Abstract
Promising therapeutic approaches for eradicating HIV include transcriptional activation of provirus from latently infected cells using latency-reversing agents (LRAs) and immune-mediated clearance to purge reservoirs. Accurate detection of cells capable of producing viral antigens and virions, and the measurement of clearance of infected cells, is essential to assessing therapeutic efficacy. Here, we apply enhanced methodology extending the sensitivity limits for the rapid detection of subfemtomolar HIV gag p24 capsid protein in CD4+ T cells from ART-suppressed HIV+ individuals, and we show viral protein induction following treatment with LRAs. Importantly, we demonstrate that clinical administration of histone deacetylase inhibitors (HDACis; vorinostat and panobinostat) induced HIV gag p24, and ex vivo stimulation produced sufficient viral antigen to elicit immune-mediated cell killing using anti-gp120/CD3 bispecific antibody. These findings extend beyond classical nucleic acid endpoints, which are confounded by the predominance of mutated, defective proviruses and, of paramount importance, enable assessment of cells making HIV protein that can now be targeted by immunological approaches.
Keywords: AIDS/HIV, Infectious disease
Clinical administration of HDAC inhibitors induces HIV protein expression and elicits immune-based clearance ex vivo.
Introduction
HIV persistence in latently infected, resting CD4+ T cells is broadly recognized as a barrier to eradicating HIV (1, 2). To eliminate the latent reservoir, infected cells must be recognized as harboring virus and then effectively destroyed and cleared. Latency disruption is under intense investigation, and several strategies are being evaluated (3–6). Recent therapeutic approaches have focused on the use of latency-reversing agents (LRAs) to induce viral transcription, initiate viral translation, and subsequently elicit death through viral cytopathic effects or immune-mediated cell killing (“shock and kill”). A class of small molecules known as histone deacetylase inhibitors (HDACis) has been broadly examined as LRAs, and clinical studies have shown that single and multiple administration results in induced viral RNA transcription in CD4+ T cells from antiretroviral therapy–suppressed (ART-suppressed) HIV+ subjects (7–11). However, direct quantitation of cell-associated viral protein following clinical administration of HDACis has not been possible. Other agents, such as bromodomain inhibitors and protein kinase C (PKC) agonists, have also been shown to induce viral transcription in vitro and in preclinical models and are under further evaluation as potential clinical HIV “shock” agents (6, 12–16). Worth noting, the induction of viral protein expression is a prerequisite for several investigational immune-mediated “kill” strategies (3–5). Broadly neutralizing antibodies are already being used in clinical trials, and other strategies involving the use of other forms of antibodies that target viral protein are currently being developed, highlighting the need for approaches that can demonstrate low-level detection of viral antigens and report on the efficacy of such therapeutic approaches (5, 17, 18).
The exploration of cure strategies targeting the latent HIV reservoir highlights a need for new assays and research tools to quantify viral burden in tissue and various body fluids.
The rare incidence of latently infected cells for which robust phenotypic markers are still needed presents a significant challenge for identifying these cells and measuring changes in viral expression (3, 19, 20). In addition, latency reversal may induce proviral transcription in only a fraction of this population, and the observed increases in cellular HIV-1 RNA may represent defective proviruses and could also result from alternative splicing or read-through transcripts, which may not give rise to efficient splicing, translation, and protein expression (19–22). Quantifying virus production has been difficult, and traditional cell culture–based assays, such as viral outgrowth assays (VOA), which measure HIV gag p24 antigen in culture medium after expansion in culture for 2–3 weeks, require large sample volumes and have limited throughput and clinical application (23, 24). TILDA, as an example of many cell-associated HIV RNA assays described, may offer some advantages to quantitative VOA (qVOA) such as increased throughput and decreased sample requirements, but still only measures RNA transcripts (25). An alternative approach that directly measures protein production and also affords increased sensitivity as a component of an existing ex vivo assay for measuring HIV reservoir and/or can be leveraged to assess protein levels needed to induce viral cytopathicity or promote immune-mediated clearance is an important advancement for HIV cure research (26–28). Toward this goal, we report and apply enhanced methodology for rapid and sensitive detection of HIV gag p24 protein in latently infected CD4+ T cells from ART-suppressed HIV+ individuals and demonstrate that latency reversal of HIV can produce sufficient viral antigen to enable immune-targeted clearance.
Results
Direct measurement of HIV gag p24 in ART-suppressed HIV+ CD4+ T cells.
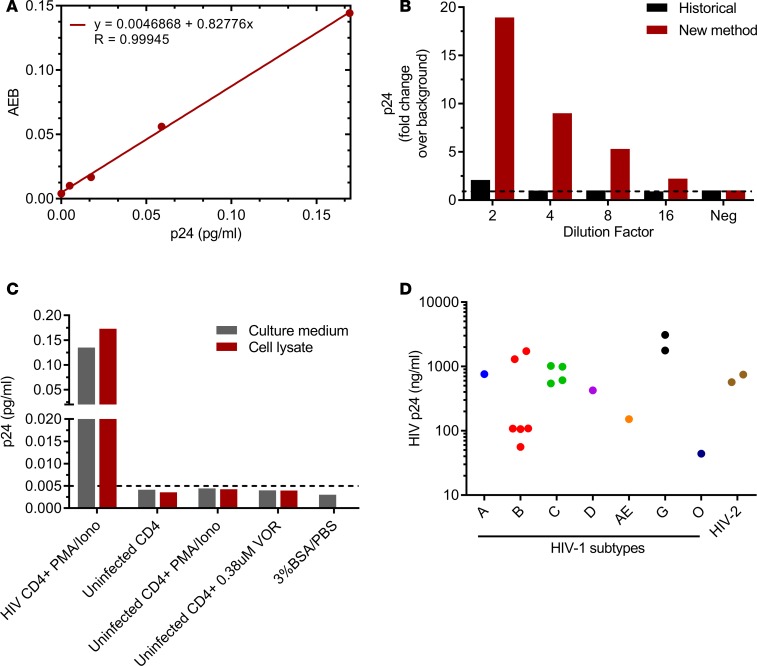
Recent studies have reported the use of a digital immunoassay to measure HIV gag p24 protein from serum or plasma of acutely infected HIV+ individuals or cultured media of HIV-infected CD4+ T cells (29–31). Despite these advances, commercial methods encounter matrix issues and lack the same degree of sensitivity in quantifying cell-associated viral p24 protein from aviremic HIV+ individuals. To overcome this limitation, we optimized the digital immunoassay protocol, detailed in Methods, for the rapid and sensitive detection of HIV gag p24 protein in latently infected CD4+ T cell lysates prepared from ART-suppressed HIV+ individuals and applied this approach to assess proviral reactivation and immune-mediated cell kill. Robust linearity in detecting p24 in CD4+ T cell lysates from aviremic HIV+ individuals was demonstrated with an analytical limit of detection (LOD) of 5 fg/ml (Figure 1A), corresponding to 125,000 p24 copies/ml or 18,000 copies/144 μl assay input and, thus, about 9 viruses/144 μl input (29, 30). The optimized methods extended the sensitivity LODs for HIV p24 in cell lysates (Figure 1B). Additional studies in the HIV-1–infected MOLTIIIB cell line also suggested HIV gag protein quantification is feasible in 0.125 cells/ml lysate (~2% cell/144 μl) in this cell model (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.92901DS1),
Figure 1. Ultrasensitive detection of HIV gag p24.
(A) Linear dilution and low fg/ml detection of recombinant p24 spiked into CD4+ T cell lysates from uninfected donors, representative of n = 4. AEB, average enzyme per bead. (B) Representative data of n = 3 of robust p24 quantitation in cell lysates from ART-suppressed HIV+ CD4 T cells using new method versus historical (29, 30). (C) Measurable p24 in cell lysate and culture medium from HIV-infected CD4+ cells stimulated with PMA/ionomycin but not in uninfected CD4 cells stimulated either with PMA/ionomycin or SAHA, or in BSA/PBS buffer, n = 1. Dashed line represents analytical LOD, 5 fg/ml. (D) Detection of p24 in viral lysates from 18 clinically diverse HIV-1/2 subtypes.
To demonstrate assay specificity and breadth, we first evaluated p24 levels in BSA/PBS buffer alone and CD4+ T cells isolated from uninfected or ART-suppressed HIV+ donors and treated ex vivo with either PMA/Ionomycin or the HDACi vorinostat (VOR). Detectable p24 levels were observed only in HIV+ cell lysates (Figure 1C). Additionally, magnetic bead-based immunodepletion studies were performed using either p24 ELISA antibodies or mouse IgG incubated with CD4+ T cell lysates from HIV+ viremic donors. HIV gag p24 was absent in the flow-through (FT) of p24 antibody–depleted samples but measurable in the IgG control FT, demonstrating specificity of the assay for HIV gag protein (Supplemental Figure 1B). To further assess whether the assay also recognized unprocessed gag polyprotein, recombinant proteins p55 and p24 were diluted 2-fold from 20–0.3 fM in 3% BSA/PBS buffer and measured simultaneously. Linear dilution and low fg/ml detection of recombinant p55 was observed, suggesting recombinant HIV gag p55 polyprotein is also measurable in vitro (Supplemental Figure 1C). It is unclear, however, how efficiently the p24 antibody recognizes gag p55 in vivo. Assay breadth was demonstrated by evaluating viral lysates from 18 genetically diverse clinical isolates, including 8 HIV-1 subtypes spanning groups M and O, and 1 HIV-2 isolate. As shown in Figure 1D, viral HIV gag protein was detected in all 18 clinical isolates, and protein values correlated significantly with RNA copies/μl (r = 0.837; P < 0.001; data not shown).
Latency-reversing agents induce viral p24 expression.
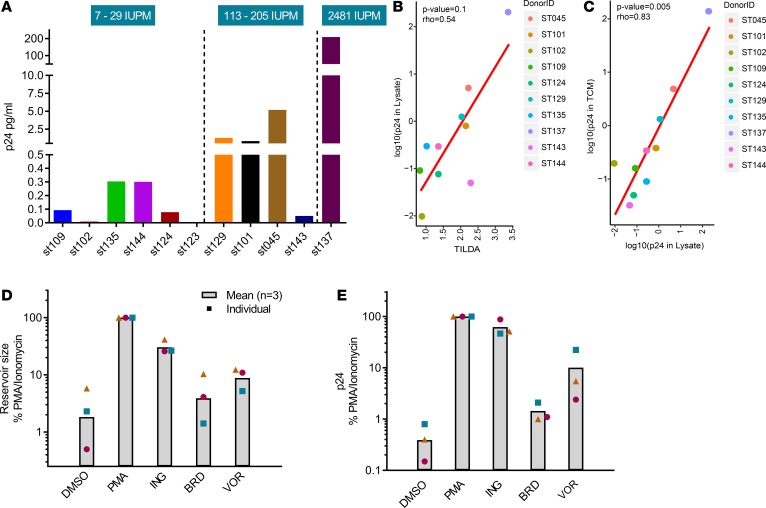
Viral protein production is critically important to investigational immunotherapies (5, 17, 18, 32, 33), and thus, it is equally important to assess viral protein production following latency reversal and, subsequently, to determine if the level of viral protein induction on a reactivated cell is sufficient to enable immune-mediated clearance. Classical measures of the HIV reservoir leverage nucleic acid readouts to estimate the burden of disease or viral transcription, but they often overestimate reservoir size due to the presence of defective proviruses. Techniques that quantify the frequency or percentage of cells that become transcriptionally active following viral reactivation have recently been developed, including the Tat/Rev-induced msRNA limiting dilution assay (TILDA); however, assays like TILDA do not quantify translationally competent proviruses. Toward the initial goal of detecting viral protein upon latent cell reactivation, we assessed the relationship between HIV-1 gag protein induction and inducible reservoir size, as measured by TILDA, in 11 ART-suppressed subjects following 48-hour PMA/Ionomycin stimulation. TILDA measurements allowed the segregation of subjects into 3 categories of small, moderate, and large inducible reservoirs (Figure 2A). The majority of subjects displayed inducible p24 levels that trended positively with the TILDA readout; however, a statistically significant correlation between TILDA and p24 was lacking (r = 0.54, P < 0.1; Figure 2B). Notably, a significant correlation was observed between p24 measured in cell lysates and culture medium (r = 0.83, P = 0.005; Figure 2C), and extended studies are underway to assess virus-associated p24 in the culture medium versus contribution, if any, from cellular leakage. Next, we evaluated HIV reactivation in cells treated ex vivo with either 10 nM ingenol-3-angelate, 5 μM bromosporine, or 750 nM VOR and observed comparable trends between TILDA and p24 following 48-hour stimulation of CD4+ T cells from ART-suppressed HIV+ subjects (Figure 2, D and E). Measurable induction was also observed at shorter incubation periods (6–24 hours; data not shown). Importantly, we demonstrate that HDACis and other LRAs can elicit both viral RNA and protein expression ex vivo.
Figure 2. Comparison of TILDA and p24 protein expression following latency reversal in ART-suppressed CD4+ T cells.
(A) Comparison of cell-associated p24 and inducible reservoir sizes, as measured by TILDA, from 11 ART-suppressed HIV+ subjects, n = 1 per patient. IUPM represents the number of tat/rev msRNA–expressing cells per million, and IUPM values are ranked left to right. (B) Correlation study of TILDA and cell-associated p24; r = 0.54, P = 0.1, Spearman correlation test. (C) Significant correlation observed between p24 in cell lysates and tissue culture medium (TCM); r = 0.83, P = 0.005, Spearman correlation method. (D and E) Quantitation of HIV reactivation by TILDA (D) and p24 digital ELISA (E) following 48-hr ex vivo stimulation with latency-reversing agents in CD4+ T cells from ART-suppressed HIV+ subjects. n = 3 subjects per condition. PMA, PMA/ionomycin; ING, ingenol-3-angelate; BRD, bromosporine; VOR, vorinostat.
VOR and panobinostat induce HIV gag p24 following clinical HDACi administration.
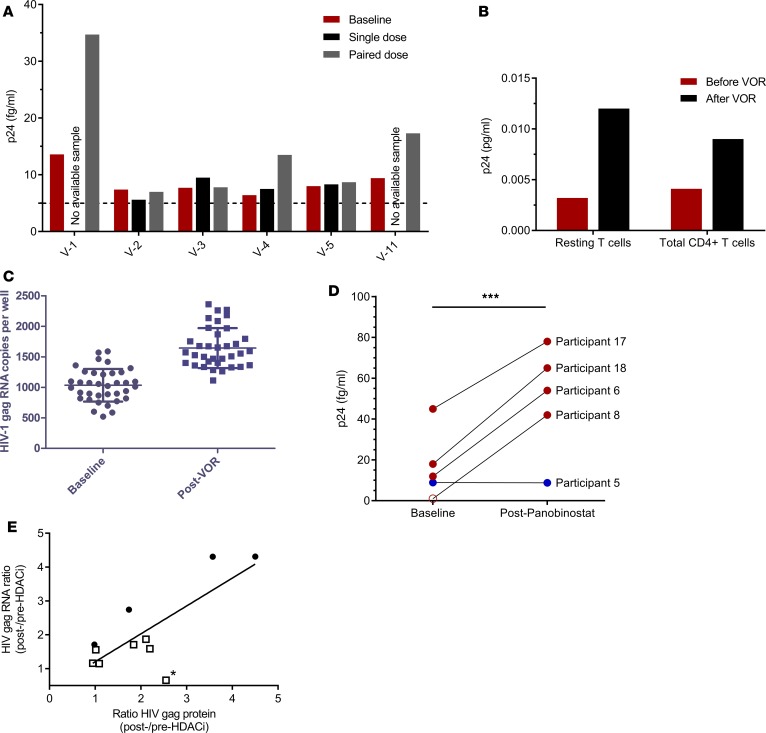
HDACis are the most studied latency-reversing agents and have shown induction of cell-associated HIV RNA in vivo using single or multiple dosing paradigms (7–11, 34); however, no study to date, to our knowledge, has demonstrated the direct induction of viral protein after clinical administration of HDACis. To determine if in vivo dosing of HDACis can induce viral protein expression, we examined changes in cell-associated p24 from peripherally isolated CD4+ T cells following clinical administration of 2 HDACis, either VOR or panobinostat. A limited number of available samples were obtained from previously published or recently completed clinical trials (9, 10, 34). Briefly, aviremic HIV+ individuals received either VOR (400 mg) at 72-hour intervals or panobinostat (20 mg) 3 times per week every other week for 8 weeks while maintaining combination ART (9, 10, 34). We quantified changes in p24 from baseline following administration of 1, 2, or 10 doses of VOR (9, 34), as well as multiple doses (6 – 10) of panobinostat (Supplemental Table 1), for subjects in whom remaining samples were available for p24 assessment. In the VOR study, PBMCs were collected from HIV+ subjects by leukapheresis 3–6 hours after VOR, and total CD4+ T cells were subsequently isolated and cultured for 48 hours with 1 μM raltegravir to prevent further rounds of infection. Peripheral blood cells were obtained by venipuncture on panobinostat-treated subjects at baseline and 8 hours after dose. Similarly, CD4+ T cells were isolated and cultured with 1 μM raltegravir for 48 hours. Cells were cultured to allow sufficient time for p24 translation to occur, since collection was performed within a few hours of drug administration to enable the primary clinical study endpoint measurements of RNA quantitation. Applying this approach, 3 of 6 VOR-treated subjects showed measurable increases in p24 protein above baseline, primarily after the second dose, and dilution of donor V-1 sample demonstrated linear detection of p24 within the analytical range of the assay (Figure 3A and Supplemental Figure 2). An additional sample was available for a patient receiving 10 doses of VOR and showed p24 induction above baseline in both total and resting memory CD4+ T cell populations (Figure 3B). Both VOR dosing paradigms results in increased cell-associated HIV-1 gag RNA in the subjects studied (Figure 3, C and E; refs. 9 and 34). For the participant receiving 10 doses of VOR, p24 was assessed immediately after thawing cells and demonstrates the ability to also detect protein without time in culture, although it is unknown whether or not dependencies exist between the numbers of clinically administered doses, the time of sample collection, and the detection of p24. In the panobinostat study, p24 measurements were also made on available samples from 5 of 15 participants from the Rasmussen et al. study (10), as shown in Supplemental Table 1. Briefly, 4 participants had matched baseline and post-panobinostat samples, with the panobinostat samples collected the same day as drug administration (participants 6, 8, 17, and 18; Supplemental Table 1). To rule out selection bias, we have included data comparing the fold change in cell-associated HIV RNA at the relevant time points in the panobinostat study for the 5 individuals contributing samples for the present study versus the remaining 10 study participants (Supplemental Figure 3). Four of 4 subjects showed increased p24 relative to baseline following either the seventh panobinostat dose (participants 8, 17, 18) or ninth dose (participant 6; Figure 3D), with the increase being statistically significant (P < 0.001). Participant 5 was examined 5 days after the sixth dose of panobinostat (i.e., during the rest period; Supplemental Table 1) and showed no detectable change relative to baseline (Figure 3D). Of note, this patient displayed very low levels of cell-associated RNA during panobinostat treatment and, like only 1 of 15 participants in the original study, plasma HIV RNA from this individual remained continuously undetectable throughout the clinical study (10). Thus, it remains unclear if this represents a return to baseline 5 days after drug administration or if it represents very low p24 levels. Comparison of post-/pre-HDACi ratios of mean cell-associated HIV gag RNA and protein following clinical administration of HDACis showed statistical significant correlations (Figure 3E, P < 0.01). Collectively, these data demonstrate that clinical administration of HDACis increase viral RNA transcription and viral protein expression.
Figure 3. Clinical administration of HDACis induces HIV protein expression in ART-suppressed subject CD4+ T cells.
(A–C) In vivo vorinostat administration induces p24 expression in CD4+ T cells following 1, 2 (A), or 10 (B) doses of vorinostat administered clinically every 72 hrs (refs. 7, 9; see ref. 34 for cell-associated HIV RNA levels corresponding to single and paired VOR dosing), and (C) for 10-dose VOR. (D) Clinical administration of panobinostat resulted in statistically significant increase in p24 above baseline, ***P < 0.001 student’s t test (see ref. 10 for RNA data; open circle represents 1 fg/ml p24, a value below analytical limit of detection [LOD]). (E) Comparison of post-/pre-HDACi ratios of mean cell-associated (ca) HIV gag RNA copies/ml and gag protein fg/ml following clinical administration of vorinostat (open squares) or panobinostat (closed circles), with statistical significance determined using linear regression (P < 0.01). n = 11. Note, subject V-1 was the only participant who was not observed to have an induction of ca-RNA (40) after VOR administration, although an increase of cell-associated p24 was detected. If this data point was excluded (asterisk), the correlation between induction of cell-associated p24 and ca-HIV RNA would be even more significant.
Latency reversal induces sufficient viral protein to enable immune-based clearance.
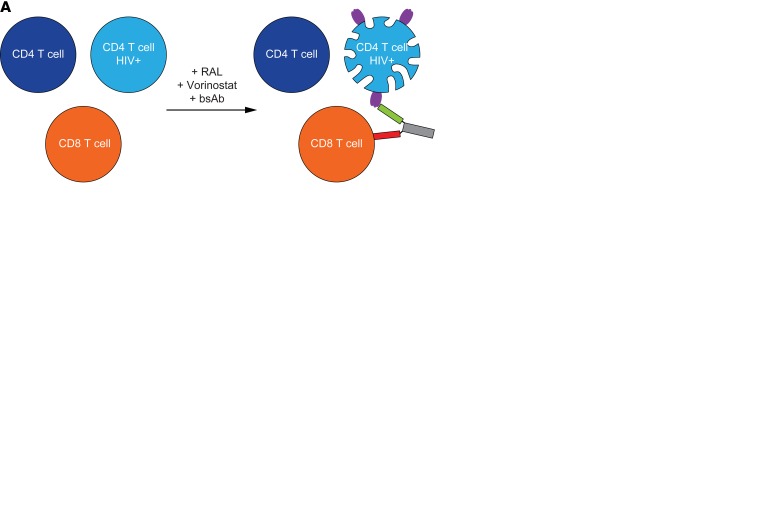
As highlighted above, several current immunological strategies are aimed at engaging the immune system to clear HIV+-expressing cells (5, 17, 18, 32, 33). We hypothesized that in vivo and ex vivo treatment with latency-reversing agents could result in sufficient viral protein expression to enable redirected CD8+ cell killing and that changes in viral levels could be assessed directly by measuring a reduction in HIV gag p24. To explore this hypothesis, autologous CD4+ and CD8+ T cells were isolated from ART-suppressed HIV+ subjects and coincubated at endogenous levels with either a low concentration of PMA/ionomycin (10 ng/ml PMA + 0.1 μg/ml ionomycin) or 380 nM VOR for 72 hours in the presence or absence of gp120/CD3 or RSV/CD3 bispecific antibodies (BsAbs; Figure 4A). As shown in Figure 4, statistically significant increases in p24 protein were observed over DMSO controls following ex vivo stimulation of CD4+ T cells with either latency-reversing agent (P < 0.01). The addition of gp120/CD3 BsAbs to the culture of HIV+ reactivated cells resulted in reduced p24 protein, comparable with DMSO levels, whereas the addition of the control RSV/CD3 BsAb had no appreciable effect and showed comparable p24 levels with PMA/ionomycin (Figure 4B) or VOR (Figure 4C) treatment alone. We also found that shorter incubations of latency-reversing agents (<24 hrs) in isolated cells from ART-suppressed HIV+ individuals produced sufficient protein for immune-mediated clearance (data not shown). Interestingly, the extent of p24 protein reduction in the presence of HIV+ envelope (ENV) BsAb and latency reversal is ~1.5-fold that of induced protein following latency reversal alone and significantly greater than respiratory syncytial virus (RSV) BsAb controls (P < 0.01), thereby suggesting efficient clearance of reactivated latent cells, as well as cells persistently expressing HIV antigen (Figure 4D). CD4+ T cell viability remained similar across treatment conditions (Supplemental Figure 4). These proof-of-concept studies illustrate HDACis, as well as other latency-reversing agents, can elicit viral protein production to facilitate redirected cell killing and support immune clearance approaches under evaluation (5, 17, 18, 35–39).
Figure 4. Immune-mediated clearance of HIV+ CD4+ T cells following HIV reactivation and redirected CD8+ T cell killing.
(A) Schematic of immune-mediated targeting of HIV+ CD4+ T cells by autologous CD8+ T cells using gp120/CD3 bispecific antibodies. (B and C) Quantitation of p24 in cell lysates following 72-hr ex vivo HIV reactivation of ART-suppressed CD4+ T cells with either PMA/ionomycin, n = 3 subjects (B), or vorinostat, n = 4 subjects (C). p24 concentrations are reduced significantly in the presence of gp120/CD3 (ENV) but not RSV/CD3 (RSV) bispecific antibodies. (D) Ratio HIV gag protein changes following latency reactivation with either VOR or PMA/ionomycin relative to unstimulated (LRA/DMSO) or in the presence of either HIV ENV (LRA/LRA+ENV BsAb) or RSV (LRA/LRA+RSV BsAb) bispecific antibodies. Input numbers are shown for PMA/ionomycin- or VOR-treated cells. Ratios were excluded when 1 comparator was below analytical LOD. Significance was determined using ANOVA. **P < 0.01. Error bars indicate mean ± SEM.
Discussion
The investment in HIV cure research and the ability to monitor the effectiveness of therapeutic interventions will depend largely on accurate and sensitive quantitation of viral burden in different tissues and body fluids. New ELISA technologies support increased detection of HIV gag p24 in plasma, serum, and cultured cell medium; however, these methods show decreased sensitivity in cell lysates due to matrix interference and in plasma and serum of chronically infected HIV+ subjects due to immune complex formation after seroconversion (29, 30). Here, we report optimized methods that enable sensitive and rapid detection of subfemtomolar cell–associated HIV gag p24 protein in HIV-infected CD4+ T cells from ART-suppressed individuals. Quantifying very small amounts of cell-associated viral antigen produced by low numbers of cells from aviremic/low-reservoir HIV+ individuals is an important advancement, as viral protein expression is believed to contribute to tissue inflammation and persistent immune activation, and induction is a prerequisite for several investigational immune-mediated “kill” strategies (3–5, 40). The assay can be completed in approximately 1 hour, typically requires 1–2 million peripheral CD4+ T cells from ART-suppressed HIV+ individuals, and can be applied to quantify viral protein in cell lysates or culture medium.
It is well established that the majority of HIV proviruses harbor defective genomes and, thus, fail to produce replication-competent virus (20). Traditional assays aimed at reservoir characterization measure HIV DNA or RNA and are sensitive and scalable but have limitations in distinguishing defective from translation- or replication-competent proviruses. This inherently leads to an overestimation of the size of the reservoir (20). In our study, we compared the relationship between HIV-1 gag protein production and inducible reservoir size, as measured by tat/rev multiply spliced RNA (msRNA) (TILDA). Although a positive trend was observed, this did not reach statistical significance, which may be attributable, in part, to poor PCR probe recognition due to mismatches in HIV proviral sequences or to the inability of inducible RNA to make protein. Conversely, the presence of transcriptionally competent “defective” proviruses capable of producing aberrant gag protein that may be poorly recognized by the p24 antibody is also a possible explanation (40). Worth noting, significant correlations were observed between HDACi-induced cell-associated RNA and protein from blood CD4+ T cells in subjects treated clinically with HDAC inhibitors, and further work is ongoing to inform on the correlations between p24 immunoassay and other viral endpoints, including qVOA. Overall, assessment of HIV gag protein provides a tool for measuring protein production following latency reversal or protein levels needed to induce viral cytopathicity or promote immune-mediated clearance; it also affords increased sensitivity as a component of an existing ex vivo assay for measuring HIV reservoir (26–28).
In this study, we show the first direct evidence to our knowledge that clinical administration of HDACis results in measurable increases in HIV-1 gag p24 protein in CD4+ T cells. Additionally, we show ex vivo HDACi treatment of autologous CD4+ and CD8+ T cells from ART-suppressed HIV+ individuals can induce reactivation of HIV and produce sufficient viral antigen to enable redirected immune cell killing, mediated by gp120/CD3 BsAbs. These observations have implications for evaluating HIV-1 eradication strategies, as they highlight application of an ultrasensitive, rapid tool for monitoring the rarer population of translation-competent virus that must be effectively reactivated and cleared. Future studies geared at assessing other viral antigens, sequencing viral genomes, and conducting viral outgrowth studies to assess relationships between defective proviruses producing RNA-competent protein, replication-competent virus, and HIV protein production will be informative to understanding how the generation of viral antigens also contributes to persistent immune activation. Furthermore, in light of the clinical studies using broadly neutralizing antibodies and antibody-based approaches to engage the immune system, this approach becomes increasingly important for assessing the reduction of HIV-expressing cells and the effectiveness of “shock and kill” strategies in preclinical or clinical settings.
Methods
Reagents.
PMA, ionomycin, ingenol-3-angelate, and bromosporine were purchased from Sigma-Aldrich. Raltegravir and VOR were synthesized at Merck. Recombinant HIV gag p24 protein was obtained from US Biological.
Isolation of T cells.
Leukapheresis was performed on VOR subjects over a period spanning 3–6 hours after dose, and venipuncture was performed on panobinostat subjects at 8 hours after dose. PBMCs were isolated either from leukopaks or from whole blood via Ficoll-gradient. Resting CD4+ T cells, total CD4+ T cells, and total CD3+ T cells were isolated from PBMCs using Stemcell Technologies kits, as previously described (7, 9, 10, 25).
Ex vivo HIV reactivation.
Enriched CD4+ T cells (1 × 106 to 5 × 106 cells/ml) were cultured in complete RPMI 1640 media (cRPMI) containing 10% FBS, 2 mM glutamine, and the antibiotics penicillin and streptomycin. Cells were treated ex vivo for indicated times with either 0.1% DMSO, 100 ng/ml PMA/1 μg/ml ionomycin, 10 ng/ml PMA/0.1 μg/ml ionomycin, 10 nM ingenol-3-angelate, 5 μM bromosporine, 750 nM VOR, or 380 nM VOR. For TILDA studies, cells were collected and put through a 35-μm cell strainer (BD Falcon; Thermo Fisher Scientific) to remove cell clumps, and cell yield following titration was greater than 90%. For p24 studies, cells and culture medium were recovered after the treatment period by centrifugation at 1500 g for 5 minutes at room temperature. Cell density and viability were determined with a Vi-cell XR instrument (Beckman Coulter). Samples were processed for p24 digital ELISA and TILDA as described below.
HIV p24 digital immunoassay.
The fully automated Quanterix HD-1 analyzer for single molecule detection has been reported previously (29–31). In this study, cell lysates were prepared by adding a final concentration of 1% Triton X-100/PBS to 2 × 106 cells. For culture medium supernatants, Triton X-100 was added to a final concentration of 1% (10% volume of 10% Triton x-100 prepared in PBS). Samples were stored frozen at –80°C for more than 1 hour or until analysis. Upon thaw, cell lysates were diluted 2- to 5-fold with dilution buffer (50% of blocker casein in PBS [Thermo Fisher Scientific, catalog 37528] and 50% of FBS (Thermo Fisher Scientific, catalog 10438018]). For culture medium supernatants, samples were diluted with an equal volume of dilution buffer and frozen at –80°C until analysis or minimally > 1 hour. Samples were then centrifuged for 5 minutes at 10,000 g at room temperature to remove insoluble material prior to p24 measurement on the Quanterix analyzer. All other assay reagents and assay reaction conditions followed the manufacturer’s p24 kit protocol (29, 30) (Quanterix). p24 concentration was calculated based on a p24 calibration curve with 4-parameter curve fitting.
p24 assay specificity.
p24 was quantified in BSA/PBS buffer alone or in lysates from uninfected or HIV+ CD4+ T cells in the presence and absence of either PMA/ionomycin or VOR. Additionally, HIV+ CD4+ T cells from viremic donors were lysed (1 million cells/ml) in 1% Triton x-100/PBS and divided into 4 aliquots (1 ml each) in eppendorf tubes. Simoa capture antibody beads (100 μl), or beads with detection antibody or normal mouse IgG (biotin-labeled simoa detection antibody or mouse IgG) were incubated with Dynabeads with streptavidin (M-280) at 4°C for 2 hours. Unbound antibody was washed away, and magnets were added into each tube, mixed, and incubated at 4°C overnight. The supernatant FT after IP was collected by magnet, and p24 in FT was measured in Simoa (Quanterix). BSA/PBS (3%) was negative in the assay, and mock IP lysate was used as positive sample. Immunodepletion results are shown in Supplemental Figure 1B.
In a separate study, MoltIIIB cell line (AIDS Research Program) with 100% integrated HIV was resuspended initially at 1 × 106 cells/ml culture medium and further diluted to 100 cells/ml. 40 μl (4 cells) was added to 960 μl of 1% Triton x-100 in PBS to become 4 cells/ml lysate. Lysate was further diluted in 3% BSA/PBS from 4–0.125 cells/ml, and p24 was quantified using digital ELISA. Results are shown in Supplemental Figure 1A.
To assess whether the assay also recognized unprocessed Gag polyprotein, recombinant protein p55 (Invitrogen, catalog RP4920) and p24 (US Biological, catalog H6003-25) were measured simultaneously in Quanterix p24 assay. Both proteins were diluted with 3% BSA/PBS starting at 20, 10, 5, 2.5, 1.25, 0.62, 0.3, and 0 fM. Both samples were measured in p24 digital ELISA, and the data were graphed based on the spiking in p24 or p55 concentration (fM) and the signal average enzyme per bead (AEB). Results are shown in Supplemental Figure 1C.
HIV virus cultures.
HIV isolates representing HIV-1 group M (subtype A, B, C, D, AE, G) and O and HIV-2 were obtained from the NIH AIDS Reference and Reagent Repository (Germantown, Maryland, USA) or isolated from CD8-depleted PBMCs by coculture with PHA-activated T cells from healthy donors. Virus isolates evaluated included 93RW034, JrFl, Bal, QZ4589, ASM57, 92TH594, 93BR029, BRH84155, BRH95436, ZA/97/003, 93MW959, 92UG024, THA/92/006, RU570, JV1083, 301342, 301340, and BCF03. Working stocks were amplified by infecting PHA-activated PBMCs or MT-4 cells stably expressing the CCR5 coreceptor and GFP virus. Viral isolates were harvested from the culture supernatant without additional purification. Diluted virus was inactivated with addition of final 1% Triton x-100 in PBS and incubated at room temperature for 30 minutes and then frozen at –80°C until analysis. The viral lysate was diluted at 1:100, 1:1,000, 1:10,000, and 1:100,000 with 3% BSA/PBS before assay.
TILDA.
TILDA analysis was performed as previously described (25) with modifications in quantitative PCR (qPCR) in which TaqMan Fast Advanced Master Mix instead of LightCycler buffer was used and performed on a Quantstudio 12K flex instrument (Thermo Fisher Scientific).
Measurement of p24 from clinical samples.
Cryopreserved cells were thawed, and CD4+ T cells were isolated as described above. 2 × 106/ml CD4+ T cells per sample were cultured in the presence of 1 μM raltegravir for 48 hours in cRPMI media and then harvested and prepared for p24 quantification, as described above.
BsAbs and ex vivo shock and kill.
Antibodies with dual specificity for HIV gp120 and CD3 were generated using published sequences. Antibodies containing a RSV-specific antibody (Synagis) arm and anti-CD3 were also constructed and used as nonspecific control. CD3-enriched cells from HIV -infected ART-suppressed donors were prepared as described above and cultured in cRPMI. Cells were seeded at a density of 2 × 106 cells/ml and were incubated with either 0.1% DMSO, 10 ng/ml PMA and 0.1 μg/ml ionomycin, or 380 nM VOR in the presence or absence of 100 ng/ml BsAbs. Raltegravir was added at a final concentration of 1 μM to prevent secondary rounds of replication. Following a 72-hour incubation period, cell lysates were prepared and p24 was quantified, as described above.
Statistics.
HIV gag protein concentration was calculated based on a calibration curve with linear regression 4-parameter curve fitting. A Spearman correlation test was used for analysis of p24 in cell lysates versus culture medium as well as comparison with TILDA, and P < 0.05 was considered significant. ANOVA or two-parameter student’s t tests were used for statistical significance for all other statistical tests, and the method used is noted in the figure legends.
Study approval.
HIV-infected subjects receiving stable, standard-of-care ART with plasma HIV-1 RNA fewer than 50 copies per ml and a CD4 count of more than 300 μl-1 for at least 6 months were enrolled in the VOR clinical studies following informed consent (7, 9, 34); HIV-infected adults receiving ART with virological suppression (<50 copies per ml, at least 2 measurements per year) for at least 2 years and CD4 counts above 500 cells per μl were enrolled in the panobinostat study following informed consent (10). VOR studies were approved by the UNC institutional biomedical review board and the FDA (7, 9, 34); panobinostat clinical study was conducted at Aarhus University Hospital, Denmark, and approved by the ethics committee in accordance with the principles of the Declaration of Helsinki (10). For ex vivo studies, additional samples were obtained from HIV+ donors with suppressed viremia (<50 copies per ml) from Philadelphia Fight and Sanguine Biosciences under IRB approval and patient informed consent.
Author contributions
GW, MS, D. Graham, JS, NMA, and BJH performed the experiments and analyzed the data. GW, D. Gorman, RJOB, WB, DJH, and BJH provided input on research design. GW, MS, and BJH wrote the manuscript. TAR, OSS, MT, LØ, DMM, NMA, and RPS oversaw clinical studies, provided clinical samples, and provided cell-associated gag RNA measurements. GW and AT performed statistical analyses. All authors reviewed and provided comments on the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Yanyan Zheng and Laurence Fayadat-Dilman for contributing to thedesign, generation, and characterization of the BsAb reagents. We’d also like to thank the HIV+ patients for donating biological samples to support cure research. Partial research funding was provided by NIH grant U01 AI095052 (to DMM) and The Danish Council for Strategic Research and Aarhus University (to TAR, OSS, MT, and LØ). Research reported in this publication was also supported (in part) by CARE, a Martin Delaney Collaboratory, the National Institute of Allergy and Infectious Diseases, National Institute of Neurological Disorders And Stroke (NINDS), National Institute On Drug Abuse (NIDA) and the National Institute Of Mental Health (NIMH) of the National Institutes of Health, grant number 1UM1AI126619. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Version 1. 08/08/2017
Electronic publication
Footnotes
WB’s present address is: Gritstone Oncology, Emeryville, California, USA.
Conflict of interest: DMM reports personal fees from Merck & Co. Inc. and from Gilead Sciences outside the submitted work.
Reference information: JCI Insight. 2017;2(16):e92901. https://doi.org/10.1172/jci.insight.92901.
Contributor Information
Guoxin Wu, Email: Guoxin_wu@merck.com.
Michael Swanson, Email: michael.swanson@merck.com.
Aarthi Talla, Email: axt427@case.edu.
Donald Graham, Email: donald_graham@merck.com.
Julie Strizki, Email: julie.strizki@merck.com.
Daniel Gorman, Email: dan.gorman@merck.com.
Wade Blair, Email: wblair@gritstone.com.
Martin Tolstrup, Email: marttols@rm.dk.
David M. Margolis, Email: david_margolis@med.unc.edu.
References
- 1.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Siliciano JM, Siliciano RF. The Remarkable Stability of the Latent Reservoir for HIV-1 in Resting Memory CD4+ T Cells. J Infect Dis. 2015;212(9):1345–1347. doi: 10.1093/infdis/jiv219. [DOI] [PubMed] [Google Scholar]
- 3.Martin AR, Siliciano RF. Progress Toward HIV Eradication: Case Reports, Current Efforts, and the Challenges Associated with Cure. Annu Rev Med. 2016;67:215–228. doi: 10.1146/annurev-med-011514-023043. [DOI] [PubMed] [Google Scholar]
- 4.Spivak AM, Planelles V. HIV-1 Eradication: Early Trials (and Tribulations) Trends Mol Med. 2016;22(1):10–27. doi: 10.1016/j.molmed.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cillo AR, Mellors JW. Which therapeutic strategy will achieve a cure for HIV-1? Curr Opin Virol. 2016;18:14–19. doi: 10.1016/j.coviro.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Rasmussen TA, Anderson JL, Wightman F, Lewin SR. Cancer therapies in HIV cure research. Curr Opin HIV AIDS. 2017;12(1):96–104. doi: 10.1097/COH.0000000000000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487(7408):482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott JH, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014;10(10):e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archin NM, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis. 2014;210(5):728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmussen TA, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV. 2014;1(1):e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 11.Søgaard OS, et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS Pathog. 2015;11(9):e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20(4):425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird GM, et al. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest. 2015;125(5):1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darcis G, et al. An In-Depth Comparison of Latency-Reversing Agent Combinations in Various In Vitro and Ex Vivo HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression. PLoS Pathog. 2015;11(7):e1005063. doi: 10.1371/journal.ppat.1005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang G, et al. Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation. PLoS Pathog. 2015;11(7):e1005066. doi: 10.1371/journal.ppat.1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez C, et al. Bryostatin-1 for latent virus reactivation in HIV-infected patients on antiretroviral therapy. AIDS. 2016;30(9):1385–1392. doi: 10.1097/QAD.0000000000001064. [DOI] [PubMed] [Google Scholar]
- 17.Pantaleo G, Levy Y. Therapeutic vaccines and immunological intervention in HIV infection: a paradigm change. Curr Opin HIV AIDS. 2016;11(6):576–584. doi: 10.1097/COH.0000000000000324. [DOI] [PubMed] [Google Scholar]
- 18.Stephenson KE, Barouch DH. Broadly Neutralizing Antibodies for HIV Eradication. Curr HIV/AIDS Rep. 2016;13(1):31–37. doi: 10.1007/s11904-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crooks AM, et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J Infect Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton K, et al. Broad activation of latent HIV-1 in vivo. Nat Commun. 2016;7:12731. doi: 10.1038/ncomms12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 24.Cillo AR, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci USA. 2014;111(19):7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Procopio FA, et al. A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine. 2015;2(8):874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trautmann L. Kill: boosting HIV-specific immune responses. Curr Opin HIV AIDS. 2016;11(4):409–416. doi: 10.1097/COH.0000000000000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banga R, Procopio FA, Perreau M. Current approaches to assess HIV-1 persistence. Curr Opin HIV AIDS. 2016;11(4):424–431. doi: 10.1097/COH.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 28.Henrich TJ, Deeks SG, Pillai SK. Measuring the Size of the Latent Human Immunodeficiency Virus Reservoir: The Present and Future of Evaluating Eradication Strategies. J Infect Dis. 2017;215(suppl_3):S134–S141. doi: 10.1093/infdis/jiw648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang L, et al. Simple diffusion-constrained immunoassay for p24 protein with the sensitivity of nucleic acid amplification for detecting acute HIV infection. J Virol Methods. 2013;188(1-2):153–160. doi: 10.1016/j.jviromet.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Cabrera C, Chang L, Stone M, Busch M, Wilson DH. Rapid, Fully Automated Digital Immunoassay for p24 Protein with the Sensitivity of Nucleic Acid Amplification for Detecting Acute HIV Infection. Clin Chem. 2015;61(11):1372–1380. doi: 10.1373/clinchem.2015.243287. [DOI] [PubMed] [Google Scholar]
- 31.Passaes CP, et al. Ultrasensitive HIV-1 p24 Assay Detects Single Infected Cells and Differences in Reservoir Induction by Latency Reversal Agents. J Virol. 2017;91(6):e02296–16. doi: 10.1128/JVI.02296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richman DD. HIV: Cure by killing. Nature. 2015;528(7581):198–199. doi: 10.1038/nature16321. [DOI] [PubMed] [Google Scholar]
- 33.Martrus G, Altfeld M. Immunological strategies to target HIV persistence. Curr Opin HIV AIDS. 2016;11(4):402–408. doi: 10.1097/COH.0000000000000289. [DOI] [PubMed] [Google Scholar]
- 34.Archin MN, et al. Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J Clin Invest. 2017;127(8):e 92684. doi: 10.1172/JCI92684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung JA, et al. Expanded cytotoxic T-cell lymphocytes target the latent HIV reservoir. J Infect Dis. 2015;212(2):258–263. doi: 10.1093/infdis/jiv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pegu A, et al. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun. 2015;6:8447. doi: 10.1038/ncomms9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung JA, et al. Dual-Affinity Re-Targeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. J Clin Invest. 2015;125(11):4077–4090. doi: 10.1172/JCI82314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloan DD, et al. Targeting HIV Reservoir in Infected CD4 T Cells by Dual-Affinity Re-targeting Molecules (DARTs) that Bind HIV Envelope and Recruit Cytotoxic T Cells. PLoS Pathog. 2015;11(11):e1005233. doi: 10.1371/journal.ppat.1005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu CL, et al. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science. 2016;352(6288):1001–1004. doi: 10.1126/science.aaf1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamichi H, et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci USA. 2016;113(31):8783–8788. doi: 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.