Abstract
Human endogenous retroviruses (HERVs), remnants of ancestral viral genomic insertions, are known to represent 8% of the human genome and are associated with several pathologies. In particular, the envelope protein of HERV-W family (HERV-W-Env) has been involved in multiple sclerosis pathogenesis. Investigations to detect HERV-W-Env in a few other autoimmune diseases were negative, except in type-1 diabetes (T1D). In patients suffering from T1D, HERV-W-Env protein was detected in 70% of sera, and its corresponding RNA was detected in 57% of peripheral blood mononuclear cells. While studies on human Langerhans islets evidenced the inhibition of insulin secretion by HERV-W-Env, this endogenous protein was found to be expressed by acinar cells in 75% of human T1D pancreata. An extensive immunohistological analysis further revealed a significant correlation between HERV-W-Env expression and macrophage infiltrates in the exocrine part of human pancreata. Such findings were corroborated by in vivo studies on transgenic mice expressing HERV-W-env gene, which displayed hyperglycemia and decreased levels of insulin, along with immune cell infiltrates in their pancreas. Altogether, these results strongly suggest an involvement of HERV-W-Env in T1D pathogenesis. They also provide potentially novel therapeutic perspectives, since unveiling a pathogenic target in T1D.
Keywords: Autoimmunity, Endocrinology
Keywords: Beta cells, Diabetes, Innate immunity
The envelope protein of a human endogenous retrovirus is expressed in the pancreas of T1D patients and is associated with immune cell recruitment.
Introduction
Type-1 diabetes (T1D) is a complex autoimmune disease resulting from a poorly understood combination of genetic and environmental factors that drive immune responses against pancreatic β cells, eventually leading to their destruction. The pathogenic process begins months to years ahead of clinical symptoms and may become serologically detected by the presence of autoantibodies against one or several β cell antigens (1). Although T1D is characterized by T cell–associated depletion of β cells, studies show that both innate and adaptive immunity are involved in the disease progression (2). Additionally, while the endocrine pancreas constitutes the primary focus of research efforts, it was recently evidenced that the exocrine pancreas is also affected in T1D pathogenesis (3, 4). This complexity is illustrated by the heterogeneity of T1D progression among patients, including age at diagnosis and rate of β cell loss. Reflecting this complexity, the disease progression model has been subjected to debates — from a linear model of β cell disappearance to a relapsing-remitting model, similar to the one described in multiple sclerosis (MS), another autoimmune disease (5, 6).
A wide range of environmental factors have been studied in a global search for T1D etiology, including dietary factors, toxins, stress, viral infections, and gut microbiome. Most particularly, enteroviruses have been associated with T1D in many instances, but this still raises questions (7–11). However, a meta-analysis based on molecular studies showed a significant association between enteroviral infection and T1D (12). Despite decades of research, no causative link between viral infections and T1D pathogenesis has been confirmed (6). A new type of pathogenic element is emerging in the etiology of autoimmune diseases, the human endogenous retroviruses (HERVs), which are remnants of ancient viral infections that occurred millions of years ago (13). HERVs were generated by a process called endogenization, which happened when environmental retroviruses infected the germinal cells of hosts during evolution, leading to their transmission into the genome of the progeny and to next generations. Owing to molecular properties of retroviral ancestors, HERVs are subjected to intragenomic dissemination through retrotransposition and recombination, thus creating multiple copies of HERVs genes (14). As a consequence, HERVs represent approximately 8% of the human genome, although most HERV copies are inactivated by mutations or deletions, or are silenced by epigenetic modifications (15). Within HERVs, the HERV-K family had once been presented to be associated with T1D in humans (16), but this finding has later been disproved by several studies (17–19). Nonetheless, murine endogenous retrovirus antigens have been associated with T1D phenotype in NOD mice, in which the disease onset is related to the presence of an envelope protein (20–22).
Among the various families of HERVs, the HERV-W family appears clearly involved in MS pathogenesis (23–25). Virus-like particles containing RNA of a human retrovirus named MS-associated retrovirus (MSRV) were first isolated in cultured cells from MS patients. It subsequently turned out that this RNA belonged to a previously unknown HERV family, HERV-W (26–28). Following the initial characterization, successive studies have confirmed the association of HERV-W expression with MS. Particularly, the envelope protein of HERV-W, HERV-W-Env, also named MSRV-Env, has been found elevated in sera of MS patients and its corresponding RNA in human peripheral blood mononuclear cells (PBMC) (24, 29). HERV-W-Env has also been detected in infiltrated and resident macrophages in MS brain lesions (24, 30). To date, 2 full-length envelope proteins from the HERV-W family have been described, a pathogenic protein named MSRV-Env from first MS isolates or HERV-W-Env and a physiologic protein named Syncytin-1, which is specifically involved in syncytiotrophoblast formation during placentation. Syncytin-1 was revealed to be expressed by a modified HERV-W gene after domestication throughout evolution (31).
Interestingly, environmental viruses such as EBV can trigger the reactivation of otherwise silenced HERV-W-env gene copies, thus suggesting that HERV-W may be the missing link between environmental agents and MS onset (32, 33). Proinflammatory and autoimmune properties of HERV-W-Env have been characterized in vivo and in vitro (23, 34–37) and appeared to be mediated by the interaction with TLR4, a receptor involved in the initiation of innate immune response against microbial infection (35). The consistent results of preclinical studies have led to the development of a monoclonal antibody targeting HERV-W-Env, named GNbAC1, which is currently tested in clinical trials for an MS indication (38). As part of MS studies, cohorts of patients suffering from other autoimmune disorders have been assessed for the presence of HERV-W-Env protein in their sera. No significant associations were found (e.g., in patients with systemic lupus erythematosus or rheumatoid arthritis; refs. 24 and 39), whereas T1D patients displayed significant HERV-W-Env expression in a preliminary study, prompting further investigations into HERV-W-Env expression in larger cohorts of T1D patients.
In the present study, we show a significant HERV-W-Env expression in serum, PBMCs, and pancreata from T1D patients. A correlation was found between HERV-W-Env and macrophage infiltrates in T1D pancreata. These findings are corroborated by an HERV-W-env transgenic mouse model displaying hyperglycemia and decreased levels of insulin, and by in vitro experiments demonstrating that HERV-W-Env directly inhibits insulin secretion in human Langerhans islets.
Results
HERV-W-Env expression is associated with T1D in humans.
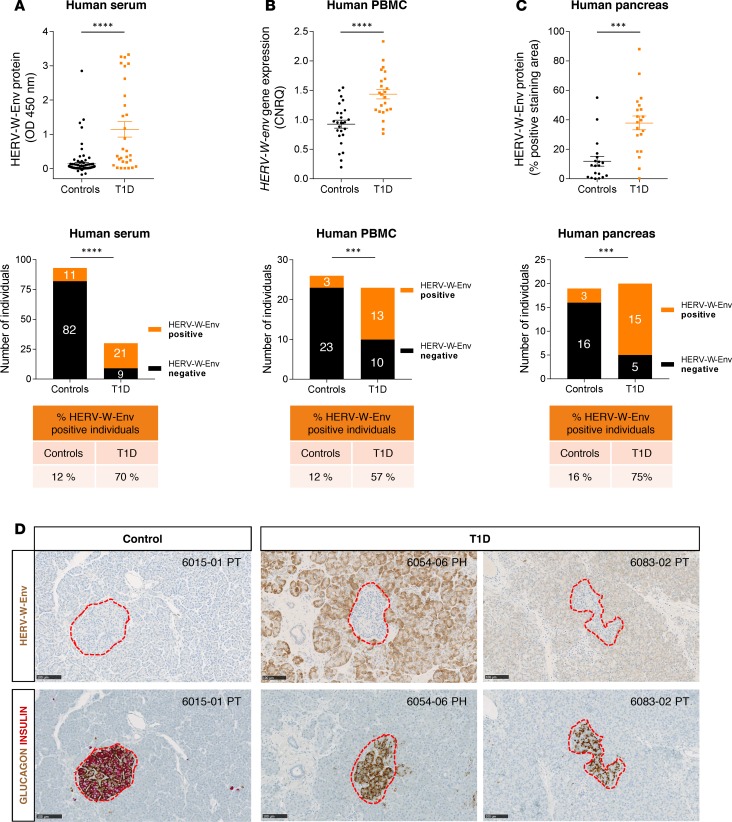
We initially assessed the presence of the HERV-W-Env protein in sera of T1D patients using an ELISA assay developed with monoclonal antibodies raised against HERV-W-Env. We found that HERV-W-Env was significantly detected in T1D compared with control individuals (P < 0.0001; Figure 1A). In this cohort, 21/30 (70%) T1D patients were positive for HERV-W-Env, whereas 11/93 (12%) control individuals were positive. We then tested HERV-W-env RNA expression in PBMC and detected HERV-W-env RNA by quantitative reverse transcription PCR (qRT-PCR) in PBMC of 13/23 (57%) T1D patients, a highly significant difference compared with control individuals 3/26 (12%) (P < 0.0001; Figure 1B). Receiver operating characteristic (ROC) curve analysis demonstrated that both ELISA and qRT-PCR tests correctly discriminate between controls and T1D patients (P < 0.0001; Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.94387DS1).
Figure 1. HERV-W-Env is expressed in human T1D patients.
(A) Serum from controls (n = 93) and T1D (n = 30) were analyzed by sandwich ELISA using monoclonal antibodies raised against HERV-W-Env. Each sample has been tested in triplicate. Results are presented as mean of the triplicate for each donor and as mean ± SEM for each group. Significance determined by Mann Whitney U test. (B) PBMC RNA was extracted from controls (n = 26) and T1D (n = 23) and analyzed by qRT-PCR assay. HERV-W-env expression was normalized to housekeeping genes B2M and YWHAZ using qbase+ and is expressed as calibrated normalized relative quantities (CNRQ). Each qPCR was done in duplicate, individual CNRQ values are plotted, and mean ± SEM for each group is shown. Significance determined by unpaired t test. (C) HERV-W-Env expression was assessed on pancreas slices from controls (n = 19) and T1D (n = 20) stained using GN_mAb_Env03 and automatically quantified. The HERV-W-Env positive area was reported to the total pancreatic tissue area. Two slides per individuals were quantified — one in the head and one in the tail. Results are presented as mean of the 2 slides for each donor and as mean ± SEM for each group. Significance determined by Mann Whitney U test. Individuals from cohorts presented in A–C were classified into either positive or negative groups for HERV-W-Env based on thresholds set at mean + 2 SD of control group. Cutoffs were set at 0.231 OD450nm for ELISA, at 1.356 CNRQ for qRT-PCR, and at 21.43% for pancreas IHC. Results are presented as contingency plots representing the number of individuals positive (in orange) or negative (in black). Significance determined by χ2 test. (D) Pancreas slices of three nPOD donors stained with GN_mAb_Env03 and adjacent slides coimmunostained with insulin (red) and glucagon (brown) are presented. Pancreatic Langerhans islets are highlighted with red dotted line. nPOD case ID is indicated in the upper right corner, with block number and pancreas zone (PH, pancreas head; PT, pancreas tail). Scale bars: 100 μm. ***P < 0.001, ****P < 0.0001.
These concordant results between protein and RNA expression in blood prompted us to seek the pathogenic protein in the pancreas. For this purpose, the network for pancreatic organ donors with diabetes (nPOD) organization provided us with a collection of pancreas slices from 20 T1D and 19 nondiabetic donors. These pancreas sections were immunostained with GN_mAb_Env03, a monoclonal antibody specifically targeting HERV-W-Env (Supplemental Figure 2), and the stained area was then quantified in an automated and blinded manner. The pathogenic protein was found significantly expressed in pancreata from T1D patients, compared with non-T1D donors (P < 0.001; Figure 1C). Within this cohort, 15/20 (75%) T1D patients displayed specific HERV-W-Env+ staining, whereas 3/19 (16%) of controls displayed positivity. Importantly, this histologic study uncovered that HERV-W-Env protein was mainly expressed in acinar cells, in the vicinity of Langerhans islets (Figure 1D). This IHC study confirmed initial observations of HERV-W-Env expression in pancreata of T1D patients, using GN_mAb_Env04, another monoclonal antibody specific for HERV-W-Env but targeting another epitope than GN_mAb_Env03 (Supplemental Figure 3).
In the present cohort, we further analyzed the characteristics of HERV-W-Env protein expression within the pancreas. The positive staining observed in T1D slides could be variable, as exemplified in Figure 2A, from clusters of acinar cells expressing HERV-W-Env (Figure 2A, upper panels) to widespread detection with variable intensity (Figure 2A, middle and lower panels). Because these patterns could represent early clusters of HERV-W activation, followed by spreading and increasing intensity, as observed for donor 6054 displaying 30 years of T1D duration (Figure 2A, lower panels), we analyzed the correlation between T1D duration and HERV-W-Env expression. In the 3 cohorts and sample types tested, we observed that elevated HERV-W-Env was found from short to long disease duration (Figure 2, B–D), suggesting possible fluctuations or continuous expression in the course of T1D pathology.
Figure 2. Characteristics of HERV-W-Env expression in pancreas and during the course of the disease.
(A) Three nPOD T1D cases stained with GN_mAb_Env03 are presented at 0.5× original magnification, and corresponding higher magnification 3× and 10×. nPOD case ID, block number, and pancreas zone are indicated on the left, and disease duration is indicated on the right. Scale bars: 0.5 ×, 5mm; 3×, 500 μm; 10×, 250 μm. (B–D) For each of the 3 cohorts presented in Figure 1, A–C, HERV-W-Env expression is presented as a function of the disease duration, the dotted line representing the positivity cutoff. T1D duration is presented as function of HERV-W-Env protein expression in serum as determined by ELISA (B), as a function of HERV-W-env gene expression in PBMC (C), or as a function of HERV-W-Env expression in pancreas as determined by IHC (D).
Pancreatic HERV-W-Env expression is associated with macrophage recruitment in T1D patients.
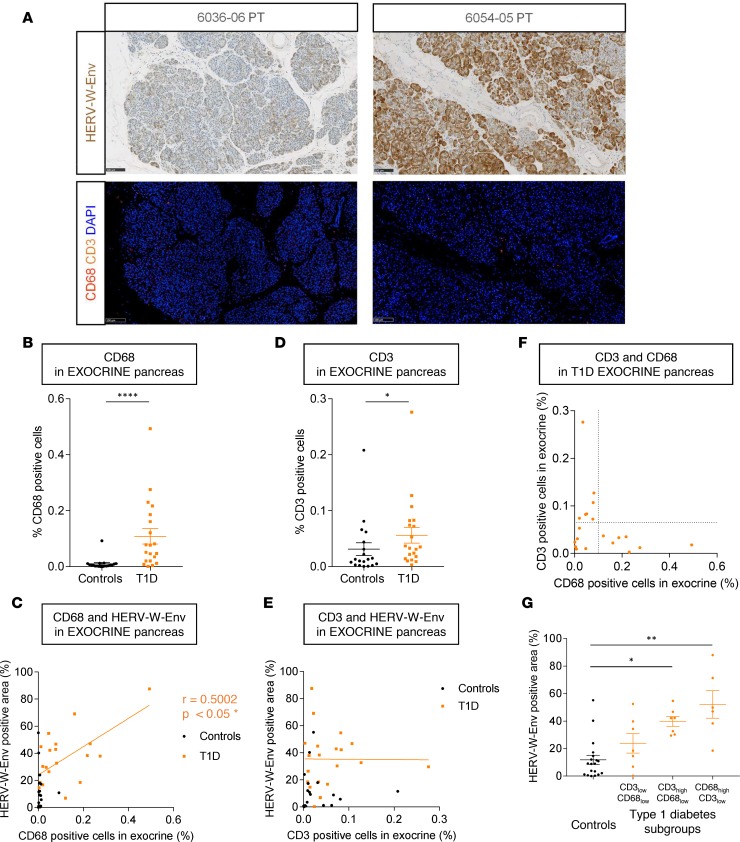
Since T1D pathogenesis is characterized by pancreatic infiltrates of immune cells, we analyzed this feature in parallel to HERV-W-Env expression. For this purpose, slides adjacent to those stained for HERV-W-Env were immunostained for CD3, CD68, and glucagon. Representative observations are presented in Figure 3A. CD3+ and CD68+ cells were quantified in the exocrine and endocrine pancreas, in automated and blinded conditions. Since HERV-W-Env is expressed by acinar cells (Figure 1D), we first focused on the exocrine pancreas. We observed a significant increase of CD68 infiltrates within T1D exocrine pancreas compared with controls (P < 0.0001; Figure 3B). The amount of infiltrated macrophages was correlated with HERV-W-Env expression (P < 0.05; Figure 3C), suggesting that HERV-W-Env protein is associated with macrophage recruitment in the exocrine pancreas of T1D patients. Analyzing T-lymphocyte infiltrates, we found that CD3+ cell infiltrates were also increased in the exocrine pancreas from T1D patients compared with controls (P < 0.05; Figure 3D). However, no correlation was found between HERV-W-Env expression and T-lymphocyte infiltrates in the exocrine pancreas (Figure 3E), though an increased number of CD3+ cells are observed in some instances within clusters of acinar cells overexpressing HERV-W-Env (Supplemental Figure 4A). In pancreatic islets, no difference in CD68+ or CD3+ cell infiltrates was found between T1D and controls (Supplemental Figure 4, B and C). Moreover, no correlations between HERV-W-Env expression and CD68+ or CD3+ cell infiltrates in the endocrine pancreas were observed (Supplemental Figure 4, D and E).
Figure 3. Proinflammatory features associated with HERV-W-Env expression within the pancreas.
(A) Pancreas of nPOD T1D cases 6036 and 6054 were immunostained with GN_mAb_Env03 (upper panels), and adjacent slides were immunostained with anti-CD3 (orange) and anti-CD68 (red) (lower panels). Lower panels display the same areas as upper panels. Scale bars: 100 μm. (B–G) Percentages of CD68+ (B) and CD3+ (D) cells in the exocrine pancreas are presented for controls (n = 19) and T1D patients (n = 20) and as a function of HERV-W-Env+ area (C and E, respectively) (Figure 1C). The relationship between CD3+ and CD68+ cells for each T1D donor is presented in F, which allowed to determine 3 groups: (i) CD3low/CD68low, (ii) CD3high/CD68low, and (iii) CD68high/CD3low. Based on this plot, the cutoff for CD3+ cells is set at 0.065%, and for CD68+ cells, it is set at 0.1%. The HERV-W-Env+ area presented in Figure 1C was reanalyzed in these 3 groups and was presented as mean for each individual and mean ± SEM for each group (G). Significance determined by Mann Whitney U test (B and D), by Spearman test (C), and by Kruskal-Wallis test followed by Dunn multiple comparison test (G). *P < 0.05, **P < 0.01, ****P < 0.0001.
Since macrophages have been shown to promote differentiation of CD8+ cytotoxic T cells in T1D pathogenesis (40) and to stimulate CD4+ T cell proliferation (41), a correlation between macrophages and CD3+ cells was analyzed. Surprisingly, we found no correlation between CD68+ and CD3+ cells (Figure 3F). The presence of macrophages or T lymphocytes appeared to be mutually exclusive within a given individual, thus suggesting the presence of 3 different histological phenotypes in T1D pancreata, (i) CD3low/CD68low, (ii) CD3high/CD68low, and (iii) CD68high/CD3low. Given this observation, we reanalyzed HERV-W-Env expression in these 3 populations of T1D donors and in controls donors (Figure 3G). Interestingly, T1D pancreata with low immune infiltrates (CD3low/CD68low) displayed low levels of HERV-W-Env, similar to control individuals, whereas T1D CD68high/CD3low presented the strongest difference compared with controls (P < 0.01). Nonetheless, T1D CD3high/CD68low pancreata also displayed a significant but smaller difference (P < 0.05).
HERV-W-env expression in transgenic mice promotes hyperglycemia, decreased levels of insulin, and pancreatic immune cell infiltrates.
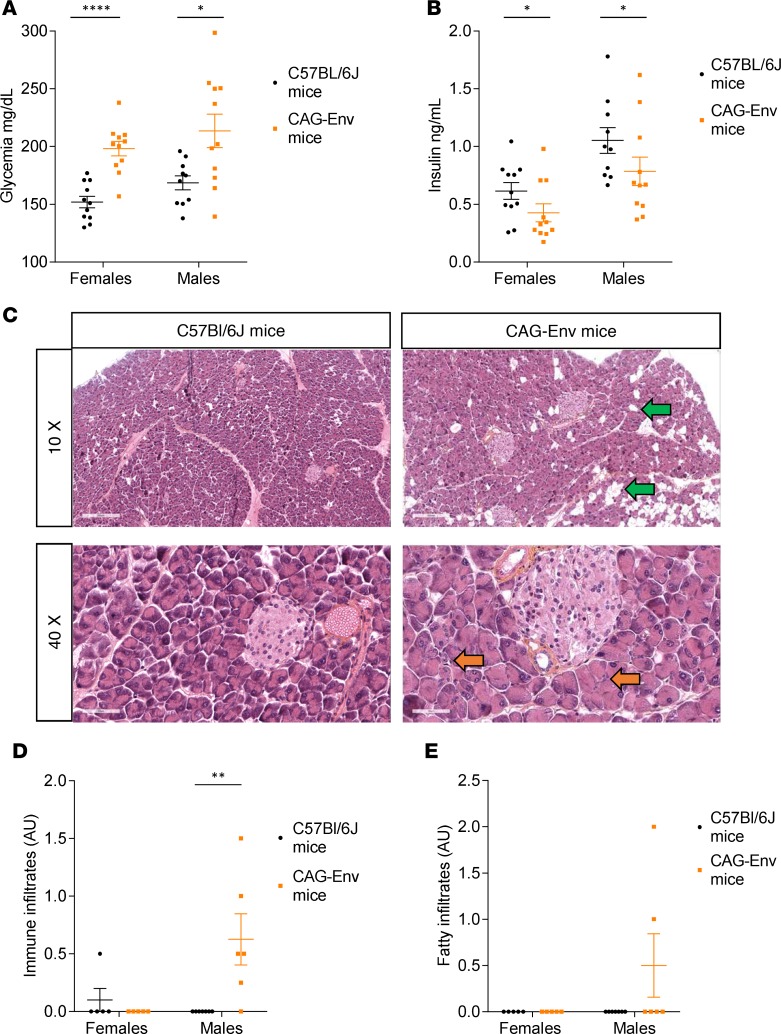
To investigate HERV-W-Env effects in mice, we developed a transgenic mouse model with a C57BL/6J background, in which the HERV-W-env transgene is expressed under the control of the ubiquitous CAG promoter but regulated by HERV-W 3’LTR. HERV-W-env transgene is inserted in the HPRT locus of the murine X-chromosome, and these transgenic mice, named CAG-Env mice, are viable and fertile. We first measured fasting glycemia in 7-week-old CAG-Env mice and found that both sexes were hyperglycemic, though this phenotype was more homogenous and significant in females (Figure 4A). CAG-Env female mice displayed a glycemia on average 30% higher compared with their nontransgenic C57BL/6J counterparts (198 ± 6 mg/dl vs. 152 ± 6 mg/dl; P < 0.0001). A comparable difference was seen in CAG-Env males, accounting for 27% (214 ± 15 mg/dl vs. 169 ± 6 mg/dl; P < 0.05). This hyperglycemia was associated with significantly lower levels of insulin in both sexes, with more homogeneity in females (Figure 4B). CAG-Env males displayed a 25% lower insulinemia than their C57BL/6J counterparts (0.79 ± 0.12 ng/ml vs. 1.05 ± 0.11 ng/ml; P < 0.05). A comparable reduction of 31% in insulinemia was observed in CAG-Env females (0.43 ± 0.08 ng/ml vs. 0.62 ± 0.08 ng/ml; P < 0.05). Since HERV-W-Env expression is associated with macrophage infiltrates in the exocrine pancreas of T1D patients (Figure 3C), we searched for immune cell infiltrates within the exocrine pancreas of these mice. Representative images of pancreata from 12-week-old CAG-Env transgenic mice, and WT mice are presented in Figure 4C. Immune cell infiltrates were observed in both sexes in CAG-Env mice but to a higher degree in males (orange arrows). Quantification revealed that CAG-Env males had statistically significant immune cell infiltrates within their exocrine pancreas (Figure 4D). Interestingly, we also observed fatty infiltrates of variable importance within the exocrine pancreas of CAG-Env mice, as pointed out by green arrows, although this observation was not statistically significant (Figure 4E). We did not observe significant insulitis in CAG-Env mice compared with WT mice (data not shown).
Figure 4. Transgenic mice expressing HERV-W-env display hyperglycemia and pancreatic abnormalities.
(A and B) Seven-week-old CAG-Env transgenic mice expressing of HERV-W-env and control C57Bl/6J mice of the same age were fasted for 5 hours before measuring glycemia (A) and insulinemia (B). Results are presented as individual values and as mean ± SEM. n = 10 or 11 mice in each group. Significance determined by unpaired t test (glycemia) or by Mann-Whitney U test, one-tailed (insulinemia). (C) Pancreas sections of 12-month-old CAG-Env mice (n = 5 females and n = 6 males) and C57BL/6J mice (n = 5 females and n = 7 males) were stained with HES (Hematoxylin Eosin Saffran). Representative images of C57BL/6J (left panels) and CAG-Env (right panels) mouse pancreata were presented at 10× (upper panels) and 40× (lower panels) original magnification. Fatty infiltrates are highlighted with green arrows and immune cells infiltrates with orange arrows. Scale bars: upper panels, 200 μm; lower panels, 50 μm. (D and E) Pancreas sections from 12-month-old CAG-Env mice (n = 5 females and n = 6 males) and C57BL/6J mice (n = 5 females and n = 7 males) were analyzed for immune cell infiltration (D) and fat infiltration (E). A gradation from 0–3 was affected to each slide, 0 corresponding to no infiltration of fat or immune cells and 3 corresponding to high level of infiltrations. Two sections distant by 500 μm were analyzed for each mouse, blindly and independently by 2 people, and their evaluations were averaged for each slide. Results are presented as mean of the 2 slides for each mouse and as mean ± SEM for each group. Significance was determined by Mann Whitney U test. *P < 0.05; **P < 0.01, ****P < 0.0001.
HERV-W-Env directly impairs insulin secretion by pancreatic β cells in vitro.
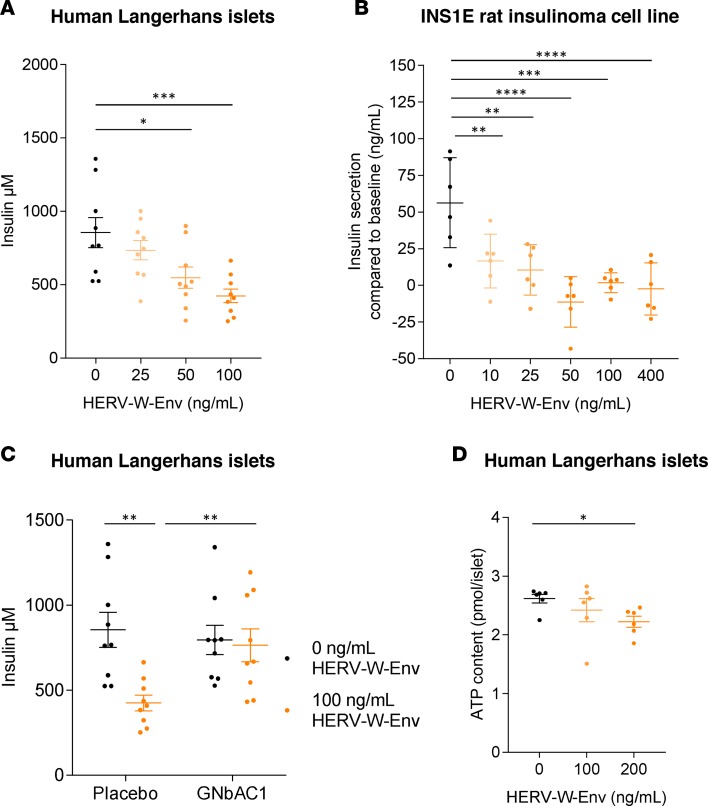
Immunohistological analysis of human pancreata revealed that acinar cells are permissive cells for the production of HERV-W-Env (Figure 1D). This suggests that a secretion of HERV-W-Env by acinar cells onto β cells may occur, raising the question of direct cytotoxic effects of HERV-W-Env. This protein could indeed directly affect β cell physiology, as pancreatic β cells express TLR4 (42–44), the receptor through which HERV-W-Env exerts its pathogenic effects. To test this hypothesis in vitro, human pancreatic Langerhans islets, isolated postmortem from healthy individuals, were exposed to increasing recombinant HERV-W-Env concentrations, ranging from 25 ng/ml to 100 ng/ml (400 pM to 1.6 nM). Our results evidence that insulin secretion was inhibited by HERV-W-Env in a dose-dependent manner (Figure 5A). At the highest concentration of HERV-W-Env (100 ng/ml), we observed 50% inhibition in insulin secretion compared with the control condition (P < 0.001). In control condition, human β cells secreted 855 ± 103 μM of insulin, and this secretion was progressively inhibited by increasing HERV-W-Env concentration, to reach a minimum of 424 ± 46 μM. The same inhibitory profile was observed using a rat insulinoma cell line, INS1E (45) (Figure 5B). In response to 10 mM of glucose, secretion of insulin by INS1E was 56.3 ± 13 ng/ml in the control condition, while it dropped below the baseline at 50 ng/ml of HERV-W-Env (P < 0.001). At higher doses of HERV-W-Env (100 and 400 ng/ml), insulin secretion fluctuated around the baseline. We further evaluated the specificity of the observed pathogenic effect toward human β cells using GNbAC1, a neutralizing monoclonal antibody raised against HERV-W-Env. We demonstrate that GNbAC1 allowed human β cells exposed to HERV-W-Env to maintain insulin secretion. Indeed, in the presence of GNbAC1, insulin secretion remained stable above 750 μM, despite 100 ng/ml of HERV-W-Env, whereas it dropped significantly in the absence of GNbAC1 (P < 0.01; Figure 5C). The inhibition of insulin secretion observed was not related to HERV-W-Env toxicity affecting β cell viability, as it did not affect ATP content at 100 ng/ml (Figure 5D). However, at higher HERV-W-Env concentration (200 ng/ml), a slight decrease of 15% in viability was observed (P < 0.05), which may indicate some impact on β cell viability at elevated concentrations but cannot explain the observed dose-response inhibition of insulin secretion.
Figure 5. Direct pathogenic effects of HERV-W-Env toward pancreatic β cells.
(A) Human Langerhans islets have been exposed for 48 hours to HERV-W-Env (0, 25, 50, 100 ng/ml), and insulin secretion has been measured in response to 20 mM glucose stimulation. Results are presented as individual values and as mean ± SEM. Each condition has been performed in triplicate and in 3 independent experiments. Significance determined by Bonferroni’s test. (B) INS1E cells have been exposed for 72 hours to increasing concentrations of HERV-W-Env (0, 10, 25, 50, 100, 400 ng/ml). Insulin secretion has been measured either in absence of glucose (baseline) or in response to 10 mM glucose stimulation. Results are presented as the difference in concentration between glucose stimulation and baseline. Each condition has been performed in triplicate and in 2 independent experiments, and data are presented as individual values and as mean ± SEM. Significance determined by Bonferroni’s test. (C) Human Langerhans islets have been exposed for 48 hours to HERV-W-Env (0 and 100 ng/ml) and to GNbAC1 (3 μg/ml) or placebo. Insulin secretion has been measured in response to 20 mM glucose stimulation. Results are presented as individual values and as mean ± SEM. Each condition has been performed in triplicate and in 3 independent experiments. Significance determined by unpaired t test. (D) Effect of HERV-W-Env on human Langerhans islet viability has been assessed using human islet microtissues exposed to 0, 100, and 200 ng/ml of HERV-W-Env for 48 hours before being lysed. Lysates were analyzed for ATP content using Promega CellTiter-Glo Luminescent Cell Viability Assay. Results are presented as individual values and as mean ± SEM. Each condition has been performed six times, and 1 experiment has been performed. Significance determined by Dunn’s test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
HERV and autoimmune diseases.
HERVs are remnants of viral infections of the primate germline millions of years ago, which have been transmitted to their progeny. Tens of HERV families have been described, representing 8% of the human genome — few of which have been associated with several autoimmune diseases such as systemic lupus erythematous, rheumatoid arthritis, MS, amyotrophic lateral sclerosis, and Sjögren’s syndrome (13, 24, 46–48). In particular, the HERV-W family has been involved in the pathogenesis of MS through its envelope protein HERV-W-Env, also known as MSRV-Env (23–25).
HERV-W-Env and T1D in humans.
Present results show an association between HERV-W-Env and T1D in humans, which is supported by the detection of HERV-W-Env by 3 different methods (ELISA, qRT-PCR, IHC) and in 3 different types of human samples (serum, PBMC, pancreas) (Figure 1). Indeed, the pathogenic protein was detected in the sera of 70% of T1D patients by ELISA and in the pancreas by IHC in 75% of T1D patients. HERV-W-env RNA was also detected in the PBMC of 57% of T1D patients. By comparison, 12%–16% of non-T1D individuals displayed HERV-W-Env positivity in the 3 cohorts. This observation may have various meanings, from “healthy carriers” to potential prodromal phases of HERV-W–associated diseases, including T1D or MS.
Functional effects of HERV-W-Env relevant for T1D pathogenesis.
Beyond the detection of an abnormal expression in T1D patients (Figure 1), this study showed that HERV-W-Env has a dual pathophysiological effect of relevance for T1D pathogenesis: (i) a direct effect toward β cells (Figure 5) and (ii) an indirect immune-mediated effect (Figures 3 and 4). This dual pathogenic pattern has already been observed in MS, in which HERV-W-Env affects the immune system — causing autoimmune neuroinflammation (37) — and directly blocks oligodendrocyte precursor cell differentiation, thereby impairing remyelination (25).
Direct functional effects of HERV-W-Env.
In vitro studies evidenced direct inhibition of insulin secretion by HERV-W-Env in a dose-dependent manner in primary cultures of human Langerhans islets and in the rat INS1E insulinoma cell line (Figure 5, A and B). This inhibition reached 50% at 100 ng/ml of HERV-W-Env in human β cells, without affecting cell viability (Figure 5D). The underlying mechanism of this inhibition requires further investigations, though it has already been described that HERV-W-Env exerts its pathogenic effects through an interaction with the TLR4 receptor (35, 36). From a T1D perspective, this mechanism appears relevant, as TLR4 is expressed by β cells (42–44). Moreover, the inhibition of insulin secretion following TLR4 stimulation has already been observed in human, rat, and mouse pancreatic β cells by using lipopolysaccharide (LPS), the prototypical ligand of TLR4 (42, 43). Inhibition appeared related to the repression of key transcription factors for β cell functions, PDX-1 and Maf-A, in a TLR4-dependent manner and via NF-κB signaling pathway (43).
At concentrations above 200 ng/ml, HERV-W-Env directly affects β cell viability in vitro (Figure 5D), a process that could also occur in vivo. Although we have observed that HERV-W-Env is expressed in the pancreas by acinar cells surrounding β islets (Figure 1D), the concentration of HERV-W-Env to which the β cells are exposed in the pancreas in vivo can only be estimated to be locally elevated. Notably, a decrease in human β cell viability has already been described upon TLR4 activation after exposure to LPS (42).
Interestingly, it has already been evidenced that HERV-W-Env also displays direct pathogenic effects toward endothelial cells and Schwann cells of peripheral nerves (49, 50). This raises questions regarding the comorbidities associated with T1D, such as diabetic vasculopathies and peripheral diabetic neuropathy. Since our results suggest that HERV-W-Env expression may be continuous during T1D evolution (Figure 2), it is conceivable that elevated levels of circulating HERV-W-Env protein at certain periods (Figure 2B) may also directly affect endothelial or Schwann cells, thus contributing to the development of T1D comorbidities. It would therefore be worth investigating whether the highest levels of HERV-W-Env in blood are associated with a higher risk of comorbidities in T1D patients.
Indirect immune-mediated effects of HERV-W-Env.
Present results also showed that HERV-W-Env expression impacts the immune system. Indeed, HERV-W-env RNA is significantly detected in PBMCs from T1D patients compared with control individuals (Figure 1B), and HERV-W-Env expression in acinar cells correlates with immune cell infiltrates within the exocrine pancreas of T1D patients (Figure 3C). This is further corroborated by the phenotype of transgenic mice expressing HERV-W-env, in which increased immune cell infiltrates are observed in the pancreas (Figure 4, C and D).
HERV-W-Env has already been described to affect the adaptive immunity. In previous studies, lethal brain hemorrhages were observed after injection of MSRV virus–like particles from MS cell cultures and appeared to be T cell–mediated (34). HERV-W-Env also displays a superantigen-like activity, leading to the polyclonal activation of T cells in vitro (23). In the present study, we did not observe a correlation between HERV-W-Env expression in the pancreas and CD3+ cell infiltrates (Figure 3E). However, in some instances, clusters of acinar cells overexpressing HERV-W-Env colocalized with important CD3+ cell infiltrates (Supplemental Figure 4A). This observation might reflect a recruitment of T cells that may only occur at a precise stage of the disease and which could be related to immune-mediated effects of HERV-W-Env.
However, an upstream and dominant immunopathogenic effect of HERV-W-Env relates to innate immunity, as this pathogenic protein interacts with TLR4, a receptor involved in innate immune response against microbial infections (35). HERV-W-Env was shown to promote monocyte maturation, DC differentiation, and release of proinflammatory cytokines, such as TNFα, IL1β, IL12β, and IL6 (34–36). In this respect, we have shown that HERV-W-Env affects the innate immune system of T1D patients, as the presence of this protein correlates with CD68+ cell detection in the exocrine pancreas (Figure 3C). CD68 is a marker typically used to identify macrophages, a cell type well-known to express TLR4 and mainly involved in innate immunity. The innate immune system plays an important role in the pathogenesis of T1D, in particular via TLR4 receptor, whose surface expression and mRNA are significantly increased in human T1D monocytes and are associated with microvascular complications in T1D (51, 52). Several downstream targets of TLR4 signaling, including MyD88, TRIF, and NF-κB, are also upregulated within T1D patients (51). Hyperglycemic environment also led to increased expression of TLR4 in macrophages from NOD mice (53). These reports highlight the importance of TLR4 in T1D pathogenesis and further corroborate the present findings that suggest an involvement of HERV-W-Env in this disease.
Abnormalities in T1D exocrine pancreas.
The quantitative immuno-histological study of human T1D pancreas has also evidenced the presence of immune cell infiltrates within the exocrine part of the T1D pancreas. Indeed, CD68+ and CD3+ cells are significantly increased in exocrine pancreas of T1D patients compared with control individuals (Figure 3, B and D). The abnormalities of the exocrine pancreas are increasingly recognized clinical features of T1D, although this is not a well-studied domain. Indeed, studies have evidenced that T1D patients displayed pancreatic exocrine insufficiency in 43%–80% of cases, along with reduced pancreatic weight and volume compared with nondiabetic controls (4). These clinical features have long been thought to be secondary to β cell loss, as insulin is a trophic factor for pancreatic acinar cells. However, this one-way view has recently been challenged by new clinical features. It appeared that acinar atrophy is independent of diabetes duration and of the presence of surviving β cells (4). Additionally, nondiabetic autoantibody–positive individuals were shown to display smaller pancreas size (54), and 74% of T1D patients presented autoantibodies against an exocrine pancreatic enzyme specifically expressed by acinar cells, the bile salt-dependent lipase (55). These studies support the hypothesis that T1D is a combined endocrine-exocrine disease, at least in a proportion of cases.
This hypothesis has recently been reinforced by an immunohistological study demonstrating an increase of CD4+ and CD8+ T lymphocytes in the exocrine pancreas of T1D patients (56). Our observation of an increased number of CD3+ T lymphocytes in the exocrine pancreas of T1D patients (Figure 3D) thus corroborates this previous study. Additionally, quantification of CD68+ cells reveals the presence of macrophages infiltrates within the exocrine part of the T1D pancreas (Figure 3B), a previously unknown pancreatic abnormality in T1D patients. Interestingly, we observed a mutually exclusive pattern of CD3+ and CD68+ cells in a given individual, suggesting an immunological heterogeneity in T1D patients, and/or in periods of disease, based on histological phenotypes (Figure 3F). This allowed us to describe 3 subgroups — (i) CD3low/CD68low, (ii) CD3high/CD68low, and (iii) CD68high/CD3low — each of them being characterized by a specific level of HERV-W-Env expression (Figure 3G). Immunological heterogeneity has already been described among T1D patients (57) and is possibly related to the variability observed in the disease clinical features (6), including age at diagnosis and rate of β cell loss.
Irrespective of the focus on HERV-W-Env, our results support the increasing number of evidence demonstrating that dysfunctions of the exocrine part of the pancreas is an important clinical feature of T1D. HERV-W-Env could be one of the pathogenic elements involved in dysfunction of the exocrine pancreas, due to its expression by acinar cells (Figure 1D). Interestingly, in the mouse model expressing HERV-W-env, the abnormalities in the pancreas were only detected in the exocrine part and mostly consisted of immune cell infiltrates (Figure 4, C–E).
Endogenous retroviruses in T1D etiology.
Present results demonstrate the abnormal expression of HERV-W-Env in T1D patients and are evidencing relevant biological mechanisms associated with this protein in T1D pathogenesis, thus supporting the involvement of an HERV element in the pathogenesis of T1D. So far, only the HERV-K family had been incriminated in T1D etiology in humans; however, such results have later been disproved (58). To date, only a recent publication links the low number of HERV-K copies within the C4 gene cluster to an increased risk of developing T1D. The authors suggest that the endogenous retroviral insertion could offer protection against T1D, but they did not provide any functional relationship between the low copy numbers and the T1D susceptibility (59). Conversely, present results now clearly indicate the involvement of an endogenous retrovirus, HERV-W, in the pathogenesis of T1D.
Though representing 8% of our genome, the majority of HERV elements have lost their transcriptional or protein-coding capacity. The few copies retaining such capacities are normally silenced, unless transactivated in some pathological conditions. Environmental viruses can trigger the transactivation of members of the HERV-W family, as this has been demonstrated for herpes virus simplex type 1, cytomegalovirus, or EBV (33, 60, 61). The transactivation of HERV-W by EBV is of particular interest, owing to the recent demonstration that EBV was the only virus found to be associated with T1D in an immunoproteomic array profiling antiviral antibodies of 23 viral strains (62).
Clinical perspectives and conclusions.
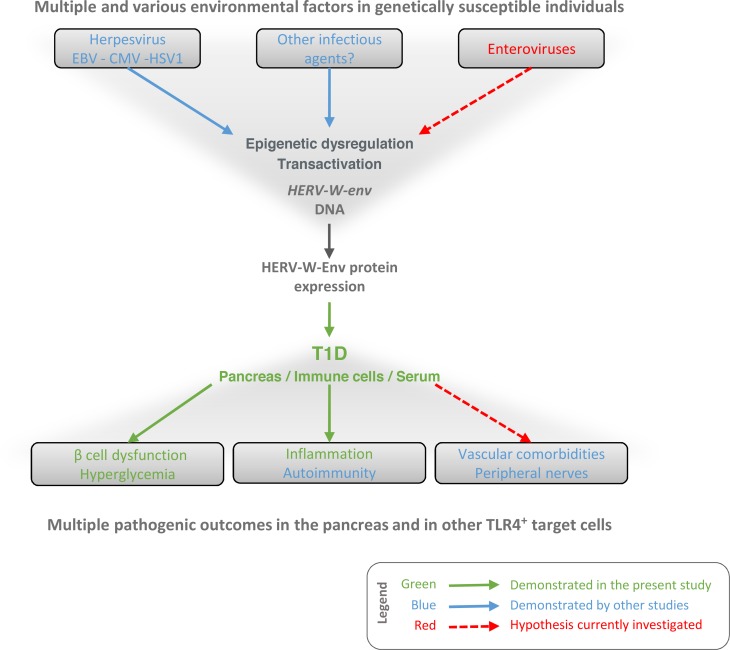
The present study, summarized in Figure 6, suggests that HERV-W-Env may be situated at a critical position in the pathogenic cascade leading to T1D, located between multiple environmental factors operating in genetically susceptible individuals and multiple pathogenic clinical outcomes. This positioning provides a rationale for the neutralization of HERV-W-Env in T1D. These therapeutic perspectives are currently tested in a phase-IIa clinical trial in T1D patients using GNbAC1, a humanized monoclonal IgG4 antibody neutralizing HERV-W-env (registered at ClinicalTrials.gov; NCT03179423).
Figure 6. Current working model.
This diagram illustrates the current working model in which the present study is integrated. Data presented demonstrate that HERV-W-Env protein is expressed in T1D patients and particularly in their pancreas (Figure 1), where it may promote β cell dysfunction (Figure 5) and macrophages recruitment (Figure 3), possibly leading to hyperglycemia and decrease levels of insulin (Figures 4 and 5). As reported by other studies, HERV-W-Env displays other pathogenic properties that may be relevant in T1D pathogenesis. In particular, HERV-W-Env promotes autoimmunity (23) and impairs TLR4+ cells such as endothelial cells (49) and Schwann cells (50), both of which are associated with major T1D comorbidities. The underlying question is why the HERV-W-env gene has been transcriptionally activated and translated into a pathogenic protein. An explanation relies upon its transactivation and epigenetic dysregulation by infectious agents, as it has already been demonstrated (33, 60, 61). In T1D, the question of such dysregulation by enterovirus is raised (12), as well as the role of EBV (62). This diagram illustrates how, in a global pathogenic cascade leading to the disease, HERV-W-Env expression is placed at a crucial position, between multiple environmental factors operating in genetically susceptible individuals and multiple clinical outcomes. This positioning provides a rationale for the neutralization of HERV-W-Env as a therapeutic target in T1D. Within this framework, GNbAC1 — a humanized monoclonal IgG4 antibody neutralizing HERV-W-env — is currently tested in a phase IIa clinical trial in T1D patients (NCT03179423).
Methods
Supplemental Methods are available online with this article.
Subjects
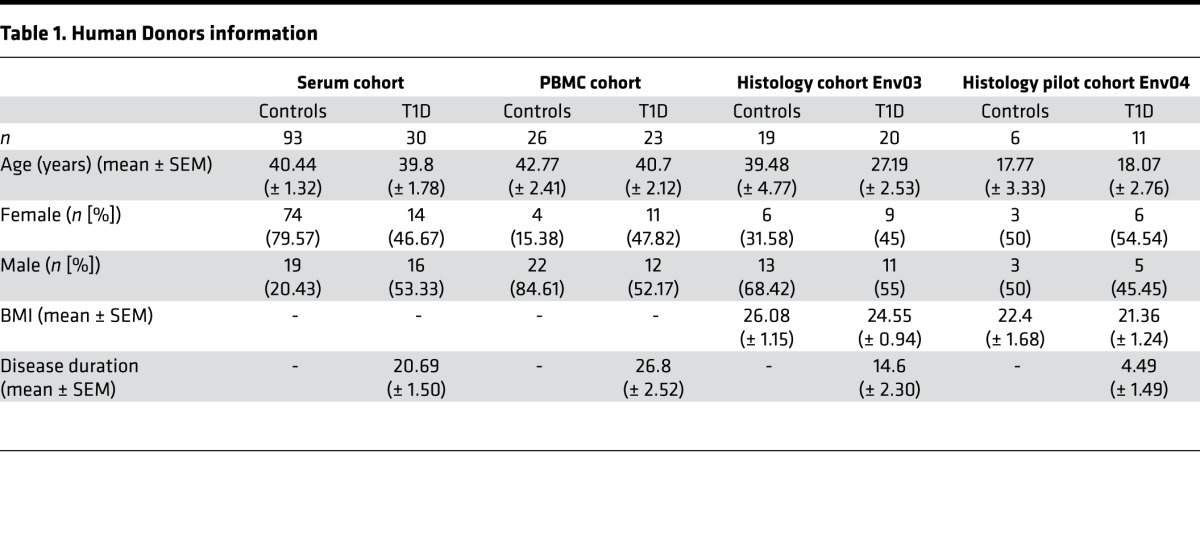
Human pancreata from T1D (n = 20) and control individuals (n = 19) were collected from organ donors through the nPOD program (nPOD investigator number 1152; Miami, Florida, USA) according to procedures previously described (63). Although not suffering from T1D, control individuals from nPOD were not all heathy individuals, as mentioned in clinical information. PBMC (n = 33) and serum (n = 93) from non-T1D blood donors were purchased from Etablissement Français du Sang (EFS, Lyon, France). PBMC (n = 25) and serum (n = 30) from T1D patients and human Langerhans islets from healthy organ donors with a BMI below 25 were purchased from TEBU BIO (Le Perray-en-Yvelines, France). Donor information is presented in Table 1.
Table 1. Human Donors information.
Antibodies
All antibody references are presented in Supplemental Table 1. Antibodies targeting HERV-W-Env were developed by GeNeuro. Mouse monoclonal antibodies targeting HERV-W-Env were obtained by immunization with purified recombinant proteins (GN_mAb_Env01, GN_mAb_Env03, GN_mAb_Env04) or with vectorized plasmids expressing the HERV-W-env gene with mouse codon–adapted sequence (GN_mAb_Env16). GNbAC1 is a humanized IgG4 antibody grafted with the 6 complementarity-determining region (CDRs) of an HERV-W-Env–neutralizing clonal hybridoma directed against the HERV-W protein encoded by MSRV-env clone (GenBank, AF331500-1; https://www.ncbi.nlm.nih.gov/genbank/), produced in CHO cells as a recombinant antibody under GMP conditions for therapeutic use. These custom antibodies have already been tested and published in peer-reviewed publications (24, 25, 30, 35, 50). GN_mAb_Env04 does not recognize the same epitope that GN_mAb_Env03.
IHC and Immunofluorescence
Paraffin sections from 39 donors (19 controls and 20 T1D) were obtained from nPOD (nPOD investigator number 1152). For each donor, paraffin sections from the head and from the tail were obtained and immunostained, in order to avoid bias that could arise from heterogeneity within a pancreas. For 1 donor, only the head of the pancreas was available. For the CD3, CD68, and glucagon study, 25 donors remained available with both head and tail sections. The results of the automated quantification are presented as the mean of the head and tail sections.
HERV-W-Env staining using GN_mAb_Env03 antibody
The HERV-W-Env IHC was performed on the automated platform Benchmark (Ventana, Roche) with the detection kit UltraView DAB (brown chromogen), without pretreatment. GN_mAb_Env03 monoclonal antibody was developed by GeNeuro and has already been validated in several publications (24, 25, 30). GN_mAb_Env03 monoclonal antibody was used at a concentration of 5 μg/ml for the 77 slides. Eight additional slides (4 T1D, 4 non-T1D) were used as controls, with mouse IgG2a isotype. Counterstaining was applied with hematoxylin II and bluing reagent. Slides were digitized with the slide scanner (Hamamatsu), objective ×20, and quantification was made using Indica Labs HALO platform. An algorithm was designed based on pattern recognition that discriminates pancreas tissue (analyzed areas) from fatty inclusions, vasculo-nervous structures, and surrounding connective tissue (excluded areas). Image analysis based on red, green, and blue (RGB) spectra was used to detect brown staining (DAB) within the positive areas (pancreas). The algorithm was designed to allow the detection of specific brown staining according to a threshold of intensity, and nonspecific edge staining of sections was not taken into account. It calculates pancreas area (mm²), staining area (mm²), and percentage (% stained area/pancreas area). Quantification was performed under blinded conditions, with anonymized slides from control and T1D donors.
Insulin and glucagon staining
The coimmunostaining for insulin and glucagon was performed on the automated platform Benchmark with the detection kits UltraView DAB (glucagon labeling) and Ultraview Red (insulin labeling). Following a CC2 short pretreatment, anti-glucagon (G2654, Sigma-Aldrich) antibody diluted 1/64,000 was incubated for 1 hour. Detection kit UltraView DAB was then applied followed by an 8-minute heat denaturation at 85°C. The anti-insulin antibody (3014, Cell Signaling Technology) was then incubated for 1 hour at 1/400, before using UltraView Red detection kit. Counterstaining with hematoxylin II was eventually applied for 4 minutes. The isotype control slides were incubated with a rabbit monoclonal isotype antibody (ab172730, Abcam) at 10 μg/ml and a mouse IgG1 isotype antibody (ab18447, Abcam) at 1.16 μg/ml. The slides were then digitalized with the slide scanner Nanozoomer (Hamamatsu), objective ×20.
CD3, CD68, and glucagon staining
The coimmunostaining for CD3, CD68, and glucagon was performed on the automated platform Discovery XT-VENTANA. After deparaffinization and antigen retrieval using CC1 solution, slides were incubated 1 hour at 37°C with a mix of anti-CD3 (A0452, Dako) and anti-CD68 (M0876, Dako) antibodies, before incubating anti-mouse (FP-SC 4110, Interchim) and anti-rabbit (FP-SB 5110, Interchim) secondary antibodies for 45 minutes at 37°C. Anti–glucagon-FITC antibody (BS-3796-A488, Interchim) was subsequently incubated for 45 minutes at 37°C. Slides were digitalized with the slide scanner NANOZOOMER 2.0RS/C10730-12 (Hamamatsu; objective ×20, resolution 0.46 μm/pixel). Image analysis was performed on the whole pancreas slide by using an algorithm from HALO platform combined with a tissue classifier (Indica Labs). Segmentation between exocrine/endocrine pancreas and other tissue types (connective tissues) were processed using Tissue Classifier based on color, texture, and contextual features. Islet regions were identified as a contiguous area of glucagon. CD3 and CD68 cells were detected according to thresholds of intensity within the endocrine and exocrine pancreas. The algorithm calculates the total number of cells within each pancreas part, the number of CD3+ cells, the number of CD68+ cells, and the areas of endocrine and exocrine parts of the pancreas. Quantification was performed under blinded conditions, with anonymized slides from control and T1D donors.
ELISA on serum
Serum from 93 controls and 30 T1D patients were used in this study. HERV-W-Env ELISA was performed twice in 2 independent facilities by 2 different experimenters, and each ELISA was performed in triplicate. Ninety-six–well plates were coated with GN_mAb_Env04 and GN_mAb_Env16 antibodies mix (2.5 μg/ml each in 0.05 M carbonate-bicarbonate coating buffer) or with coating buffer only (uncoated wells) overnight at 4°C. Plates were then washed and blocked (1X PBS + 0.5% Tween 20 + 2% BSA) for 1 hour at 37°C. Nondiluted serum samples were incubated in triplicates for 1 hour at 37°C. Plates were then washed and incubated with HRP-conjugated GN_mAb_Env01 antibody (0.5 μg/ml in blocking buffer) for 1 hour at 37°C. Plates were washed before revelation with tetramethylbenzidine (TMB) substrate solution. After addition of stop solution, OD was measured at 450 nm. The OD450nm of each sample was corrected by subtracting the OD450nm of the corresponding uncoated well. The results are presented as the mean value of each sample obtained in the 2 independent laboratories.
qRT-PCR on PBMC
PBMC from 33 non-T1D and 25 T1D blood donors were used in this study. RNA was isolated using miRNeasy micro kit (Qiagen) following the manufacturer instructions. The RNA concentration was determined using Nanodrop 2000 UV-VIS Spectrophotometer (Thermo Fisher Scientific). RNA quality control (RIN) was performed using 2100 Bioanalyzer microfluidic gel electrophoresis system (Agilent). On-column DNase digestion was performed during RNA extraction, and DNAse treatment in solution was performed using the Heat&Run gDNA removal kit (Arcticzymes). cDNA synthesis was performed using iScript Advanced cDNA Synthesis kit (Bio-Rad), with 120 ng of total RNA as input. Samples were excluded according to 2 exclusions criteria: RIN value below 6 or RNA concentration after extraction below 10 ng/μl. Following these criteria, 7 non-T1D and 2 T1D samples have been excluded from the analysis. Each PCR reaction was made in duplicate, with 2.5 ng cDNA in each PCR reaction. All measurements have been performed in 384-wells plates (CFX384, Bio-Rad) in a reaction volume of 5 μl, using SybrGreen I. Primer sets are described in Supplemental Table 2. HERV-W-env RNA expression analysis was performed with Biogazelle’s qbase+ (http://www.qbaseplus.com), which uses an improved model of the ΔΔCt approach. Gene expression is expressed as calibrated normalized relative quantities (CNRQ). CNRQ values are calculated using multiple reference genes by combining PCR replicate errors, amplification efficiency, and normalization variances.
Mouse model
CAG-Env transgenic mice expressing HERV-W-env were developed in a C57BL/6J background. In these mice, the MSRV-pV14-env transgene (GenBank, AF331500-1) is expressed under the control of the ubiquitous CAG promoter and is inserted in the HPRT locus of the murine X-chromosome. The HERV-W-env 3’LTR region is present in the transgene, allowing specific regulation of its expression in tissues. Mouse genotyping is performed using primer sets detailed in Supplemental Table 2. Pathogen-free C57BL/6J mice from Charles River Laboratories and CAG-Env mice from Plateforme de Haute Technologie Animale (PHTA, Grenoble, France) were received after weaning. After a 1-month period of acclimation, blood was collected at the saphenous vein after a 5-hour fasting period. Blood glucose was measured using glycemia reader FreeStyle Optium (Abbot). Measures were performed in duplicates. Remaining blood was centrifuged at 820 g for 10 minutes at 4°C, and serum was collected and conserved at –20°C before measuring insulin using Ultrasensitive Mouse Insulin ELISA kit (MERCODIA, 10-1249-01) according to supplier recommendations. After euthanization, pancreata were dissected, included in paraffin, and sections of 4 μm were stained with Hematoxylin Eosin Safran. Slides were digitalized with the slide scanner NANOZOOMER 2.0RS/C10730-12 (Hamamatsu).
Histology
Pancreas sections from CAG-Env mice and C57BL/6J mice were analyzed for the presence of insulitis and for immune cell and fat infiltration in the exocrine pancreas. A gradation from 0–3 was affected to each slide, 0 corresponding to no infiltration of fat or immune cells and 3 corresponding to high levels of infiltrations. Two sections separated by 500 μm were analyzed for each mouse, independently and by 2 persons unaware of mice genotype, and the mean of their evaluations was made for each slide. Final results are presented as the mean of the 2 sections for each mouse and as mean ± SEM for each group.
Cell culture
Human Langerhans islets.
Human Langerhans Islets purchased from Tebu-Bio were cultured at 37°C, 5% CO2, in complete sterile medium composed of proprietary islet media (PIM[S]) (Prodo Labs, PIM-S002GMP) supplemented with 5% PIM(ABS) (Prodo Labs, PIM-ABS001GMP), 1% PIM(G) (Prodo Labs, PIM-G001GMP), 15 μM Ciprofloxacin (Sigma-Aldrich, 17850-5G-F), 1.4 μM Amphotericin (Sigma-Aldrich, A2942, 50 ml), and 10 μM Gentamycin (Sigma-Aldrich, G1397, 10 ml). Islets were first seeded at a density of 1,000 islet equivalents (IEQ) in 25-cm² flasks with 12 ml of complete medium. Four days after flask seeding, islets were seeded at 75 IEQ/ml into 24-wells plates (1 ml/well). The following day, cells were treated with recombinant HERV-W-Env at 25, 50, and 100 ng/ml (PX Therapeutics) and GNbAC1 antibody 3 μg/ml (GeNeuro). HERV-W-Env buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.5% SDS, 10 mM DTT) and GNbAC1 buffer (20 mM Histidin, 5% saccharose, 0,01% Tween20, pH 6.0) were used in control conditions and kept constant in every condition. HERV-W-Env and GNbAC1 were preincubated 45 minutes at 25°C in PIM(ABS) and subsequently diluted 1:200 in complete PIM(S) medium without serum. Islets were incubated with these mixes for 5 hours, before adding complete PIM(S) medium with 10% of PIM(ABS) to achieve a final concentration of 5% PIM(ABS). Each condition was performed in triplicate. Islets were incubated for 2 days, at 37°C, 5% CO2.
The evening before Glucose Stimulation Insulin Secretion (GSIS) assay, islets were washed and incubated with HERV-W-Env and GNbAC1 as stated previously, except that dilutions were made in serum free 3 mM glucose medium. After an overnight incubation, islets were washed with 3 mM glucose medium and preincubated 1 hour with the same medium at 37°C, 5% CO2. Cells were then incubated 3 times 1 hour in 1 ml of 3 different solutions at 37°C: i) 3 mM glucose 2, 5% BSA medium, ii) 12 mM glucose 2, 5% BSA, and iii) 20 mM glucose 2, 5% BSA. Supernatants of the last incubation were collected for insulin secretion measurement using the Insulin ELISA kit (Mercodia, catalog 10-1113-01) according to manufacturer recommendations.
Islet viability was assessed by InSphero using islet microtissues. To overcome heterogeneity of purified islets directly obtained from deceased donors, a postisolation protocol was developed by InSphero to take advantage of the inherent ability of dispersed epithelial cells to reestablish cell-cell contact and assemble into spheroids. Briefly, after a mild dissociation of primary islets yielding to a single cell suspension, cells were reaggregated in hanging drop cultures, in given numbers and proportions for each cell type (α, β, and δ cells). This scaffold-free self-assembly of dispersed cells allows reconstituted islets to produce their own extracellular matrix and yields to islets of uniform size and cellular composition. Islet microtissues were dosed for 5 hours with HERV-W-Env (0, 200, and 400 ng/ml) in media without serum. HERV-W-Env buffer concentration was kept constant in the 3 conditions. After this first incubation step, 35 μl of the media were removed and 35 μl of culture media containing 10% serum were added to obtain a final concentration of 5%. Islet microtissues were incubated for 48 hours and then lysed to analyze total ATP content using Promega CellTiter-Glo Luminescent Cell Viability assay. This experiment was performed once with 6 replicates.
INS1-E rat insulinoma cell line.
INS-1E cells, provided to J. Portoukalian by P. Maechler (Geneva, Switzerland) and used between passage 82–86, were cultured at 37°C, 5% CO2 in complete sterile medium composed of RPMI 1640 glutamax (Life Technologies, catalog 61870-010) supplemented with 10% heat-inactivated FCS, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, 10 mM HEPES, and 1% penicillin/streptomycin. Cells were seeded at 150,000 cells/ml into 12-wells plates (1 ml/well). The following day, cells were treated with 500 μl HERV-W-Env protein (PX Therapeutics), ranging from 20–800 ng/ml, diluted with INS-1E medium 0% serum. HERV-W-Env buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.5% SDS, 10 mM DTT) was used in control condition (0 ng/ml HERV-W-Env) and kept constant in every condition. After 6 hours of incubation, 500 μl of INS-1E medium 20% serum was added to each well. Each condition was performed in triplicate. INS-1E–treated cells were incubated for 3 days, at 37°C, 5% CO2. Before GSIS assay, cells were maintained for 2 hours in DMEM glucose-free medium at 37°C. Cells were then washed twice with glucose-free Krebs-Ringer bicarbonate HEPES buffer (KRBH) (1,350 mM NaCl, 3.6 mM KCl, 50 mM NaHCO3, 5 mM NaH2PO4, 5 mM MgCl2, 15 mM CaCl2, 10 mM HEPES and 0.1% BSA, pH7.4), and preincubated 15 minutes at 37°C in KRBH buffer. INS1E were then incubated two times 30 minutes in 1 ml of 2 different solutions: i) glucose-free KRBH 0.1% BSA and ii) KRBH 0.1% BSA with 10 mM glucose. Supernatants were collected to measure insulin secretion using the Ultrasensitive Rat Insulin ELISA kit (MERCODIA, catalog 10-1251-01) according to manufacturer recommendations.
Statistics
Normality of the distribution was assessed by Shapiro-Wilk normality test. Depending on the normality of distributions, differences between group pairs were analyzed with Student t test or Mann-Whitney U test. Group differences were analyzed either by ANOVA test followed by Bonferroni’s multiple comparisons test or by Kruskal-Wallis test followed by Dunn’s multiple comparisons test. Correlations between CD3 and CD68 density and HERV-W-Env+ staining area were analyzed using Spearman correlation. Receiver operating characteristic (ROC) curve analyses were performed on values from the 3 cohorts presented in Figure 1, A–C. Individuals were classified into either positive or negative groups for HERV-W-Env based on thresholds set at mean plus 2 SD of each control groups (without outliers above Mean plus 1 SD). Cut-offs were set at 0.231 OD450 for ELISA, at 1.356 CNRQ for qRT-PCR, and 21.43 % for pancreas IHC. Contingency tables were subsequently analyzed using χ2 test. Unless otherwise stated, tests were performed two-tailed. All analyses were performed using GraphPad Prism version 7 (GraphPad Software). Data are presented as individual values and as mean ± SEM unless otherwise indicated. Findings were assumed statistically significant at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Study approval
Mouse experiments were performed in accordance with the animal protection decree 2013-118 (1st February 2013). All experimental procedures were registered and approved by ethical committee C2EA-55, protocol DR2013-34, and by French Ministry of Research, reference number 01653.01. Human pancreata from T1D and control individuals were collected through the nPOD program according to procedures and ethical review by the University of Florida IRB as previously described (63). EFS provided PBMC and serum from non-T1D blood donors under the convention 14-1797. PBMC, serum, and human Langerhans islets from T1D patients were provided by TEBU BIO Company, which guarantees the compliance with international standards for donation and collection of human tissue, including a written informed consent received from donors prior to blood collection. Importation from the US of these tissues and cells was made with authorization IE-2013-677.
Author contributions
SL designed experiments, interpreted data and wrote the manuscript. JM designed experiments, interpreted data and revised the manuscript. JJ, AD, NQ, and KR performed in vivo and in vitro experiments. MN developed the algorithms used for staining quantification. MS performed immunostainings. JD and RG developed and performed ELISA assay. TP developed and performed qRT-PCR assay. JP and JLT performed preliminary experiments and provided expertise for the study design. HP designed experiments, interpreted data and revised the manuscript.
Supplementary Material
Acknowledgments
The authors are grateful to Irina Kusmarteva, Alberto Pugliese, and Mark Atkinson from nPOD for their help in providing pancreata specimens with histological and clinical data, and for their advice on experimental design. This research was performed with the support of the nPOD, a collaborative T1D research project sponsored by JDRF and would have not been possible without the generosity of donors and their families, as well as the dedication of clinicians, scientists and all persons who make nPOD effective. The authors thank Benjamin Charvet and Thomas Rückle for commenting on the manuscript, and Antoinette Angeloni for administrative support. This work was funded by GeNeuro Innovation.
Version 1. 08/31/2017
Electronic publication
Footnotes
Conflict of interest: SL, JM, JJ, AD, NQ, and KR receive compensation for their work at GeNeuro Innovation. MN and MS receive compensation for their work at Excilone. TP receives compensation for his work at Biogazelle. HP receives compensation for his work at GeNeuro Innovation and is an inventor on patents owned by bioMérieux, INSERM, and GeNeuro but has transferred all his rights to bioMérieux or to GeNeuro under applicable laws for employed inventors.
Reference information: JCI Insight. 2017;2(17):e94387. https://doi.org/10.1172/jci.insight.94387.
Contributor Information
Sandrine Levet, Email: sl@geneuro.com.
Julie Medina, Email: jme@geneuro.com.
Julie Joanou, Email: jj@geneuro.com.
Amandine Demolder, Email: ade@geneuro.com.
Nelly Queruel, Email: nq@geneuro.com.
Matthieu Normand, Email: matthieu.normand@excilone.com.
Marine Seffals, Email: marine.seffals@excilone.com.
Julie Dimier, Email: julie.dimier@gmail.com.
Thomas Piofczyk, Email: thomas.piofczyk@biogazelle.com.
Jacques Portoukalian, Email: jacques.portoukalian@univ-lyon1.fr.
Jean-Louis Touraine, Email: jean-louis.touraine@chu-lyon.fr.
Hervé Perron, Email: hperron@geneuro.com.
References
- 1.Atkinson MA. The pathogenesis and natural history of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;211 doi: 10.1101/cshperspect.a007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tai N, Wong FS, Wen L. The role of the innate immune system in destruction of pancreatic beta cells in NOD mice and humans with type I diabetes. J Autoimmun. 2016;71:26–34. doi: 10.1016/j.jaut.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell-Thompson M, et al. Insulitis and β-Cell Mass in the Natural History of Type 1 Diabetes. Diabetes. 2016;65(3):719–731. doi: 10.2337/db15-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell-Thompson M, Rodriguez-Calvo T, Battaglia M. Abnormalities of the Exocrine Pancreas in Type 1 Diabetes. Curr Diab Rep. 2015;15(10):79. doi: 10.1007/s11892-015-0653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Herrath M, Sanda S, Herold K. Type 1 diabetes as a relapsing-remitting disease? Nat Rev Immunol. 2007;7(12):988–994. doi: 10.1038/nri2192. [DOI] [PubMed] [Google Scholar]
- 6.Christoffersson G, Rodriguez-Calvo T, von Herrath M. Recent advances in understanding Type 1 Diabetes. F1000Res. 2016:5. doi: 10.12688/f1000research.7356.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson SJ, et al. Evaluation of the fidelity of immunolabelling obtained with clone 5D8/1, a monoclonal antibody directed against the enteroviral capsid protein, VP1, in human pancreas. Diabetologia. 2014;57(2):392–401. doi: 10.1007/s00125-013-3094-7. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Calvo T, von Herrath MG. Enterovirus infection and type 1 diabetes: closing in on a link? Diabetes. 2015;64(5):1503–1505. doi: 10.2337/db14-1931. [DOI] [PubMed] [Google Scholar]
- 9.Richardson SJ, et al. Detection of enterovirus in the islet cells of patients with type 1 diabetes: what do we learn from immunohistochemistry? Reply to Hansson SF, Korsgren S, Pontén F et al [letter] Diabetologia. 2014;57(3):647–649. doi: 10.1007/s00125-014-3167-2. [DOI] [PubMed] [Google Scholar]
- 10.Hober D, Sauter P. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat Rev Endocrinol. 2010;6(5):279–289. doi: 10.1038/nrendo.2010.27. [DOI] [PubMed] [Google Scholar]
- 11.Hansson SF, Korsgren S, Pontén F, Korsgren O. Detection of enterovirus in the islet cells of patients with type 1 diabetes: what do we learn from immunohistochemistry? Diabetologia. 2014;57(3):645–646. doi: 10.1007/s00125-013-3138-z. [DOI] [PubMed] [Google Scholar]
- 12.Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu P. The potential role of retroviruses in autoimmunity. Immunol Rev. 2016;269(1):85–99. doi: 10.1111/imr.12371. [DOI] [PubMed] [Google Scholar]
- 14.Belshaw R, et al. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci USA. 2004;101(14):4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 2012;13(4):283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 16.Conrad B, Weissmahr RN, Böni J, Arcari R, Schüpbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90(2):303–313. doi: 10.1016/S0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 17.Jaeckel E, Heringlake S, Berger D, Brabant G, Hunsmann G, Manns MP. No evidence for association between IDDMK(1,2)22, a novel isolated retrovirus, and IDDM. Diabetes. 1999;48(1):209–214. doi: 10.2337/diabetes.48.1.209. [DOI] [PubMed] [Google Scholar]
- 18.Kim A, et al. Human endogenous retrovirus with a high genomic sequence homology with IDDMK(1,2)22 is not specific for Type I (insulin-dependent) diabetic patients but ubiquitous. Diabetologia. 1999;42(4):413–418. doi: 10.1007/s001250051173. [DOI] [PubMed] [Google Scholar]
- 19.Muir A, Ruan QG, Marron MP, She JX. The IDDMK(1,2)22 retrovirus is not detectable in either mRNA or genomic DNA from patients with type 1 diabetes. Diabetes. 1999;48(1):219–222. doi: 10.2337/diabetes.48.1.219. [DOI] [PubMed] [Google Scholar]
- 20.Gaskins HR, Prochazka M, Hamaguchi K, Serreze DV, Leiter EH. Beta cell expression of endogenous xenotropic retrovirus distinguishes diabetes-susceptible NOD/Lt from resistant NON/Lt mice. J Clin Invest. 1992;90(6):2220–2227. doi: 10.1172/JCI116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsumura H, Miyazawa M, Ogawa S, Wang JZ, Ito Y, Shimura K. Detection of endogenous retrovirus antigens in NOD mouse pancreatic beta-cells. Lab Anim. 1998;32(1):86–94. doi: 10.1258/002367798780559464. [DOI] [PubMed] [Google Scholar]
- 22.Bashratyan R, et al. Type 1 diabetes pathogenesis is modulated by spontaneous autoimmune responses to endogenous retrovirus antigens in NOD mice. Eur J Immunol. 2017;47(3):575–584. doi: 10.1002/eji.201646755. [DOI] [PubMed] [Google Scholar]
- 23.Perron H, et al. Multiple sclerosis retrovirus particles and recombinant envelope trigger an abnormal immune response in vitro, by inducing polyclonal Vbeta16 T-lymphocyte activation. Virology. 2001;287(2):321–332. doi: 10.1006/viro.2001.1045. [DOI] [PubMed] [Google Scholar]
- 24.Perron H, et al. Human endogenous retrovirus type W envelope expression in blood and brain cells provides new insights into multiple sclerosis disease. Mult Scler. 2012;18(12):1721–1736. doi: 10.1177/1352458512441381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kremer D, et al. Human endogenous retrovirus type W envelope protein inhibits oligodendroglial precursor cell differentiation. Ann Neurol. 2013;74(5):721–732. doi: 10.1002/ana.23970. [DOI] [PubMed] [Google Scholar]
- 26.Perron H, et al. Isolation of retrovirus from patients with multiple sclerosis. Lancet. 1991;337(8745):862–863. doi: 10.1016/0140-6736(91)92579-Q. [DOI] [PubMed] [Google Scholar]
- 27.Perron H, et al. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc Natl Acad Sci USA. 1997;94(14):7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blond JL, et al. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol. 1999;73(2):1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mameli G, et al. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not Human herpesvirus 6. J Gen Virol. 2007;88(Pt 1):264–274. doi: 10.1099/vir.0.81890-0. [DOI] [PubMed] [Google Scholar]
- 30.van Horssen J, van der Pol S, Nijland P, Amor S, Perron H. Human endogenous retrovirus W in brain lesions: Rationale for targeted therapy in multiple sclerosis. Mult Scler Relat Disord. 2016;8:11–18. doi: 10.1016/j.msard.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 32.Perron H, Lang A. The human endogenous retrovirus link between genes and environment in multiple sclerosis and in multifactorial diseases associating neuroinflammation. Clin Rev Allergy Immunol. 2010;39(1):51–61. doi: 10.1007/s12016-009-8170-x. [DOI] [PubMed] [Google Scholar]
- 33.Mameli G, et al. Expression and activation by Epstein Barr virus of human endogenous retroviruses-W in blood cells and astrocytes: inference for multiple sclerosis. PLoS One. 2012;7(9):e44991. doi: 10.1371/journal.pone.0044991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firouzi R, et al. Multiple sclerosis-associated retrovirus particles cause T lymphocyte-dependent death with brain hemorrhage in humanized SCID mice model. J Neurovirol. 2003;9(1):79–93. doi: 10.1080/13550280390173328. [DOI] [PubMed] [Google Scholar]
- 35.Rolland A, Jouvin-Marche E, Viret C, Faure M, Perron H, Marche PN. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J Immunol. 2006;176(12):7636–7644. doi: 10.4049/jimmunol.176.12.7636. [DOI] [PubMed] [Google Scholar]
- 36.Madeira A, Burgelin I, Perron H, Curtin F, Lang AB, Faucard R. MSRV envelope protein is a potent, endogenous and pathogenic agonist of human toll-like receptor 4: Relevance of GNbAC1 in multiple sclerosis treatment. J Neuroimmunol. 2016;291:29–38. doi: 10.1016/j.jneuroim.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Perron H, et al. Human endogenous retrovirus protein activates innate immunity and promotes experimental allergic encephalomyelitis in mice. PLoS One. 2013;8(12):e80128. doi: 10.1371/journal.pone.0080128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curtin F, et al. A placebo randomized controlled study to test the efficacy and safety of GNbAC1, a monoclonal antibody for the treatment of multiple sclerosis - Rationale and design. Mult Scler Relat Disord. 2016;9:95–100. doi: 10.1016/j.msard.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Gaudin P, et al. Infrequency of detection of particle-associated MSRV/HERV-W RNA in the synovial fluid of patients with rheumatoid arthritis. Rheumatology (Oxford) 2000;39(9):950–954. doi: 10.1093/rheumatology/39.9.950. [DOI] [PubMed] [Google Scholar]
- 40.Lehuen A, Diana J, Zaccone P, Cooke A. Immune cell crosstalk in type 1 diabetes. Nat Rev Immunol. 2010;10(7):501–513. doi: 10.1038/nri2787. [DOI] [PubMed] [Google Scholar]
- 41.Walker LS, von Herrath M. CD4 T cell differentiation in type 1 diabetes. Clin Exp Immunol. 2016;183(1):16–29. doi: 10.1111/cei.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garay-Malpartida HM, Mourão RF, Mantovani M, Santos IA, Sogayar MC, Goldberg AC. Toll-like receptor 4 (TLR4) expression in human and murine pancreatic beta-cells affects cell viability and insulin homeostasis. BMC Immunol. 2011;12:18. doi: 10.1186/1471-2172-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amyot J, Semache M, Ferdaoussi M, Fontés G, Poitout V. Lipopolysaccharides impair insulin gene expression in isolated islets of Langerhans via Toll-Like Receptor-4 and NF-κB signalling. PLoS One. 2012;7(4):e36200. doi: 10.1371/journal.pone.0036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, Song L, Gao X, Chang W, Qin X. Toll-like receptor 4 on islet β cells senses expression changes in high-mobility group box 1 and contributes to the initiation of type 1 diabetes. Exp Mol Med. 2012;44(4):260–267. doi: 10.3858/emm.2012.44.4.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merglen A, Theander S, Rubi B, Chaffard G, Wollheim CB, Maechler P. Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology. 2004;145(2):667–678. doi: 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- 46.Le Dantec C, Vallet S, Brooks WH, Renaudineau Y. Human endogenous retrovirus group E and its involvement in diseases. Viruses. 2015;7(3):1238–1257. doi: 10.3390/v7031238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trela M, Nelson PN, Rylance PB. The role of molecular mimicry and other factors in the association of Human Endogenous Retroviruses and autoimmunity. APMIS. 2016;124(1-2):88–104. doi: 10.1111/apm.12487. [DOI] [PubMed] [Google Scholar]
- 48.Li W, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7(307):307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duperray A, et al. Inflammatory response of endothelial cells to a human endogenous retrovirus associated with multiple sclerosis is mediated by TLR4. Int Immunol. 2015;27(11):545–553. doi: 10.1093/intimm/dxv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faucard R, et al. Human Endogenous Retrovirus and Neuroinflammation in Chronic Inflammatory Demyelinating Polyradiculoneuropathy. EBioMedicine. 2016;6:190–198. doi: 10.1016/j.ebiom.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Devaraj S, Dasu MR, Rockwood J, Winter W, Griffen SC, Jialal I. Increased toll-like receptor (TLR) 2 and TLR4 expression in monocytes from patients with type 1 diabetes: further evidence of a proinflammatory state. J Clin Endocrinol Metab. 2008;93(2):578–583. doi: 10.1210/jc.2007-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devaraj S, Jialal I, Yun JM, Bremer A. Demonstration of increased toll-like receptor 2 and toll-like receptor 4 expression in monocytes of type 1 diabetes mellitus patients with microvascular complications. Metab Clin Exp. 2011;60(2):256–259. doi: 10.1016/j.metabol.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohammad MK, et al. Dysregulated Toll-like receptor expression and signaling in bone marrow-derived macrophages at the onset of diabetes in the non-obese diabetic mouse. Int Immunol. 2006;18(7):1101–1113. doi: 10.1093/intimm/dxl045. [DOI] [PubMed] [Google Scholar]
- 54.Campbell-Thompson M, Wasserfall C, Montgomery EL, Atkinson MA, Kaddis JS. Pancreas organ weight in individuals with disease-associated autoantibodies at risk for type 1 diabetes. JAMA. 2012;308(22):2337–2339. doi: 10.1001/jama.2012.15008. [DOI] [PubMed] [Google Scholar]
- 55.Panicot L, Mas E, Thivolet C, Lombardo D. Circulating antibodies against an exocrine pancreatic enzyme in type 1 diabetes. Diabetes. 1999;48(12):2316–2323. doi: 10.2337/diabetes.48.12.2316. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez-Calvo T, Ekwall O, Amirian N, Zapardiel-Gonzalo J, von Herrath MG. Increased immune cell infiltration of the exocrine pancreas: a possible contribution to the pathogenesis of type 1 diabetes. Diabetes. 2014;63(11):3880–3890. doi: 10.2337/db14-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arif S, et al. Blood and islet phenotypes indicate immunological heterogeneity in type 1 diabetes. Diabetes. 2014;63(11):3835–3845. doi: 10.2337/db14-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coppieters KT, Boettler T, von Herrath M. Virus infections in type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2(1):a007682. doi: 10.1101/cshperspect.a007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mason MJ, et al. Low HERV-K(C4) copy number is associated with type 1 diabetes. Diabetes. 2014;63(5):1789–1795. doi: 10.2337/db13-1382. [DOI] [PubMed] [Google Scholar]
- 60.Ruprecht K, et al. Regulation of human endogenous retrovirus W protein expression by herpes simplex virus type 1: implications for multiple sclerosis. J Neurovirol. 2006;12(1):65–71. doi: 10.1080/13550280600614973. [DOI] [PubMed] [Google Scholar]
- 61.Assinger A, Yaiw KC, Göttesdorfer I, Leib-Mösch C, Söderberg-Nauclér C. Human cytomegalovirus (HCMV) induces human endogenous retrovirus (HERV) transcription. Retrovirology. 2013;10:132. doi: 10.1186/1742-4690-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bian X, et al. Immunoproteomic Profiling of Antiviral Antibodies in New-Onset Type 1 Diabetes Using Protein Arrays. Diabetes. 2016;65(1):285–296. doi: 10.2337/db15-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pugliese A, et al. The Juvenile Diabetes Research Foundation Network for Pancreatic Organ Donors with Diabetes (nPOD) Program: goals, operational model and emerging findings. Pediatr Diabetes. 2014;15(1):1–9. doi: 10.1111/pedi.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.