Abstract
Mutations in over 30 genes are known to result in impairment of the adaptive immune system, causing a group of disorders collectively known as severe combined immunodeficiency (SCID). SCID disorders are split into groups based on their presence and/or functionality of B, T, and NK cells. Piglets from a line of Yorkshire pigs at Iowa State University were shown to be affected by T− B− NK+ SCID, representing the first example of naturally occurring SCID in pigs. Here, we present evidence for two spontaneous mutations as the molecular basis for this SCID phenotype. Flow cytometry analysis of thymocytes showed an increased frequency of immature T cells in SCID pigs. Fibroblasts from these pigs were more sensitive to ionizing radiation than non-SCID piglets, eliminating the RAG1 and RAG2 genes. Genetic and molecular analyses showed two mutations were present in the Artemis gene, which in homozygous or compound heterozygous state cause the immunodeficient phenotype. Rescue of SCID fibroblast radiosensitivity by human Artemis protein demonstrated that the identified Artemis mutations are the direct cause of this cellular phenotype. The work presented here reveals two mutations in the Artemis gene that cause T− B− NK+ SCID in pigs. The SCID pig can be an important biomedical model, but these mutations would be undesirable in commercial pig populations. The identified mutations and associated genetic tests can be used to address both of these issues.
Introduction
Severe Combined Immunodeficiency (SCID) is a diverse group of primary immunodeficiencies that involve impaired development or function of both B and T cells(1). Phenotypes causing SCID can be classified based on the presence or absence of B, T, and Natural Killer (NK) cells and further categorized by the genetic defect that causes the disease. Mutations in genes necessary for V(D)J recombination at the T cell receptor (TCR) or Immunoglobulin (IG) clusters, which are required for receptor maturation and T and B cell survival, respectively, lead to T− B− NK+ SCID(2). T− B− NK+ SCID defects can be split into two groups based on sensitivity of affected cells to ionizing radiation. Lesions in the RAG1 and RAG2 genes result in immunodeficient animals that are resistant to radiation(3). Cells with defects in DNA-PKcs, DNA ligase IV, Cernunnos, or Artemis are radiosensitive, as they are unable to repair damage to DNA caused by ionizing radiation(4).
SCID animals are valuable biomedical models and have been used to study many aspects of human immune function, disease progression, as well as vaccine and drug development and testing. SCID mice, in particular, have been widely used as a vehicle to study the human immune system and play important roles in pre-clinical studies of the stability and function of human stem cells and their differentiated counterparts as potential therapies(5, 6). In order to improve SCID mice as biomedical models, genetic engineering has been utilized to overcome some of the differences between the immune environment of mice and humans(7). Much progress has been made in creating immunodeficient mice models used in biomedical research(7, 8) through selection of appropriate mice strains and conditioning regimens(9). However, regardless of these advances, the translatability of rodent immunology and inflammatory studies to humans is imperfect(10, 11) and other models may be beneficial(12, 13).
Anatomical, genetic, and immunological similarities underpin the usefulness of swine as a large animal model for humans(13, 14). Recently, transgenic methods have been used to create SCID pigs, including mutations in the X-linked IL2RG gene(15, 16) and mutations in RAG1 or in both RAG1 and RAG2(17, 18). Recently, our group(19) reported the first identification of a naturally occurring form of SCID in a line of purebred Yorkshire pigs that had been selected for increased feed efficiency at Iowa State University(20). Necropsy of four piglets that died abnormally early in a viral challenge study showed they lacked functional adaptive immune systems. In subsequent work, we(21) demonstrated that these immunodeficient pigs failed to reject human cancer cells, and Ewen et al.(22) reported minimal circulating B and T cells but normal amounts of NK cells in a preliminary analysis of these SCID pigs. We present in this paper the first report of the characterization of the genetic lesions that cause the SCID phenotype in this pig population. Quantitative real-time PCR (qRT-PCR) and flow cytometry assays confirmed that the SCID pigs lack B and T cells, but do have NK cells; we also demonstrate T cell maturation defects in the thymus. Irradiation of fibroblasts from these SCID pigs and normal controls showed that the genetic defect is in the group of genes involved in DNA damage repair. Genetic mapping, phasing of genetic markers, and targeted sequencing revealed two independent mutations in the Artemis gene that cause SCID in this line of pigs. We confirmed that mutations in Artemis are responsible for the radiosensitivity of fibroblasts by rescuing this phenotype with expression of human Artemis in fibroblasts derived from the SCID pigs. These Artemis deficient pigs provide a large animal model to study various aspects of human disease and immune function.
Materials and Methods
All animal experiments were performed under a protocol approved by the Iowa State University Institutional Animal Care and Use Committee (Ames, Iowa).
Flow cytometry to determine cellular phenotype
Thymic tissues were collected into ice-cold Minimal Essential Media, supplemented with HEPES, and penicillin/streptomycin. Tissues were minced into 2×2 cm2 pieces and treated with 100 U/ml collagenase (collagenase I; Life Technologies, Grand Island, New York) in HBSS (+calcium and magnesium; Life Technologies) for 30 min at 37°C, with gentle agitation. Cells were washed in PBS, counted, and 1,000,000 cells were stained using the following fluorescently conjugated antibodies: Alexa Fluor647 conjugated anti-pig CD8a (clone 76-2-11; BD Biosciences, San Diego, California), FITC conjugated anti-pig CD4 (clone 74-12-4; BD Biosciences), PE-conjugated anti-pig γδ (clone MAC320; BD Biosciences), or the appropriate isotype controls (BD Biosciences), or a with FITC-conjugated mouse anti-pig CD3ε (clone PPT3; Southern Biotech, Birmingham, Alabama). Cells were washed and resuspended in 400 μL 0.2% BSA/PBS solution prior to acquisition on an EC800 flow cytometer (Sony Biotechnology, Champaign, Illinois). Data was analyzed using FCS Express 4 software (DeNovo Software, Los Angeles, California).
For samples used in the homozygous h12 and h16 comparison, an aliquot of 100 μL of whole blood, collected in ACD, was transferred to a 12×75 mm polystyrene tube. Fc receptors were blocked with 10 μL of normal mouse serum (Equitech-Bio, Inc, Kerrville, Texas). Directly-conjugated antibodies to CD3ε (FITC conjugated, clone PPT3; Southern Biotech, Birmingham, Alabama), CD16 (PE-conjugated, clone G7; Serotec, Raleigh, North Carolina), and CD21 (APC-conjugated, clone B-ly4; BD Biosciences) were added and incubated for 30 minutes. Red blood cells were lysed using Multi-species RBC Lysis Buffer (eBiosciences, San Diego, California), and then washed twice in PBS-1% BSA (Fraction V bovine serum albumin; GE Healthcare Life Sciences). Cells were acquired on a Sony EC800 cytometer, equipped with counts/μL capabilities, and those detected within the lymphocyte gate were analyzed for the expression of CD3ε, CD21, or CD16 lineage-specific markers.
Radiosensitivity testing of fibroblasts from SCID and non-SCID piglets
Tissue processing and fibroblast collection was performed similarly as described by Ross et al.(23). Ear snip tissue of 20 young piglets from five litters from carrier-by-carrier matings was collected into PBS with Gentamycin and stored at 4°C until processing, three to 24 hours. Tissue was then soaked in 100% ethanol for 2–3 minutes, washed twice with PBS, then transferred to a dry dish and minced. Trypsin was added to the minced tissue and incubated at 37°C for 30 minutes. Trypsin was deactivated with cell culture medium, containing low glucose DMEM, 15% Fetal Bovine Serum (FBS), and Gentamycin, then centrifuged for 10 minutes at 600 × g. Cell culture media was removed from tissue and replaced with fresh media and cultured in the same medium to allow fibroblasts to migrate from tissue pieces. Fibroblasts were passaged to purify the culture, then stored in freezing media made up of FBS with 10% DMSO in liquid nitrogen. Frozen cultures were thawed and cultured in cell culture medium for approximately 7 days prior to further analysis.
Radiosensitivity assay
For irradiation, 150,000 fibroblast cells were suspended in 15 mL fresh cell culture medium with four tubes per biological replicate. Samples were irradiated using a gamma irradiation machine (Mary Greeley Medical Center, Ames, Iowa) at 0, 2, 4, or 8 gamma rays (Gy). Each sample was then plated in triplicate at a density of 1,000 cells per plate and cultured for two weeks, with one media change 7 days post irradiation. After 14 days of culture, plates were washed with PBS, stained with methyl blue, and the number of individual colonies with a diameter greater than 2 mm determined by visual inspection were counted(24).
Statistical analysis
The average number of colonies of three replicates for each animal and radiation dose was used as observations. Percentage survival for each animal was calculated by dividing the average number of colonies at each radiation dose (2, 4, and 8 Gy) by the average number of colonies at 0 Gy irradiation for that animal. Survival percentage was analyzed using Proc Mixed in SAS 9.3 (SAS, Inc.; Cary, North Carolina), with the fixed effects of SCID status, radiation dose, and their interaction. Litter and animal within litter were fitted as random effects. Different error variances were allowed for each radiation dose.
Data from irradiation of fibroblasts from pigs homozygous for each haplotype, and from compound heterozygous and normal pigs were also analyzed as described above, with the additional fixed effects of haplotype within SCID status and the interaction of haplotype and radiation dose.
Genome wide association analysis
Subjects
Genomic DNA from 20 SCID affected pigs, 50 unaffected littermates, their six parents, and 96 ancestors from the previous seven generations of the experimental residual feed intake (RFI) selection line at Iowa State University(20) (ISU) was isolated from tail or ear tissue using the Qiagen DNeasy Blood and Tissue Kit (Hilden, Germany). DNA samples were sent to GeneSeek Inc. (Lincoln, Nebraska) and genotyped using the Illumina Porcine SNP60 Beadchip(25). SCID status was determined by plotting lymphocyte against white blood cell numbers obtained from complete blood cell analysis of whole blood from each piglet (data not shown).
Statistical analysis
Initial attempts to map the genomic region harboring the causative mutation using runs of homozygosity(26) in the affected piglets were unsuccessful. Associations of SNPs with the binary phenotype of affected versus unaffected SCID status were then analyzed with the dfam, disease within family, option of Plink(27) to determine the genomic region most strongly associated with the categorical disease phenotype. The dfam option examines differential transmission of alleles from parents to affected versus unaffected offspring. This analysis assumes homogeneity of the associated allele within each family, but not between families.
Haplotype analysis
Following the results from the association study, genotypes of subjects used for the GWAS for 21 SNPs located in a 1-Mb region surrounding Artemis from the SNP60 panel were separated into haplotypes using PHASE 2.1.1(28). This revealed two haplotypes that segregated with SCID status when present in a homozygous or compound heterozygous state.
Candidate gene sequencing
Total RNA was extracted from either ear tissue or fibroblasts cultured from ear tissue using the TriZol extraction method (Gibco BRL, Life Technologies, New York). mRNA was reverse transcribed using SuperScript VILO (Invitrogen, Carlsbad, California). To examine the entire Artemis coding sequence for variation, the full coding region of the Artemis cDNA was PCR amplified using primers in the first and last exon: forward 5′-GGATCCGTGTTCGCCAACGCT-3′ and reverse 5′-GCGGCCGCAGAGCTGCCTTTTAGGTTAT-3′, using an annealing temperature of 60°C and an elongation time of 2 minutes and 30 seconds. Artemis cDNA PCR products were cloned into a TOPO vector and grown in E. coli bacteria on LB plates with Ampicillin. Individual colonies were then picked into LB-Ampicillin media and PCR amplified to ensure that each colony contained a vector with Artemis cDNA. Positive PCR products were cleaned using Exo-Sap and sent to the ISU DNA Facility for sequencing.
To examine the genomic region for sequence variation, primers were designed to amplify targeted exons of Artemis and portions of each surrounding intron. The genomic region containing exons 7 and 8 was PCR amplified using primers: forward 5′-CTCAGTGGGTTAGGGACCTG-3′ and reverse 5′-GCCATCTGATAGGGTTTCCA-3′, using an annealing temperature of 54°C and an elongation time of 60 seconds. The genomic region containing exons 10 and 11 was PCR amplified using primers: forward 5′-GCTAAAGTCCAGGCCAGTTG-3′ and reverse 5′-CAAGAGTCCCCACCAGTCTT-3′, using an annealing temperature of 56°C and an elongation time of 60 seconds. PCR products were cleaned using Exo-Sap and sent to the ISU DNA Facility for sequencing. Sequences for each haplotype were compared to those of normal littermates and to the reference sequence obtained from Sus scrofa build 10.2.
Rescue of Radiosensitive Phenotype
Fibroblasts for h12/h16 compound heterozygous SCID (n=2) and normal animals (n=3) from 3 litters were cultured in the same manner as described above. Fibroblasts were transfected with either 5 μg of the human Artemis plasmid(29) (pExodus CMV.Artemis, which was procured through deposition to Addgene [plasmid #40211] by Andrew Scharenberg), with 3.45 μg of the empty pExodus vector construct (also deposited by A. Scharenberg. Addgene; plasmid #39991), or shocked with no plasmid added. Cells were electroporated using electroporation media and conditions described by Ross et al.(23). Briefly, 1,000,000 cells in 200 μL were electroporated using three, 1 ms square-wave pulses of 300 volts in a 2 mm gap cuvette using a BTX ECM 2001 electroporator. Transfected cells were incubated overnight in cell culture media. One day after transfection, fibroblasts were subjected to 4 Gy radiation and plated in triplicate as described above. Cells were cultured for 14 days, with one media change seven days post irradiation, stained with methyl blue, and colonies determined by visual inspection and counted.
Statistical Analysis
The average number of colonies of three replicates for each animal and plasmid was used as observations. Data were analyzed using Proc Mixed in SAS 9.3 (SAS, Inc.; Cary, North Carolina), including the fixed effects of SCID status, plasmid, and their interaction. Litter and animal within litter were fitted as random effects. Different error variances were allowed for data from normal versus SCID animals.
Results
SCID phenotype segregates as an autosomal recessive trait in Yorkshire pig population
Weaned piglets of the eighth generation of a line of purebred Yorkshires selected for increased feed efficiency (residual feed intake; RFI) at Iowa State University (ISU)(20) were sent to Kansas State University (KSU) to undergo experimental infection with the Porcine Reproductive and Respiratory Syndrome virus to evaluate their ability to respond to an infectious disease stressor(30). Necropsy of four of these piglets that died soon after arrival at KSU showed that they had very low numbers of circulating B and T cells, and their lymph nodes and thymus were small(19). Pedigree analysis of the four litters that these piglets were from showed that the six parents of these four litters were related. To explore the genetic basis of this phenotype, matings between these six parents were repeated to produce a total of 13 litters (data not shown), each of which produced at least one affected piglet, as diagnosed by numbers of white blood cells and lymphocytes based on complete blood cell (CBC) counts of whole blood; plots of lymphocyte to white blood cell counts within litter distinguished suspected SCID piglets from non-SCID littermates (data not shown). A total of 176 piglets were born, 12 of which died before their SCID status could be determined. Of the remaining 164 piglets, 31 (19%) were confirmed to have SCID based on very low lymphocyte counts as measured by standard CBC or flow cytometry. This frequency was lower (p=0.07) than the expected 25% if the immunodeficiency phenotype was caused by an autosomal recessive mutation. However, it is possible that a portion of the 12 pigs with unknown SCID phenotype in these litters were actually SCID, thus the SCID phenotype may be under-reported in this population. Based on these results, we hypothesized that the immunodeficiency phenotype was caused by an autosomal recessive mutation.
T− B− NK+ SCID pigs are defective in thymocyte maturation
Flow cytometry analysis, recently published by Ewen et al.(22), demonstrated that concentrations of circulating leukocytes in peripheral blood of normal and affected pigs were similar among granulocytes, monocytes, and NK cell populations; however, B and T cell populations were significantly reduced in SCID piglets. Consistent findings were produced when quantitative real-time PCR (qRT-PCR) was used to measure the relative expression of B, T, and NK cell marker genes in whole blood of SCID and non-SCID piglets; non-SCID piglets were shown to have significantly higher levels of B and T cell marker gene expression compared to SCID littermates (p<0.01; data not shown).
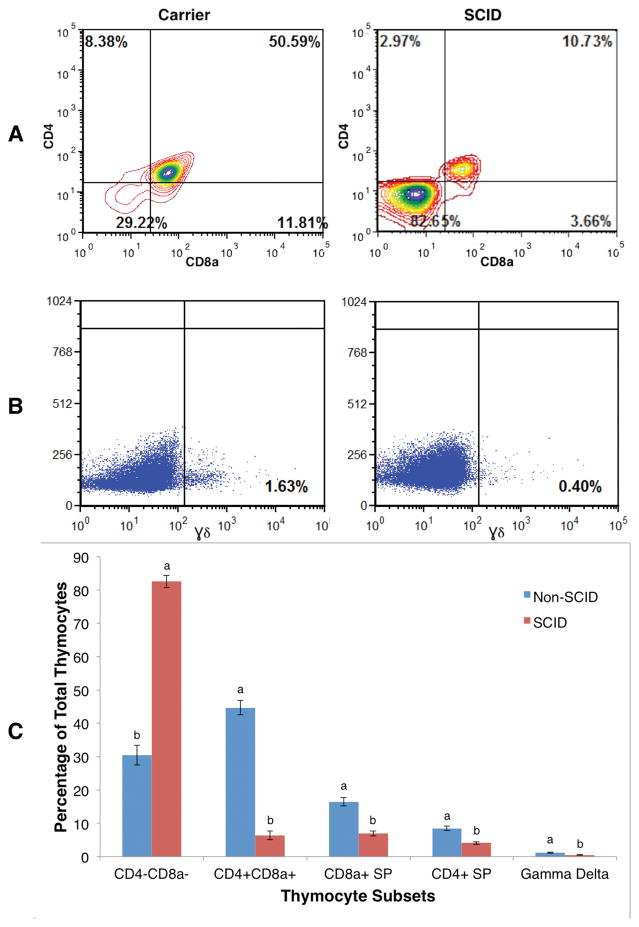
To help ascertain the mechanism underlying the marked T cell paucity in the SCID pigs, thymocytes were prepared from thymic tissues and stained with CD3ε, CD8a, CD4, and γδ lineage markers to monitor thymocyte development in these animals. Flow cytometric analysis revealed that the frequency of immature CD4− CD8a− double negative thymocytes was significantly greater in the SCID pigs, 82.5% (±2.94%), compared to their non-SCID littermates, 30.4% (±1.8%; p=0.001; Fig. 1), indicating an arrest of differentiation at or prior to TCR rearrangement(31). Furthermore, the frequencies of CD4+ CD8a+ double positive and CD4+ and CD8+ single positive cells, which are usually observed following TCR rearrangement(32, 33), were also significantly reduced in thymocytes derived from SCID piglets (p<0.01). These cells were almost exclusively CD3ε negative, as CD3+ cells are approximately10-fold reduced in abundance in SCID thymocytes compared to those in non-SCID (Fig. 1C). Aberrant thymocyte development was not restricted to αβTCR–bearing cells, as γδ+ thymocytes were also significantly decreased in SCID affected pigs compared to non-SCID pigs (p=0.0054; Fig. 1). These findings from flow cytometry and qRT-PCR of lymphocyte marker expression indicated that the SCID phenotype may be caused by a lesion early in the lymphocyte developmental pathway, as both B and T cells were absent or low in number, but late enough that NK cells were unaffected, resulting in T− B− NK+ SCID.
Figure 1. Flow cytometry shows very few CD4+ CD8+ and γδ+ cells in SCID pig thymus.
The number in each diagram is the percentage of each cell type in thymocyte populations of a SCID pig or a non-SCID carrier littermate (panels A and B). CD8a (x-axis) and CD4 (y-axis; panel A) and γδ (panel B) are T-cell surface markers. The least square means of percentages of each thymocyte sub-population are shown in panel C. Error bars represent the standard error of the estimates. Bars with different letters (a and b) indicate statistically significant (p<0.01) differences between SCID and non-SCID expression within cellular subset.
SCID fibroblasts are sensitive to ionizing radiation
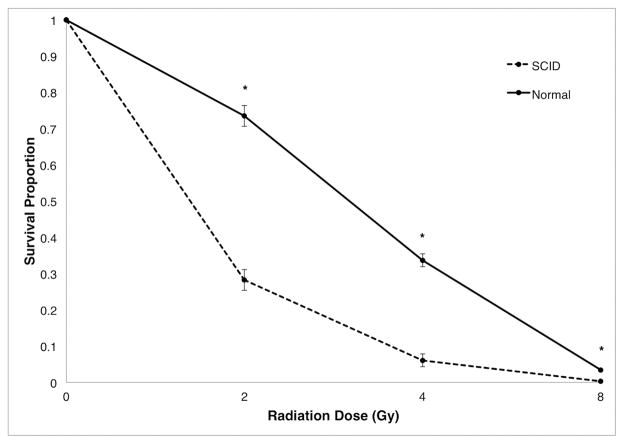
Human SCID patients with a T− B− NK+ phenotype have defects in genes in the somatic recombination pathway required for TCR and IG maturation. This involves two groups of genes; the first group includes RAG1 and RAG2, whose proteins catalyze the first step of TCR and IG maturation, creating a double-strand DNA break (DSDB) in pre-T and pre-B cells(34). The second group of genes repairs these breaks in the process of forming functional TCR and IG receptors(35). Defects in RAG1 and RAG2 affect somatic DNA recombination, which occurs only in B and T lymphocytes. However, the ubiquitously expressed genes in the second group are involved in the non-homologous end joining (NHEJ) DNA damage repair pathway, which is important in all cells(36). Cells with lesions in DNA damage repair genes show increased sensitivity to ionizing radiation(37). To determine if the genetic defect was in the first or second group of genes, we tested the ability of fibroblasts from SCID and normal pigs to repair DSDB caused by ionizing radiation. Fibroblasts cultured from skin tissue of SCID and non-SCID littermates from five litters were gamma-irradiated. Fibroblast survival was significantly decreased for cells from the SCID pigs compared to cells from normal littermates at all radiation doses tested (p<0.0001; Fig. 2). Even at the lowest dose of radiation, 2 gamma-rays (Gy), the proportion of fibroblasts that survived and developed colonies was significantly lower for those derived from SCID pigs, 0.28 ± 0.03, compared to those of normal pigs, 0.73 ± 0.03 (p<0.0001). This radiosensitivity in fibroblasts suggested that the mutation causing SCID in these pigs was in one of the ubiquitously expressed genes involved in NHEJ repair of DSDB.
Figure 2. Effect of ionizing radiation on fibroblasts from SCID and normal pigs.
Fibroblasts from SCID piglets (n=10) and normal littermates (n=10) were exposed to increasing doses of gamma-rays. Colonies (≥ 2 mm) were stained and counted after 14 days. Survival proportion was calculated as the average number of surviving colonies for three replicates at each radiation dose divided by the number of colonies from non-irradiated cells for each animal. Error bars represent the standard error of the least squares means. * indicates statistically significant (p<0.0001) difference between SCID and normal pigs within radiation dose.
Genetic mapping of the locus mutated in SCID pigs
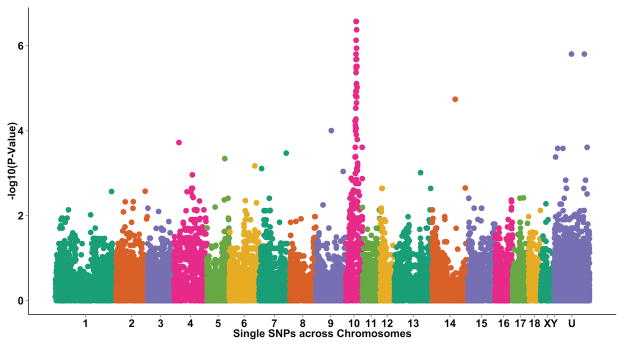
Genotypes for over 60,000 single nucleotide polymorphisms (SNP) represented on the Illumina Porcine SNP60 panel(25), covering the entire porcine genome, were obtained on the six carrier parents, 20 SCID affected piglets, 50 unaffected littermates of the SCID piglets, and 96 ancestors of these animals. A genome wide association study was performed to identify the region harboring the causative mutation. We identified a 5.6 Mb region on Sus scrofa chromosome 10 that was differentially transmitted (p=2.7 × 10−7 for SNP with strongest association) to SCID affected versus unaffected piglets (Fig. 3). This region was found to contain the porcine Artemis gene, which, when mutated in humans(38) or mice(39), causes a T− B− NK+ immunodeficiency phenotype. Lesions in Artemis have also been shown to cause increased sensitivity to ionizing radiation(40).
Figure 3. Manhattan plot of the Genome Wide Association Study for SCID status.
Results show the –log(p-value) of the association of ordered SNPs on Sus scrofa chromosomes 1 through 18, X, Y, and unknown (UNK) with SCID status based on the dfam option in PLINK.
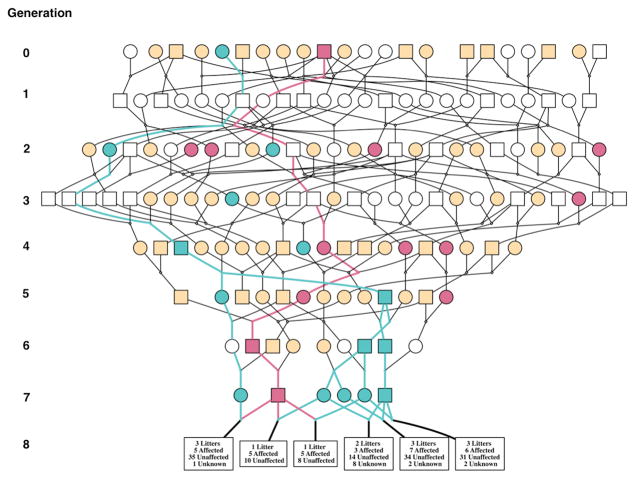
To confirm the suspected autosomal recessive manner of transmission, a haplotype analysis was performed using 21 SNPs from the SNP60 panel in the 1-Mb region surrounding Artemis. Results showed two distinct haplotypes (h12 and h16), which when present in a compound heterozygous or homozygous recessive state, were associated with the SCID phenotype. Both of these SNP haplotypes could be traced back to the founder generation of the RFI selection line(20), which was sourced from commercial Yorkshire populations in the Midwest region of the USA (Fig. 4).
Figure 4. Pedigree of SCID ancestors showing carriers of mutated haplotypes based on the Illumina Porcine SNP60 panel.
Circles represent females and squares represent males. Green symbols are h16 haplotype carriers and pink symbols are h12 haplotype carriers, with lines of each color tracking the respective SCID haplotype through the pedigree to the founder generation. Beige symbols are pigs that were genotyped with the SNP60 panel and did not carry either SCID haplotype. White symbols in generations 0 through 7 represent non-genotyped individuals. Boxes in generation 8 give information on the numbers of SCID (Affected) and non-SCID (Unaffected) piglets and the number of piglets that died before their SCID phenotype was determined (Unknown) from each parent pair.
Candidate gene sequencing identifies independent mutations in Artemis
To identify the causal mutation, the coding portion of the Artemis cDNA from fibroblast mRNA was sequenced for affected piglets and normal littermates (Fig. 5). Selected exons and flanking portions of introns were also amplified and sequenced from genomic DNA. Multiple examples of alternative splicing events were seen in cDNA of both SCID and normal pigs (S1 Fig.). The most abundant transcript observed in cDNA from normal pigs contained all exons present in the human Artemis cDNA, but is missing the second exon of an Artemis porcine cDNA sequence isolated from alveolar macrophages that was reported by others (NM_001258445.1). This exon was not present in any of the Artemis transcripts sequenced in this study, although it exists in the pig genome (Sus scrofa 10.2) between exon 1 and what we refer to as exon 2 and may represent a bona fide exon. However, based on BLAST analysis of sequences that are present in dbEST (17 February 2015), cDNAs with the top six alignments did not contain this exon. In addition, this exon did not align to any portion of human Artemis cDNA sequences (NM_001033855.2). Both genome-guided transcriptome assembly and de novo transcriptome assembly using whole blood RNAseq data from 31 pigs in an unrelated project was found to contain only one Artemis transcript, which did not contain this exon (H. Liu and C.K. Tuggle, unpublished data). Based on these findings, we believe the swine reference transcript that contains this extra exon (NM_001258445.1) is a minor transcript. For clarity and consistency with the human literature, we denoted this extra exon as exon 1A and considered the second exon that is present in the transcripts observed in our study as exon 2. Translation of the longest transcript we sequenced from non-SCID pigs would produce a protein containing 712 amino acids, with 81% sequence identity to the human encoded Artemis protein.
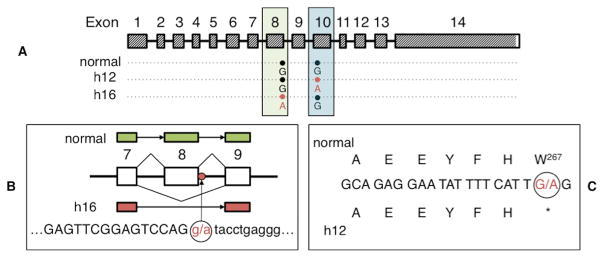
Figure 5. Two independent mutations were found in Artemis.
(Panel A) The coding region of the Artemis transcript is indicated by slanted lines and genotypes at mutated positions are shown for chromosomes that carry normal, h12, and h16 haplotypes. Mutant alleles that cause SCID are shown in pink lettering. (Panel B) Genomic sequence of the h16 haplotype shows a splice donor site mutation (g.51578763 G→A) responsible for the lack of exon 8 in all h16 transcripts. Capital letters denote exonic sequence while lower case letters denote intronic sequence. (Panel C) A nonsense point mutation (g.51584489 G→A) in exon 10 changes the Tryptophan at position 267 to a stop codon in the h12 haplotype.
Haplotype 16 has a lesion causing a predicted splice defect
The majority of cDNAs sequenced from normal pigs contained exons 1–14. The most complete transcript sequenced from cDNA from the h16 SCID haplotype contained all these exons except for exon 8, resulting in the loss of 141 nucleotides (S1 Fig.). In fact, none of the cDNA sequences obtained from h16 chromosomes contained exon 8. Genomic sequencing of exon 8 and a portion of the surrounding introns revealed a splice donor site mutation in intron 8 (g.51578763 G→A; Fig. 5b). This G→A mutation was only seen in the h16 genomic sequence, as a G nucleotide at this position was seen in both the h12 and normal sequences; to distinguish h16 from h12 transcripts in compound heterozygous animals, we used a synonymous point mutation in exon 13 that we identified to be present only in h16 transcripts (position g.51587796). This G→A nucleotide conversion disrupts the signals required to correctly splice exon 8 to exon 9, which explains the observed lack of exon 8 in cDNAs expressed from h16 haplotype chromosomes. An Artemis transcript with no exon 8 would retain the normal reading frame but would be expected to produce a protein missing 47 amino acids of the predicted full length 712-amino acid Artemis protein.
Haplotype 12 has a nonsense point mutation and a predicted severely truncated protein
Multiple examples of alternative splicing, including aberrant splicing within exons and one intron, were observed for transcripts from chromosomes that carried the h12 haplotype (S1 Fig.). All transcripts sequenced from homozygous h12 animals were found to lack the 137 base pair long exon 10, which would cause a frameshift that results in a stop codon shortly after the missing exon. The predicted protein translated from transcripts missing exon 10 would be severely truncated; at 277 amino acids long, it would be < 50% of normal length. To investigate the cause of this lack of exon 10 in all h12 transcripts, exons 10 and 11 and portions of the surrounding introns were amplified from genomic DNA of homozygous h12 animals. Signal sequences required for splicing of exon 10 were identical to those seen in the sequence of normal pigs, as well as in the reference sequence (Sscrofa 10.2, www.ensembl.org/Sus_scrofa/Info/Index). However, sequence analysis of the exon 10 genomic region in homozygous h12 pigs identified a G→A nonsense point mutation at g.51584489 that changes the Tryptophan amino acid codon at position 267 to a stop codon (Fig. 5c). Our interpretation of these data is that any transcript that contained exon 10 was not stable enough for cDNA analysis to detect. Further, while not identified as part of any cDNA from homozygous h12 pigs, the predicted translation of this presumably unstable transcript containing exon 10 with the g.51584489 G→A mutation would also produce a severely truncated protein of 266 amino acids.
Animals homozygous for either mutation are not different in numbers of specific lymphocytes or in radiosensitivity
The flow cytometry and radiosensitivity assays described earlier were performed on a combination of compound heterozygous and homozygous h12 or h16 SCID animals. To determine if homozygosity for either mutation would result in altered severity of phenotypes, flow cytometry of circulating blood and assays of fibroblast irradiation were performed. Peripheral blood from SCID piglets that were homozygous for either mutation or compound heterozygous had greatly decreased percentages of CD21+ and CD3+ cells compared to carrier littermates (p<0.01), but no differences were seen among pigs with different SCID genotypes (p>0.10; S2 Fig.). Fibroblasts from each SCID genotype were equally sensitive to ionizing radiation at all radiation doses (p>0.10; S3 Fig.), and fibroblasts from each SCID genotype were significantly more sensitive than fibroblasts from normal pigs at 2 and 4 Gy (p<0.05) and tended to be more sensitive at 8 Gy (p=0.087; S3 Fig.).
Transfection with human Artemis rescues radiosensitivity of fibroblasts
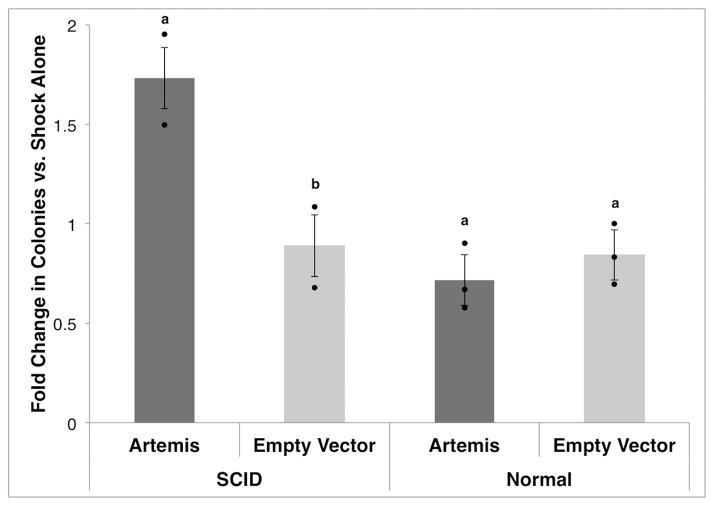
To further confirm the role of Artemis in the observed SCID phenotype, we assessed the complementation of the radiosensitivity of SCID pig fibroblasts by expression of human Artemis. Fibroblasts cultured from compound heterozygous SCID (n=2) and normal (n=3) piglets from three litters were transfected with a human Artemis containing plasmid (pArt), with the empty vector construct (pExo), or shocked without the addition of either plasmid. Cells were then subjected to 4 Gy radiation and survival measured. Addition of pArt increased the radioresistance of fibroblasts from SCID pigs (p=0.001), while cells from normal littermates were unaffected by the addition of pArt (p=0.11; Fig. 6), and pExo had no effect on the radiosensitivity of fibroblasts of SCID or non-SCID genotypes.
Figure 6. Rescue of sensitivity to ionizing radiation.
Fibroblasts from compound heterozygous SCID (n=2) and normal (n=3) littermates were transfected with human Artemis-expressing plasmid (5 μg, Artemis), with a molar equivalent of pExodus plasmid without the Artemis gene (3.45 μg, Empty Vector), or shocked without plasmid added. Fibroblasts were exposed to a 4 Gy radiation dose 24 hours after transfection. Colonies (≥ 2 mm) were counted after 14 days of growth. The average number of surviving colonies for three replicates for a given plasmid was divided by the number of colonies from shock only cells for each animal. Error bars represent the standard error of the least squares means. Dots show individual observations. Bars with different letters within affected status (a and b) represent statistical differences between means with p<0.01.
Discussion
Through immunophenotyping, genetic mapping, cDNA and genomic DNA sequencing, fibroblast radiation sensitivity testing, and cDNA rescue of radiosensitivity phenotypes of fibroblasts, we provide substantial evidence that a novel T− B− NK+ immunodeficiency trait in pigs is caused by two independent mutations in the same gene that codes for a DNA repair enzyme, which is known to cause this type of SCID phenotype in human patients. We document that the identified mutations in the Artemis gene are necessary to observe a SCID phenotype in directed matings and are sufficient to cause the radiosensitivity phenotype observed in SCID fibroblasts from this population.
Novel immunodeficiency phenotype in pigs is a recessive Mendelian trait that shows defects in thymocyte maturation consistent with flawed somatic recombination
Cino Ozuno, et al.(19) reported the initial analysis of the first cases of naturally occurring SCID in pigs, which was serendipitously found within a line selected for a commercially relevant trait at ISU(20). In our expanded analysis of litters born within the pedigree of this line, a Mendelian and non-sex-linked segregation of SCID affected status in litters born to carrier-by-carrier matings was observed, indicating that the SCID phenotype was an autosomal recessive trait(2). Findings from qRT-PCR of lymphocyte marker expression in whole blood showed that the SCID pigs had very low numbers of B and T cells, while NK cells were present. These results suggested that the ability of B and T lymphocytes to recombine genetic material to form functional receptors was compromised in the SCID affected piglets. Flow cytometry of thymus tissue showed an increased population of CD4− CD8a− and γδ− T cells in SCID pigs compared to non-SCID littermates. This suggested an arrest in early thymocyte development and TCR rearrangement in the SCID pigs(31, 33). The increased sensitivity to ionizing radiation in fibroblasts of SCID pigs further narrowed the list of candidate genes to be evaluated for mutations to those involved in the NHEJ DNA damage repair system.
Two distinct mutations in the Artemis gene found in SCID pigs
Genomic association analysis of the population segregating the SCID phenotype pointed to a region containing the Artemis gene, while SNP genotype phasing results indicated that there were two distinct haplotypes that each carried a mutation postulated to cause the SCID phenotype. Both of these haplotypes could be traced back to the founders of this population, which suggests that these deleterious mutations may be present in commercial Yorkshire pigs in the US. This is an important implication for the swine industry, as SCID piglets thrive while suckling from their dam, but succumb to opportunistic infections after weaning. Parentage information is generally not tracked into the nursery, at least in part due to cross fostering practices, the lack of permanent individual identification and the frequent use of mixed semen from more than one boar for artificial insemination. Therefore, although the death of several littermates represents a concentrated loss for that litter, this clue to a genetic defect is not likely to be recognized on commercial farms. In addition, if the frequency of the SCID mutations is low, carrier-by-carrier matings may be rare in industry practice.
Sequencing of Artemis cDNA from non-SCID and SCID pigs carrying each haplotype identified multiple examples of alternative splicing. The cause for absence of specific exons in sequences of each SCID haplotype was explored using targeted genomic sequencing. A point mutation at the splice donor site for intron 8 in the h16 genomic sequence explained the lack of exon 8 in all cDNA sequences from the h16 haplotype. A compendium of 48 different causative mutations in Artemis in human SCID patients has cataloged six splice site mutations, although none involve exon 8(41). Translation of the most complete h16 transcript would produce a protein of 665 amino acids, lacking a 47 amino acid long portion of the beta-CASP region, which is required for Artemis function(42, 43). Genomic deletions of Artemis exons 5 through 8(38) and exons 7 and 8(40) have been shown to cause SCID in human patients. For h12, no cDNAs were identified that contained exon 10, but the most complete cDNAs from homozygous h12 cells contained all other exons that were present in the non-SCID cDNA sequence. Because the lack of exon 10 causes a frameshift and the open reading frame stops in exon 11, the longest protein that is predicted to be encoded by the observed cDNAs from haplotype h12 is 277 amino acids long, containing only 39% of the normal protein. Interestingly, genomic sequence of the h12 haplotype identified a nonsense point mutation in exon 10, which would result in a protein of only 266 amino acids if this exon 10 were present in mRNA. This severely truncated protein would lack over half of the total amino acid sequence, including a highly conserved part of the beta-CASP motif(42). Therefore, in both observed cDNAs and RNA transcripts predicted from genomic sequence, h12 can encode only a greatly shortened protein, with 277 or 266 amino acids, respectively. In humans, an 8 base pair insertion in exon 14 of Artemis that changed a cysteine to a stop codon at position 330 of the protein was also shown to cause SCID(44). Pannicke et al.(41) lists several small deletions causing SCID, the majority of which lead to a frameshift that introduces novel nonsense codons that cause truncations that are distal to exon 10, in exons 12, 13, and 14. This suggests that either version of the h12 haplotype protein is highly unlikely to be functional in DNA damage repair.
We thus hypothesized that the h12 mutation may cause a more severe immunodeficient phenotype than the h16 mutation, in which only 47 internal amino acids are predicted to be lost. However, flow cytometry and fibroblast irradiation showed no phenotypic differences between SCID pigs that were homozygous for either mutation or compound heterozygous. While single missense mutations, as well as deletions of single amino acids without a frameshift, can lead to a SCID or Omenn syndrome phenotype in humans(41), examples exist for frameshift mutations producing a truncated protein with partial activity. For example, the D451fsX10 mutation is a large truncation of >200 amino acids that retains partial nuclease activity(45). In a mouse model, the hypomorphic D451fsX10 mutation is associated with aberrant DNA joining that causes chromosomal rearrangements(46). Thus, underlying cellular differences in DNA repair activity between the h12 and h16 encoded proteins may exist, and further studies should be conducted to determine the functional differences between these two mutations in pigs.
Value of Artemis SCID pigs in preclinical testing
Similarities between humans and pigs emphasize the value of the pig as a biomedical model(13), and pigs as large animal models for many diseases are being created(47). Body, tissue and organ size make them physiologically very similar, allowing surgical and imaging techniques to translate well(48). Size similarity also makes the pig a more suitable model for testing delivery of cells (i.e., heart) and for measuring location/stability of transplanted cells under orthopedic stress (i.e., joint tissue). However, the use of pig models in regenerative medicine has been minimal, due to the heretofore lack of a porcine immuno-compromised model that could be used as an xenogenic transplant model. As recently demonstrated for human cancer cells, the SCID pigs we describe here cannot reject a xenograft(21). Immunodeficient porcine models have long been sought, as is evident by the recently reported transgenic SCID pigs with targeted IL2RG(15, 16) and RAG1 and/or RAG2 mutations(17, 18). Here, we have described two naturally occurring genetic defects in Artemis that cause SCID in pigs. These porcine models will be valuable in studies involving cellular and organ transplantation, human cancer progression and treatment, vaccine development, and many other immunological questions. Specifically, defects in Artemis (as well as all genes encoding proteins needed for VDJ recombination) have been shown to be associated with poor outcome of stem cell transplantation in humans (reviewed in 2). In the case of Artemis, affected patients have substantially higher risk of late toxicity following hematopoietic cell transplants (HCT) as compared to patients with RAG lesions (49). This was attributed to the use of alkylating agents in conditioning regimens. A separate study also found Artemis defects were associated with negative clinical events following HCT, which included graft vs host disease and a range of infections(50). While Xiao et al.(51) generated an Artemis deficient mouse model that replicates the phenotype observed in human Artemis deficient SCID patients, these Artemis deficient pigs provide an additional and perhaps more relevant preclinical model to test and improve clinical therapies used in treating these patients. Future studies will focus on defining optimal husbandry practices to increase the lifespan of SCID pigs so that such modeling can be performed.
Supplementary Material
Acknowledgments
We would like to sincerely thank G. Kuper and the staff at the ISU swine breeding farm in Madrid, Iowa, E. Powell, the Tuggle, Dekkers, Ross, Ellinwood, and Lab Animal Resources group members at ISU, and the Rowland group at KSU for their assistance with animal care and lab work. We acknowledge B. MacPhail, Jaeger Corporation (Omaha, Nebraska), for help with irradiation of fibroblasts. We are grateful to M. Georges for valuable advice regarding genetic mapping and sequencing results of the two mutations.
Funding
E. Waide was a Fellow supported by USDA NIFA National Needs grant # 2010-38420-20328. This work was supported by the ISU Bailey Faculty Development Award and PIIR grants funded by ISU Research Foundation.
Footnotes
Author Contributions
E.H.W., J.C.M.D., C.K.T., and J.W.R. designed experiments and J.C.M.D., C.K.T., and J.W.R. supervised research. E.H.W. performed genetic mapping, genomic and cDNA sequencing, irradiation and transfection of fibroblasts, and statistical analysis of all data. R.R.R.R., C.R.W. and C.L.E. performed and supervised the flow cytometric analyses. A.B.E. performed quantitative RT-PCR assays. D.M.T. performed initial genomic mapping analyses and, along with N.J.B., assisted E.H.W. in further analyses. N.J.B. developed a method to diagnose SCID status based on CBC data. N.V.L.S. supervised statistical analyses performed by E.H.W. N.M.E. participated in project discussions and facilitated the cell irradiation work with Mary Greeley Medical Center. E.H.W., C.L.E. and C.K.T. wrote the manuscript.
Competing Interest
The authors have filed a patent that includes testing for the genetic mutations discovered in this study.
References
- 1.Notarangelo LD. Primary immunodeficiencies. J Allergy Clin Immunol. 2010;125:S182–S194. doi: 10.1016/j.jaci.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 2.Cossu F. Genetics of SCID. Ital J Pediatr. 2010;36:76. doi: 10.1186/1824-7288-36-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger Ra, Hansen-Hagge TE, Desiderio S, Lieber MR, Bartram CR. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 4.De Villartay JP. V(D)J recombination deficiencies. Adv Exp Med Biol. 2009;650:46–58. doi: 10.1007/978-1-4419-0296-2_4. [DOI] [PubMed] [Google Scholar]
- 5.Blum B, Benvenisty N. The Tumorigenicity of Human Embryonic Stem Cells. Adv Cancer Res. 2008;100:133–158. doi: 10.1016/S0065-230X(08)00005-5. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham JJ, Ulbright TM, Pera MF, Looijenga LHJ. Lessons from human teratomas to guide development of safe stem cell therapies. Nat Biotechnol. 2012;30:849–857. doi: 10.1038/nbt.2329. [DOI] [PubMed] [Google Scholar]
- 7.Shultz LD, Brehm MA, Garcia JV, Greiner DL. Humanized mice for immune system investigation: Progress, promise and challenges. Nat Rev Immunol. 2012;12:786–798. doi: 10.1038/nri3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia S, Freitas AA. Humanized mice: Current states and perspectives. Immunol Lett. 2012;146:1–7. doi: 10.1016/j.imlet.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Brehm Ma, Shultz LD. Human allograft rejection in humanized mice: a historical perspective. Cell Mol Immunol. 2012;9:225–231. doi: 10.1038/cmi.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 11.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West Ma, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plews J, Gu M, Longaker M, Wu J. Large animal induced pluripotent stem cells as pre-clinical models for studying human disease. J Cell Mol Med. 2013;16:1196–1202. doi: 10.1111/j.1582-4934.2012.01521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: A model for human infectious diseases. Trends Microbiol. 2012;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prather RS, Lorson M, Ross JW, Whyte JJ, Walters E. Genetically Engineered Pig Models for Human Diseases. Annu Rev Anim Biosci. 2013;1:203–219. doi: 10.1146/annurev-animal-031412-103715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki S, Iwamoto M, Saito Y, Fuchimoto D, Sembon S, Suzuki M, Mikawa S, Hashimoto M, Aoki Y, Najima Y, Takagi S, Suzuki N, Suzuki E, Kubo M, Mimuro J, Kashiwakura Y, Madoiwa S, Sakata Y, Perry ACF, Ishikawa F, Onishi A. Il2rg gene-targeted severe combined immunodeficiency pigs. Cell Stem Cell. 2012;10:753–758. doi: 10.1016/j.stem.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe M, Nakano K, Matsunari H, Matsuda T, Maehara M, Kanai T, Kobayashi M, Matsumura Y, Sakai R, Kuramoto M, Hayashida G, Asano Y, Takayanagi S, Arai Y, Umeyama K, Nagaya M, Hanazono Y, Nagashima H. Generation of Interleukin-2 Receptor Gamma Gene Knockout Pigs from Somatic Cells Genetically Modified by Zinc Finger Nuclease-Encoding mRNA. PLoS One. 2013;8:1–8. doi: 10.1371/journal.pone.0076478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K, Kwon DN, Ezashi T, Choi YJ, Park C, Ericsson AC, Brown AN, Samuel MS, Park KW, Walters EM, Kim DY, Kim JH, Franklin CL, Murphy CN, Roberts RM, Prather RS, Kim JH. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci U S A. 2014;111:7260–7265. doi: 10.1073/pnas.1406376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Guo X, Fan N, Song J, Zhao B, Ouyang Z, Liu Z, Zhao Y, Yan Q, Yi X, Schambach A, Frampton J, Esteban Ma, Yang D, Yang H, Lai L. RAG1/2 Knockout Pigs with Severe Combined Immunodeficiency. J Immunol. 2014;193:1496–503. doi: 10.4049/jimmunol.1400915. [DOI] [PubMed] [Google Scholar]
- 19.Cino-Ozuna A, Rowland R, Nietfeld J, Kerrigan M, Dekkers J, Wyatt C. Preliminary Findings of a Previously Unrecognized Porcine Primary Immunodeficiency Disorder. Vet Pathol. 2012;50:144–146. doi: 10.1177/0300985812457790. [DOI] [PubMed] [Google Scholar]
- 20.Cai W, Casey DS, Dekkers JCM. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J Anim Sci. 2008;86:287–298. doi: 10.2527/jas.2007-0396. [DOI] [PubMed] [Google Scholar]
- 21.Basel MT, Balivada S, Beck AP, Kerrigan MA, Pyle MM, Dekkers JCM, Wyatt CR, Rowland RRR, Anderson DE, Bossmann SH, Troyer DL. Human xenografts are not rejected in a naturally occurring immunodeficient porcine line: a human tumor model in pigs. Biores Open Access. 2012;1:63–68. doi: 10.1089/biores.2012.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewen C, Cino-Ozuna A, He H, Kerrigan M, Dekkers J, Tuggle C, Rowland R, Wyatt C. Analysis of blood leukocytes in a naturally occurring immunodeficiency of pigs shows the defect is localized to B and T cells. Vet Immunol Immunopathol. 2014;162:174–179. doi: 10.1016/j.vetimm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Ross JW, Whyte JJ, Zhao J, Samuel M, Wells KD, Prather RS. Optimization of square-wave electroporation for transfection of porcine fetal fibroblasts. Transgenic Res. 2010;19:611–620. doi: 10.1007/s11248-009-9345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darroudi F, Wiegant W, Meijers M, Friedl AA, van der Burg M, Fomina J, van Dongen JJM, van Gent DC, Zdzienicka MZ. Role of Artemis in DSB repair and guarding chromosomal stability following exposure to ionizing radiation at different stages of cell cycle. Mutat Res - Fundam Mol Mech Mutagen. 2007;615:111–124. doi: 10.1016/j.mrfmmm.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 25.Ramos AM, Crooijmans RPMA, Affara NA, Amaral AJ, Alan L, Beever JE, Bendixen C, Churcher C, Clark R, Dehais P, Hansen MS, Hedegaard J, Hu Z, Kerstens HH, Law AS, Milan D, Nonneman DJ, Rohrer GA, Rothschild MF, Smith TPL. Design of a High Density SNP Genotyping Assay in the Pig Using SNPs Identified and Characterized by Next Generation Sequencing Technology. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drögemüller C, Reichart U, Seuberlich T, Oevermann A, Baumgartner M, Boghenbor KK, Stoffel MH, Syring C, Meylan M, Müller S, Müller M, Gredler B, Sölkner J, Leeb T. An unusual splice defect in the mitofusin 2 gene (mfn2) is associated with degenerative axonopathy in tyrolean grey cattle. PLoS One. 2011;6:e18931. doi: 10.1371/journal.pone.0018931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Certo MT, Gwiazda KS, Kuhar R, Sather B, Curinga G, Mandt T, Brault M, Lambert AR, Baxter SK, Jacoby K, Ryu BY, Kiem HP, Gouble A, Paques F, Rawlings DJ, Scharenberg AM. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods. 2012;9:973–975. doi: 10.1038/nmeth.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunkelberger J, Boddicker N, Serão N, Young J, Rowland R, Dekkers J. Response of pigs divergently selected for residual feed intake to experimental infection with the PRRS virus. Livest Sci. 2015;177:132–141. [Google Scholar]
- 31.Šinkora M, Butler JE. The ontogeny of the porcine immune system. Dev Comp Immunol. 2009;33:273–283. doi: 10.1016/j.dci.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinkora M, Sinkora J, Reháková Z, Butler JE. Early ontogeny of thymocytes in pigs: sequential colonization of the thymus by T cell progenitors. J Immunol. 2000;165:1832–1839. doi: 10.4049/jimmunol.165.4.1832. [DOI] [PubMed] [Google Scholar]
- 33.Sinkora M, Sinkorová J, Cimburek Z, Holtmeier W. Two groups of porcine TCRgammadelta+ thymocytes behave and diverge differently. J Immunol. 2007;178:711–719. doi: 10.4049/jimmunol.178.2.711. [DOI] [PubMed] [Google Scholar]
- 34.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y. DNA damage: a trigger of innate immunity but a requirement for adaptive immune homeostasis. Nat Rev Immunol. 2006;6:261–270. doi: 10.1038/nri1804. [DOI] [PubMed] [Google Scholar]
- 36.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109:45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 37.Dvorak CC, Cowan MJ. Radiosensitive Severe Combined Immunodeficiency Disease. Immunol Allergy Clin North Am. 2010;30:125–142. doi: 10.1016/j.iac.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moshous D, Callebaut I, De Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, De Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 39.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid Phenotype Associated with Defective V(D)J Coding End Processing in Artemis-Deficient Mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Moshous D, Zhou Y, Wang J, Xie G, Salido E, Hu D, de Villartay JP, Cowan MJ. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J Immunol. 2002;168:6323–6329. doi: 10.4049/jimmunol.168.12.6323. [DOI] [PubMed] [Google Scholar]
- 41.Pannicke U, Hönig M, Schulze I, Rohr J, Heinz GA, Braun S, Janz I, Rump EM, Seidel MG, Matthes-Martin S, Soerensen J, Greil J, Stachel DK, Belohradsky BH, Albert MH, Schulz A, Ehl S, Friedrich W, Schwarz K. The most frequent DCLRE1C (ARTEMIS) mutations are based on homologous recombination events. Hum Mutat. 2010;31:197–207. doi: 10.1002/humu.21168. [DOI] [PubMed] [Google Scholar]
- 42.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poinsignon C, Moshous D, Callebaut I, de Chasseval R, Villey I, de Villartay JP. The metallo-beta-lactamase/beta-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J Exp Med. 2004;199:315–321. doi: 10.1084/jem.20031142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomashov-Matar R, Biran G, Lagovsky I, Kotler N, Stein A, Fisch B, Sapir O, Shohat M. Severe combined immunodeficiency (SCID): From the detection of a new mutation to preimplantation genetic diagnosis. J Assist Reprod Genet. 2012;29:687–692. doi: 10.1007/s10815-012-9765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Giblin W, Kubec M, Westfield G, St Charles J, Chadde L, Kraftson S, Sekiguchi J. Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability. J Exp Med. 2009;206:893–908. doi: 10.1084/jem.20082396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs C, Huang Y, Masud T, Lu W, Westfield G, Giblin W, Sekiguchi JM. A hypomorphic Artemis human disease allele causes aberrant chromosomal rearrangements and tumorigenesis. Hum Mol Genet. 2011;20:806–819. doi: 10.1093/hmg/ddq524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walters EM, Wolf E, Whyte JJ, Mao J, Renner S, Nagashima H, Kobayashi E, Zhao J, Wells KD, Critser JK, Riley LK, Prather RS. Completion of the swine genome will simplify the production of swine as a large animal biomedical model. BMC Med Genomics. 2012;5:55. doi: 10.1186/1755-8794-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swindle MM. Swine in the laboratory: Surgery, anesthesia, imaging, and experimental techniques. 2. CRC Press; Boca Raton, Florida: 2007. [Google Scholar]
- 49.Schuetz C, Neven B, Dvorak CC, Leroy S, Ege MJ, Pannicke U, Schwarz K, Schulz AS, Hoenig M, Sparber-Sauer M, Gatz Sa, Denzer C, Blanche S, Moshous D, Picard C, Horn BN, De Villartay JP, Cavazzana M, Debatin KM, Friedrich W, Fischer A, Cowan MJ. SCID patients with ARTEMIS vs RAG deficiencies following HCT: Increased risk of late toxicity in ARTEMIS-deficient SCID. Blood. 2014;123:281–289. doi: 10.1182/blood-2013-01-476432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neven B, Leroy S, Decaluwe H, Le Deist F, Picard C, Moshous D, Mahlaoui N, Debré M, Casanova JL, Dal Cortivo L, Madec Y, Hacein-Bey-Abina S, De Saint Basile G, De Villartay JP, Blanche S, Cavazzana-Calvo M, Fischer A. Long-term outcome after hematopoietic stem cell transplantation of a single-center cohort of 90 patients with severe combined immunodeficiency. Blood. 2009;113:4114–4124. doi: 10.1182/blood-2008-09-177923. [DOI] [PubMed] [Google Scholar]
- 51.Xiao Z, Dunn E, Singh K, Khan IS, Yannone SM, Cowan MJ. A non leaky Artemis-deficient mouse that accurately models the human SCID phenotype including resistance to hematopoietic stem cell transplantation. Biol Blood Marrow Transpl. 2009;15:1–11. doi: 10.1016/j.bbmt.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.