BMD and eBMD, three also showed strong associations with height in the GIANT consortium in the same direction of DXA BMD23, suggesting that the latter relationships may partly have reflected size effects (although it must be noted that several other concordant eBMD and DXA BMD associated loci also showed associations with height). Whereas bone size and bone mass generally show a strong positive correlation, genetic influences leading to greater bone size might be inversely related to trabecular bone density at certain sites, due to reduced mechanical strain as a consequence of a larger and hence stronger skeleton. However, despite these few discrepancies, overall there was a strong positive correlation between estimated effect size for the genome-wide significant heel eBMD SNPs in the present UK Biobank Study and estimated effect sizes for DXA-derived BMD at other skeletal sites in our previous GEFOS-Seq study (femoral neck: Pearson’s r = 0.64 (0.57 – 0.71); lumbar spine: r = 0.69 (0.62 – 0.75); forearm: r = 0.49 (0.39 – 0.58)) (Fig. 1)4. Adjusting for weight had little effect on the genome-wide significant results, save for partially attenuating the strength of association between eBMD and known adiposity variants (Supplementary Table 5).
Figure 1. Effect size in standard deviations for heel eBMD (y-axis) from the current UK Biobank Study plotted against effect size in standard deviations from the previous GEFOS-Seq paper for BMD at the (A) femoral neck, (B) lumbar spine, and (C) forearm (x-axis).
Only conditionally independent variants that reach genome-wide significance (P < 6.6 x10-9) for eBMD in the UK Biobank Study are plotted. Minus log10P-value for the (any) fracture analysis in UK Biobank is represented by the shading of the data points (black for robust evidence of association with fracture and white for poor evidence of association). SNPs that reach Bonferroni corrected significance for fracture (P < 1.6 x10-4) are labelled in the diagram. The blue dashed trend line shows a strong correlation between estimated effect sizes at the heel and at other sites of the body. SNPs at SLC8A1 and AQP1 were significantly related with fracture after correction for multiple testing (P < 1.6 x 10-4) and have not previously been reported associated with BMD or fracture although they both reached nominal significance (P < 0.05) in the previous GEFOS-Seq scan.
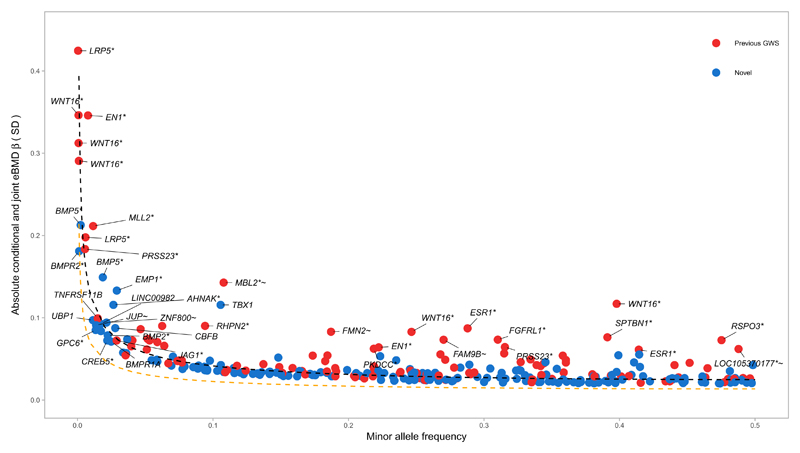
Since we employed a large sample size and genotyped and/or imputed low frequency variants (MAF <1%), we next assessed the allelic architecture of eBMD (Fig 2). We found a strong relationship between MAF and effect size, which in general followed the statistical power of our study design. The variants of largest effect (where each allele increased eBMD by 0.44 standard deviations [SD], P = 5 x 10-11) were in the gene IGHMBP2 (within 0.5Mb of known variants in LRP5) and the known EN1 and WNT16/CPED1 loci. We also detected several rare (MAF <1%) and low frequency variants (1% < MAF < 5%) in novel loci including rare variants near the genes BMP5 and BMPR2. Comparing the mean absolute effect sizes of genome-wide significant variants, we found a 6.5-fold difference in effects attributed to rare compared to common variants.
Figure 2. Relationship between absolute conditional and joint analysis effect size in standard deviations (y-axis) and minor allele frequency (x-axis) for 307 conditionally independent SNPs.
Red circles represent SNPs at previously reported BMD loci. Blue circles denote SNPs at novel loci. The black dashed line shows the effect size required for 80% power to detect association at a given minor allele frequency at genome-wide significance (α = 6.6x10-9) in the present study. The orange dashed line shows the effect size required for 80% power to detect association at a given minor allele frequency at genome-wide significance (α = 6.6x10-9) assuming N = 483,230 individuals when the full UKBiobank Study becomes available.
Sex-specific analyses across the genome and tests of sex heterogeneity at genome-wide significant SNPs revealed a single variant rs17307280 at FAM9B on the X chromosome that was significantly associated with eBMD in men only (Supplementary Fig. 4, Supplementary Table 6) (heterogeneity P = 1.4 x 10-11), replicating previous results from Estrada et al.3
Effects on Fracture
We tested the relationship between eBMD-associated SNPs and fracture. We identified 14,492 individuals (58% women) in UK Biobank reporting a previous fracture, irrespective of the trauma mechanism, since high-trauma fractures are predicted by low BMD and are predictive of future low-trauma fracture, suggesting a shared aetiology 24,25. In total, we observed that twelve eBMD SNPs were associated with fracture, after control for multiple testing (P ≤ 1.6 x 10-4). Sensitivity analyses, including only 8,540 individuals (69% women) reporting a fracture resulting from a simple fall (i.e. from standing height), were consistent with these findings (Table 1). Of these twelve loci, variants at AQP1 and SLC8A1 have not been associated with BMD or risk of fracture previously (although both SNPs show nominal association (P < 0.01) with DXA derived measures of BMD in the previous GEFOS-Seq study4 (Fig. 1, Supplementary Table 3)). Genome-wide significant eBMD variants showed an inverse relationship between their effect on eBMD and odds of fracture (Supplementary Fig. 5).
Table 1.
Genome-wide significant eBMD associated SNPs that show significant association with risk of fracture (P < 1.6 x 10-4)
| ANY FRACTURE | FALL FRACTURE | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RSID | CHR | BP | C.GENE | EA | NEA | EAF | OR | CI95%-L | CI95%-U | P | OR | CI95%-L | CI95%-U | P | STATUS |
| rs10490046 | 2 | 40630678 | SLC8A1 | A | C | 0.78 | 0.94 | 0.92 | 0.97 | 6.8x10-6 | 0.94 | 0.91 | 0.98 | 1.4x10-3 | NOVEL |
| rs112069922 | 4 | 1034997 | IDUA | C | T | 0.95 | 0.89 | 0.84 | 0.93 | 4.8x10-6 | 0.90 | 0.84 | 0.96 | 2.2x10-3 | KNOWN |
| rs9491689 | 6 | 127398595 | RSPO3 | C | A | 0.72 | 1.05 | 1.03 | 1.08 | 5.0x10-5 | 1.05 | 1.02 | 1.09 | 2.0x10-3 | KNOWN |
| rs7741021 | 6 | 127468274 | RSPO3 | A | C | 0.52 | 1.07 | 1.04 | 1.09 | 1.5x10-8 | 1.07 | 1.04 | 1.10 | 4.8x10-6 | KNOWN |
| rs4869744 | 6 | 151908012 | ESR1 | T | C | 0.71 | 0.95 | 0.93 | 0.98 | 1.3x10-4 | 0.95 | 0.92 | 0.98 | 8.0x10-4 | KNOWN |
| rs2941741 | 6 | 152008982 | ESR1 | G | A | 0.58 | 1.05 | 1.03 | 1.08 | 6.5x10-6 | 1.07 | 1.04 | 1.11 | 2.4x10-6 | KNOWN |
| rs10276670 | 7 | 30956489 | AQP1 | A | G | 0.77 | 0.95 | 0.92 | 0.97 | 4.1x10-5 | 0.94 | 0.91 | 0.97 | 3.5x10-4 | NOVEL |
| rs2536195 | 7 | 120959155 | WNT16 | A | G | 0.6 | 1.10 | 1.07 | 1.12 | 2.6x10-15 | 1.13 | 1.10 | 1.16 | 1.6x10-15 | KNOWN |
| rs10668066 | 7 | 120965464 | WNT16 | G | GCACC | 0.75 | 1.09 | 1.07 | 1.12 | 1.5x10-11 | 1.13 | 1.09 | 1.17 | 2.5x10-12 | KNOWN |
| rs7099953 | 10 | 54426489 | MBL2 | G | T | 0.89 | 0.90 | 0.87 | 0.93 | 4.9x10-9 | 0.89 | 0.84 | 0.93 | 5.0x10-7 | KNOWN |
| rs7209826 | 17 | 41796406 | SOST | A | G | 0.62 | 1.05 | 1.03 | 1.07 | 3.6x10-5 | 1.06 | 1.03 | 1.10 | 7.1x10-5 | KNOWN |
| rs188810925 | 17 | 41798194 | SOST | G | A | 0.92 | 1.09 | 1.04 | 1.14 | 9.2x10-5 | 1.11 | 1.05 | 1.17 | 3.3x10-4 | KNOWN |
*Beta (β) and standard errors (SE) from BOLT-LMM were transformed using the following formula: (β or SE) / (μ *(1- μ)), where μ = number of cases/number of controls. Approximate odds ratios (OR) and 95% confidence intervals (CI95%) were calculated from the transformed beta and standard error. RSID = Reference SNP cluster ID, CHR = Chromosome, BP = Base pair position of the variant according to human reference sequence (Hg19/GRCh37), C.GENE = closest gene, EA = Effect allele, NEA = Non-effect allele, EAF = Effect allele frequency, and P = Strength of evidence against the null hypothesis of no association between variant and self-reported fracture (i.e. P-value), ANY FRACTURE = any self-reported fracture within the last five years (N = 14,492 cases / 130,563 controls) and FALL FRACTURE = self-reported fracture within the last five years that occurred as the result of a simple fall (N = 8,540 cases / 131,333 controls).
Shared Genetic Factors
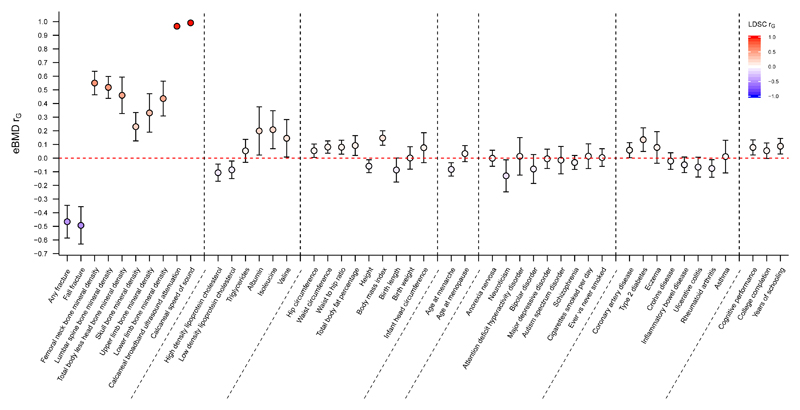
We tested whether eBMD had a shared genetic aetiology with 247 other diseases and biomedically relevant traits using LD score regression26 as implemented in LDHub27. This method estimates the degree of shared genetic risk factors between two diseases or traits, although it says nothing about how this shared genetic aetiology arises (i.e. whether one variable causes the other or whether the relationship between eBMD and the other variable is mediated by an underlying variable like body mass index which is itself partially genetic). Fig. 3 shows that genetically increased eBMD was strongly and negatively correlated with fracture (rg = -0.47, 95% CI: -0.59, -0.35). Further, measures of BMD at other skeletal sites showed moderate positive genetic correlations with eBMD (Fig. 3) consistent with the concordant directions observed at the genome-wide significant loci (Fig. 1).
Figure 3. Genetic correlations between eBMD as measured in the UK Biobank Study (y-axis) and other traits and diseases (x-axis) estimated by LD score regression implemented in LDHub.
Genetic correlation (rG) and corresponding 95% confidence intervals (error bars) between eBMD and traits were estimated using linkage-disequilibrium score regression. The genetic correlation estimates (rG) are shaded according to their magnitude and direction (red for positive and blue for negative correlation).
We also tested to see whether eBMD was genetically correlated with a range of other complex traits and diseases (Supplementary Table 7; Fig. 3). We observed that eBMD was weakly and negatively correlated with HDL, LDL, height, age at menarche and rheumatoid arthritis (Fig. 3). On the other hand, eBMD was weakly positively genetically correlated with body mass index (BMI), waist circumference, waist-hip ratio, coronary heart disease and type 2 diabetes. These findings support a shared genetic etiology of several common traits and diseases with eBMD, as has been shown previously between BMD, adiposity and type 2 diabetes through Mendelian randomization28,29.
Gene Prioritization
Strategy One: Bioinformatic, Statistical and Functional Genomics in Humans
We used several bioinformatics and statistical genetics tools to prioritize likely candidate genes and variants. These included the Variant Effect Predictor software30 to identify deleterious coding variation at genome-wide significant loci (Supplementary Table 8), the FINEMAP software to create configurations of plausible causal SNPs around each conditionally independent lead SNP (Supplementary Table 9), ENCODE maps of DNase I hypersensitivity sites (DHS) 31,32 and contextual analysis of transcription factor occupancy (CATO) scores4 to identify SNPs perturbing transcription factor activity, and cis-eQTL evidence in human osteoblasts 33 (Supplementary Table 10). These results are fully described in the Supplementary Note.
Strategy Two: Data-Driven Expression-Prioritized Integration
The second gene prioritization approach applied the DEPICT computational tool34. We identified 273 genes as most likely to be driving the eBMD association signals (false discovery rate (FDR) < 0.05). Among these 273 genes are several with an established role in bone metabolism including BMP2, LRP5, EN1, RUNX2, JAG1, ESR1, COL21A1 and SOST (Supplementary Table 11).
We next tested the DEPICT prioritized genes for enriched expression in any of 209 Medical Subject Heading (MeSH) tissue and cell type annotations34. 62 tissue or cell type annotations were identified (at a FDR of 5%) among the entries defined from the medical subject heading tissue and cell annotations (Supplementary Table 12, Supplementary Fig. 6). The strongest evidence of enriched expression of the genes mapping to eBMD loci came from chondrocytes and cartilage, although systems other than the musculoskeletal system were also overrepresented (i.e., cardiovascular system [7/12 significant entries], membranes tissue [6/7 significant entries] and connective tissue cells [5/7 significant entries]).
The DEPICT prioritized genes were also tested for enriched gene sets, which identified more than 1,000 significantly enriched (FDR 5%) gene sets. After clustering in 35 ‘meta gene-sets’, most clusters were related to skeletal growth (e.g. regulation of mineralized tissue development, vertebral fusion, abnormal craniofacial development, cartilage development) or to signalling pathways involved in bone biology (e.g. mesenchymal stem cell differentiation, BMP or WNT signalling). More global biological processes were also highlighted (e.g. transcription factor binding and regulation, chromatin remodelling complex or cell development) (Supplementary Fig. 7).
Meta-analysis gene-set enrichment of variant associations (MAGENTA) software produced similar results implicating gene sets involved in bone mineralization and development, Cadherin, WNT and Hedgehog signalling pathways, as well as other pathways worthy of further investigation (oncogenic pathways, melanogenesis etc) (Supplementary Table 13).
All genes prioritized by DEPICT were tested for expression in mouse osteoblasts, osteoclasts and osteocytes. Among the 273 genes prioritized, 241 had mouse homologs (the majority that did not have a known homologue were long non-coding RNAs), 92% were expressed in osteoblasts, 66% in osteoclasts, and 83% in osteocytes (Supplementary Table 14). Taken together, 95.4% of these genes were expressed in at least one of the three cell types. This represents a substantial enrichment of genes expressed in osteoblast, osteocyte and osteoclasts (P < 0.0001 for each of osteoblasts, osteocytes and osteoclasts).
We then investigated whether a skeletal phenotype had been reported in the International Mouse Phenotyping Consortium (IMPC: http://www.mousephenotype.org) or Mouse Genome Informatics (MGI: http://www.informatics.jax.org) databases in knockout mice harbouring a deletion of any of the prioritized genes. 189 (78%) of the 241 DEPICT-prioritized genes had mouse knockout phenotype data available and 62 (33%) of these had skeletal abnormalities (Supplementary Table 14).
Strategy Three: Deep Phenotyping of Murine Knockouts of Selected Genes within 1MB of Lead SNPs
The third gene prioritization approach identified all genes within 1MB of lead SNPs at associated eBMD loci. These genes were compared with knockout mice generated at the Wellcome Trust Sanger Institute for the IMPC 35. Knockout mice had been generated for 120 of the prioritized genes and bespoke skeletal phenotyping was undertaken as part of the Origins of Bone and Cartilage Disease (OBCD) Program 36. Specifically, we performed both structural and functional analysis of skeletal samples using digital x-ray microradiography, micro-CT and biomechanical testing. Results were compared with normal reference data from >250 controls with identical C57BL/6 genetic background. 43 (36%) of these 120 prioritized genes had significantly abnormal bone structure, representing a 2-fold enrichment compared to previous analysis of 100 unselected knockout mice 36 (χ2= 8.359, P = 0.0038) (Supplementary Table 15).
GPC6 Findings
These parallel strategies identified 100 genes with an abnormal skeletal phenotype when disrupted in mutant mice (Supplementary Table 14, Supplementary Table 15). However, all three gene prioritization strategies identified GPC6 and, therefore this gene was selected for further study (Supplementary Table 16).
GPC6 encodes a member of the glycosylphosphatidylinositol-anchored, membrane-bound heparan sulfate proteoglycan protein family. Loss of function mutations in GPC6 result in omodysplasia-1 (MIM 258315) a rare autosomal recessive skeletal dysplasia characterized by short limbed dwarfism with craniofacial dysmorphism, indicating a role for GPC6 in skeletal biology37, although the gene has not previously been implicated in osteoporosis.
Our bioinformatics pipeline provided evidence for a functional association at the GPC6 locus. A single SNP in GPC6, rs1933784, in high linkage disequilibrium with the conditionally independent lead SNP rs147720516 at this locus (r2 > 0.9), is a plausible causal and functional variant. We observed that rs1933784 is a low frequency SNP (MAF = 0.05) significantly associated with eBMD (P = 2.3x10-10), with a high causal probability (log10 Bayes factor = 2.4), and lies within DHS in several cell types (Supplementary Table 16). The rs1933784 variant also shows some evidence of association with expression of GPC6 in osteoblasts (P = 4.7x10-3) (Supplementary Table 16).
GPC6 was identified by DEPICT as the most likely gene responsible for the association at this locus. Gpc6 is expressed in osteoblasts and osteocytes in mice (Supplementary Table 14). Gpc6 had a similar level of enrichment (1.76 log fold-enrichment) in osteocytes when compared to genes known to play a key role in the skeleton, such as Lrp5 (1.95 log fold-enrichment) (Supplementary Fig. 8), an important receptor that influences bone mass through canonical Wnt-signalling, and Runx2 (1.73 log fold-enrichment), a key transcription factor in osteoblast differentiation.
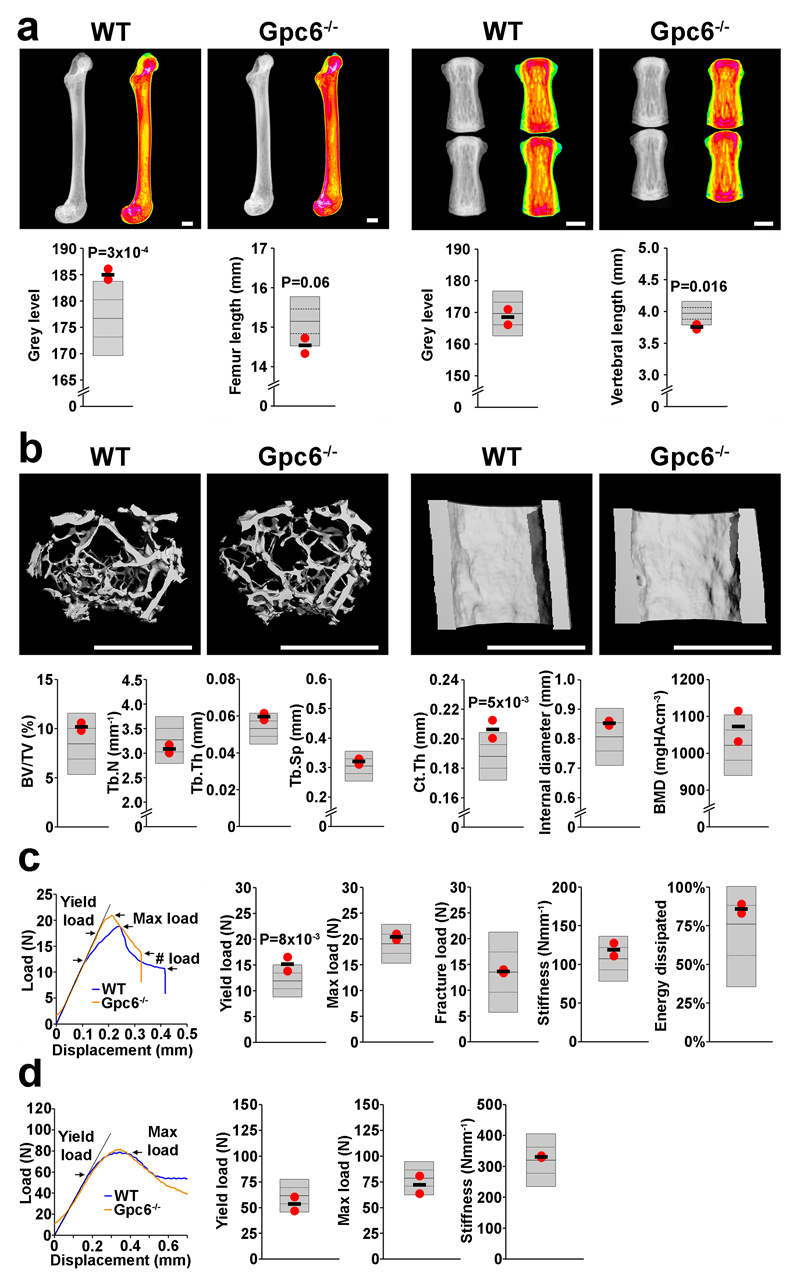
Adult female Gpc6-/- mice were analyzed and results compared to >250 wild type controls of identical C57BL/6 background. Consistent with the phenotype of omodysplasia-1, Gpc6-/-mice, had femurs and vertebrae that were shorter than wild type (-1.95 and -2.17 SD, permuted P = 0.06, P = 0.016 respectively). Gpc6-/- mice also had increased femoral bone mineral content (+2.4 SD, permuted P = 3x10-4), and increased cortical thickness (+2.3 SD, permuted P = 5x10-3). The biomechanical consequence of these structural abnormalities was an increase in yield load (+2.1SD, permuted P = 8x10-3) which reflects an increased material elasticity (Fig. 4). While the phenotype of Gpc6-/- mice is consistent with human omodysplasia-1, no information is available regarding adult manifestations of the condition. Thus further studies in Gpc6-/- mice are required to characterize the cellular and molecular mechanisms underlying the role of GPC6 in the pathogenesis of osteoporosis.
Figure 4. Increased bone mass and strength in adult Glypican 6 knockout mice (Gpc6-/-).
(a) X-ray microradiography images of femur and caudal vertebrae from female wild-type (WT) and Gpc6-/- mice at postnatal day 112 (P112). Pseudocolored grey-scale images in which low bone mineral content (BMC) is green and high BMC is pink. Graphs show reference ranges derived from >250 WT mice of identical age, sex and genetic background (C57BL/6), mean (solid line), 1.0SD (dotted lines) and 2.0SD (grey box). Values for parameters from individual Gpc6-/- mice are shown as red dots and mean values as a black line (n=2 animals). (b) Micro-CT images of proximal femur trabecular bone (left) and mid-diaphysis cortical bone (right) from WT and Gpc6-/- mice. Graphs showing trabecular bone volume/tissue volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), trabecular spacing (Tb.Sp), cortical thickness (Ct.Th), internal cortical diameter and cortical bone mineral density (BMD). (c) Representative load displacement curves from destructive 3-point bend testing of WT and Gpc6-/- femurs showing yield load, maximum load, fracture load and gradient of the linear elastic phase (stiffness). Graphs showing yield load, maximum load, fracture load, stiffness and energy dissipated prior to fracture (Toughness) (d) Representative load displacement curves from destructive compression testing of WT and Gpc6-/- caudal vertebra showing yield load, maximum load and stiffness. P values generated by permutation analysis as described in the methods. Scale bars: a,b, 1mm.
Finally, we queried 87 separate GWAS using the web-utility “PhenoScanner”, where full genome-wide summary statistics were available for the conditionally independent genome-wide significant SNPs for eBMD (rs72635657, rs147720516) at the GPC6 locus, for any associations with a p-value <0.0538. We identified one association at a p-value <0.05, for rs72635657 with femoral neck BMD (P = 0.015). We also searched the NHGRI-EBI catalogue39 of published genome-wide association studies for the gene GPC6 (accessed 22nd March 2017). SNPs in the region of GPC6 had previously shown evidence of association with Attention Deficit Hyperactivity Disorder, FEV1 following bronchodilation, alzheimer’s disease, neuroticism and lower facial height although the lead SNPs reported in these scans were not in appreciable linkage disequilibrium with the lead conditionally independent SNPs in the present study (all r2 < 0.1).
Discussion
In this study, we have increased the number of genetic loci associated with BMD in humans almost threefold and doubled the amount of variance explained for this trait. Further, we have demonstrated that several BMD associated variants also influence risk of fracture. We have prioritized genes for future study and provided functional evidence that GPC6 plays a role in BMD and the pathophysiology of osteoporosis.
Our findings provide clear evidence that the genetic architecture of BMD is highly polygenic. The effect sizes observed follow a close relationship with MAF within the limits of the statistical power of the study. This suggests that further low-frequency and rare variants of moderate to large effect will be identified in future studies, which will likely be helpful to understand cellular and molecular mechanisms. Drug targets supported by evidence from human genetics are more likely to result in clinically useful therapies in general, and this has been demonstrated for musculoskeletal conditions 13,14. Thus, our findings will be helpful in identifying pathways and proteins amenable to pharmacologic manipulation to decrease the burden of fracture in the population.
GPC6 encodes a glypican that may serve as a novel drug target for osteoporosis care since it is a cell-surface protein involved in signalling, whose loss of function leads to increased bone mineral content, likely due to increased cortical bone and a resultant increased elasticity. GPC6 is a member of the glypican family (GPC1-6) of glycosylphosphatidylinositol-anchored, membrane-bound heparan sulfate proteoglycan core proteins that are involved in cellular growth control and differentiation. Mutations of GPC3, GPC4 and GPC6 result in developmental skeletal abnormalities but limited or no information is available from affected adults (OMIM 312870, OMIM 258315). The heparan sulfate proteoglycans attached to the GPC6 core protein regulate skeletal signalling pathways involved in bone formation and mineralization, including those mediated by fibroblast growth factors, vascular endothelial growth factor, Hedgehog, and bone morphogenetic protein pathways 40. In addition, the adult high bone mass phenotype and increased cortical bone thickness identified in Gpc6-/- mice in these studies is consistent with the recently identified direct role for GPC6 in the modulation of Wnt signalling 41,42, which is the key regulator of osteoblastic bone formation and is associated with BMD in humans. Overall, these findings suggest a number of possible new pharmacological targets that include not only the core protein GPC6, but also the heparan sulphate synthetic (EXT1-2) and modification enzymes (NDST1-4, GLCE, HS2ST, and HS6ST1-3) that specifically regulate growth factor binding and activity. The availability of global and tissue-specific Gpc6-/- mice 35 now provides the opportunity to test these possibilities directly. However we caution that whilst GPC6 and associated proteins appear to be promising targets for pharmacotherapy, other factors will need to be considered before these molecules are confirmed to be suitable candidates for pharmacological manipulation (e.g. likelihood of unintended side effects etc).
There are several limitations to our study. First, despite the high concordance between the loci identified using ultrasound derived measures of BMD and previous studies utilizing DXA derived BMD, there were some notable differences. Our study did not replicate associations at 18 known BMD loci from previous studies. Also, our list of genome-wide significant variants included some that were strongly related to eBMD at the heel, but have not been found in previous studies utilizing DXA derived BMD measures at other body sites in considerably smaller sample sizes. For some of these loci such as TBX1, this may simply be a consequence of the associated variants having neither been genotyped nor tagged well in previous studies. For other loci it may reflect genetic influences that are specific to the heel (e.g. genetic responses of the heel to ground reaction forces) that are not present at other body sites. Interestingly, we identified variants at six loci where the direction of effect was opposite between heel eBMD and DXA derived BMD at other sites, although notably at CPED1 the variants also showed association with risk of fracture in the direction consistent with the heel eBMD association. Whilst the reason behind these differences is unclear, the implication is that ultrasound measures of the heel capture aspects of bone structure beyond those obtained by central DXA, and is consistent with previous observations that ultrasound measures of the heel predict risk of osteoporotic fracture over and above hip BMD43.
Second, our study does not provide a definitive biological mechanism through which variants at genome-wide significant loci causally affect eBMD. Our eQTL analyses were not consistent with SNP effects being mediated through osteoblast expression at a majority of loci. This is likely because at least some of the identified eBMD-associated SNPs may act on cell types other than osteoblasts, such as osteocytes and osteoclasts. Further, the relatively small sample size of 95 individuals in the osteoblast eQTL experiment may have led to uncertain estimates. Last, the expression of genes in culture may reflect different biological processes than those in vivo. Whilst differences in gene expression are not the only mechanism through which the functional effects of an association can be mediated, we expect that large scale genetical genomics studies investigating the pattern of genetic association in osteoblasts, osteocytes and osteoclasts will facilitate answers to how these eBMD associations are mediated in the not too distant future.
Third, our study had limited ability to detect very rare variants (i.e. MAF <0.1%) or rare variants of small effect (MAF <1% and effect size <0.05 SD). Finally, our study has only investigated the genetic aetiology of osteoporosis in European individuals. It is likely that the study of populations of different ancestry will reveal novel loci important in the regulation of BMD, as has been shown in the study of other conditions44.
In summary, our findings shed light on the pathophysiologic mechanisms that underlie BMD and fracture risk in humans. Proteins identified and prioritized by these studies have identified signalling pathways that represent new drug targets for the prevention and treatment of osteoporosis- a major health care priority.
Online Methods
Measurement of eBMD, fracture and weight in UKBB
In 2006-2010, the UK Biobank recruited 502,647 individuals aged between 37-76 years (99.5% were 40-69 years) from across the country. Each participant provided information regarding their health and lifestyle using touch screen questionnaires, physical measurements and agreement to have their health followed and they also provided blood, urine and saliva samples for future analysis. UK Biobank has ethical approval from the Northwest Multi-centre Research Ethics Committee (MREC) and informed consent was obtained from all participants. Quantitative ultrasound assessment of calcanei was performed in UK Biobank participants using a Sahara Clinical Bone Sonometer [Hologic Corporation (Bedford, Massachusetts, USA)]. Details of the complete protocol employed are publicly available on the UK Biobank website (https://biobank.ctsu.ox.ac.uk/crystal/docs/Ultrasoundbonedensitometry.pdf). Participants were initially measured at baseline (N = 487,428) and had either their left calcaneus (N = 317,815), right calcaneus (N = 4,102) or both calcanei measured (N = 165,511). A subset of these subjects were followed up at two further time points (N = 20,104) and (N = 7,988), during which both heels were measured. A detailed description of the ascertainment procedure is described in the Supplementary Fig 2. Prior to quality control, ultrasound data were available for 488,683 individuals at either baseline and/or follow-up assessment. Estimated bone mineral density [eBMD, (g/cm2)] was derived as a linear combination of speed of sound (SOS) and bone ultrasound attenuation (BUA) (i.e. eBMD = 0.002592 * (BUA + SOS) − 3.687). To reduce the impact of outlying measurements, quality control was applied to male and female subjects separately using the following exclusion thresholds: Speed of sound [SOS; Male: (≤ 1,450 and ≥ 1,700 m/s), Female (≤ 1,455 and ≥ 1,700 m/s)] and broadband ultrasound attenuation [BUA; male: (≤ 27 and ≥ 138 dB/MHz), female (≤ 22 and ≥ 138 dB/MHz)]. Individuals exceeding the following thresholds for eBMD were excluded: [Male: (≤ 0.18 and ≥ 1.06 g/cm2), Female (≤ 0.12 and ≥ 1.025 g/cm2)]. Bivariate scatter plots of eBMD, BUA and SOS were visually inspected and any measurements not clustering with the others were removed, leaving a total of 483,230 valid measures (476,618 left and 6,612 right calcaneus) for SOS, BUA and BMD (265,057 females and 218,173 males). Please see Supplementary Fig. 2 for a detailed description of the QC pipeline and Supplementary Table 1 for an overview of descriptive statistics of the cohort after quality control.
We defined 14,492 individuals (8,439 female and 6,053 male) as having a fracture, based on answering yes to the question “Have you fractured/broken any bones in the last 5 years?” at either baseline or first follow-up. Individuals were coded as missing if they responded “Do not know” or “Prefer not to answer” at both the baseline and first follow-up, otherwise they were coded as controls (N=130,563). Self-reported fractures have low false positive and false negative rates.45 Individuals who responded yes to having a fracture were also asked “Did the fracture result from a simple fall (i.e. from standing height)?” We created a second variable using this question, where 8,540 individuals (5,853 female and 2,687 male) had a fracture from a simple fall and 131,333 individuals did not report a fracture. Weight was measured using the Tanita BC418MA body composition analyser.
Preparation, quality control and genetic analysis in UK Biobank samples
Genotype data from the interim May 2015 release of UK Biobank were available for a subset of 152,729 participants. Data was imputed centrally by UK Biobank using IMPUTE2 46 to a 1000G (Oct 2014) and UK10K reference panel. In addition to the quality control metrics performed centrally by UK Biobank (UK Biobank document #155580 http://biobank.ctsu.ox.ac.uk/crystal/docs/genotyping_qc.pdf), we defined a subset of “white European” ancestry samples using a K-means (K=4) clustering approach based on the first 4 genetically determined principal components. A maximum of 142,487 individuals (76,067 females and 66,420 males) with genotype and valid QUS measures were available for the present analyses. We tested genetic variants for association with eBMD assuming an additive allelic effect, in a linear mixed non-infinitesimal model implemented in BOLT-LMM 47 to account for cryptic population structure and relatedness. Genotyping array, age and sex were included as covariates in all models. We also included weight as a covariate in a sensitivity analysis to investigate whether power to detect association was increased or if weight mediated the relationship between genotype and eBMD (i.e. some variants may be primarily associated with weight and their effect on eBMD mediated through a causal effect of weight on eBMD 29). Only SNPs down to a minor allele frequency of 0.1% and with an info-score threshold of > 0.4 were analysed. We additionally analysed the association between eBMD and directly genotyped SNPs on the X chromosome, adjusting for genotyping array, age, sex and the first 4 ancestry principal components, using Plink v1.09 beta 3.38 (7 Jun 2016) software48 and a nested sample of unrelated subjects (N= 135,729). As the analyses for the X chromosome data were based upon observed genotypes, our quality control was slightly different. We excluded SNPs with evidence of deviation from Hardy-Weinberg Equilibrium (1×10-6), minor allele frequency < 0.1% and overall missing rate > 5%, resulting in 15,552 X chromosome SNPs for analysis. Heterogeneity between sexes in effect size coefficients was tested using EasyStrata 49. Manhattan and Miami plots of our genome-wide association scans were generated by EasyStrata version 15.3. Regional association plots were generated using LocusZoom (v1.3) 50, using linkage-disequilibrium information estimated from our reference UKBiobank sample, together with the December 2016 release of the NHGRI-EBI Catalog of published genome-wide association studies. SNPs that were associated with eBMD at genome-wide significant levels were additionally tested for association with fracture using BOLT-LMM including age, sex, BMI and the time of reporting the fracture as fixed effects 47.
Estimation of genome-wide significance threshold
Traditional estimates of the genome-wide significance threshold for common variants (MAF >5%) in European populations (i.e. α = 5 x 10-8) are based on a Bonferroni correction of α = 0.05/106, since there are an estimated 1 million statistically independent SNPs above this MAF threshold. However, in the case of UK Biobank, we have assessed SNPs down to a minor allele frequency of 0.1% in 142,487 individuals and applied an info-score threshold of > 0.4, resulting in 17.17M SNPs. Therefore, we defined a new and more conservative threshold to declare genome-wide significance accounting for the number of independent statistical tests performed in our data. To do so, we applied the method we previously used within the UK10K sequencing consortium 4 which assesses the correlation between nearby test statistics empirically. Analysis of permuted data derived from a small proportion of all tested variants allows assessment of the correlation patterns. Hence, we can estimate, in subsets of the genome of varying size, the relationship between the Bonferroni significance threshold and the empirical significance threshold that corrects for correlations, and thereby extrapolate to the whole genome. Specifically, when assessing all 740,018 variants that met our filtering criteria across chromosome nine (Supplementary Fig. 9), we saw a good linear fit between family wise error rate (α = 0.05), divided by the number of tests and the empirical significance thresholds. Our estimated genome-wide significance threshold then, accounting for all SNPs MAF ≥ 0.1% and info-score > 0.4 was α = 6.6 x 10-9.
Approximate conditional association analysis
In order to detect multiple independent association signals at each of the genome-wide significant eBMD loci, we applied approximate conditional and joint genome-wide association analysis using the software package GCTA 51. SNPs with high collinearity (multiple regression R2 > 0.9) were ignored and those situated more than 20 MB away were assumed to be in complete linkage equilibrium. A reference sample of 15,000 unrelated (pairwise relatedness < 0.025) individuals of white British origin, randomly selected from UK Biobank was used to model patterns of linkage disequilibrium (LD) between variants. The reference genotyping dataset consisted of the same 17M variants assessed in our GWAS, but with an additional QC step excluding SNPs that deviated from Hardy-Weinberg Equilibrium (1×10-6). Conditionally independent variants reaching GWAS significance were annotated to the physically closest gene using bedtools52 v2.26.0 and the Hg19 Gene range list available from https://www.cog-genomics.org/plink2/.
Estimation of variance explained by significant variants and SNP heritability
We estimated the proportion of phenotypic variance tagged by all SNPs on the genotyping array (i.e. the SNP heritability) using BOLT-REML53. In order to calculate the variance explained by all genome-wide significant SNPs we first employed the method of Bigdeli et al. (2016) to shrink the effect sizes of SNPs likely to suffer from “winner’s curse”54. Briefly the method works by shrinking the effect sizes of SNPs that just pass significance whilst having a negligible effect on SNPs that are more strongly significant (and consequently more accurately and precisely estimated). After calculating the corrected effect sizes we then removed the combined effect of the SNPs on individual’s eBMD and recalculated the total expected variance in BOLT-LMM. The difference between this estimate and the total expected variance calculated on the original data without the SNP correction is an estimate of the variance explained by all SNPs.
LD Score regression
To estimate the amount of genomic inflation present in the data that was due to residual population stratification, cryptic relatedness, and other latent sources of bias, we used LD score regression15. LD scores were calculated for all high quality SNPs (i.e. INFO score > 0.9 and MAF > 0.1%) from a dataset consisting of 15,000 unrelated individuals from the UK Biobank. To estimate the genetic correlation between eBMD and other complex traits and diseases including those related to osteoporosis, we used a relatively new method based on LD Score regression as implemented in the online web utility LDHub 26,27. This method uses the cross products of summary test statistics from two GWAS and regresses them against a measure of how much variation each SNP tags (its “LD Score”). Variants with high LD Scores are more likely to contain more true signals and hence provide more chance of overlap with genuine signals between GWAS. The LD score regression method uses summary statistics from the GWAS meta-analysis of eBMD and the other traits of interest, calculates the cross-product of test statistics at each SNP, and then regresses the cross-product on the LD Score. The slope of the regression is a function of the genetic covariance between traits:
where Ni is the sample size for study i, ρg is the genetic covariance, M is the number of SNPs in the reference panel with MAF between 5% and 50%, lj is the LD score for SNP j, Ns quantifies the number of individuals that overlap both studies, and ρ is the phenotypic correlation amongst the Ns overlapping samples. Thus, if there is sample overlap (or cryptic relatedness between samples), it will only affect the intercept from the regression (i.e. the term ) and not the slope, and hence estimates of the genetic covariance will not be biased by sample overlap. Likewise, population stratification will affect the intercept but will have minimal impact on the slope (i.e. intuitively since population stratification does not correlate with linkage disequilibrium between nearby markers).
Gene prioritization and pathway analysis
To establish functional connections we conducted three different analyses implemented in the DEPICT v1 tool34. First, to prioritize genes with relevant biological roles in the eBMD associated loci functional similarities among genes from different associated regions were tested, where genes with high functional similarity across regions obtained lower prioritization P values. Second, we analysed expression enrichment across particular tissues or cell types, by testing whether genes in the associated eBMD loci were seen highly expressed in any of the 209 Medical Subject Heading (MeSH) annotations using data from 37,427 expression arrays. Third, we performed a gene set enrichment analysis, which tests if the genes in the associated eBMD loci are enriched in reconstituted gene-sets. The 10,968 gene-sets tested have been generated from diverse databases, i.e. Gene Ontology, Kyoto encyclopedia of genes and genomes, REACTOME, InWeb database (high confidence protein-protein interaction), and the Mouse Genetics Initiative (phenotype-genotype relationships). All three analyses used False Discovery Rate (FDR) to adjust for multiple testing, significance was defined at FDR 5%.
The DEPICT analyses were based on independent lead SNPs (r2<0.1, European populations 1000 genomes reference panel) with P-values below the genome-wide significant threshold (P < 6.64x10-9). As many of the Gene-sets tested come from different repositories, they overlap, hence significantly enriched gene-sets were further grouped into ‘meta gene sets’ by similarity clustering, as previously described34. The visualization of these ‘meta gene-sets’ was performed in cytoscape 55, filtering for ‘meta gene sets’ at FDR <1%.
We also compared the DEPICT gene set enrichment results to analyses using the Meta-analysis gene-set enrichment of variant associations (MAGENTA) software56. Briefly, MAGENTA maps each gene in the genome to a single index SNP with the lowest p value within a 110 kb upstream and 40 kb downstream window (excluding genes in the HLA region due to complex patterns of linkage disequilibrium). This P value is then corrected for confounding factors (e.g. SNP density, gene size etc.) in a linear regression model and each gene is ranked by its adjusted gene score. The observed number of gene scores in a given pathway, with a ranked score above a specified threshold (i.e. 95th and 75th percentiles of all gene scores) is then calculated. This observed statistic is then compared to 1,000,000 randomly permuted pathways of identical size. This generates an empirical GSEA P value for each gene set. A gene set was declared significant when an individual pathway reached a FDR < 0.05 in either analysis. We tested 3217 pre-specified gene sets from Gene Ontology, Ingenuity, Kyoto Encyclopedia of Genes and Genomes (KEGG), Protein Analysis Through Evolutionary Relationships (PANTHER), BioCarta and Reactome databases.
Prioritising candidate genes and possible causal variants at each eBMD locus
We combined a number of approaches to identify possible causal SNPs at each eBMD signal (defined here as all SNPs within 500 kb of conditionally independent lead SNP that met genome-wide significance). First, we annotated all SNPs within a locus (defined as +/- 500 kb from a conditionally independent lead SNP) for deleterious coding variation annotation using the Variant Effect Predictor (VEP)30 if they were significantly associated with eBMD (P < 6.6x10-9). Deleterious SNPs were classified as such if they had one of the following sequence ontology terms: frameshift_variant, inframe_deletion, inframe_insertion, initator_codon_variant, missense_variant, splice_acceptor_variant, splice_donor_variant, stop_gained, or stop_lost.
Next, we identified 305 autosomal lead SNPs and further defined sets of plausible causal SNPs within each locus using FINEMAP57. For each locus, FINEMAP implements a shotgun stochastic search algorithm to test multiple causal configurations of SNPs, calculating within a Bayesian framework the posterior probabilities of each configuration to identify the number of likely causal SNPs. We note that this approach assumes that the true causal variant(s) have been included in the analysis and have been well imputed. We also emphasize that approaches such as this that are based solely on association test statistics and linkage disequilibrium are unlikely to be definitive with respect to the identification of causal variants/genes. Thus, we regard these fine mapping analyses as one of several approaches that can be used to implicate specific variants/genes in osteoporosis aetiology. When the same variant/gene is implicated using multiple independent approaches (e.g. mouse knockout, human knockout, gene expression and eQTL studies etc.) then we can be increasingly confident of the identity of the gene/variant(s) underlying the statistical association.
For a given number of plausible causal SNPs, FINEMAP will calculate for each SNP their Bayes factor which quantifies the evidence that the particular SNP is causal. We retained only SNPs with Bayes factors greater than 100, or log10 Bayes factors greater than 2, as our plausible causal SNPs for each locus.
Each set of plausible causal SNPs was then annotated for overlap with DNase I hypersensitive sites (DHS) using a master list derived from 115 cell types4. DHS are focal sites of open chromatin comprising the collective transcription factor (TF) binding sites in a given cell type. We further annotated each SNP inhabiting a DHS using Contextual Analysis of Transcription Factor Occupancy (CATO) scores. CATO, previously described by Maurano et al. 4, scores the likelihood of a variant causing allelic imbalance of a DHS by modelling both local sequence context and direct effects on the TF recognition sequences for 44 TF motif families. CATO scores range between 0 and 1 and we considered SNPs with CATO scores greater than 0.1 as having very strong functional evidence (corresponding to a 51% positive predictive rate in the initial training set 4).
Genetically modified animals used for functional validation
The International Mouse Phenotyping Consortium (IMPC) together with the International Knockout Mouse Consortium (IKMC) are generating null alleles for all protein coding genes on a C57BL/6 genetic background. These mice are phenotyped through a broad-based phenotyping screen 58. This approach can be used for functional investigation of candidate genes identified by GWAS analysis of human disease traits, and studies have already ascribed novel functions for poorly annotated or previously unpublished genes. The Origins of Bone and Cartilage Disease (OBCD) study (http://www.boneandcartilage.com) is undertaking a validated multi-parameter skeletal phenotype screen 36 of mutant mouse lines generated by Wellcome Trust Sanger Institute as part of the IKMC and IMPC effort.
OBCD methods
Samples from 16 week-old female wild-type and knockout mice are stored in 70% ethanol, anonymised and randomly assigned to batches for rapid throughput analysis in an unselected fashion. The relative bone mineral content and length of the femur and caudal vertebrae are determined at 10μm pixel resolution by digital X-ray microradiography (Faxitron MX20). Micro-CT (Scanco uCT50, 70kV, 200μA, 0.5mm aluminium filter) is used to determine cortical bone parameters (thickness Ct.Th, BMD, medullary diameter) at 10μm voxel resolution in a 1.5mm region centred in the mid-shaft region 56% along the length of the femur distal to the femoral head, and trabecular parameters (bone volume BV/TV, trabecular number Tb.N, thickness Tb.Th, spacing Tb.Sp) at 5μm voxel resolution in a 1mm region beginning 100μm proximal to the distal growth plate. Biomechanical variables of bone strength and toughness (yield load, maximum load, fracture load, % energy dissipated prior to fracture) are derived from destructive 3-point bend testing of the femur and compression testing of caudal vertebra 6 and 7 (Instron 5543 load frame, 100N load cell) 36. Overall, 19 skeletal parameters are reported for each individual mouse studied and compared to reference data obtained from >250 16 week-old wild-type C57BL/6 female mice. Coefficients of Variation (CV) for each skeletal parameter were as follows:
femur BMC (2.0%) and length (2.1%); vertebra BMC (2.1%) and length (2.3%); trabecular bone BV/TV (18.5%), Tb.N (7.3%), Tb.Th (7.9%) and Tb.Sp (8.3%); cortical bone Ct.Th (4.3%), Int.Diam (6.0%) and BMD (4.0%); femur yield load (13.2%), maximum load (10.0%), fracture load (29.0%), stiffness (13.7%) and energy dissipated prior to fracture (26.7%); and vertebra yield load (13.0%), maximum load (10.3%) and stiffness (13.3%).
In Supplementary Table 15, we have highlighted knockout mice with phenotypes greater than 2 SDs away from the mean of wild type mice. We generated P-values for the reported Gpc6-/- mouse phenotypes through permutation. To do so we first identified the least extreme phenotype for the Gpc6-/- mice tested. We then permuted the knockout labels 100,000 times to observe the number of times we observed two knockout animals with both phenotypes as extreme as the least extreme Gpc6-/- mouse phenotype. The P-value was then calculated as the number of extreme permutations divided by 100,000. All mouse studies were undertaken by Wellcome Trust Sanger Institute Mouse Genetics Project as part of the International Knockout Mouse Consortium and licensed by the UK Home Office in accordance with the Animals (Scientific Procedures) Act 1986 and the recommendations of the Weatherall report.
Gene expression in primary human and murine osteoblasts
To study human osteoblasts, we undertook cis-eQTL analyses of plausible causal regulatory SNPs in 95 primary human osteoblasts as previously described by Grundberg et al33, performed with an updated imputation panel, the combined UK10K and 1000 Genomes Phase 1 v3 reference panel59. We used an alpha level of α=0.05 to identify possible gene targets of plausible causal SNPs.
We investigated the possibility that heel eBMD associations and cis-eQTL effects in osteoblasts may represent different signals (i.e. as opposed to a causal effect of osteoblast expression on eBMD) by performing two sample summary Mendelian randomization analysis on osteoblast eQTL and heel eBMD GWAS results60,61. A HEIDI (heterogeneity in dependent instruments) test was used to identify situations in which the lead cis-eQTL was likely to be in linkage disequilibrium with two distinct causal variants (i.e. one affecting gene expression and the other affecting eBMD variation), as opposed to expression of the relevant gene mediating the relationship between the SNP and eBMD. Intuitively the test works by comparing estimates of the putative causal effect of gene expression on eBMD obtained by Mendelian randomization analysis of each variant whilst taking into account dependencies between the SNPs. Under a causal model, different SNPs should produce the same causal estimate (subject to sampling error), whereas under a model of linkage (i.e. two separate signals in the region, one affecting gene expression in osteoblasts and the other affecting eBMD), the estimates from the Mendelian randomization analysis may significantly differ. In the context of our study, a significant HEIDI test suggests that expression of the relevant gene in osteoblasts does not mediate the SNP – eBMD association. We therefore performed HEIDI tests for all the probes listed in Supplementary Table 10 that were implicated in our gene expression analyses. In order to avoid weak SNP instruments potentially affecting our results we only included SNPs in the eQTL analysis that exhibited strong evidence of association (i.e. F statistic > 10)61.
Gene expression profiles of candidate genes were examined in primary mouse osteoblasts undergoing differentiation. These data have been described in detail previously 62 and are publicly available from the Gene Expression Omnibus (GSE54461). To study murine osteoblasts, pre-osteoblast-like cells were obtained from neonatal calvaria collected from C57BL/6J carrying a transgene expressing Cyan Florescent Protein (CFP) under the control of the Col 3.6Kb promoter. The cells were placed into culture for 4 days in growth media, removed from culture and cells not expressing CFP were removed by FACS sorting. The remaining pre-osteoblast cells were re-plated, were subjected to an osteoblast differentiation cocktail and RNA was collected every two days from day 2 to 18 days post differentiation. Next Generation RNA sequencing (RNAseq) was used to evaluate the transcriptome at each time point using an Illumina HiSeq 2000. Three technical replicates per samples were sequenced. The alignments for abundance estimation of transcripts was conducted using Bowtie version 0.12.9, using the NCBIm37 reference genome. Expression level per gene was calculated using RSEM version 1.2.0 with the parameters of --fragment-length-mean 280 and --fragment-length-sd 50, and the expression level for each sample was normalized relative to the per sample upper quartile.
Gene expression in murine osteocytes
Osteocyte expression was determined through an analysis of whole transcriptome sequencing derived from four different bone types from throughout the mouse skeleton – the tibia, femur, humerus and calvaria (marrow removed, n=8 per bone). A threshold of expression was determined based on the distribution of normalised gene expression for each sample modifying a statistical approach by Hart et al. (2013) 63. “Expressed” genes were above this threshold for 8 of 8 replicates in any bone type. The specificity of these genes expression in the skeleton was determined by comparing transcriptome-sequencing data from bone-samples with osteocytes isolated to those with the marrow left intact (n=5 per group) (S.E.Y, J.H.D.B, G.R.W, P.I.C manuscript in preparation).
Gene expression in mouse osteoclasts
Expression of genes in murine osteoclasts was determined using publically available data obtained using Next-Gen RNA-sequencing applied to bone marrow derived osteoclasts obtained from 6-8 week old C57BL/6 mice (GEO Accession Number: GSM1873361).
Data Availability and Accession Code Availability Statements
The human genotype and phenotype data on which the results of this study are based are available upon application from the UK Biobank Study (http://www.ukbiobank.ac.uk/). GWAS summary statistics from this study available via the GEnetic Factors for OSteoporosis Consortium website (http://www.gefos.org/). No new datasets or related accession codes were generated as part of this study. Mouse phenotype data are available online at the IMPC (http://www.mousephenotype.org) and OBCD (http://www.boneandcartilage.com).
Supplementary Material
Acknowledgements
We thank Pak Sham for helpful discussions and Marcus Schull for assistance with high performance computing.
This work was supported by a Medical Research Council Programme Grant (MC_UU_12013/4 to D.M.E); a Wellcome Trust Strategic Award (grant number 101123) and a Wellcome Trust Project Grant (grant number 094134) (awarded to G.R.W, J.H.D.B and P.I.C); the Netherlands Organization for Health Research and Development ZonMw VIDI 016.136.367 (funding F.R, C.M-G, K.T), and the mobility stimuli plan of the European Union Erasmus Mundus Action 2: ERAWEB programme funding (K.T); the NIAMS/NIH (AR060981 and AR060234 to C.L.A.B).
We thank research nurses and assistants at the Departments of Surgical and Medical Sciences, Uppsala University, Sweden for large-scale collection of bone samples and culture of primary osteoblasts. This part of the work was supported by Genome Quebec, Genome Canada, and the Canadian Institutes of Health Research (CIHR). We would like to thank Thomas Winkler for providing invaluable technical support for the EasyStrata Software used in this manuscript.
N.M.W. is supported by a National Health and Medical Research Council Early Career Fellowship (APP1104818). E.G. was supported by the Swedish Research Council. J.A.M is supported by funding from the Réseau de Médecine Génétique Appliquée (RMGA) and the Fonds de Recherche du Québec - Santé (FRQS). C.M.T.G is supported by the Natural Sciences and Engineering Research Council of Canada. P.I.C is supported by Mrs Janice Gibson and the Ernest Heine Family Foundation. C.L.G is funded by Arthritis Research UK (ref 20000). J.B.R. is funded by the Canadian Institutes of Health Research, the Fonds du Recherche Québec Santé and the Jewish General Hospital. D.M.E. is funded by an Australian Research Council Future Fellowship (FT130101709).
This research has been conducted using the UK Biobank Resource (Application number 12703). Access to the UK Biobank study data was funded by University of Queensland Early Career Researcher Grant (2014002959).
Footnotes
URLS
Mouse Phenotyping Consortium (IMPC: http://www.mousephenotype.org)
Mouse Genome Informatics (MGI: http://www.informatics.jax.org)
The Origins of Bone and Cartilage Disease study (OBCD: http://www.boneandcartilage.com)
UK Biobank (UKBB: http://www.ukbiobank.ac.uk/)
GEnetic Factors for OSteoporosis Consortium (GEFOS: http://www.gefos.org/)
Author Contributions
Conceived and designed experiments: S.K., F.R., J.H.T., P.I.C., C.L.A-B., J.H.D.B., G.R.W., J.B.R., D.M.E.
Performed statistical Analysis: J.P.K., J.A.M., C.M-G., V.F., N.M.W., S.E.Y., J.Z., K.T., K.G., C.X., C.M.T.G., C.L.A-B., J.D.B., G.R.W.
Wrote the paper: J.P.K., J.A.M., C.M-G., V.F., N.M.W., S.E.Y, C.L.G., K.T., C.M.T.G., M.T.M, S.K., F.R., J.H.T., P.I.C., C.L.A-B., J.H.D.B., G.R.W., J.B.R., D.M.E.
Generation of mouse models and/or functional experiments: S.E.Y., E.J.G., J.G.L., A.S.P., P.C.S., R.A., V.D.L., N.C.B., D.K-E., A-T.A., K.F.C., J.K.W., F.K., D.J.A, P.I.C, C.L.A-B, J.H.D.B, G.R.W.
Generation of heel eBMD Data: N.C.H., C.C.
Lead analysts: J.P.K., J.A.M., C.M.G.
Jointly supervised research: J.B.R., D.M.E
Revised and reviewed the paper: All authors.
Competing Financial Interests Statement
Authors declare no competing financial interests.
References
- 1.Cauley JA, et al. Long-term risk of incident vertebral fractures. JAMA. 2007;298:2761–7. doi: 10.1001/jama.298.23.2761. [DOI] [PubMed] [Google Scholar]
- 2.Liu CT, et al. Heritability of prevalent vertebral fracture and volumetric bone mineral density and geometry at the lumbar spine in three generations of the Framingham study. J Bone Miner Res. 2012;27:954–8. doi: 10.1002/jbmr.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng HF, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–7. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arden NK, Baker J, Hogg C, Baan K, Spector TD. The heritability of bone mineral density, ultrasound of the calcaneus and hip axis length: a study of postmenopausal twins. J Bone Miner Res. 1996;11:530–4. doi: 10.1002/jbmr.5650110414. [DOI] [PubMed] [Google Scholar]
- 6.Howard GM, Nguyen TV, Harris M, Kelly PJ, Eisman JA. Genetic and environmental contributions to the association between quantitative ultrasound and bone mineral density measurements: a twin study. J Bone Miner Res. 1998;13:1318–27. doi: 10.1359/jbmr.1998.13.8.1318. [DOI] [PubMed] [Google Scholar]
- 7.Hunter DJ, et al. Genetic variation in bone mineral density and calcaneal ultrasound: a study of the influence of menopause using female twins. Osteoporos Int. 2001;12:406–11. doi: 10.1007/s001980170110. [DOI] [PubMed] [Google Scholar]
- 8.Lee M, et al. Unique and common genetic effects between bone mineral density and calcaneal quantitative ultrasound measures: the Fels Longitudinal Study. Osteoporos Int. 2006;17:865–71. doi: 10.1007/s00198-006-0075-4. [DOI] [PubMed] [Google Scholar]
- 9.Bauer DC, et al. Broadband ultrasound attenuation predicts fractures strongly and independently of densitometry in older women. A prospective study. Study of Osteoporotic Fractures Research Group. Arch Intern Med. 1997;157:629–34. [PubMed] [Google Scholar]
- 10.Bauer DC, et al. Quantitative ultrasound predicts hip and non-spine fracture in men: the MrOS study. Osteoporos Int. 2007;18:771–7. doi: 10.1007/s00198-006-0317-5. [DOI] [PubMed] [Google Scholar]
- 11.Gonnelli S, et al. Quantitative ultrasound and dual-energy X-ray absorptiometry in the prediction of fragility fracture in men. Osteoporos Int. 2005;16:963–8. doi: 10.1007/s00198-004-1771-6. [DOI] [PubMed] [Google Scholar]
- 12.Moayyeri A, et al. Genetic determinants of heel bone properties: genome-wide association meta-analysis and replication in the GEFOS/GENOMOS consortium. Hum Mol Genet. 2014;23:3054–68. doi: 10.1093/hmg/ddt675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–60. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 14.Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genet. 2012;13:576–88. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 15.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan EL, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koller DL, et al. Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. J Clin Endocrinol Metab. 2010;95:1802–9. doi: 10.1210/jc.2009-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards JB, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–12. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivadeneira F, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Styrkarsdottir U, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–65. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 21.Styrkarsdottir U, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–7. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 22.Xiong DH, et al. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet. 2009;84:388–98. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackey DC, et al. High-trauma fractures and low bone mineral density in older women and men. JAMA. 2007;298:2381–8. doi: 10.1001/jama.298.20.2381. [DOI] [PubMed] [Google Scholar]
- 25.Sanders KM, et al. The exclusion of high trauma fractures may underestimate the prevalence of bone fragility fractures in the community: the Geelong Osteoporosis Study. J Bone Miner Res. 1998;13:1337–42. doi: 10.1359/jbmr.1998.13.8.1337. [DOI] [PubMed] [Google Scholar]
- 26.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2016;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad OS, et al. A Mendelian Randomization Study of the Effect of Type-2 Diabetes and Glycemic Traits on Bone Mineral Density. J Bone Miner Res. 2017;32(5):1072–1081. doi: 10.1002/jbmr.3063. [DOI] [PubMed] [Google Scholar]
- 29.Kemp JP, Sayers A, Smith GD, Tobias JH, Evans DM. Using Mendelian randomization to investigate a possible causal relationship between adiposity and increased bone mineral density at different skeletal sites in children. Int J Epidemiol. 2016;45:1560–1572. doi: 10.1093/ije/dyw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roadmap Epigenomics, C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grundberg E, et al. Population genomics in a disease targeted primary cell model. Genome Res. 2009;19:1942–52. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassett JH, et al. Rapid-throughput skeletal phenotyping of 100 knockout mice identifies 9 new genes that determine bone strength. PLoS Genet. 2012;8:e1002858. doi: 10.1371/journal.pgen.1002858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campos-Xavier AB, et al. Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am J Hum Genet. 2009;84:760–70. doi: 10.1016/j.ajhg.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staley JR, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malinauskas T, Jones EY. Extracellular modulators of Wnt signalling. Curr Opin Struct Biol. 2014;29:77–84. doi: 10.1016/j.sbi.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Malinauskas T, Aricescu AR, Lu W, Siebold C, Jones EY. Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nat Struct Mol Biol. 2011;18:886–93. doi: 10.1038/nsmb.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakane H, Yamamoto H, Matsumoto S, Sato A, Kikuchi A. Localization of glypican-4 in different membrane microdomains is involved in the regulation of Wnt signaling. J Cell Sci. 2012;125:449–60. doi: 10.1242/jcs.091876. [DOI] [PubMed] [Google Scholar]
- 43.Moayyeri A, et al. Quantitative ultrasound of the heel and fracture risk assessment: an updated meta-analysis. Osteoporos Int. 2012;23:143–53. doi: 10.1007/s00198-011-1817-5. [DOI] [PubMed] [Google Scholar]
- 44.Paternoster L, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–56. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ismail AA, et al. Validity of self-report of fractures: results from a prospective study in men and women across Europe. EPOS Study Group. European Prospective Osteoporosis Study Group. Osteoporos Int. 2000;11:248–54. doi: 10.1007/s001980050288. [DOI] [PubMed] [Google Scholar]
- 46.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–90. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler TW, et al. EasyStrata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics. 2015;31:259–61. doi: 10.1093/bioinformatics/btu621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. doi: 10.1038/ng.2213. S1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loh PR, et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet. 2015;47:1385–92. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bigdeli TB, et al. A simple yet accurate correction for winner's curse can predict signals discovered in much larger genome scans. Bioinformatics. 2016;32:2598–603. doi: 10.1093/bioinformatics/btw303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito R, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–76. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segre AV, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benner C, et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koscielny G, et al. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42:D802–9. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.International Mouse Knockout, C. Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 60.Hrabe de Angelis M, et al. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat Genet. 2015;47:969–78. doi: 10.1038/ng.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans DM, Davey Smith G. Mendelian Randomization: New Applications in the Coming Age of Hypothesis-Free Causality. Annu Rev Genomics Hum Genet. 2015;16:327–50. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 63.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 64.Kemp JP, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014;10:e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hart T, Komori HK, LaMere S, Podshivalova K, Salomon DR. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics. 2013;14:778. doi: 10.1186/1471-2164-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmbeck K, et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 67.Evans BR, et al. Mutation of membrane type-1 metalloproteinase, MT1-MMP, causes the multicentric osteolysis and arthritis disease Winchester syndrome. Am J Hum Genet. 2012;91:572–6. doi: 10.1016/j.ajhg.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human genotype and phenotype data on which the results of this study are based are available upon application from the UK Biobank Study (http://www.ukbiobank.ac.uk/). GWAS summary statistics from this study available via the GEnetic Factors for OSteoporosis Consortium website (http://www.gefos.org/). No new datasets or related accession codes were generated as part of this study. Mouse phenotype data are available online at the IMPC (http://www.mousephenotype.org) and OBCD (http://www.boneandcartilage.com).