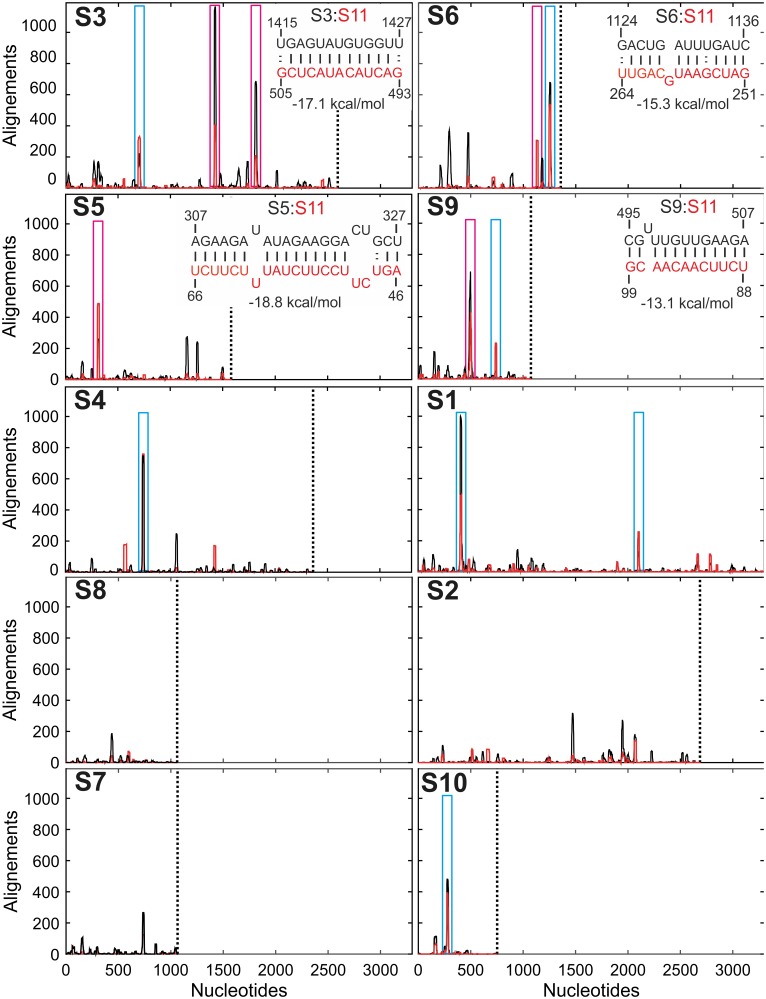
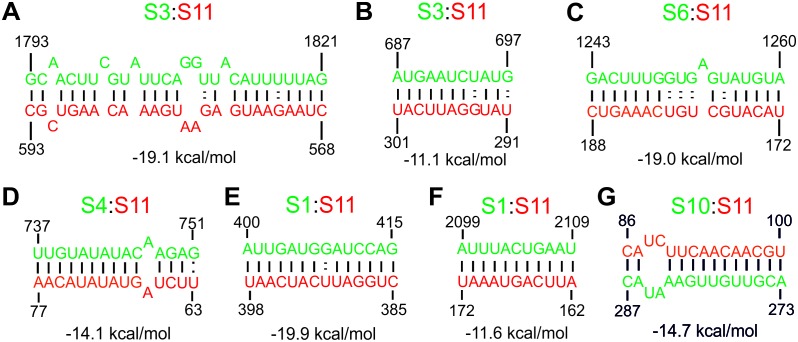
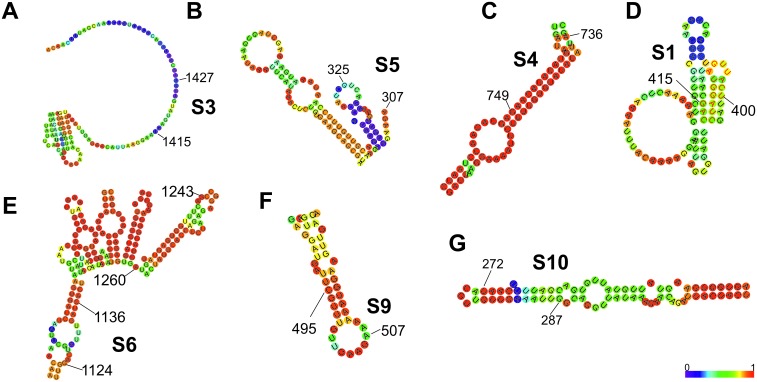
Figure 3. Inter-segment interactions between ssRNAs S11 and S1-S10, identified via RNA-RNA SELEX.
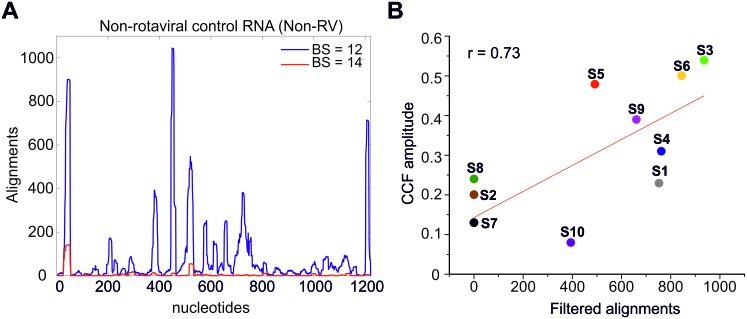
SELEX-enriched sequences were aligned against S1-S10 genomic segments using probabilistic score-based filtering (Materials and methods) to construct the resulting histogram plots. Each histogram peak corresponds to a sequence with a potential to interact with the S11 RNA alone (shown as black peaks), or in the presence of NSP2 (peaks shown in red). These histograms are plotted in the order of decreasing CCF amplitudes (see Figure 1B). Each plot is scaled to the size of the longest segment S1 (3302 nts), and the 3’-end of each RNA segment is marked with a vertical dotted line. Sequences that can stably base-pair with S11 RNA in the presence of NSP2 are highlighted (magenta boxes). A number of genomic sequences identified in the RNA-RNA SELEX experiments may be sequestered by stable local secondary structures (peaks identified by cyan boxes). Insets – RNA-RNA interaction sites between RNA S11 (bottom strand shown in red), and its interacting partners S3, S6, S5 and S9 (top strand, shown in black).