Abstract
Cardiac hypertrophy, as a response to hemodynamic stress, is associated with cardiac dysfunction and death, but whether hypertrophy itself represents a pathological process remains unclear. Hypertrophy is driven by changes in myocardial gene expression that require the MEF2 family of DNA-binding transcription factors, as well as the nuclear lysine acetyltransferase p300. Here we used genetic and small-molecule probes to determine the effects of preventing MEF2 acetylation on cardiac adaptation to stress. Both nonacetylatable MEF2 mutants and 8MI, a molecule designed to interfere with MEF2-coregulator binding, prevented hypertrophy in cultured cardiac myocytes. 8MI prevented cardiac hypertrophy in 3 distinct stress models, and reversed established hypertrophy in vivo, associated with normalization of myocardial structure and function. The effects of 8MI were reversible, and did not prevent training effects of swimming. Mechanistically, 8MI blocked stress-induced MEF2 acetylation, nuclear export of class II histone deacetylases HDAC4 and -5, and p300 induction, without impeding HDAC4 phosphorylation. Correspondingly, 8MI transformed the transcriptional response to pressure overload, normalizing almost all 232 genes dysregulated by hemodynamic stress. We conclude that MEF2 acetylation is required for development and maintenance of pathological cardiac hypertrophy, and that blocking MEF2 acetylation can permit recovery from hypertrophy without impairing physiologic adaptation.
Keywords: Cardiology, Cell Biology
Keywords: Cell stress, Epigenetics, Heart failure
Interfering with stress-induced MEF2 acetylation prevents and reverses established cardiac hypertrophy and normalizes contractile function by remodeling the transcriptional response to stress.
Introduction
After birth, the size of the heart is tightly coupled to the size and metabolic requirements of the body. Hypertrophy is the normal mechanism for increasing heart size during postnatal growth and athletic training, but it also develops in response to sustained increases in workload caused by diseases such as hypertension and valve dysfunction (1). For reasons that remain unclear, these disorders elicit a dysfunctional hypertrophic response characterized by contractile dysfunction, myocardial stiffening, fibrosis, circulatory failure, and an increased risk of death (2). Hypertrophy is a frequent precursor of heart failure, especially heart failure with preserved ejection fraction (HFpEF), an extremely common and lethal condition for which there is no effective therapy (3).
Accumulating evidence suggests that hypertrophy may not be essential for successful adaptation to increased cardiac workload, and may in fact be directly harmful under conditions of sustained stress (4–11). At the same time, hypertrophy is increasingly appreciated not only as a herald of cardiac morbidity and mortality but an early stage of heart failure itself (12, 13), leading to calls for earlier intervention in the process (8). However, beyond agents that mitigate hypertension and other secondary causes of hypertrophy, few molecular tools are available to study the effects of inhibiting hypertrophy directly.
Cardiac hypertrophy is the result of a coordinated gene expression program enacted largely within postmitotic myocytes. This epigenetic response to extracellular stress promotes changes in sarcomeric protein production, diastolic relaxation, metabolism, calcium handling, and collagen synthesis, and is largely under the control of transcription factors in the stress-responsive muscle enhancer factor-2 (MEF2) family (14). The 4 vertebrate MEF2 isoforms feature an amino terminal MADS domain (present in MCM1, agamous, deficiens, and serum response factor) that mediates dimerization and binding to a canonical DNA motif (CTA(A/T)4TAG/A) and a MEF2-specific domain required for coregulatory factor binding, as well as C-terminal transcriptional activation domains. Although widely expressed, MEF2 promotes highly tissue- and developmental stage–specific gene expression in response to activating signals (15). MEF2 participates in the development and function of striated muscle, as well as in cells of immune, vascular, and neuronal origin (14), and regulates a large number of the genes involved in pathological hypertrophy, both directly and indirectly through other transcription factors and microRNAs (16).
MEF2 transcriptional activity is under tight temporal and spatial control, responding to cellular context and activation of certain intracellular signaling pathways, especially those that signal through calcium. The activity of MEF2 in turn determines its gene targets in a context-dependent manner. Under resting conditions, MEF2 is constitutively associated with the class IIa histone deacetylases (HDACs) HDAC4, -5, -7 and -9 (14, 17), which maintain MEF2 in a transcriptionally inactive state (18–20). In response to extracellular signals, posttranslational formation of ϵ-N-acetyl-lysine residues on MEF2 by lysine acetyltransferases (KATs) p300 and P/CAF (17, 21) promotes MEF2 DNA binding and transcriptional activation. This acetylation is aided by the signal-dependent phosphorylation of class IIa HDACs, which are then dissociated from MEF2 and exported into the cytosol (7, 22–24), allowing levels of both acetyl-MEF2 and p300 to increase markedly under conditions of pressure overload (25).
Deletion of either Mef2D (9) or p300 (25) blunts the development of hypertrophy, while deletion of class IIa HDACs potentiates hypertrophy, consistent with a model in which KATs and HDACs act in opposition. However, the extent to which MEF2 represents the key acetylation substrate in hypertrophy remains unclear. Nonselective HDAC inhibitors such as trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA) repress hypertrophy (26) through mechanisms that do not directly involve MEF2 (27, 28). The deacetylase activity of class IIa HDACs is in fact very weak (29), and these HDACs (-4, -5 and -9) may function more as acetyl-lysine readers that recruit other silencing molecules, including class I HDACs and the N-CoR/SMRT complex (30–32). These molecules in turn appear to silence MEF2 by deacetylase-independent mechanisms (20, 33–35).
Here we demonstrate that MEF2 acetylation is markedly increased in the failing human heart, and is required for myocyte hypertrophy. We show that a recently developed small-molecule probe designed to occupy the MEF2-coregulator interface blocks myocardial MEF2 acetylation in vivo, and effectively impedes and reverses cardiac myocyte hypertrophy in multiple stress models in vivo, restoring cardiac structure and function in the face of ongoing pressure overload. Mechanistically, we show that 8MI blocks the activation-induced nuclear export of class IIa HDACs and destabilizes p300, resulting in global remodeling of the hypertrophic transcriptome.
Results
Human myocardial analysis.
The acetylation state of MEF2 was determined in a series of human left ventricular (LV) myocardial samples, representing both nonfailing and failing hearts (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.91068DS1). MEF2 acetylation was elevated in all failing heart samples relative to nonfailing controls (Figure 1A), consistent with reported increases in p300 levels and MEF2-dependent transcription in human heart failure (25, 36).
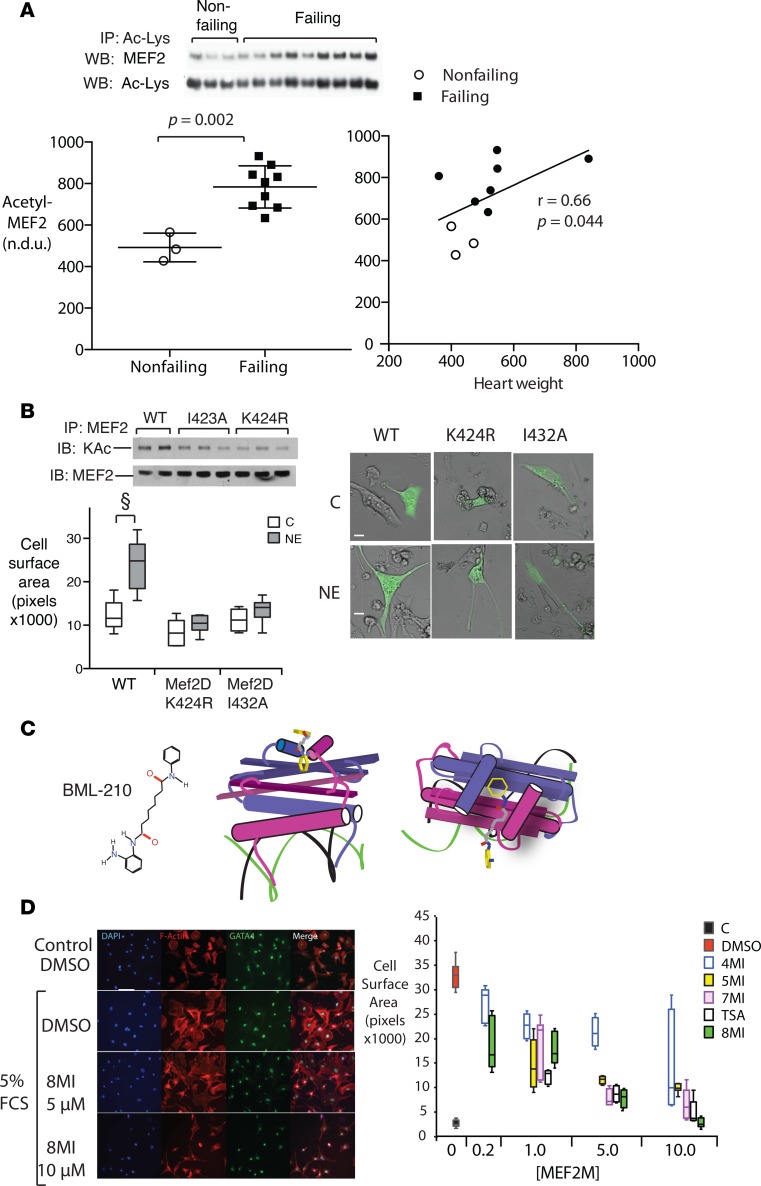
Figure 1. MEF2 acetylation is increased in human heart failure and required for cardiac myocyte hypertrophy in culture.
(A) Muscle enhancer factor-2 (MEF2) acetylation in heart failure. Human left ventricular myocardial samples were homogenized and subsequent lysates were immunoprecipitated with an anti–acetyl-lysine (anti–Ac-Lys) antibody as described (see Methods) and immunoblotted with antibodies against total MEF2 and Ac-Lys as a loading control. Above: Representative immunoblots. Below: Acetyl-MEF2 (normalized to Ac-Lys) (n = 14 subjects). Left: Individual nonfailing versus failing values plotted together with mean ± SEM. Exact 2-tailed P value was calculated using Mann-Whitney test. Right: Scatter plot of the same data showing correlation of acetyl-MEF2 with heart weight. Spearman r value and 2-tailed P were generated using Prism v.6 software. n.d.u., normalized densitometry units. (B) Acetylation-defective MEF2 mutants block endogenous MEF2 acetylation and hypertrophy in culture. Neonatal rat ventricular myocytes (NRVMs) expressing EGFP and WT MEF2 or 1 of 2 acetylation-defective MEF2D mutants (I423A or K424R) were cultured in the presence of 4 μM norepinephrine (NE) or its vehicle (C) for 48 hours. Above: Representative immunoblots showing MEF2 lysine acetylation in the presence of the indicated MEF2 expression vectors. Below left: Growth response to NE. Left: Representative merged brightfield and fluorescence images. Scale bars: 20 μm. Below right: Quantification of myocyte surface area. n = 3 independent experiments. Graph displays interquartile range ± SEM. (C) Chemical probe of MEF2-coactivator interface. Left: Chemical structure of parent molecule BML-210. Center and right: 3D structure views of BML-210 interaction with the MEF2-coregulator interface, side and top views, respectively. Green and black wires: DNA helices. Purple and magenta: MEF2 monomers. An interactive 3D version of these figures is provided in Supplemental File 1. (D) MEF2 modulation prevents serum-induced myocyte hypertrophy in culture. NRVMs were exposed to 5% fetal calf serum (FCS) in the presence of a series of BML-210 derivatives at the indicated concentrations (40) or their vehicle (DMSO). Left: Representative images. Scale bar: 50 μm. Right: Myocyte surface area was quantified using NIH ImageJ. Graph summarizes 3 separate experiments and displays interquartile range and SEM.
Dependence of myocyte hypertrophy on MEF2 acetylation.
Neonatal rat ventricular myocytes (NRVMs) were transiently transfected with WT MEF2 or 1 of 2 mutants: MEF2D K424R, which eliminates a lysine substrate of p300/CBP, and I423A, which targets an adjacent residue (20). Both MEF2 mutants significantly impaired MEF acetylation compared with WT MEF2 (Figure 1B) in response to norepinephrine, a potent hypertrophic stimulus (25, 37). Similarly, norepinephrine induced a near-doubling in size in myocytes expressing WT MEF2, but not in cells expressing either MEF2 mutant (Figure 1B), indicating a requirement for MEF2 acetylation.
MEF2-coregulator interaction is required for myocyte hypertrophy.
To dynamically modulate MEF2 acetylation, we exploited a series of molecules derived from BML-210, a pimeloylanilide o-aminoanilide (PAOA) compound, that were developed to bind the MADS-box/MEF2S domain of MEF2 competitively with p300 and HDAC4 on the MEF2 MADS domain (38–42) (Figure 1, C and D). BML-210 and its analogs were originally described as HDAC inhibitors, but have properties that are not reproduced by class I and II HDAC inhibitors such as TSA and SAHA (43), including the ability to block MEF2-dependent transcription by direct binding to MEF2 (40). Serum-starved NRVMs were placed in 5% fetal calf serum (FCS) in the presence of a series of BML-210 derivatives, the nonselective HDAC inhibitor TSA, or their vehicle DMSO, and myocyte size was determined at 48 hours. NRVMs increased substantially in surface area within 48 hours of FCS treatment (Figure 1E) in the presence of DMSO. The BML-210 analogs inhibited FCS-induced myocyte growth in a dose-dependent manner (Figure 1, E and F), with an order of potency resembling that of the same compounds in a MEF2 reporter assay (40). TSA also inhibited myocyte growth.
8MI prevents cardiac hypertrophy in vivo.
The most effective of the MEF2-binding compounds from the cell-based assay (designated 8MI) was tested in a model of moderate pressure overload. 8MI, or its vehicle as control, was administered over a range of concentrations immediately prior to and for 28 days following transverse aortic coarctation (TAC). At the end of 28 days TAC induced a 50.0% increase in normalized heart weight in control animals (heart weight/tibia length [HW/TL]) compared with mice undergoing a sham operation (Figure 2A). 8MI blunted this increase in a dose-dependent manner, essentially normalizing heart weight at the highest dose used (40 mg/kg). Similarly, echocardiographic wall thickness was increased by 35.9% ± 1.0% in control animals but only 6.9% ± 1.4% in mice at the highest dose of 8MI (Figure 2B). Treatment with 8MI also prevented ventricular remodeling (Figure 2C). TAC induced substantial renal and/or hepatic dysfunction in 2 of 3 DMSO-treated mice, but only in 1 of 9 mice receiving any dose of 8MI (Supplemental Table 2).
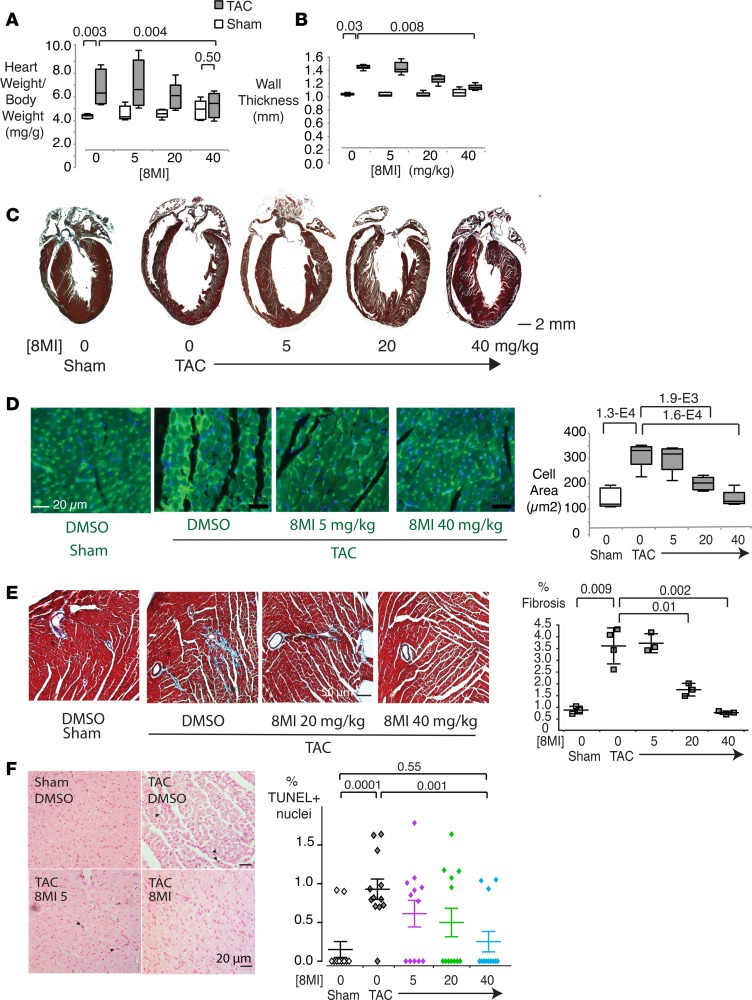
Figure 2. A muscle enhancer factor-2 (MEF2) modulator prevents cardiac hypertrophy in vivo.
(A) Normalization of cardiac mass following transverse aortic coarctation (TAC) in 8MI-treated mice. Heart weight to tibia length ratio (HW/TL) was determined in mice 21 days after TAC or a sham operation and receiving 8MI at the indicated doses (n = 4–5 per group). (B) Normalization of echocardiographic posterior wall thickness by 8MI. Measurements were taken in living mice treated as indicated between 20 and 21 days after TAC. (C) Normalization of cardiac geometry after TAC in 8MI-treated mice. Representative Masson’s trichrome–stained 4-chamber cross sections of hearts from mice treated as in A. Original magnification, ×10. (D) Normalization of myocyte size in vivo. Left: Representative wheat germ agglutinin (WGA)–stained sections of myocardium from mice treated as indicated. Right: Quantification of cell size in WGA-stained sections. N = 4 sections × 3 hearts per group. Graphs in A, B, and D display interquartile range and SEM. (E) Reduced fibrosis associated with pressure overload in 8MI-treated mice. Left: Masson’s trichrome stain of myocardium as in D. Original magnification, ×200. Right: Fibrotic area was quantified and expressed relative to total tissue area as described in Methods. n = 4 sections × 3 mice per group. (F) 8MI reduces apoptosis during pressure overload. Left: Myocardial sections at 21 days after TAC or sham operation. Arrows = TUNEL-stained nuclei. Right: Quantification. n = 20 sections × 3 mice per group. For graphs in E and F, scatter plots of individual measures are shown with mean ± SEM superimposed. P values are shown above the graphs. Two-tailed P values were computed by first performing 1-way ANOVA followed by the Dunn’s multiple comparison test, with significance set to 0.05.
Myocyte cross-sectional area was significantly induced by TAC (sham, 146 ± 17 vs. TAC, 319 ± 22 μm2) but this growth was eliminated by treatment with 8MI (Figure 2D). Ventricular fibrosis, prominent in mice 28 days after TAC, was eliminated by escalating doses of 8MI (Figure 2E). Pressure load–induced apoptosis was also reduced (Figure 2F). Similarly, a core group of cardiac hypertrophy–associated gene transcripts encoding collagen (Col1 and Col3), atrial natriuretic peptide (Nppa), B-type natriuretic peptide (Nppb), sarco-endoplasmic reticulum ATPase 2 (Atp2a2), and α- and β-myosin heavy chains (Myh6 and Myh7) were increased by TAC, and normalized in a dose-dependent manner by 8MI treatment (Supplemental Figure 1, A–F).
MEF2 modulation prevents contractile dysfunction.
Echocardiographic studies revealed normal cardiac function in all mice at baseline, and in sham-operated mice treated with either DMSO or 8MI at all doses (Supplemental Figure 2). As expected, TAC induced a 37% fall in ejection fraction (EF) at 28 days (sham, 82.4% ± 1.8% vs. TAC, 51.7% ± 5.1%, Supplemental Figure 2A). Treatment with 8MI treatment preserved systolic function in the face of sustained pressure overload, in a dose-dependent manner, with mice maintaining a near-normal EF of 75.4% at the highest dose of 8MI (Supplemental Figure 2A). Fractional shortening (FS) and stroke volume were depressed by TAC and were similarly restored in the presence of 8MI, while LV end-diastolic diameter (LVIDd), LV end-systolic diameter (LVIDs), and LV systolic and diastolic volumes diminished (Supplemental Figure 2, A–F, H), consistent with reduced remodeling as noted in Figure 2B. Heart rate was unaffected in all mice (Supplemental Figure 2G).
8MI reversibly blocks myocardial hypertrophy and dysfunction in a swimming stress model.
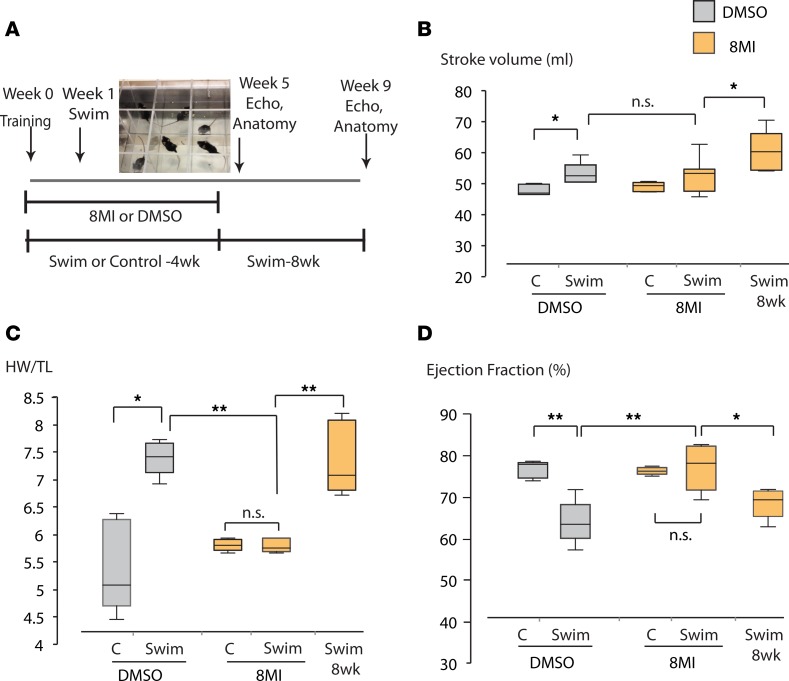
In a different model of stress-induced hypertrophy, forced swimming (Figure 3A), all swimming mice developed increased resting stroke volume over the course of the experiment, demonstrating a positive physical conditioning effect (44) that was unaffected by 8MI (Figure 3B). While swimming is often viewed as a pure model of exercise-induced hypertrophy, evidence from rat models suggests that it causes less increase in skeletal muscle blood flow, total cardiac output, and metabolic activity than treadmill exercise (45), while evoking a significant stress response reflected in elevated blood pressure (46), ACTH and cortisol (47), and plasma and tissue norepinephrine levels (48) compared with running or sedentary animals. Consistent with this, cardiac hypertrophy developed in swimming mice relative to briefly immersed controls (Figure 3C and Supplemental Table 1), accompanied by a substantial reduction in EF (Figure 3D). These pathological changes were completely prevented by 8MI, but subsequently developed after discontinuation of 8MI in mice that continued to swim (Figure 3, C and D).
Figure 3. Prevention of swimming stress–induced hypertrophy by 8MI requires continued treatment.
(A) Protocol outline. (B) Stroke volume increases with time. No difference is seen between vehicle- and 8MI-treated groups at 4 weeks. (C) Cardiac hypertrophy does not develop during treatment with 8MI, but proceeds after discontinuation of treatment at 4 weeks. HW/TL = left ventricular weight indexed to tibia length. (D) Systolic dysfunction associated with hypertrophy is prevented by 8MI. In a separate group, ejection fraction deteriorates over the 4 weeks after 8MI discontinuation. Graphs in B, C, and D display interquartile range and median. n = 5 mice per treatment group. *P < 0.05; **P < 0.01, calculated using the Mann-Whitney test. n.s., not significant.
8MI reverses established hypertrophy.
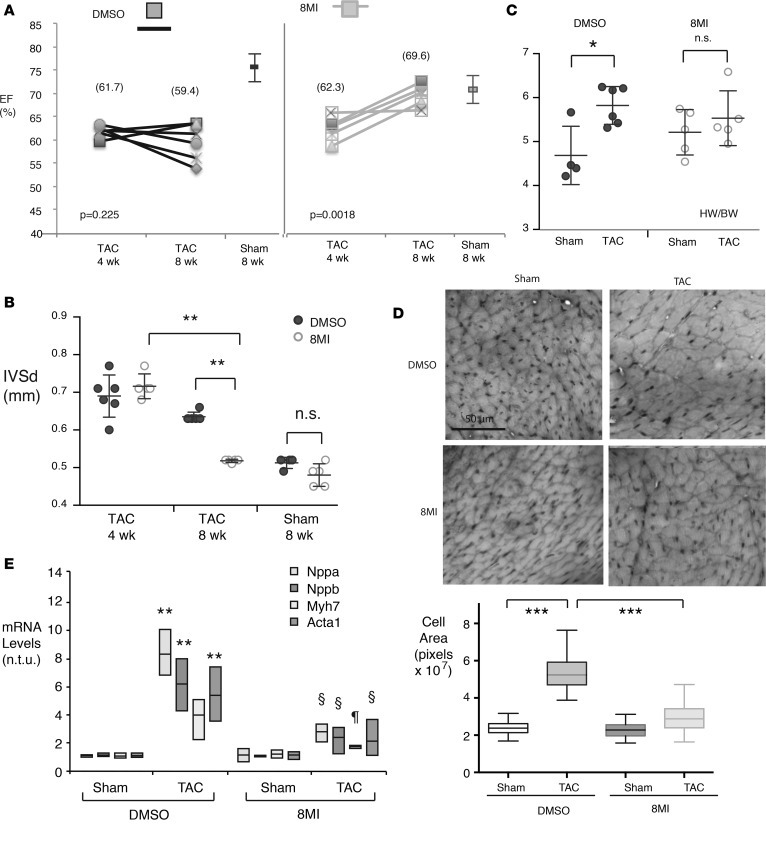
In a third experiment, mice underwent either a sham operation or aortic banding, and hypertrophy and contractile dysfunction were confirmed and quantified 4 weeks later by echocardiographic EF (Figure 4A), LV diastolic septal thickness (LVSd), and LVIDs (Figure 4B and Supplemental Figure 1). Beginning at postoperative week 5, 8MI or its vehicle was administered daily and continued until week 9. 8MI effectively normalized contractility and LV geometry relative to sham-operated mice, with no effect on heart rate (Figure 4, A and B, and Supplemental Figure 3). TAC mice receiving DMSO did not improve, and most exhibited further contractile dysfunction and remodeling (Figure 4, A and B, and Supplemental Figure 3). Normalized heart weights for 8MI-treated TAC and sham-operated animals were identical at 8 weeks, while in the DMSO group, TAC hearts were 20% larger than sham (Figure 4C). As expected, TAC was accompanied by a doubling of myocyte cross-sectional area in animals receiving DMSO; in contrast, after 4 weeks of 8MI, myocytes from TAC animals were only 29% larger than sham (Figure 4D). Interrogation of a panel of hypertrophy-associated genes revealed a marked blunting of the transcriptional response to TAC in animals after 4 weeks of 8MI. These observations collectively support the substantial reversal of established maladaptive hypertrophy after 4 weeks of 8MI administration.
Figure 4. 8MI reverses hypertrophy and associated contractile dysfunction.
Mice were subjected to transverse aortic coarctation (TAC) or a sham operation and treated with 8MI or its vehicle beginning 4 weeks later as described in Methods. n = 5 per group. (A) Contractile dysfunction is reversed by 8MI. Graph compares percentage ejection fraction (EF) for each TAC mouse at 4 and 8 weeks, with mean and SEM for sham-operated mice at 8 weeks, separated by treatment group. Note that EF remains low in TAC mice receiving DMSO, but improves in mice receiving 8MI. (B) 8MI reduces TAC-mediated heart weight increase. IVSd, interventricular septal thickness in diastole. (C) Septal hypertrophy is reversed by 8MI. Septal thickness was determined by echocardiography at 4 and 8 weeks. For B and C, values for individual mice are plotted together with mean ± SEM; n = 6. (D) Myocyte size is reduced in TAC mice receiving 8MI. Above: Masson’s trichrome–stained sections were obtained from the left ventricular free wall of mice from the indicated treatment groups. Scale bar: 50 μm. Below: Myocyte cross-sectional area was estimated using ImageJ. n = 100 cells × 3 fields for each condition. Graph shows interquartile range and SEM. ***P < 0.001. (E) 8MI normalizes hypertrophy-associated gene expression in mice after TAC. Data range and mean are plotted for each condition, n = 3 mice per group. For all graphs, *P < 0.05, **P < 0.01 for sham versus TAC, §P < 0.05 for TAC mice, 8MI versus DMSO (2-way ANOVA with post-hoc testing for multiple comparisons). n.t.u., normalized transcript units.
8MI treatment inhibits myocardial MEF2 acetylation in vivo.
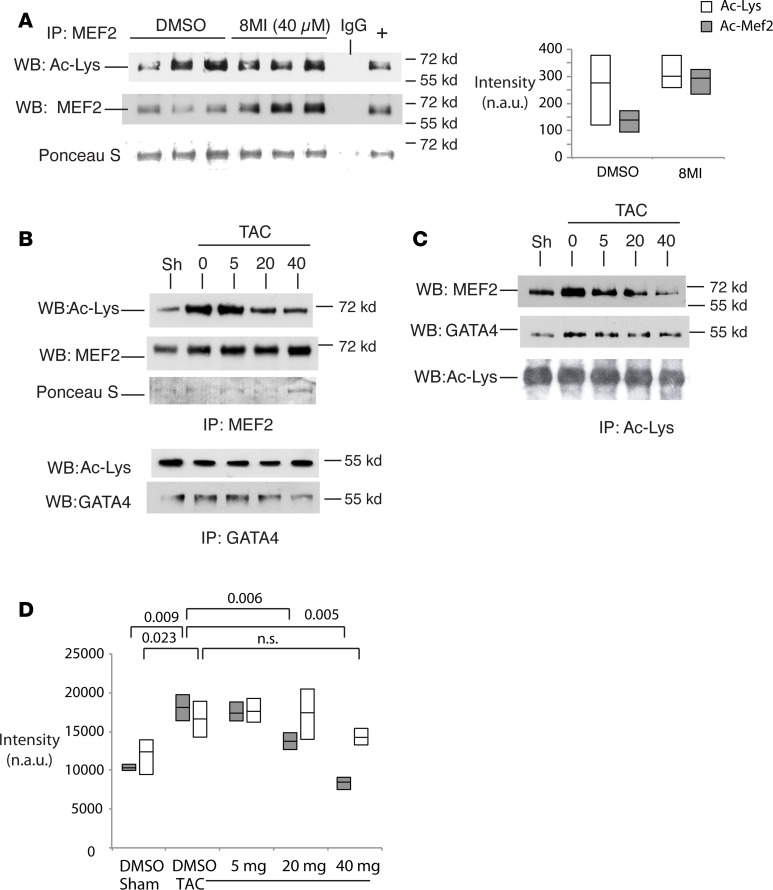
MEF2 acetylation was assessed in myocardial tissue from sham- and TAC-operated mice in the presence of vehicle or 8MI. 8MI did not affect total acetyl-MEF2 under sham conditions, although fractional MEF2 acetylation was reduced (Figure 5A). TAC increased both fractional (Figure 5B) and total (Figure 5C) acetyl-MEF2 levels, and this increase was blocked by 8MI in a dose-dependent manner. In contrast, TAC induction of total and acetyl-GATA4 (Figure 5, B and C) was not affected by 8MI (Figure 5, C and D).
Figure 5. 8MI decreases stress-induced muscle enhancer factor-2 (MEF2) acetylation.
(A) Acetyl-Mef2 (Ac-Mef2) levels in nonstressed murine hearts with and without 8MI. Mice were administered 8MI 40 mg/kg or its vehicle (DMSO) by i.p. injection for 4 weeks. Myocardial lysates were immunoprecipitated with anti–acetyl-lysine (anti–Ac-Lys) antibody and immunoblotted with antibodies against total Ac-Lys or MEF2. Ponceau S stain was used to confirm loading. Left: Representative Western blots. Right: Quantification. Graph displays full data range with mean. Open bars: Total Ac-Lys. Filled bars: Ac-Mef2. n = 3 per treatment group. (B and C) Mef2 acetylation is increased by transverse aortic coarctation (TAC) and reduced by 8MI. Mice were subjected to TAC or a sham operation, as described in Methods. Myocardial lysates were immunoprecipitated with (B) anti–pan-Mef2 and -Gata4 antibodies, or (C) anti–Ac-Lys antibody and probed with antibodies against Mef2, Gata4, or Ac-Lys as shown. (D) Quantification of data as in C normalized to total Ac-Lys. Dark bars: Ac-Mef2. Light bars: Ac-Gata4. Graph displays full data range with mean. n = at least 3 biological replicates; exact P values are indicated (2-way ANOVA with post-hoc testing for multiple comparisons). n.s., not significant; n.a.u., normalized arbitrary units.
8MI promotes MEF2 silencing through loss of p300 and nuclear retention of class IIa HDACs.
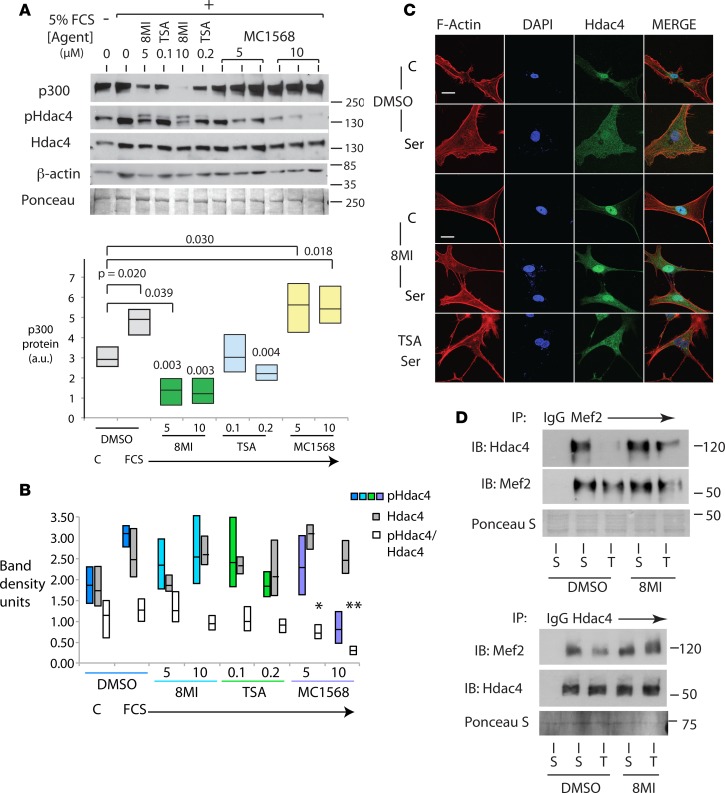
We next asked whether the effects of 8MI on hypertrophy were related to its action on MEF2-coregulator interactions, as predicted by our earlier data (40), and compared the effects of broad-spectrum (TSA) and class IIa–selective (MC1568) HDAC inhibitors, using experimentally validated concentrations sufficient to block hypertrophy in a cell-based assay (ref. 49 and data not shown). In cultured NRVMs stimulated with FCS, 8MI prevented the expected rise in p300 levels, and in fact p300 protein levels fell significantly compared with those in resting cells (Figure 6A). Both TSA and MC1568 blocked hypertrophy, but MC1568 had no effect on p300 levels. TSA prevented p300 induction, but did not affect baseline p300 levels (Figure 6A).
Figure 6. 8MI and HDAC inhibition prevent cardiac hypertrophy by different mechanisms.
(A) 8MI blocks induction of p300 by serum. Neonatal rat ventricular myocytes (NRVMs) were exposed to 5% fetal calf serum (FCS) in the presence of 8MI (5 and 10 μM), trichostatin A (TSA; 0.1 and 0.2 μM), MC1568 (5 and 10 μM), or their vehicle (0) as indicated. Control cells received fresh serum-free media (TIB). Protein levels of p300 and β-actin were assayed by immunoblot. Above: Representative immunoblots. Below: Quantification of 3 independent experiments. n = at least 3 biological replicates; exact P values are indicated (2-way ANOVA with post-hoc testing for multiple comparisons). (B) 8MI does not prevent serum-induced phosphorylation of histone deacetylase 4 (Hdac4). Quantification of p-Ser-Hdac4 (p-Hdac4) and Hdac4 immunoblots as shown in A. Graph displays full data range with mean. n = 3; *P < 0.05, **P < 0.01 (2-way ANOVA with post-hoc testing for multiple comparisons). (C) 8MI blocks nuclear export of Hdac4. Serum-starved NRVMs were fed with 5% FCS or fresh serum-free media in the presence of 8MI, TSA, or their vehicle DMSO as indicated. Hdac4, F-actin (Actin), and nuclear DNA (DAPI) were visualized as described in Methods. Scale bars: 20 μm. Representative images shown; n = 3 independent experiments. (D) 8MI reverses effects of stress on Hdac-Mef2 interaction. Myocardial lysates from sham-operated mice (S) and mice subjected to transverse aortic coarctation (T) were immunoprecipitated with anti-Mef2 (upper panel) or anti-Hdac4 (lower panel) antibodies and immunoblotted with Hdac4 and Mef2 as indicated. Anti-IgG was used as a control (IgG). Ponceau S staining was used to confirm protein loading. Representative blots of 3 independent experiments.
In response to growth signaling, class IIa HDACs undergo phosphorylation that promotes their export into the cytosol (50). Levels of both total and p-Ser498-HDAC4 increased in response to FCS (Figure 6, A and B). MC1568 significantly reduced both total and relative HDAC4 phosphorylation (Figure 6, A and B). In 8MI-treated cells, total p-HDAC4 was not reduced, and a second, more slowly migrating p-HDAC4 band was observed (Figure 6A), suggesting that 8MI may induce additional posttranslational modifications in p-HDAC4 that alter its protein-protein interactions or subcellular location. Consistent with this, immunofluorescence studies demonstrated that serum-induced translocation of both Hdac4 and Hdac5 did not occur in 8MI-treated cells (Figure 6C and Supplemental Figure 3), while TSA had no effect at an equivalent antihypertrophic concentration (10 nM, Figure 6C).
We next examined the interaction between MEF2 and HDAC4 in reciprocal coimmunoprecipitation assays of whole-cell lysates from LV myocardium (Figure 6D). Under these conditions, the nuclear membrane is disrupted and does not bar HDAC4-MEF2 reassociation in vitro. In lysates from pressure-overloaded myocardium, HDAC4 binding to MEF2 was sharply impaired relative to sham-operated myocardium (Figure 6D). By contrast, HDAC4 from 8MI-treated myocardium largely retained the ability to associate with MEF2. Thus, it appears that 8MI blocks HDAC4 posttranslational modifications or protein associations that normally preclude its interaction with MEF2 under stress.
8MI transforms the transcriptional response to pressure overload.
To determine the extent to which 8MI produced selective silencing of MEF2-dependent transcription, we used RNA sequencing (RNA-Seq) followed by gene set enrichment analysis (GSEA). We analyzed LV muscle from mice 4 weeks after TAC or a sham operation, accompanied by daily oral administration of 8MI or its vehicle (blank) by gavage.
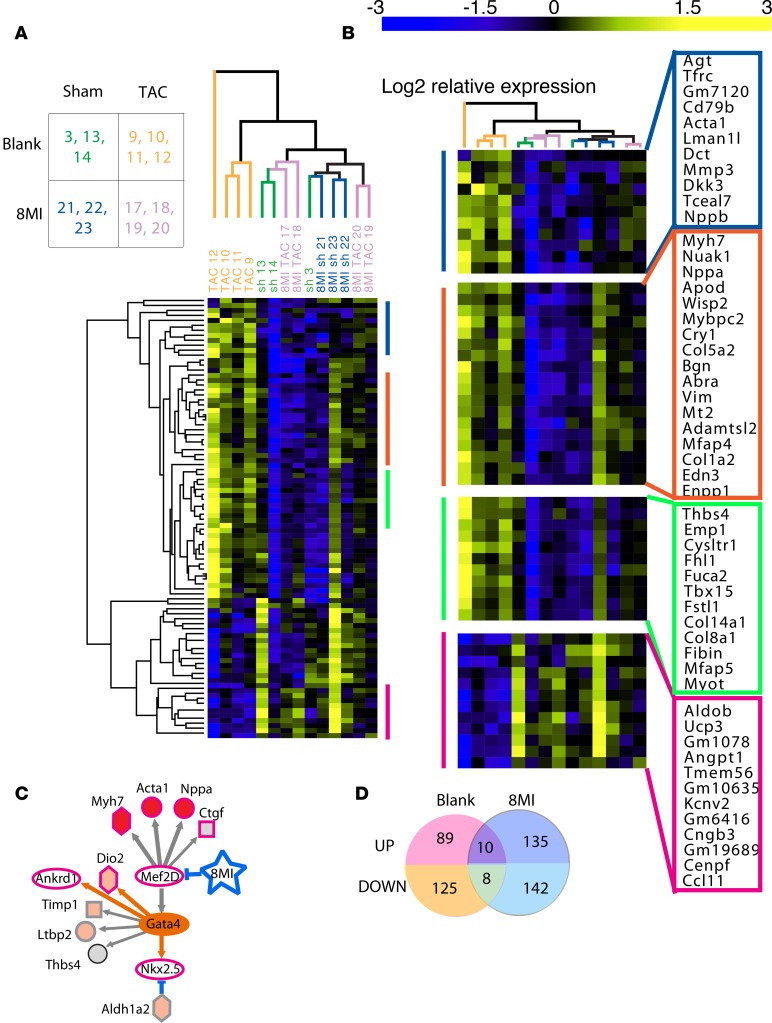
Unsupervised hierarchical clustering demonstrated that only the TAC-operated animals treated with the blank solution were clearly identifiable as a separate group based on gene expression (Figure 7A). Sham and TAC animals receiving 8MI sorted indiscriminately, because 8MI treatment caused a widespread reversal of the hypertrophic gene expression profile. In addition to genes previously surveyed individually by reverse transcription-PCR (RT-PCR) (Supplemental Figure 1), we observed a number of previously reported and potentially novel hypertrophy-associated transcripts (Figure 7B). Four subclusters are annotated in Supplemental Table 3. Cluster 1 includes Acta1 and Nppb; cluster 2 contains Nppa and Col1a1; both are normalized by 8MI. Cluster 4 contains transcripts that are repressed in hypertrophy, and normalized by 8MI. Cluster 3 contains some genes, including Fstl1, that are upregulated both in the presence and absence of 8MI.
Figure 7. RNA-Seq reveals widespread reversal of hypertrophic transcription by muscle enhancer factor-2 (MEF2) silencing.
Analysis pipeline is described in Methods. (A) Unsupervised hierarchical cluster analysis of the cardiac transcriptome after transverse aortic coarctation (TAC) or a sham operation followed by 4 weeks of daily treatment with 8MI or its vehicle (blank) by gavage. Differentially up- and downregulated genes in left ventricle are shown; 1 column = 1 mouse; n = 3 per group. Visualization and analysis were done in GENE-E (https://software.broadinstitute.org/GENE-E/index.html). The full data set is provided as Supplemental File 2. (B) Expanded view of 4 selected clusters. See gene annotations in Supplemental Table 3. (C) Ingenuity Network analysis implicates MEF2 as 8MI target. See Supplemental Table 4 for complete set of identified networks. (D) 8MI transforms the transcriptional response to stress. Note little overlap of stress-regulated transcripts between 8MI and blank-treated mice. Venn diagrams generated using tools at VIB, Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Pathway analysis of the genes in clusters 1–3 showed that of 27 regulatory pathways with P less than 0.005, Tgfb1 was the largest, and the highest-ranked was Gata4, consistent with its global role in cardiac gene expression. Five pathways pointed to MAPK signaling and effectors (Raf1, Map3k7, Map2k3, Map2k6, and Mapk8). However, the largest number of pathways (seven) implicated the MEF2 network putatively interrupted by 8MI (Hdac5, Hdac9, Camk2d, Mef2D, Mef2C, Mef2A, and Crebbp/p300) (Supplemental Table 4).
GSEA was used to compare promoter features of the stress-induced, 8MI-normalized genes with Motif gene sets in the Molecular Signatures Database (MSigDB) (51, 52). This analysis revealed significant overlap with MEF2 motif gene sets (MEF2, MEF2A, each FDR q value = 0.005, Supplemental Table 5) along with a motif (TGGNNNNNNKCCAR) complementary to a binding site for MEF2 in the human ACTA1 promoter (FDR q = 4.73 × 10–5; see ref. 53). We also identified Jun/AP1 and TEF1 motifs previously associated with cardiac myocyte hypertrophy (54, 55), confirming the validity of the approach. These motifs were absent from the genes upregulated by 8MI (Supplemental Table 5).
Discussion
Epigenetic modification of MEF2 as a key to hypertrophy.
Epigenetic modifiers that write, erase, or read acetyl-lysine marks have emerged as important regulators of cardiac hypertrophy (25, 56–59), although the substrates for these modifications are not well established. Acetyl-lysines are binding sites for bromodomain-containing proteins such as p300 (60) and the bromodomain and extraterminal (BET) proteins (Brd 2–4). Acetylation of MEF2 likely stabilizes its interaction with these acetyl-lysine receptors (57–59). The BET inhibitor JQ-1 has recently been shown to inhibit pressure overload hypertrophy (58, 59), suggesting that BET proteins act as hypertrophy mediators downstream of the MEF2 acetylation signal.
A small MEF2-binding molecule enforces MEF2 deacetylation.
The acetylation state of MEF2 is determined by dynamic interactions with coregulators that have either intrinsic or associated acetylation or deacetylation activities, including p300, class IIa HDACs, and cabin1, which share a common cofactor binding site on the MADS-box/MEF2S domain of MEF2 (38, 39, 41, 42). Our data indicate that by binding to this site, 8MI locks MEF2 in a deacetylated state, through a mechanism that depletes the cell of p300 and disrupts activation-induced nuclear export of class IIa HDACs. The decoupled HDACs appear to undergo additional posttranslational modifications in the nucleus, and likely retain the ability to bind MEF2 in a dynamic equilibrium with 8MI, thereby maintaining MEF2 in a deacetylated state.
p300 depletion by 8MI.
By blocking the stress-mediated induction of p300, and reducing basal p300 content, 8MI further tilts the balance in favor of MEF2 deacetylation. We previously showed that myocyte p300 is stabilized by autoacetylation, allowing it to accumulate rapidly during oxidant stress (61). Autoacetylation is normally kept in check by p300-associated HDACs; accordingly, both class 1– and class 2–selective as well as non–class-selective HDAC inhibitors have been shown to increase p300 acetylation (61–63) and accumulation (61, 62). Depletion of p300 is a unique property of 8MI that is inconsistent with catalytic HDAC inhibition and likely attributable instead to retention of nuclear HDAC activity.
It remains unclear how 8MI leads to HDAC4 nuclear retention. A number of lines of evidence suggest that class I/II HDAC–mediated transcriptional repression is linked to higher-order chromatin organization, which controls gene silencing in specific subnuclear domains. In mammalian cells, silenced chromatin localizes to a zone of hypoacetylation at the nuclear periphery (64), and class II HDACs have been shown to bind to inner nuclear membrane proteins involved in gene silencing (65, 66). We previously provided structural data that imply that HDAC4-MEF2 repressor complexes influence higher-order chromatin organization (67). It is interesting to speculate that this higher-order structure, which may be sensitive to 8MI, efficiently couples HDAC4-MEF2 complexes to nuclear export machinery.
Dispensability of the hypertrophic program.
The near-complete remodeling of the transcriptional response to stress in animals receiving 8MI demonstrates the important role of MEF2 in this process. Of the 232 genes responding to pressure overload, 214 became unresponsive in the presence of 8MI, are enriched in MEF2 motifs, and participate in MEF2-regulated networks. These genes include the canonical hypertrophy-associated Acta1, Myh7, Nppa, and Nppb. Only 18 were similarly regulated in the presence and absence of 8MI, notably including follistatin-like 1 (Fstl1), an injury-induced glycoprotein with potent cytoprotective and regenerative properties in the myocardium (68, 69). Our findings demonstrate that appropriate targeting of the harmful stress pathway mediated by MEF2 activation can spare essential components of the adaptive response and enhance cardiac function under adverse conditions.
While 8MI is not predicted to be MEF2 isoform–selective, it is likely to be selective for stress-activated MEF2 transcription. All MEF2 isoforms can target the same enhancer elements, and hence important functional differences must be conferred through cellular context– and stage-specific differences in timing of expression, and more importantly in binding affinity, activity, and availability of their coregulators (15, 19, 32, 70–73). Modulation of cell-specific MEF2-coregulator interactions could confer localized action of 8MI in tissues under stress such as the failing heart (36, 74) and in hypertrophic cardiomyopathy (75), where MEF2-dependent transcription may be excessively and pathologically activated.
We show here that blocking MEF2 acetylation both interrupts hypertrophy and preserves cardiac function, adding to the weight of evidence that stress-induced MEF2 activation in the heart is not only dispensable but also potentially injurious. Our observation that 8MI can reverse pathological hypertrophy in the face of sustained mechanical loading suggests that stress-induced damage is ongoing and dynamic, and a potential target for therapeutic intervention. When stress signaling is turned off, either by removal of the original stress, or by molecular intervention as illustrated here, the myocardium may have considerable ability to repair itself. Since hypertrophy is associated with many conditions that are poorly amenable to mechanical correction, such as diabetes, hypertension, and aging, the availability of chemical probes provides a unique opportunity to explore targeting the hypertrophic process as a way to treat cardiac dysfunction, as well as to study the role of MEF2 activity in these diseases.
Methods
Please also refer to Supplement 1, Extended Experimental Procedures.
Reagents.
Expression vectors encoding acetylation-defective or WT MEF2D were the gifts of Tso-Pang Yao (Duke University, Durham, North Carolina, USA) and have been previously described (20). 8MI and analogs were obtained through a Materials Transfer Agreement with C&C BioPharma, LTD. A full list of the antibodies and other reagents used for this study is provided in Extended Experimental Procedures.
Human tissue samples.
Anonymized LV myocardial samples and associated clinical information were harvested within 4 hours after demise or explantation and obtained through the Cooperative Human Tissue Network under a protocol approved by the University of Miami Institutional Review Board. Samples were maintained at –80°C until use.
Myocyte cell culture.
Primary neonatal rat ventricular cardiomyocyte cultures were prepared from the hearts of 1- to 3-day-old neonatal rat pups and transfected using a calcium phosphate method as previously described (37). Final cultures were greater than 90% myocytes.
Murine surgery and imaging.
Cardiac hypertrophy was induced by TAC as described previously, with minor variations (25). In the TAC prevention protocol, 8MI was administered via i.p. injection daily for 35 days beginning 1 week prior to surgical intervention. In the reversal protocol, daily i.p. injection of 8MI was initiated beginning 4 weeks after TAC and continued for 4 more weeks. Cardiac function and morphology were evaluated using a Visual Sonics 770 High Resolution Imaging System.
Swimming stress model.
We used a previously established protocol for induction of hypertrophy with swimming exercise, with minor variations (76). Mice were required to swim twice daily for 90 minutes, 5 hours apart. Control mice were immersed briefly. Mice received DMSO or 8MI (40 mg/kg i.p.) daily, beginning 1 week prior to swimming. At the end of 4 weeks, mice underwent functional and anatomical analysis as above. A subgroup of 8MI-treated mice was continued on the swimming protocol for an additional 4 weeks without further administration of the compound.
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and visualized with antibodies against HDAC-4 and- 5 and an Alexa Fluor 488–conjugated secondary antibody using a Zeiss LSM 700 confocal microscope. Images were acquired using Zen 2009 Ver. 5.5, SP2 (Zeiss).
Western blot and qPCR analysis.
Protein lysates were resolved in SDS-PAGE and transferred to nitrocellulose membranes. Proteins were detected by incubation with appropriate primary antibodies and HRP-conjugated secondary antibodies, and visualized by chemiluminescence. For quantitative PCR (qPCR), total RNA was analyzed on an ABI 7900HT Fast Real-Time PCR System using TaqMan primers (Thermo Fisher Scientific), and expression levels were normalized to 18S rRNA. All samples were run in duplicate.
RNA-Seq.
Total RNA samples from mouse LVs (n = 3–4 per treatment group) were prepared for sequencing using Illumina TruSeq total strand-specific RNA-Seq library preps and sequenced as 75-bp single reads on Illumina NextSeq V2 Flow Cells. Alignment of RNA-Seq reads to the mouse genome (Mus musculus UCSC mm10) was accomplished with TopHat and Cufflinks 2.1.1. Analysis of differential gene expression, GSEA, and heatmap visualization were performed using DESeq2, GSEA, and GENE-E software, respectively, as described in Extended Experimental Procedures. Gene Ontology and Network analysis was performed using Ingenuity software (http://www.ingenuity.com). All sequencing data have been submitted to NCBI under BioProject ID PRJNA393500.
Statistical and bioinformatics analysis.
Repeated-measures 2-way analysis of variance (ANOVA), with correction for multiple comparisons, and Mann-Whitney P values were determined using Prism (GraphPad) v.6 software. R values were determined using nonparametric Spearman correlation in Prism. Acceptance of significance was set at P less than 0.05 using 2-tailed parameters. All experimental groups had at least 3 animals. Differentially regulated transcripts were quantified and ranked for nominal significance, with 2-tailed P cutoffs of less than 0.01 for transcripts with fold change of greater than 0.70 or less than 1.5, and less than 0.05 for transcripts with fold change of 0.07 or less or 1.5 or higher. Details of RNA-Seq statistical analysis are provided in Extended Methods in the supplement.
Human studies.
All human studies were performed on anonymized tissues under protocols approved by the University of Miami Institutional Review Board.
Animal studies.
All experiments were performed on 2- to 3-month-old WT C57BL/6 mice under protocols approved by the Institutional Animal Care and Use Committee of the University of Miami.
Author contributions
LC and NHB conceptualized the study. LC, JW, S. Jain, SS, YW, NJ, XL, and NHB developed the methodologies. JW, SS, CC, GY, S. Joshi, MH, and YW conducted the investigation. NHB and S. Joshi wrote the original draft of the manuscript. NHB, S. Joshi, JW, and LC reviewed and edited the manuscript. NHB and LC acquired funding. LC and DG provided chemical probes used for this study. NHB and XL performed bioinformatic data analyses. NHB supervised the study.
Supplementary Material
Acknowledgments
We are indebted to Tso-Pang Yao for provision of critical reagents for this project, and to Claes Wahlestedt for helpful comments and advice. We thank Jing Liu for excellent technical assistance during the initiation of the project. This work was supported by grants from the NHLBI (NIH R01 HL071094, to N.H.B.) and the Miami Heart Research Institute (to N.H.B.). Shaurya Joshi is the recipient of an American Heart Association Greater Southeastern Affiliate Predoctoral Award.
Version 1. 08/29/2017
Electronic publication
Footnotes
NJ’s present address is: Department of Chemistry Faculty of Science University of Kelaniya Kelaniya, Sri Lanka.
Conflict of interest: L. Chen is the scientific founder, and N.H. Bishopric and L. Chen are scientific advisors and board members, of C&C BioPharma, Ltd., which is investigating drug-like derivatives of the 8MI chemical probe of the MEF2 coactivator interface used in this study. L. Chen, D. Gai, and N.H. Bishopric have been granted minority interests in C&C BioPharma, and the University of Miami and C&C BioPharma have patents pending on the potential of these probes in the treatment of cardiovascular diseases.
Reference information: JCI Insight. 2017;2(16):e91068. https://doi.org/10.1172/jci.insight.91068.
Contributor Information
Shaurya Joshi, Email: joshishaurya@yahoo.com.
Svetlana Speransky, Email: sxs713@med.miami.edu.
Christopher Crowley, Email: christopher.n.crowley@gmail.com.
Nimanthi Jayathilaka, Email: njayathi@gmail.com.
Xiao Lei, Email: xiaolei@usc.edu.
Yongqing Wu, Email: yongqingwu8@gmail.com.
David Gai, Email: gdh9999@gmail.com.
Sumit Jain, Email: sumitjain1484@gmail.com.
Michael Hoosien, Email: HOOSIEM@ccf.org.
Yan Gao, Email: gaoyan_0222@163.com.
Lin Chen, Email: linchen@usc.edu.
Nanette H. Bishopric, Email: nbishopric@med.miami.edu.
References
- 1.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358(13):1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.Cohn JN, Ferrari R, Sharpe N. Cardiac remodeling--concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. Behalf of an International Forum on Cardiac Remodeling. J Am Coll Cardiol. 2000;35(3):569–582. doi: 10.1016/S0735-1097(99)00630-0. [DOI] [PubMed] [Google Scholar]
- 3.Shah AM, et al. Cardiac structure and function in heart failure with preserved ejection fraction: baseline findings from the echocardiographic study of the Treatment of Preserved Cardiac Function Heart Failure with an Aldosterone Antagonist trial. Circ Heart Fail. 2014;7(1):104–115. doi: 10.1161/CIRCHEARTFAILURE.113.000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heineke J, et al. CIB1 is a regulator of pathological cardiac hypertrophy. Nat Med. 2010;16(8):872–879. doi: 10.1038/nm.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, et al. GDF15/MIC-1 functions as a protective and antihypertrophic factor released from the myocardium in association with SMAD protein activation. Circ Res. 2006;98(3):342–350. doi: 10.1161/01.RES.0000202804.84885.d0. [DOI] [PubMed] [Google Scholar]
- 6.Hill JA, et al. Cardiac hypertrophy is not a required compensatory response to short-term pressure overload. Circulation. 2000;101(24):2863–2869. doi: 10.1161/01.CIR.101.24.2863. [DOI] [PubMed] [Google Scholar]
- 7.Backs J, et al. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci U S A. 2009;106(7):2342–2347. doi: 10.1073/pnas.0813013106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109(13):1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y, et al. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest. 2008;118(1):124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antos CL, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278(31):28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 11.Meijs MF, et al. Left ventricular hypertrophy: a shift in paradigm. Curr Med Chem. 2007;14(2):157–171. doi: 10.2174/092986707779313354. [DOI] [PubMed] [Google Scholar]
- 12.Peterson GE, et al. Relationship of left ventricular hypertrophy and diastolic function with cardiovascular and renal outcomes in African Americans with hypertensive chronic kidney disease. Hypertension. 2013;62(3):518–525. doi: 10.1161/HYPERTENSIONAHA.111.00904. [DOI] [PubMed] [Google Scholar]
- 13.Kannel WB. Incidence and epidemiology of heart failure. Heart Fail Rev. 2000;5(2):167–173. doi: 10.1023/A:1009884820941. [DOI] [PubMed] [Google Scholar]
- 14.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134(23):4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 15.Naya FJ, Wu C, Richardson JA, Overbeek P, Olson EN. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development. 1999;126(10):2045–2052. doi: 10.1242/dev.126.10.2045. [DOI] [PubMed] [Google Scholar]
- 16.Liu N, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104(52):20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17(2):1010–1026. doi: 10.1128/MCB.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18(18):5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol Cell Biol. 2005;25(9):3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25(19):8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10(19):2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 22.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97(8):4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001;21(18):6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vega RB, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24(19):8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei JQ, et al. Quantitative control of adaptive cardiac hypertrophy by acetyltransferase p300. Circulation. 2008;118(9):934–946. doi: 10.1161/CIRCULATIONAHA.107.760488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Y, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113(22):2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao DJ, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108(10):4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13(3):324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 29.Bradner JE, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6(3):238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21(18):6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grégoire S, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27(4):1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang H, Choi DE, Kim H, Cho EJ, Youn HD. Cabin1 represses MEF2 transcriptional activity by association with a methyltransferase, SUV39H1. J Biol Chem. 2007;282(15):11172–11179. doi: 10.1074/jbc.M611199200. [DOI] [PubMed] [Google Scholar]
- 33.Lomonte P, Thomas J, Texier P, Caron C, Khochbin S, Epstein AL. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J Virol. 2004;78(13):6744–6757. doi: 10.1128/JVI.78.13.6744-6757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52(6):769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14(1):55–66. [PMC free article] [PubMed] [Google Scholar]
- 36.Putt ME, et al. Evidence for coregulation of myocardial gene expression by MEF2 and NFAT in human heart failure. Circ Cardiovasc Genet. 2009;2(3):212–219. doi: 10.1161/CIRCGENETICS.108.816686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishopric NH, Kedes L. Adrenergic regulation of the skeletal alpha-actin gene promoter during myocardial cell hypertrophy. Proc Natl Acad Sci U S A. 1991;88(6):2132–2136. doi: 10.1073/pnas.88.6.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, et al. Structure of the MADS-box/MEF2 domain of MEF2A bound to DNA and its implication for myocardin recruitment. J Mol Biol. 2010;397(2):520–533. doi: 10.1016/j.jmb.2010.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han A, Pan F, Stroud JC, Youn HD, Liu JO, Chen L. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature. 2003;422(6933):730–734. doi: 10.1038/nature01555. [DOI] [PubMed] [Google Scholar]
- 40.Jayathilaka N, et al. Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2. Nucleic Acids Res. 2012;40(12):5378–5388. doi: 10.1093/nar/gks189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han A, He J, Wu Y, Liu JO, Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J Mol Biol. 2005;345(1):91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 42.He J, et al. Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly. Nucleic Acids Res. 2011;39(10):4464–4474. doi: 10.1093/nar/gkr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat Chem Biol. 2006;2(10):551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 44.Schaible TF, Scheuer J. Effects of physical training by running or swimming on ventricular performance of rat hearts. J Appl Physiol Respir Environ Exerc Physiol. 1979;46(4):854–860. doi: 10.1152/jappl.1979.46.4.854. [DOI] [PubMed] [Google Scholar]
- 45.Flaim SF, Minteer WJ, Clark DP, Zelis R. Cardiovascular response to acute aquatic and treadmill exercise in the untrained rat. J Appl Physiol Respir Environ Exerc Physiol. 1979;46(2):302–308. doi: 10.1152/jappl.1979.46.2.302. [DOI] [PubMed] [Google Scholar]
- 46.Cox RH. Exercise training and response to stress: insights from an animal model. Med Sci Sports Exerc. 1991;23(7):853–859. [PubMed] [Google Scholar]
- 47.Contarteze RV, Manchado Fde B, Gobatto CA, De Mello MA. Stress biomarkers in rats submitted to swimming and treadmill running exercises. Comp Biochem Physiol, Part A Mol Integr Physiol. 2008;151(3):415–422. doi: 10.1016/j.cbpa.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Geenen D, Buttrick P, Scheuer J. Cardiovascular and hormonal responses to swimming and running in the rat. J Appl Physiol. 1988;65(1):116–123. doi: 10.1152/jappl.1988.65.1.116. [DOI] [PubMed] [Google Scholar]
- 49.Nebbioso A, et al. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009;10(7):776–782. doi: 10.1038/embor.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97(26):14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slepak TI, et al. Control of cardiac-specific transcription by p300 through myocyte enhancer factor-2D. J Biol Chem. 2001;276(10):7575–7585. doi: 10.1074/jbc.M004625200. [DOI] [PubMed] [Google Scholar]
- 54.Bishopric NH, Jayasena V, Webster KA. Positive regulation of the skeletal alpha-actin gene by Fos and Jun in cardiac myocytes. J Biol Chem. 1992;267(35):25535–25540. [PubMed] [Google Scholar]
- 55.MacLellan WR, Lee TC, Schwartz RJ, Schneider MD. Transforming growth factor-beta response elements of the skeletal alpha-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J Biol Chem. 1994;269(24):16754–16760. [PubMed] [Google Scholar]
- 56.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hang CT, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spiltoir JI, et al. BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J Mol Cell Cardiol. 2013;63:175–179. doi: 10.1016/j.yjmcc.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anand P, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154(3):569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eckner R, et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8(8):869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 61.Jain S, Wei J, Mitrani LR, Bishopric NH. Auto-acetylation stabilizes p300 in cardiac myocytes during acute oxidative stress, promoting STAT3 accumulation and cell survival. Breast Cancer Res Treat. 2012;135(1):103–114. doi: 10.1007/s10549-012-2069-6. [DOI] [PubMed] [Google Scholar]
- 62.Kim SH, Kang HJ, Na H, Lee MO. Trichostatin A enhances acetylation as well as protein stability of ERalpha through induction of p300 protein. Breast Cancer Res. 2010;12(2):R22. doi: 10.1186/bcr2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baertschi S, Baur N, Lueders-Lefevre V, Voshol J, Keller H. Class I and IIa histone deacetylases have opposite effects on sclerostin gene regulation. J Biol Chem. 2014;289(36):24995–25009. doi: 10.1074/jbc.M114.564997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- 65.Somech R, et al. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118(Pt 17):4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 66.Zhang CL, McKinsey TA, Olson EN. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc Natl Acad Sci U S A. 2001;98(13):7354–7359. doi: 10.1073/pnas.131198498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo L, Han A, Bates DL, Cao J, Chen L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc Natl Acad Sci U S A. 2007;104(11):4297–4302. doi: 10.1073/pnas.0608041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oshima Y, Ouchi N, Sato K, Izumiya Y, Pimentel DR, Walsh K. Follistatin-like 1 is an Akt-regulated cardioprotective factor that is secreted by the heart. Circulation. 2008;117(24):3099–3108. doi: 10.1161/CIRCULATIONAHA.108.767673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei K, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525(7570):479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu J, Gong NL, Bodi I, Aronow BJ, Backx PH, Molkentin JD. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J Biol Chem. 2006;281(14):9152–9162. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 71.Grégoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25(6):2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ginnan R, Sun LY, Schwarz JJ, Singer HA. MEF2 is regulated by CaMKIIδ2 and a HDAC4-HDAC5 heterodimer in vascular smooth muscle cells. Biochem J. 2012;444(1):105–114. doi: 10.1042/BJ20120152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6(2):233–244. doi: 10.1016/S1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 74.Cortés R, et al. Differences in MEF2 and NFAT transcriptional pathways according to human heart failure aetiology. PLoS One. 2012;7(2):e30915. doi: 10.1371/journal.pone.0030915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Konno T, et al. Heterogeneous myocyte enhancer factor-2 (Mef2) activation in myocytes predicts focal scarring in hypertrophic cardiomyopathy. Proc Natl Acad Sci U S A. 2010;107(42):18097–18102. doi: 10.1073/pnas.1012826107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaplan ML, et al. Cardiac adaptations to chronic exercise in mice. Am J Physiol. 1994;267(3 Pt 2):H1167–H1173. doi: 10.1152/ajpheart.1994.267.3.H1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.