Abstract
Oxidative stress is important in the pathogenesis of allergic asthma. Extracellular superoxide dismutase (EC-SOD; SOD3) is the major antioxidant in lungs, but its role in allergic asthma is unknown. Here we report that asthmatics have increased SOD3 transcript levels in sputum and that a single nucleotide polymorphism (SNP) in SOD3 (R213G; rs1799895) changes lung distribution of EC-SOD, and decreases likelihood of asthma-related symptoms. Knockin mice analogous to the human R213G SNP had lower airway hyperresponsiveness, inflammation, and mucus hypersecretion with decreased interleukin-33 (IL-33) in bronchoalveolar lavage fluid and reduced type II innate lymphoid cells (ILC2s) in lungs. SOD mimetic (Mn (III) tetrakis (N-ethylpyridinium-2-yl) porphyrin) attenuated Alternaria-induced expression of IL-33 and IL-8 release in BEAS-2B cells. These results suggest that R213G SNP potentially benefits its carriers by resulting in high EC-SOD in airway-lining fluid, which ameliorates allergic airway inflammation by dampening the innate immune response, including IL-33/ST2–mediated changes in ILC2s.
Keywords: Inflammation, Pulmonology
Keywords: Asthma, Drug therapy, Innate immunity
A single nucleotide polymorphism (rs1799895;R213G) in the extracellular superoxide dismutase changes its distribution in the lungs and protects against allergic asthma.
Introduction
A single nucleotide polymorphism (SNP) in extracellular superoxide dismutase (EC-SOD; SOD3) changes arginine to glycine at position 213 (R213G; rs1799895), resulting in redistribution of EC-SOD from lung tissues to the extracellular fluids. Involvement of oxidative stress in allergic asthma is well known; still, antioxidant therapies have not been successful. This study could provide the missing link of an effective antioxidant therapy against allergic asthma to improve asthmatic symptoms and to protect against the development of allergic asthma.
Allergic asthma is a heterogeneous disease characterized by airway hyperresponsiveness, inflammation, and remodeling (1, 2). Oxidative stress is implicated in all of these features (1) and is characterized by an imbalance of reactive oxygen species (ROS) such as superoxide (O2–), hydrogen peroxide (H2O2), hydroxyl radical (OH.), and antioxidants such as SODs, catalases, and peroxidases, which scavenge these reactive species (1, 3). Allergen exposure drives oxidative stress by causing airway epithelium and smooth muscle cells to produce ROS, particularly O2–, through activation of NADPH oxidase (4, 5). Other sources of ROS include inflammatory cells like eosinophils (6), neutrophils (7), and monocytes/macrophages (8). Asthmatics have increased hydrogen peroxide (9) and nitric oxide (10) as exhaled gases supporting the role of oxidative stress in asthma.
One of the lung’s major defenses against oxidative stress is EC-SOD. It catalyzes the dismutation of superoxide radicals into hydrogen peroxide and oxygen (11). EC-SOD is highly expressed in arteries (3, 12), lungs, and airways (13, 14), with high levels in alveolar macrophages (15). Human (16) and mouse studies (17–19) have shown that EC-SOD plays a key role in attenuating lung injury by blocking oxidative stress–mediated lung inflammation; however, little is known about the role of EC-SOD in modulating allergic airway disease. A key feature of EC-SOD is its carboxy-terminus, which has 6 positively charged amino acids that anchor it to the negatively charged extracellular matrix (ECM) (20). A SNP in the coding region of the ECM binding region of SOD3 (rs1799895) results in an arginine to glycine amino acid change at position 213 (R213G), thereby disrupting the positive charge and releasing the protein from the ECM (21) and redistributing EC-SOD to extracellular fluids such as plasma and epithelial lining fluids (22, 23). People who carry the R213G SNP have attenuated risk of exacerbations of chronic obstructive pulmonary disease (COPD) (24), but the role of the rs1799895 SNP in asthma is unknown. Two recent publications support a potential role for EC-SOD in asthma: first, EC-SOD–knockout mice have increased ovalbumin-induced (OVA-induced) allergic airway inflammation and supplementation of EC-SOD alleviates OVA-induced allergic asthma (25); second, the SOD mimetic, Mn (III) tetrakis (N-ethylpyridinium-2-yl) porphyrin (MnTE-2-PyP), has been shown to suppress CD25 expression and reduce the Th2 response in OVA-induced allergic airway inflammation in mice (26). MnTE-2-PyP is a potent SOD mimetic with a dismutation constant log Kcat (O2–) of 7.76 compared with the SOD enzymes with log Kcat (O2–) of 8.84–9.2 (27, 28).
A potential link between oxidative stress and inflammation is IL-33, which is one of the most potent alarmins that activate type II innate lymphoid cells (ILC2s) (29). Allergen exposure, specifically Alternaria, induces oxidative stress in bronchial epithelial cells that causes extracellular secretion of ATP and increases intracellular calcium concentrations preceding IL-33 release from the airway epithelium (30). IL-33 is stored in the nucleus and is released upon cellular activation (29). It binds to the ST2 receptors on ILC2s and stimulates them to secrete massive amounts of IL-5, IL-9, and IL-13 that polarize the immune response to type II (31). In this study, we hypothesized that EC-SOD plays a role in modulating allergic asthma. To test this hypothesis we investigated EC-SOD expression and genetics in 4 human populations and used 2 allergic asthma models to study mice genetically manipulated to overexpress human SOD3 or carry a knockin for the rs1799895 SNP.
Results
SOD3 transcripts are increased in sputum of severe asthmatics.
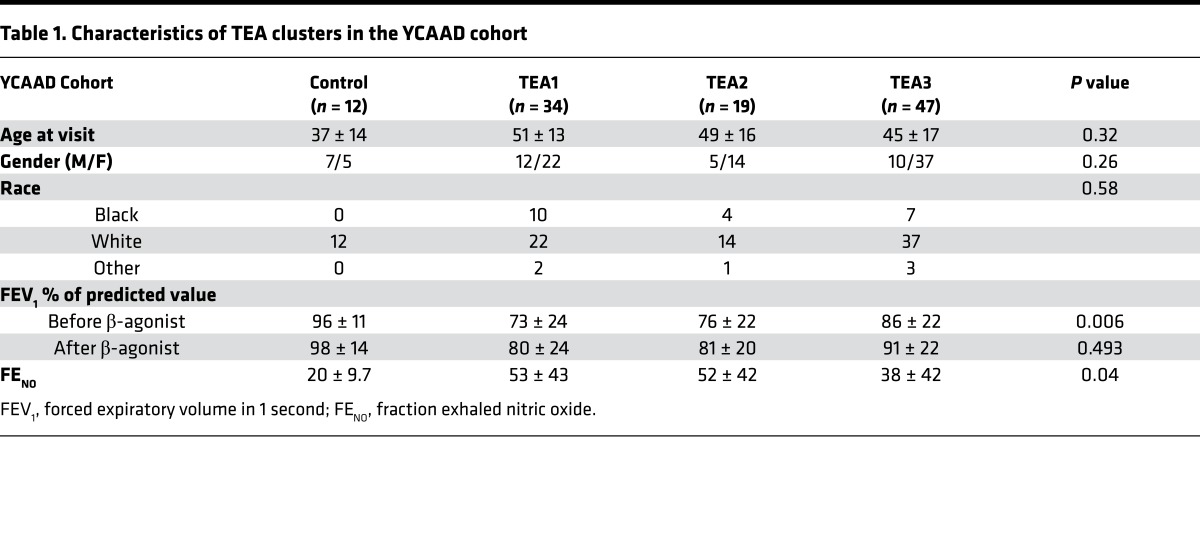
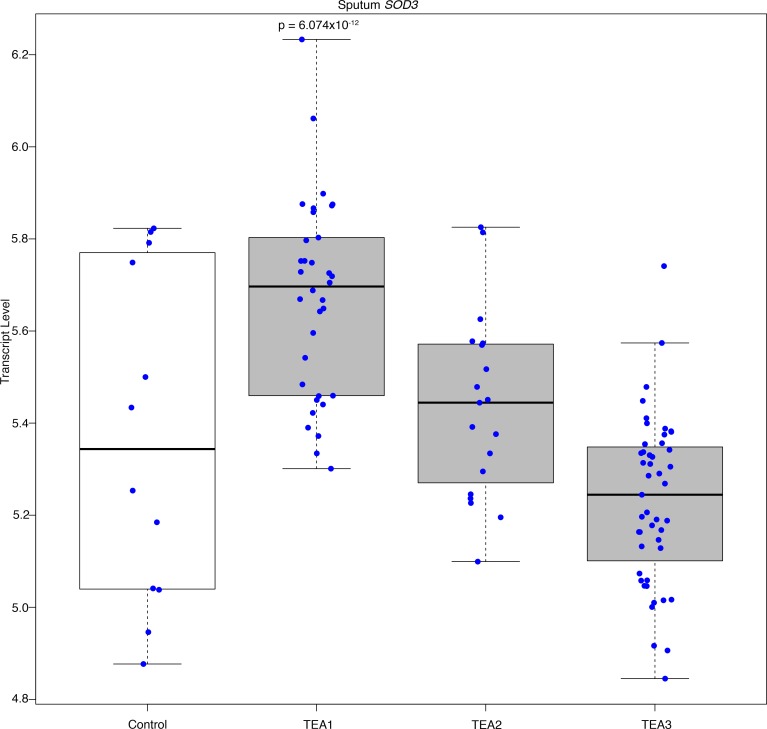
Sputum samples from the Yale Center for Asthma and Airway Diseases (YCAAD) cohort were divided into 3 groups based on transcriptomic endotypes of asthma (TEA) (32). TEA1 subjects had the highest percentage of near-fatal asthma and lowest baseline lung function with highest fraction exhaled nitric oxide (FENO) levels (Table 1). Additionally, TEA1 had the highest SOD3 mRNA expression in sputum compared with controls and less severe asthmatics (Figure 1 and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.95072DS1). Although there was evidence for an association between asthma severity and SOD3 expression, there was no difference in EC-SOD protein in the sputum (Supplemental Figure 1, B and C).
Table 1. Characteristics of TEA clusters in the YCAAD cohort.
Figure 1. Sputum SOD3 transcript increases with asthma severity.
Sputum samples were collected from the Yale Center for Asthma and Airway Diseases (YCAAD) cohort consisting of asthmatic patients (n = 112) that were divided into 3 groups based on transcriptomic endotypes of asthma (TEA) as described in (32). TEA1 (n = 34) is the most severe endotype followed by TEA2 (n = 19) and TEA3 (n = 47). CTRL (n = 12) is the non-atopic healthy control. Graph shows distribution of each individual represented by a dot in respective groups. RNA sequencing was performed to find that the SOD3 transcript was highest in the TEA1 cluster (P = 6.0737 × 10–12) and decreased with asthma severity to control levels in TEA3. The y axis represents the transcript level. Bottom and top of the boxes are the 25th and 75th percentile and the band near the middle of the box is the 50th percentile (the median).
R213G (rs1799895) SNP decreases odds of asthma and asthma-related symptoms.
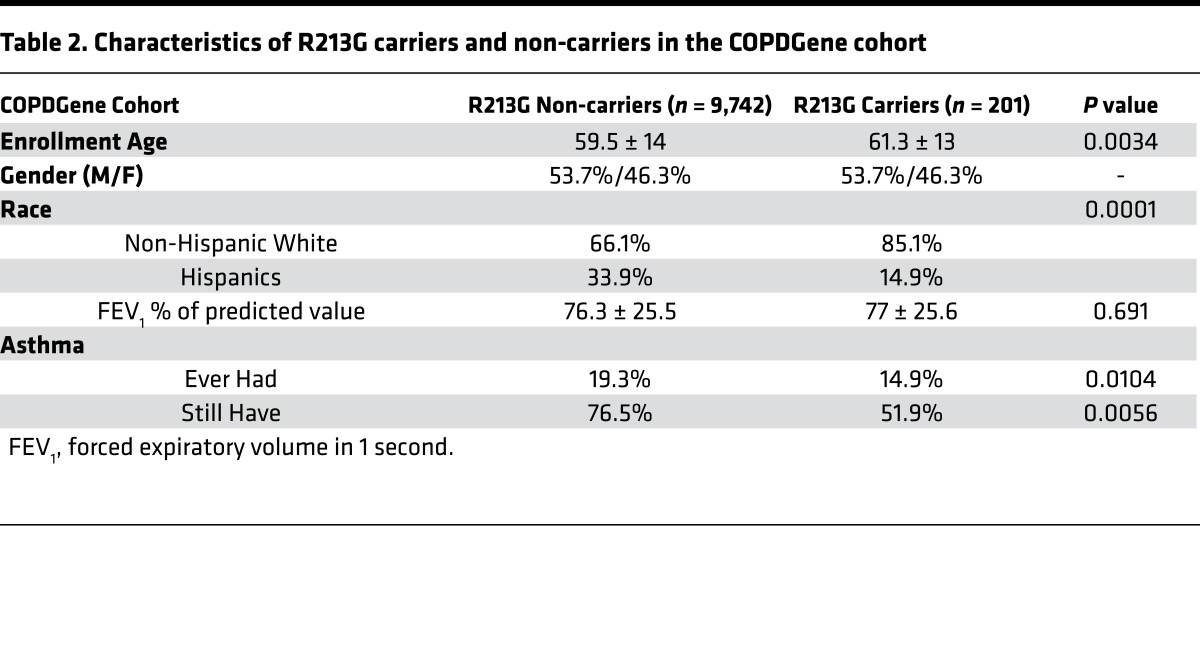
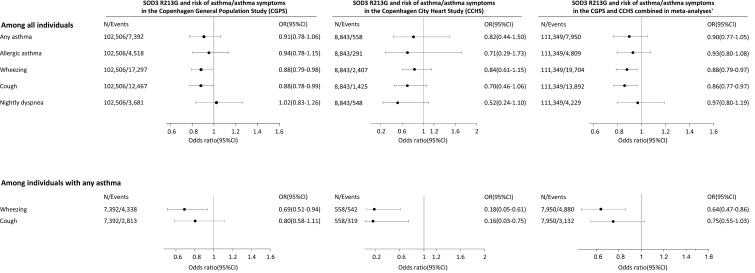
Results from the YCAAD cohort suggested an association between SOD3 expression and asthma severity, but there were no subjects who carried the R213G SNP, most likely because the cohort contained few ethnic northern Europeans, who have higher R213G carrier rates (2%–3%) compared with other populations. To examine the association between R213G carriers and asthma, we investigated 3 cohorts with large numbers of northern European ancestry subjects: the Copenhagen General Population Study (CGPS) with 102,506 subjects; the Copenhagen City Heart Study (CCHS) with 8,843 subjects; and the COPDGene study with 9,943 (Table 2). In the CGPS cohort 7,392 subjects (7%) were diagnosed with asthma and in the CCHS cohort 558 subjects (6%) were diagnosed with asthma. Among these asthmatics, R213G carriers had lower risk of wheezing and cough (odds ratios [95% CI] of 0.69 [0.51, 0.94] and 0.80 [0.58, 1.11] in CGPS, and 0.18 [0.05, 0.61] and 0.16 [0.03, 0.75] in CCHS) (Figure 2), suggesting that the R213G polymorphism also decreases the risk of asthma-related symptoms. In a third cohort (COPDGene), which contained only smokers, subjects who had childhood asthma were 69% less likely (odds ratio of 0.31 [0.14, 0.70]) to continue to have asthma if they had one allele of R213G SNP compared with the non-carriers (Table 2). These 4 population studies suggested that EC-SOD and the R213G polymorphism might be important in attenuating asthma symptoms; however, the mechanism underlying how EC-SOD attenuates this risk was not clear.
Table 2. Characteristics of R213G carriers and non-carriers in the COPDGene cohort.
Figure 2. The association between SOD3 R213G genotype and asthma phenotypes in logistic regression and in single- and multiple-failures-per-subject Cox regression in the Copenhagen General Population Study (CGPS) and the Copenhagen City Heart Study (CCHS) separately and combined.
In all analyses, SOD3 R213G is used as a continuous variable for maximal power. Cox regression was with entry at 1977, when the Danish Patient Registry began, thus including both incident and prevalent cases. Any asthma: either self-reported asthma, a hospital diagnosis of asthma, or medication for asthma among never-smokers. Allergic asthma: individuals with any asthma who also reported allergy triggers such as food, grass, flowers, animals, or others. *Combined in a fixed effect meta-analysis using the metan command in STATA. OR, odds ratio from logistic regression; CI, confidence interval. In the CGPS, a total of 99,958 R213G non-carriers (97.5%), 2,533 R213G heterozygotes (2.5%), and 15 R213G homozygotes (0.01%) were identified. In the CCHS, 8,634 were R213G non-carriers (97.6%), 207 were R213G heterozygotes (2.3%), and 2 (0.02%) were R213G homozygotes.
R213G protects against OVA-induced allergic airway hyperresponsiveness, inflammatory cell infiltration, and protein leakage.
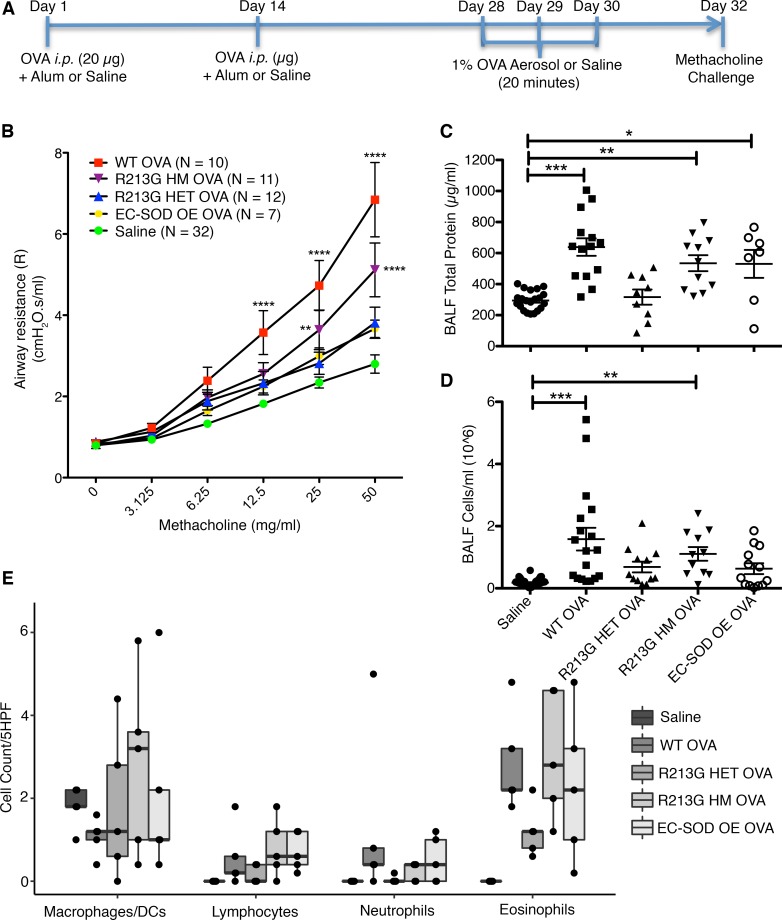
We used 4 different C57BL/6 mice strains: wild-type (WT) control mice, heterozygous (HET), and homozygous (HM) mice that were genetically modified with an R213G knockin to mimic the human R213G SNP (22), and mice that overexpressed lung SOD3 via an SPC promoter (OE) (33). Mice were sensitized and challenged with OVA (Figure 3A), and it had no effect on the EC-SOD activity among the treatment groups (data not shown). The WT mice exposed to OVA had higher airway hyperresponsiveness (AHR) measured as airway resistance, compared with the saline control (Figure 3B), but not in OE or HET OVA groups. Although the HM OVA group was protected from OVA-induced airway resistance compared with the WT OVA, it was still higher than the saline control.
Figure 3. R213G protects mice from airway hyperresponsiveness and inflammatory cell infiltration.
(A) C57BL/6 WT, R213G HET, R213G HM, and EC-SOD OE mice were sensitized and challenged with ovalbumin or saline as indicated in Figure 2A. Saline control contained all the genotypes that were included in OVA. (B) On day 32, methacholine challenge was performed on FlexiVent to measure airway resistance (R) with doses ranging from 0 to 50 mg/ml in saline. Two-way ANOVA was used with Bonferroni multiple comparisons. (C) Total protein was measured in BALF to estimate protein leakage into the airway lumen (Saline = 21, WT OVA = 14, R213G HET OVA = 9, R213G HM OVA = 11, EC-SOD OE OVA = 7). One-way ANOVA was used with Tukey’s multiple comparison test. (D) Inflammatory cell infiltration was measured as total cell count in the BALF (Saline = 47, WT OVA = 18, R213G HET OVA = 12, R213G HM OVA = 11, EC-SOD OE OVA = 13). One-way ANOVA was used with Tukey’s multiple comparison test. (E) Cytospin slides (n = 5/group), stained with H&E were manually counted to identify cell types/5 high-power fields (5HPF). Investigators were blinded to the sample group allocation for cell counts. Bottom and top of the boxes are the 25th and 75th percentile and the band near the middle of the box is the 50th percentile (the median). Two-way ANOVA was used with Bonferroni multiple comparisons. Error bars represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Protein leakage into bronchoalveolar lavage fluid (BALF) was significantly higher in WT OVA compared with saline controls (Figure 3C). HET OVA and OE OVA mice showed no change in protein leakage compared with saline, whereas HM OVA mice had higher protein content in BALF compared with the saline control.
The R213G HET, R213G HM, and OE mice exposed to OVA had significantly fewer inflammatory cells in BALF compared with the WT OVA mice (Figure 3D). Eosinophil, neutrophil, and lymphocyte counts were higher in WT OVA mice compared with the saline controls (Figure 3E), but not in the HET OVA group. Eosinophil and lymphocyte counts were higher in the HM and OE OVA groups compared with the saline group, but macrophages/DC-like-cell counts were not different among groups.
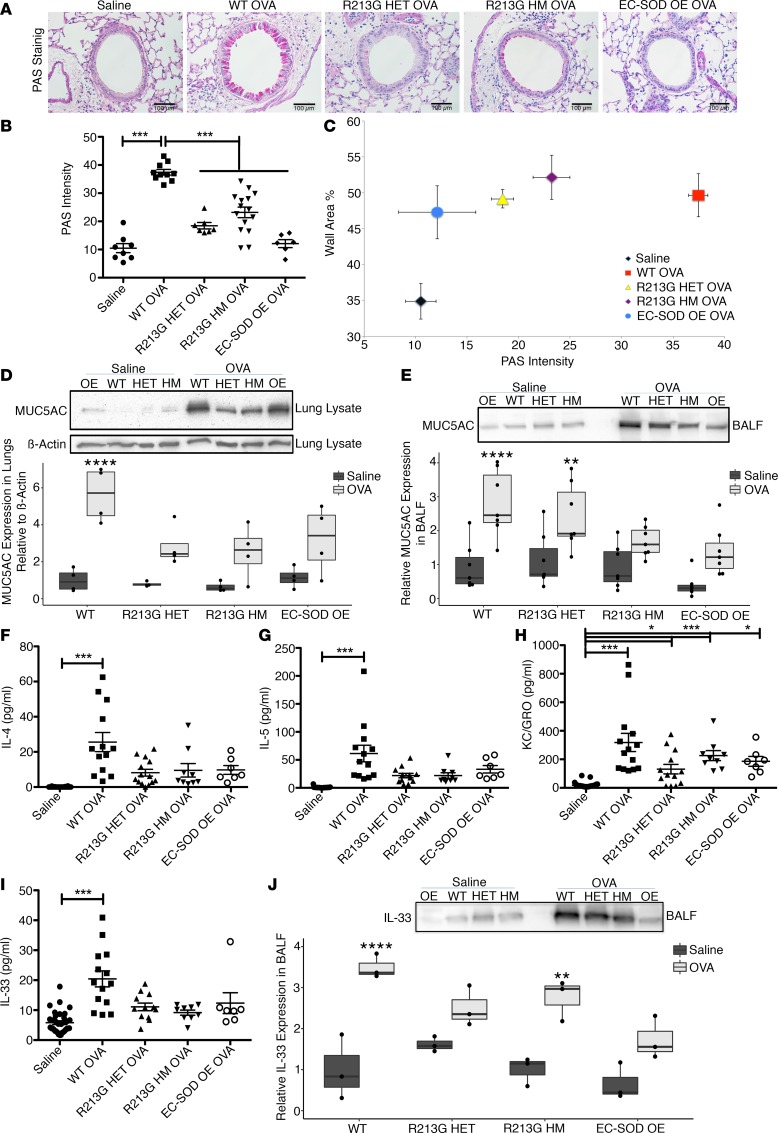
Increased EC-SOD in epithelial lining fluid protects against OVA-induced mucus hyperproduction and hypersecretion.
WT OVA-treated mice showed high periodic acid–Schiff (PAS) staining around the airway lumen in goblet cells, but HET, HM, or OE mice did not (Figure 4A). The WT OVA mice also had significantly higher mucus production (Figure 4B) and wall area thickness compared with the saline control, HET, HM, and OE OVA groups (Figure 4C).
Figure 4. R213G reduces mucus hyperproduction and secretion in OVA-exposed mice.
(A) Images (magnification, ×20) of left lung stained with PAS showing effect of OVA on different genotypes. Scale bars: 100 μm. (B) Graph shows PAS intensity in the airway epithelium, measured by FIJI software. One-way ANOVA was used with Tukey’s multiple comparison test (Saline = 8, WT OVA = 10, R213G HET = 7, R213G HM = 15, EC-SOD OE = 7). PAS intensity was also plotted against wall area percentage in Figure 4C. Mucus hyperproduction and secretion were measured with MUC5AC densitometric analysis of the Western blots in lung lysate (D) and BALF (E), n = 4–7/group. An equal volume of BALF was loaded in each well to control for variations in Western blots. Equal volumes of BALF samples were run on MSD and plotted against the standard curves to obtain concentrations of IL-4 (F), IL-5 (G), KC/GRO (H), and IL-33 (I) in pg/ml. One-way ANOVA was used with Tukey’s multiple comparison test. Saline = 32, WT OVA = 14, R213G HET OVA = 13, R213G HM OVA = 9, EC-SOD OE OVA = 7. (J) Representative of 3 samples/group and densitometric analysis of volume-controlled BALF. Bottom and top of the boxes are the 25th and 75th percentile and the band near the middle of box is the 50th percentile (the median). Analysis was done by 2-way ANOVA with Bonferroni multiple comparisons. Error bars represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To assess the changes in production and secretion of mucus, MUC5AC expression was measured in tissues and BALF. In lung tissues, mucus hyperproduction was marked by an increase in MUC5AC protein expression in WT OVA mice (Figure 4D). Mucus secretion followed a similar pattern, as measured by MUC5AC expression in BALF, even though the HET OVA group showed higher MUC5AC in BALF than the saline control (Figure 4E). Although the treatment groups had measurable differences in collagen deposition among different treatment groups or genotypes, they were not statistically significant, consistent with an acute model (Supplemental Figure 2).
R213G protects against OVA-induced cytokine release in BALF.
Th2 cytokines such as IL-4 (Figure 4F) and IL-5 (Figure 4G) were significantly increased in BALF from WT OVA mice compared with the saline controls, R213G (HET and HM), and OE OVA. However, the HM OVA group had high levels of KC/GRO (CXCL1) compared with the saline control (Figure 4H). Like most of the other inflammatory cytokines tested, IL-33 was also increased in BALF of WT, but not in R213G HET, R213G HM, or the OE mice challenged with OVA (Figure 4I). Increased IL-33 in BALF was noted using 2 independent methods with similar results: Meso Scale Discovery (MSD) assay and by Western blot (Figure 4J). Similarly, inflammatory markers such as IL-1β, IL-6, TNF-α, IFN-γ, and lymphoid cell growth factor IL-2 were increased in the BALF of WT OVA compared with the saline control, HET, HM, and OE OVA groups (Supplemental Figure 3, A and C–E). The IL-2 level was significantly higher in all OVA-exposed mice compared with the saline control, irrespective of the genotypes (Supplemental Figure 3B). There were no differences by group in IL-10, IL-12p70, or IL-25 levels (Supplemental Figure 3, F–H).
R213G also protects against Alternaria-induced innate inflammation in mice.
In a second model of allergic airway inflammation with Alternaria (Supplemental Figure 4A), WT mice showed no change in airway resistance (Supplemental Figure 4B), but an increase in inflammatory cell infiltration (Supplemental Figure 4C), predominantly due to innate immune macrophages/DC-like cells (Supplemental Figure 4D). HET and HM mice had markedly less Alternaria-induced inflammatory cell infiltration in BALF compared with WT mice exposed to Alternaria. However, macrophages/DC-like cells were higher in HM mice exposed to Alternaria, compared with the saline group (Supplemental Figure 4D).
Like the IL-33 levels in the OVA model, IL-33 was significantly increased in BALF of WT mice exposed to Alternaria compared with the saline control (Supplemental Figure 5A). Alternaria increased IL-4 and TNF-α levels in WT mice (Supplemental Figure 5, B and C), but not in HET and HM mice. No difference was detected in IL-2, IL-5, IL-10, IL-13, IL-25, KC/GRO, or IFN-γ (Supplemental Figure 5, D–J).
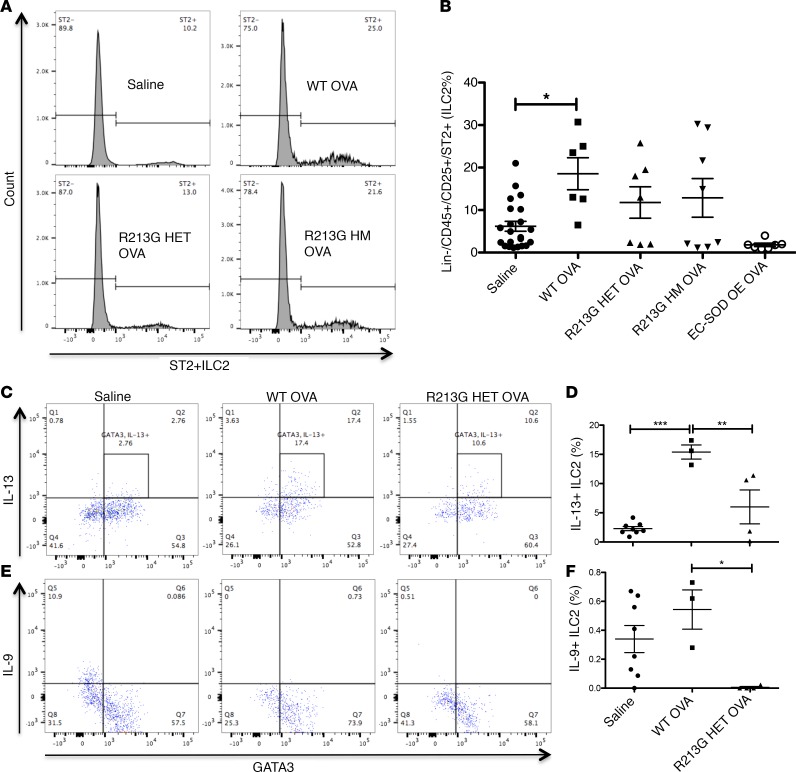
R213G reduces ILC2 numbers and activation in adaptive and innate models of allergic airway inflammation.
ILC2 percentages (after Lin–/CD45+, percentage of CD25+ cells that are ST2+) were increased in WT OVA compared with the saline control; however, R213G HET, HM, and EC-SOD OE did not show any significant differences in the number of ILC2s after OVA sensitization and challenge (Figure 5, A and B). R213G HET mice also had fewer OVA-induced ILC2s that were positive for GATA3 and expressed IL-13 (Figure 5, C and D) or IL-9 compared with WT mice (Figure 5, E and F). Similar results were seen for IL-9 in the ILC2s of Alternaria-exposed mice (Supplemental Figure 6, A–D). There were no significant differences in IL-5.
Figure 5. R213G reduces OVA-induced ILC2s in mouse lungs.
(A) Histogram showing ILC2 (Lin−, CD45+, CD25+, ST2+) differences among saline (n = 22), WT OVA (n = 6), R213G HET OVA (n = 7), R213G HM OVA (n = 8), and EC-SOD OE OVA (n = 7). ILC2% refers to the percentage of CD25+ cells that are ST2+. Analysis of ILC2% is shown in Figure 5B. One-way ANOVA was used with Tukey’s multiple comparison test. (C and D) IL-13+ ILC2s and (E and F) IL-9+ ILC2s in Saline (n = 8), WT OVA (n = 3), R213G HET OVA (n = 4). IL-13+ ILC2s were defined as Lin− (CD3, Ly-6G/Ly-6C, CD11b, CD45R/B220, TER-119), CD5−, CD49b−, TCRαβ−, TCRγδ−, CD45+, CD90+, ST2+, GATA+, and IL-13+, and IL9+ ILC2s were identified as Lin− (CD3, Ly-6G/Ly-6C, CD11b, CD45R/B220, TER-119), CD5−, CD49b−, TCRαβ−, TCRγδ−, CD45+, CD90+, ST2+, GATA+, and IL-9+. One-way ANOVA with Tukey’s multiple comparison test was used. Error bars represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
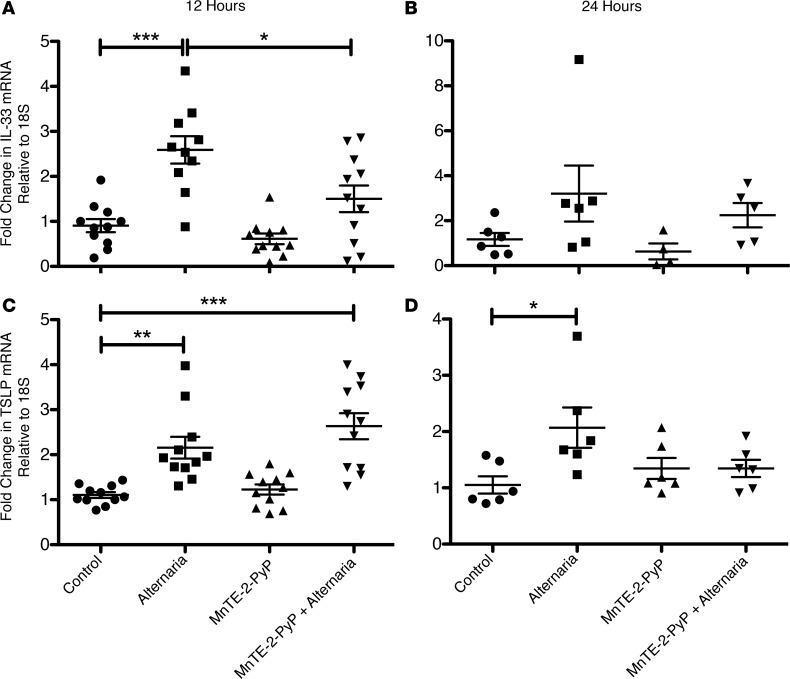
SOD attenuation of Alternaria-induced IL-33 expression in airway epithelial cells is not specific to EC-SOD.
To investigate whether a pharmacologic antioxidant could mimic the results of genetically manipulated mice, we use the SOD mimetic MnTE-2-PyP. MnTE-2-PyP significantly attenuated Alternaria-induced IL-33 mRNA expression in airway epithelium at 12 hours (Figure 6A). At 24 hours, IL-33 expression was abated in most groups (Figure 6B). The SOD mimetic did not have any effect on Alternaria-induced thymic stromal lymphopoietin (TSLP) expression at 12 hours (Figure 6C), but decreased the TSLP expression to baseline at 24 hours (Figure 6D).
Figure 6. SOD mimetic decreases Alternaria-induced IL-33 and TSLP expression in BEAS-2B cells.
Dot plot showing fold-change in IL-33 mRNA after 12 hours (A) and 24 hours (B), and in TSLP mRNA after 12 hours (C) and 24 hours (D). n = 10/group in the 12-hour time point and 6/group in the 24-hour time point. Data were generated with 4 different BEAS-2B cell passages in 3 independent experiments for the 12-hour time point and 3 different cell passages in 3 independent experiments for the 24-hour time point. One-way ANOVA was used with Tukey’s multiple comparison test. Error bars represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Alternaria significantly increased IL-8 levels in BEAS-2B cells. However, at 24 hours the IL-8 level was 100 times higher at 50,000 pg/ml than that at 12 hours (Supplemental Figure 7). MnTE-2-PyP decreased the IL-8 level to baseline at 12 hours and to insignificant differences with the control group at 24 hours.
Discussion
This study suggests a potential role for EC-SOD in human asthma. We found evidence that the SOD3 gene is differentially regulated in asthmatic sputum based on disease severity, suggesting that the increase in SOD3 expression is an adaptive response to increased oxidative stress in asthmatic airways. In 3 other independent populations we found evidence that people who carry the R213G polymorphism, which increases EC-SOD in extracellular fluids, was associated with decreased asthma-related symptoms and improved resolution of childhood asthma. We investigated these associations using both the R213G knockin SNP as well as mice that overexpressed EC-SOD in their airways. The R213G knockin and lung EC-SOD–overexpressing mouse are 2 unique approaches in which we can understand how human genetic variation and expression of SOD3 modulates the biology of asthma. As we have previously reported, R213G mice have no other changes in antioxidant expression or activity, but only in its distribution (22, 23). In general, higher airway EC-SOD, either through the R213G SNP or EC-SOD OE, resulted in lower inflammatory responses to both the OVA model and the Alternaria model. As with other human polymorphisms such as those that lead to cystic fibrosis or sickle cell disease, we found evidence that heterozygotes have better outcomes than homozygotes, which may explain why the R213G SNP is not ubiquitous (minor allele frequency is only 3%). The slightly worse outcomes in HM R213G mice may be due to near-complete depletion of EC-SOD from the ECM, associated with homozygosity. Loss of EC-SOD in tissue (i.e., with homozygosity) has important implications in vasculature and has been associated with worse cardiovascular mortality (34, 35).
Since allergic asthma is predominantly an airway disease, we are not surprised that airway EC-SOD is more important than tissue EC-SOD in the OVA and Alternaria models. Our results with both the genetically manipulated mice and SOD mimetic suggest that increased SOD activity leads to attenuated IL-33 expression and release from airway epithelium, thereby mitigating ILC2 activation in the lungs. However, based on our animal models, it is evident that the presence of EC-SOD in the extracellular airway–lining fluids and in lung tissues is crucial to achieve an optimum protection. Perhaps EC-SOD acts at 2 different stages: first, the lining-fluid EC-SOD reduces airway epithelium–derived superoxide in early allergen exposure, protecting in the innate phase; and second, the tissue-bound EC-SOD (along with the unbound EC-SOD) protects from an adaptive immune response by abating granulocyte-derived superoxide.
The increased sputum SOD3 expression in asthmatics may indicate an adaptive response to the allergen-induced oxidative stress, similar to what has been reported in subjects with a history of cigarette smoking and COPD (36). However, there was no difference in sputum EC-SOD protein. The discrepancy between RNA and protein levels in sputum may be a result of rapid degradation of EC-SOD in inflamed airways, which has been noted in a bleomycin model of lung inflammation (37) or because of a failure to translate or release the protein. EC-SOD lowers superoxide levels, which can combine with nitric oxide (also high in asthma) to form peroxynitrite and other reactive nitrogen species.
The mouse and in vitro studies here suggest several important roles for EC-SOD and the R213G SNP in allergic airway inflammation. First, high levels of EC-SOD in the airways attenuate cytokine release and cellular recruitment into the airways. This was associated with reduced mucus secretion and airway hyperresponsiveness, 2 clinical hallmarks of allergic asthma (38). EC-SOD’s attenuation of the IL-33 response was notable, given the key role of IL-33 in asthma and the importance of IL-33 in the activation of ILC2s. IL-33 regulates ST2-dependent activation in ILC2s (29, 39) and oxidative radicals affect IL-33 disulphide bond formation (39), indicating that oxidative stress might have a direct impact on the IL-33–dependent activation of ST2 in ILC2s. ILC2s play a major role in the pathogenesis of allergic diseases such as atopic dermatitis and asthma (40) by secreting massive amounts of IL-5, IL-9, and IL-13 (31). We found that high EC-SOD, either through overexpression or the R213G SNP, attenuated allergen-mediated activation of ILC2s. Perhaps EC-SOD acts directly on ILC2s by abating the oxidative stress in the microenvironment surrounding ILC2s and affecting the cellular response by decreasing NF-κB/AP-1–dependent gene activation, or through an indirect effect by decreasing epithelial expression/release of alarmins. TSLP is another alarmin that activates ILC2s and is involved in inflammatory responses in asthma (41). More studies are required to decipher the precise relationship between EC-SOD and the alarmins in inflammatory airway diseases.
Our findings that increased airway EC-SOD attenuates allergic airway inflammation are consistent with other published studies suggesting that EC-SOD attenuates lung oxidative stress–mediated lung injury from LPS, bleomycin, ozone, etc. (42–47). Allergic asthma, oxidative stress, and lung inflammation go hand-in-hand (48, 49). There are reports suggesting that EC-SOD plays a major role in determining the progression of allergic lung disease by controlling DC maturation and T lymphocyte priming, differentiation, proliferation, and activation (25). As a therapeutic intervention, EC-SOD has also been shown to reduce house dust mite–induced atopic dermatitis–like skin inflammation, mast cell activation (50), and Th2-driven allergic conjunctivitis (51), but has not been studied in asthma models. The SOD mimetic MnTE-2-PyP has also been shown to decrease OVA-induced allergic airway inflammation in mice (52) by decreasing CD25 expression on Th2 lymphocytes (26). However, the role of EC-SOD in the developmental phase of asthma and the role of EC-SOD distribution in lungs were not studied. Variability in the effect of MnTE-2-PyP on TSLP and IL-33 levels suggests that it may have different control mechanisms and perhaps work at different stages of asthma sensitization and development. Whether EC-SOD mimetics could be used as novel therapies for allergy-induced asthma remains to be shown, but airway antioxidants (ascorbate, α-tocopherol) have been reported to be low in asthma patients (53).
Overall, our data suggest that EC-SOD reduces allergen-induced superoxide generation in airway epithelial cells and thereby reduces the expression and release of IL-33, in addition to other proinflammatory cytokines. This in turn could attenuate ILC2 expansion and activation, resulting in a decreased polarization of DCs to a type II inflammation. The host response in severe asthma is apparently to increase EC-SOD expression in the airways as an adaptive response to attenuate allergen-induced airway exacerbation, to account for an increase in oxidative stress. Evolutionarily, this may also lead to increased prevalence of the R213G SNP in some populations with high asthma rates. Stable drugs such as MnTE-2-PyP, or recombinant EC-SOD could be used as therapeutic agents to boost antioxidant defenses in the airways. Identifying individuals with a genetic predisposition may guide the therapeutic approach to protect against the development of chronic inflammatory lung diseases like allergic asthma.
Study limitations.
The R213G SNP has a low minor allele frequency of 3% in non-Hispanic European and 6% in south Asian populations, but near-zero prevalence in African-Americans. Thus, the R213G findings are more relevant to these populations. It is not clear if the EC-SOD–mediated attenuation of allergic airway inflammation is via direct inhibition by IL-33 or indirectly through IL-33 mediators. We also have not yet analyzed changes in other ILC populations that may be important in determining the plasticity in allergic airway inflammation (e.g., ILC3 or ILC1). Moreover, no primary airway epithelial cells were used to replicate the BEAS-2B cell line results. We did not use recombinant or native EC-SOD in our experiments since the protein has a shorter half-life in cell culture compared with the drug used, and the activity may not stay the same during the experimental time period. These experiments may be incorporated into future experiments to expand upon this new knowledge.
Methods
Asthmatic sputum samples and gene analysis.
Sputum samples were collected from asthmatics and healthy volunteers aged greater than or equal to 12 years old, between September 2009 and June 2012, that completed the YCAAD phenotyping protocol as described in Yan et al. (32) and all subjects gave informed written consent. Briefly, the study excluded anyone who smoked within the past year, had a history chronic lung diseases other than asthma (such as COPD, allergic bronchopulmonary aspergillosis, Churg-Strauss syndrome, pulmonary vascular disease, or interstitial lung disease); had other severe chronic conditions such as congestive heart failure, chronic kidney disease, liver disease, or viral infection; or had an inability to safely tolerate the studies required for participation. The Yale University School of Medicine Human Investigation Committee approved the protocol. Samples were collected with informed consent and de-identified.
RNA was isolated from sputum samples consisting of cell pellets with an All-in-One purification kit (Norgen Biotek). The RNA quality and purity were checked on an Agilent Bioanalyzer (Agilent Technologies), and, if required, treated again to remove any DNA contamination (Qiagen). Sputum RNA (10 ng) was amplified with a WT-Ovation Pico RNA amplification System (NuGen) and processed as per the manufacturer’s instructions (Affymetrix) as described previously (54).
The data are available in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE56396.
Genotyping rs1799895 in the COPDGene cohort.
DNA was isolated from 9,943 subjects in the COPDGene cohort, all of whom gave informed written consent as previously described (55). SNP rs1799895 was genotyped using a TaqMan predesigned assay (Applied Biosystems/ThermoFisher Scientific).
The CCHS, the CGPS, and rs1799895 genotyping.
The CGPS was initiated in 2003 with ongoing enrollment, and the CCHS was initiated in 1976–1978 with followup examinations in 1981–1983, 1991–1994, and 2001–2003. For both studies, individuals aged 20–100 years were invited randomly from the Danish Civil Registration System to reflect the Danish general population. The study was approved by Herlev Hospital and Danish Ethical Committees, conducted according to the Declaration of Helsinki, and written informed consent was obtained from all participants. DNA was available in the CGPS and in the 1991–1994 and 2001–2003 examinations of the CCHS. Only individuals of Danish descent with DNA available were included. In the CGPS, individuals were genotyped for the SOD3 rs1799895 R213G variant using a TaqMan-based assay (Applied Biosystems) and Taqman GenoTyper v1.2 with a call rate of 99.3% (35). In the CCHS, individuals were genotyped using PCR followed by restriction enzyme digestion and agarose gel electrophoresis, and all R213G carriers were reanalyzed and confirmed by DNA sequencing, as described previously (34).
Animals.
WT and genetically modified C57BL/6 mice for EC-SOD as OE (overexpressors), R213G HM, and R213G HET were bred at the National Jewish Health (NJH) Biological Resource Center (BRC) under a pathogen-free condition following standard institutional guidance and approval from the IACUC at NJH. Age- and sex-matched littermates were used for experiments.
OVA sensitization and challenge.
Mice were sensitized with 20 μg OVA (Sigma-Aldrich) or saline on day 1 and day 14 by i.p. injection. Imject Alum (Thermo Fisher Scientific) was used as an adjuvant to OVA in the injections. Aerosol challenge was performed on day 28, 29, and 30 with 1% OVA through a Micro Mist Nebulizer (Hudson RCI) in a modified containment box. OVA-specific adaptive immune responses were examined 24 hours after the last allergen exposure on day 32.
Alternaria challenge.
Intranasal Alternaria (5 μg; GreerLabs) or saline was delivered in mice once per day for 3 days. At 72 hours after the last Alternaria challenge, mice were euthanized to check for lung function, inflammation, and ILCs.
Lung function.
The AHR was assessed with increasing doses of nebulized methacholine (acetyl-β-methylcholine chloride, Sigma-Aldrich) in saline from 0 to 50 mg/ml (in OVA model) or 0 to 100 mg/ml (in Alternaria model) on a flexiVent (Scireq) system by tracheostomy. The airway resistance was recorded in the flexiVent software 5.01 or FlexiWare following the methacholine dose response.
BALF and inflammatory cells.
A tracheal cannula was used to instill 1 ml PBS to retrieve BALF from the lungs of mice. The procedure was repeated 3 times to collect BALF in 2 different tubes. Both the tubes were spun at 350 g for 5 minutes at 4°C. The supernatant from the first tube was preserved for cytokine analysis in 1% BSA at –80°C. Pelleted cells were pooled in 150 μl of PBS and 10 μl of the suspension diluted 1:2 in Trypan blue was used to count the cells and check viability with a hemocytometer. Next, 50 μl was used to prepare Cytospin slides that were stained with Hema-Diff (Thermo Fisher Scientific) and then were observed under a high-power microscope for leukocyte differential counts.
Cytokine analysis.
Cytokines, including IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12p70, IL-13, IL-25, IL-33, TSLP, KC/GRO, TNF-α, and IFN-γ in different samples, were analyzed from human sputum or mouse BALF in an electrochemical based multiplex assay, V-plex or U-plex kit (MSD) according to the manufacturer’s protocol. The plate was read on a Sector Imager 2400 (MSD) immediately after adding the MSD read buffer.
Protein isolation and Western blot.
Samples from human sputum, mouse lung lysate, or mouse BALF were used for Western blotting. Frozen lung tissues were mechanically pulverized and resuspended with an ice-cold lysis buffer (50 mM Tris, 200 mM NaCl, 1% Triton X-100, pH 7.4) containing a protease inhibitor cocktail (Thermo Fisher Scientific) and phosphatase inhibitors (10 mM sodium fluoride, 1.5 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF, and 60 mM β-glycerophosphate). Validated antibodies for IL-33 (1:1,000), MUC5AC (1:1,000), and corresponding HRP-conjugated secondary antibodies were used at 1:2,000 dilution. β-Actin was used as the housekeeping genes (please see Supplemental Table 1 for all antibody information). The EC-SOD antibody was custom made by Lampire (56). Protein bands were visualized using the FluorChem system (Protein Simple) with Amersham ECL Prime (GE Healthcare). Densitometric analysis was done with FluorChem software.
Lung histology.
Left lung lobes were fixed in 4% paraformaldehyde (PFA) and sent to the histology core facility of NJH for paraffin embedding and sectioning. The lung sections were stained with hematoxylin and eosin (H&E), Masson’s trichrome, and PAS following the standards for quantitative assessment of lug structure (57). Mucus staining was measured by deconvoluting PAS-stained images with FIJI (ImageJ) and drawing lines around the basement membrane and the airway epithelium around the lumen (58, 59). The wall area percentage was calculated as (60): wall area % = (wall area/total bronchial area) × 100.
Flow cytometry.
Right-lung lobes were mechanically cut into small pieces and digested with 1 mg/ml collagenase I (Thermo Fisher Scientific) for 30 minutes at 37°C with constant inversion in RPMI 1640 complete medium (RPMI 1640 + 10% FBS + penicillin/streptomycin). The digested suspension was mechanically disrupted with 18-gauge × 1.5-inch needles (BD) in 15-ml conical tubes and filtered with 70-μm cell strainers in a fresh 50-ml conical tube. The cell suspension was centrifuged at 300 g for 10 minutes at 4°C. Red blood cells were lysed with 2 ml ACK buffer for 5 minutes at 4°C and the cells were washed with PBS before plating them in a 6-well plate in 300 μl RPMI complete medium. The cells were incubated at 37°C, 5% CO2 in a water-jacketed incubator overnight to enrich the cell population and get rid of plastic-adhered structural cells. The cell suspensions were pipetted out in fresh tubes and centrifuged at 300 g for 10 minutes at 4°C. After the cells were washed with FACS buffer (PBS + 1% BSA + 0.1% NaN3), they were plated on a round-bottom 96-well plate with 200 μl FACS buffer. The cells were blocked with Fc blocker for 20 minutes at 4°C and incubated with respective extracellular antibodies for 30 minutes at 4°C. After the cells were washed with FACS buffer, they were fixed with 4% PFA in FACS buffer for 10 minutes at 4°C. Washed cells were permeabilized with 0.1% saponin in FACS buffer for intracellular staining.
Cells were analyzed on a BD Fortessa to identify ILC2s as live lymphocytes gated with Lin− (CD3, Ly-6G/Ly-6C, CD11b, CD45R/B220, TER-119), CD5−, CD49b−, TCRαβ−, TCRγδ−, CD45+, CD90+, CD25+, ST2+, and GATA3+ (Supplemental Table 1). Intracellular staining for IL-5, IL-9, and IL-13 was also done in these ILC2s. Gating is shown in Supplemental Figure 8.
Cell culture and stimulation.
BEAS-2B cells were obtained from Rafeul Alam’s lab (NJH), cultured in complete DMEM (DMEM+ 10% FBS + 1% penicillin/streptomycin) and serum starved for 24 hours. Alternaria extract (10 μg) and/or MnTE-2-PyP (30 μM) were added to the serum-free culture media and incubated at 37°C for 12 or 24 hours. Media were preserved for cytokine assessment and RNA was extracted for quantitative PCR. MnTE-2-PyP was obtained as a gift from James Crapo (NJH).
Quantitative PCR.
RNA isolated with RNeasy (Qiagen) from the BEAS-2B cells was quantified with a Nanodrop (Thermo Fisher Scientific). Taqman assays were used for IL-33 (Hs01125943_m1) and TSLP (Hs00263639_m1) with Taqman Fast Virus 1-Step Master Mix (Thermo Fisher Scientific).
Statistics.
Data were analyzed and plotted using GraphPad Prism (version 5.0f) and R (version 3.3.1) software. One-way ANOVA was used with Tukey’s multiple comparison test to determine the statistical differences among groups. Two-way ANOVA was used with Bonferroni multiple comparisons in case of grouped comparisons. The associated P values as well as the sample size and experiment numbers are indicated throughout. Error bars represent ± SEM. Among all the data sets, P values less than 0.05 were considered significant and are denoted as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The association analysis was calculated with logistic regression and with single- and multiple-failures-per-subject Cox regression.
Study approval.
Study protocols were reviewed and approved by relevant regulatory bodies including (a) the Yale University School of Medicine Human Investigation Committee (New Haven, CT) phenotyping protocol for YCAAD, as described in Yan et al. (32), (b) the NJH Institutional Review Board (Denver, CO) for the COPDGene cohort (55), (c) Herlev Hospital and Danish Ethical Committees (Copenhagen, Denmark), and (d) The Institutional Animal Care and Use Committee (Denver, CO) for animals used under R. Bowler’s protocol. All human studies were conducted according to the Declaration of Helsinki, and written informed consent was obtained from all participants.
Author contributions
RG, HWC, RA, ENG, BGN, GLC, and RPB designed the research studies. RG, JTV, MRW, LHL, and QL conducted experiments. RG, JTV, MRW, and QL acquired data. RG, SRJ, ENG, CJK, SA, and QL analyzed data. RG, HWC, RA, ENG, GLC, BGN, and RPB wrote the manuscript.
Supplementary Material
Acknowledgments
NIH/NHLBI R01HL111288 (to R. Bowler) and R01HL086680 (to E. Nozik-Grayck). We thank James Crapo for his gift of the SOD mimetic; Mukesh Verma for his help with Alternaria and flow cytometry; Alan Copenhaver and Philip Simonian for aiding in the development of ILC flow panel; Di Jiang for help with nebulizers; Brittany C. Hartman for technical help; Josh Burbank (NJH Flow Cytometry Core) for help with flow cytometry; and the Biological Resource Center of NJH.
Version 1. 08/28/2017
Electronic publication
Funding Statement
Awarded to Dr. Russell Bowler
Awarded to Dr. Eva Nozik-Grayck
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(17):e95072. https://doi.org/10.1172/jci.insight.95072.
Contributor Information
Rohit Gaurav, Email: gauravr@njhealth.org.
Laura Hernandez-Lagunas, Email: Ana-Laura.Hernandez@ucdenver.edu.
Qing Liu, Email: qing.liu@yale.edu.
Eva Nozik-Grayck, Email: Eva.Grayck@ucdenver.edu.
Hong Wei Chu, Email: chuH@njhealth.org.
Rafeul Alam, Email: alamr@njhealth.org.
Shoaib Afzal, Email: Shoaib.Afzal@regionh.dk.
Geoffrey L. Chupp, Email: geoffrey.chupp@yale.edu.
References
- 1.Bowler RP, Crapo JD. Oxidative stress in allergic respiratory diseases. J Allergy Clin Immunol. 2002;110(3):349–356. doi: 10.1067/mai.2002.126780. [DOI] [PubMed] [Google Scholar]
- 2.Gaurav R, Agrawal DK. Clinical view on the importance of dendritic cells in asthma. Expert Rev Clin Immunol. 2013;9(10):899–919. doi: 10.1586/1744666X.2013.837260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowler RP, Crapo JD. Oxidative stress in airways: is there a role for extracellular superoxide dismutase? Am J Respir Crit Care Med. 2002;166(12 Pt 2):S38–S43. doi: 10.1164/rccm.2206014. [DOI] [PubMed] [Google Scholar]
- 4.Panday A, Sahoo MK, Osorio D, Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol. 2015;12(1):5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boldogh I, et al. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J Clin Invest. 2005;115(8):2169–2179. doi: 10.1172/JCI24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaurav R, Bewtra AK, Agrawal DK. Chloride channel 3 channels in the activation and migration of human blood eosinophils in allergic asthma. Am J Respir Cell Mol Biol. 2015;53(2):235–245. doi: 10.1165/rcmb.2014-0300OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Junger WG. Measurement of oxidative burst in neutrophils. Methods Mol Biol. 2012;844:115–124. doi: 10.1007/978-1-61779-527-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rada B, Leto TL. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib Microbiol. 2008;15:164–187. doi: 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emelyanov A, et al. Elevated concentrations of exhaled hydrogen peroxide in asthmatic patients. Chest. 2001;120(4):1136–1139. doi: 10.1378/chest.120.4.1136. [DOI] [PubMed] [Google Scholar]
- 10.Kharitonov SA, et al. Exhaled nitric oxide is increased in asthma. Chest. 1995;107(3 Suppl):156S–157S. doi: 10.1378/chest.107.3_supplement.156s. [DOI] [PubMed] [Google Scholar]
- 11.Marklund SL. Human copper-containing superoxide dismutase of high molecular weight. Proc Natl Acad Sci USA. 1982;79(24):7634–7638. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crapo JD, Day BJ. Modulation of nitric oxide responses in asthma by extracellular antioxidants. J Allergy Clin Immunol. 1999;104(4 Pt 1):743–746. doi: 10.1016/s0091-6749(99)70282-4. [DOI] [PubMed] [Google Scholar]
- 13.Oury TD, Chang LY, Marklund SL, Day BJ, Crapo JD. Immunocytochemical localization of extracellular superoxide dismutase in human lung. Lab Invest. 1994;70(6):889–898. [PubMed] [Google Scholar]
- 14.Oury TD, Day BJ, Crapo JD. Extracellular superoxide dismutase in vessels and airways of humans and baboons. Free Radic Biol Med. 1996;20(7):957–965. doi: 10.1016/0891-5849(95)02222-8. [DOI] [PubMed] [Google Scholar]
- 15.Kim H, et al. Differential expression of EC-SOD, Mn-SOD and CuZn-SOD in rat lung exposed to crystalline silica. J Occup Health. 2007;49(3):242–248. doi: 10.1539/joh.49.242. [DOI] [PubMed] [Google Scholar]
- 16.Comhair SA, Erzurum SC. Redox control of asthma: molecular mechanisms and therapeutic opportunities. Antioxid Redox Signal. 2010;12(1):93–124. doi: 10.1089/ars.2008.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao H, et al. Extracellular superoxide dismutase protects against pulmonary emphysema by attenuating oxidative fragmentation of ECM. Proc Natl Acad Sci USA. 2010;107(35):15571–15576. doi: 10.1073/pnas.1007625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gongora MC, et al. Loss of extracellular superoxide dismutase leads to acute lung damage in the presence of ambient air: a potential mechanism underlying adult respiratory distress syndrome. Am J Pathol. 2008;173(4):915–926. doi: 10.2353/ajpath.2008.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folz RJ, Abushamaa AM, Suliman HB. Extracellular superoxide dismutase in the airways of transgenic mice reduces inflammation and attenuates lung toxicity following hyperoxia. J Clin Invest. 1999;103(7):1055–1066. doi: 10.1172/JCI3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med. 2003;35(3):236–256. doi: 10.1016/S0891-5849(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 21.Petersen SV, et al. The high concentration of Arg213-->Gly extracellular superoxide dismutase (EC-SOD) in plasma is caused by a reduction of both heparin and collagen affinities. Biochem J. 2005;385(Pt 2):427–432. doi: 10.1042/BJ20041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartney JM, et al. A common polymorphism in extracellular superoxide dismutase affects cardiopulmonary disease risk by altering protein distribution. Circ Cardiovasc Genet. 2014;7(5):659–666. doi: 10.1161/CIRCGENETICS.113.000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouradian GC, et al. Superoxide dismutase 3 R213G single-nucleotide polymorphism blocks murine bleomycin-induced fibrosis and promotes resolution of inflammation. Am J Respir Cell Mol Biol. 2017;56(3):362–371. doi: 10.1165/rcmb.2016-0153OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(8):858–864. doi: 10.1164/rccm.200509-1387OC. [DOI] [PubMed] [Google Scholar]
- 25.Kwon MJ, Jeon YJ, Lee KY, Kim TY. Superoxide dismutase 3 controls adaptive immune responses and contributes to the inhibition of ovalbumin-induced allergic airway inflammation in mice. Antioxid Redox Signal. 2012;17(10):1376–1392. doi: 10.1089/ars.2012.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jungsuwadee P, Weaver MR, Gally F, Oberley-Deegan RE. The metalloporphyrin antioxidant, MnTE-2-PyP, inhibits Th2 cell immune responses in an asthma model. Int J Mol Sci. 2012;13(8):9785–9797. doi: 10.3390/ijms13089785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Archambeau JO, Tovmasyan A, Pearlstein RD, Crapo JD, Batinic-Haberle I. Superoxide dismutase mimic, MnTE-2-PyP(5+) ameliorates acute and chronic proctitis following focal proton irradiation of the rat rectum. Redox Biol. 2013;1:599–607. doi: 10.1016/j.redox.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forman HJ, Fridovich I. Superoxide dismutase: a comparison of rate constants. Arch Biochem Biophys. 1973;158(1):396–400. doi: 10.1016/0003-9861(73)90636-X. [DOI] [PubMed] [Google Scholar]
- 29.Liew FY, Girard JP, Turnquist HR. Interleukin-33 in health and disease. Nat Rev Immunol. 2016;16(11):676–689. doi: 10.1038/nri.2016.95. [DOI] [PubMed] [Google Scholar]
- 30.Uchida M, et al. Oxidative stress serves as a key checkpoint for IL-33 release by airway epithelium. Allergy. doi: 10.1111/all. [published online ahead of print March 8, 2017]. https://doi.org/10.1111/all.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halim TY, et al. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat Immunol. 2016;17(1):57–64. doi: 10.1038/ni.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan X, et al. Noninvasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Am J Respir Crit Care Med. 2015;191(10):1116–1125. doi: 10.1164/rccm.201408-1440OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auten RL, O’Reilly MA, Oury TD, Nozik-Grayck E, Whorton MH. Transgenic extracellular superoxide dismutase protects postnatal alveolar epithelial proliferation and development during hyperoxia. Am J Physiol Lung Cell Mol Physiol. 2006;290(1):L32–L40. doi: 10.1152/ajplung.00133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juul K, et al. Genetically reduced antioxidative protection and increased ischemic heart disease risk: The Copenhagen City Heart Study. Circulation. 2004;109(1):59–65. doi: 10.1161/01.CIR.0000105720.28086.6C. [DOI] [PubMed] [Google Scholar]
- 35.Kobylecki CJ, Afzal S, Nordestgaard BG. Does SOD3 R213G homozygosity influence morbidity, mortality, and lung function in the general population? Antioxid Redox Signal. 2016;24(15):884–891. doi: 10.1089/ars.2016.6629. [DOI] [PubMed] [Google Scholar]
- 36.Regan EA, et al. Smoking and COPD increase sputum levels of extracellular superoxide dismutase. Free Radic Biol Med. 2011;51(3):726–732. doi: 10.1016/j.freeradbiomed.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Fattman CL, Chu CT, Kulich SM, Enghild JJ, Oury TD. Altered expression of extracellular superoxide dismutase in mouse lung after bleomycin treatment. Free Radic Biol Med. 2001;31(10):1198–1207. doi: 10.1016/S0891-5849(01)00699-2. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, et al. Lyn kinase represses mucus hypersecretion by regulating IL-13-induced endoplasmic reticulum stress in asthma. EBioMedicine. 2017;15:137–149. doi: 10.1016/j.ebiom.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen ES, et al. Oxidation of the alarmin IL-33 regulates ST2-dependent inflammation. Nat Commun. 2015;6:8327. doi: 10.1038/ncomms9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenzie AN. Type-2 innate lymphoid cells in asthma and allergy. Ann Am Thorac Soc. 2014;11 Suppl 5:S263–S270. doi: 10.1513/AnnalsATS.201403-097AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, et al. Central role of cellular senescence in TSLP-induced airway remodeling in asthma. PLoS ONE. 2013;8(10):e77795. doi: 10.1371/journal.pone.0077795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowler RP, Arcaroli J, Abraham E, Patel M, Chang LY, Crapo JD. Evidence for extracellular superoxide dismutase as a mediator of hemorrhage-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2003;284(4):L680–L687. doi: 10.1152/ajplung.00191.2002. [DOI] [PubMed] [Google Scholar]
- 43.Bowler RP, Arcaroli J, Crapo JD, Ross A, Slot JW, Abraham E. Extracellular superoxide dismutase attenuates lung injury after hemorrhage. Am J Respir Crit Care Med. 2001;164(2):290–294. doi: 10.1164/ajrccm.164.2.2011054. [DOI] [PubMed] [Google Scholar]
- 44.Bowler RP, et al. Extracellular superoxide dismutase attenuates lipopolysaccharide-induced neutrophilic inflammation. Am J Respir Cell Mol Biol. 2004;31(4):432–439. doi: 10.1165/rcmb.2004-0057OC. [DOI] [PubMed] [Google Scholar]
- 45.Bowler RP, Nicks M, Warnick K, Crapo JD. Role of extracellular superoxide dismutase in bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2002;282(4):L719–L726. doi: 10.1152/ajplung.00058.2001. [DOI] [PubMed] [Google Scholar]
- 46.Tollefson AK, et al. Endogenous enzymes (NOX and ECSOD) regulate smoke-induced oxidative stress. Free Radic Biol Med. 2010;49(12):1937–1946. doi: 10.1016/j.freeradbiomed.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottfredsen RH, Goldstrohm DA, Hartney JM, Larsen UG, Bowler RP, Petersen SV. The cellular distribution of extracellular superoxide dismutase in macrophages is altered by cellular activation but unaffected by the naturally occurring R213G substitution. Free Radic Biol Med. 2014;69:348–356. doi: 10.1016/j.freeradbiomed.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan N, et al. 25-Hydroxyvitamin D3-deficiency enhances oxidative stress and corticosteroid resistance in severe asthma exacerbation. PLoS ONE. 2014;9(11):e111599. doi: 10.1371/journal.pone.0111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li G, et al. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic Biol Med. 2012;52(2):392–401. doi: 10.1016/j.freeradbiomed.2011.10.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YS, et al. Extracellular superoxide dismutase ameliorates house dust mite-induced atopic dermatitis-like skin inflammation and inhibits mast cell activation in mice. Exp Dermatol. 2016;25(8):630–635. doi: 10.1111/exd.13028. [DOI] [PubMed] [Google Scholar]
- 51.Lee HJ, Kim BM, Shin S, Kim TY, Chung SH. Superoxide dismutase 3 attenuates experimental Th2-driven allergic conjunctivitis. Clin Immunol. 2017;176:49–54. doi: 10.1016/j.clim.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 52.Terziev L, Dancheva V, Shopova V, Stavreva G. Antioxidant effect of MnTE-2-PyP on lung in asthma mice model. ScientificWorldJournal. 2012;2012:379360. doi: 10.1100/2012/379360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larkin EK, et al. New risk factors for adult-onset incident asthma. A nested case-control study of host antioxidant defense. Am J Respir Crit Care Med. 2015;191(1):45–53. doi: 10.1164/rccm.201405-0948OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho MH, et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2(3):214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen SV, et al. Extracellular superoxide dismutase (EC-SOD) binds to type i collagen and protects against oxidative fragmentation. J Biol Chem. 2004;279(14):13705–13710. doi: 10.1074/jbc.M310217200. [DOI] [PubMed] [Google Scholar]
- 57.Hsia CC, Hyde DM, Ochs M, Weibel ER, ATS/ERS Joint Task Force on Quantitative Assessment of Lung Structure An official research policy statement of the American Thoracic Society/European Respiratory Society: standards for quantitative assessment of lung structure. Am J Respir Crit Care Med. 2010;181(4):394–418. doi: 10.1164/rccm.200809-1522ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7–8):518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim V, et al. Airway wall thickness is increased in COPD patients with bronchodilator responsiveness. Respir Res. 2014;15:84. doi: 10.1186/s12931-014-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.