Abstract
Objective The objective of this study was to characterize the bladder microbiota in pregnancy.
Methods A prospective observational study of 51 pregnant women, admitted to a tertiary care hospital, who underwent straight catheterization urine collection or transurethral Foley catheter placement. 16S rRNA gene sequencing and enhanced quantitative urine culture assessed the maternal bladder microbiota with comparisons made to standard urine culture results.
Results Enhanced quantitative urine culture and 16S rRNA gene sequencing detected bacteria in the majority of participants. Lactobacillus and Gardnerella were the most commonly detected microbes. In contrast, standard urine culture had a 100% false-negative rate and failed to detect several known or emerging urinary pathogens.
Conclusion There are live bacteria in the bladders of most pregnant women. This challenges the definition of asymptomatic bacteriuria.
Keywords: asymptomatic bacteriuria, microbiome, bladder, pregnancy
Asymptomatic bacteriuria (ASB) is defined as the presence of a uropathogen in the absence of physical symptoms. 1 ASB strains have long been thought to be nonpathogenic variants of pathogenic species, especially Escherichia coli or Streptococcus agalactiae (also known as group B streptococcus [GBS]). However, in certain populations, such as the general obstetric population, untreated ASB is associated with subsequent urinary tract infection (UTI). This includes pyelonephritis, 2 which is associated with adverse pregnancy outcomes, maternal sepsis, and preterm labor. 2 3 Thus, routine screening and treatment of ASB are recommended during pregnancy. 4 5
However, the current approach to ASB screening is limited by the assumption that the female bladder naturally exists in a sterile state 6 7 ; thus, the presence of bacteria in urine is currently considered abnormal. Yet, several recent studies have revealed the existence of bacterial communities in the bladders of nonpregnant perimenopausal women. 8 9 10 11 12 One of the earliest studies compared multiple urine collection methods, 13 including suprapubic aspiration (SPA), which bypasses vulvovaginal contamination. This study noted that the bacterial DNA (microbiome) detected in urine obtained by transurethral catheter closely resembled those obtained by SPA, and diverged substantially from the microbiomes in vaginal swabs and mid-stream voided urine. Thus, transurethral catheterized urine specimens accurately sample the bladder and are thus the appropriate urine collection method for investigation of the bacteria present in the urinary bladders of women (the female bladder microbiota). 13
To determine whether bacterial DNA detected by 16S rRNA gene sequencing originated from live bacteria within the bladder, our group developed an enhanced quantitative urine culture protocol that detects bacteria in the vast majority of catheterized urine samples deemed “no growth” by standard clinical microbiology urine culture methods. 14 15 Thus, standard urine culture does not detect the presence of a large number of bacterial species, including most uropathogens. 16
Given compelling enhanced culture and DNA sequence evidence that the female urinary bladder of perimenopausal women is not sterile, we questioned the sterility of urine obtained from the bladders of younger pregnant women. Specifically, we used the complementary approaches of enhanced quantitative urine culture and 16S rRNA gene sequencing to determine if urine obtained by transurethral catheterization contains microbiota and then compared those results to a standard clinical microbiology urine culture protocol.
Materials and Methods
Participant Recruitment
Following Loyola Institutional Review Board approval, we prospectively invited pregnant patients presenting to labor and delivery to participate in the study if they were ≥18 years of age, proficient in English and undergoing straight catheterization urine collection or placement of a transurethral Foley catheter as part of their clinical care (e.g., for preterm labor [PTL], labor w/epidural, or preeclampsia). We excluded patients who had used antibiotics within the past 4 weeks for any reason. History of ASB was not part of the exclusion criteria and knowledge of previous urine dip was not required for participation nor collected for purposes of this study. Participants gave written informed research consent. At enrollment, participants provided demographic information, UTI history, medication (including antibiotic) use, obstetric history, tobacco use, and other comorbidities. As part of standard obstetric care, this cohort had undergone screening for ASB. For all patients, we recorded gestational age at the time of sample collection.
Sample Collection and Urine Culture
Urine was collected aseptically via the transurethral catheter into a sterile collection cup. As per clinical protocol, a portion of the urine sample was placed into a BD Vacutainer Plus C&S preservative tube and sent to the research laboratory for standard urine culture and enhanced quantitative urine culture assessment. Due to the lack of appropriate refrigeration facilities in the clinic, the remainder of the urine was stored at room temperature for 1 to 8 hours before being processed and stored down at −80°C with 10% AssayAssure (Sierra Molecular; Incline Village, NV) for subsequent sequencing.
Within 24 hours of urine collection, both standard and enhanced urine cultures were set up. Both protocols are provided in Table 1 and have been described previously. 14 The detection limit for the standard protocol was 1,000 CFU/mL. For enhanced quantitative urine culture, it was 10 CFU/mL. If enhanced quantitative urine culture was unable to detect bacteria, the sample was considered to be below the detection threshold. For samples with bacteria detected, α diversities were measured, the Bray–Curtis dissimilarity was calculated, 17 and the complete method was used for hierarchical clustering using RStudio. 18 Bray–Curtis dissimilarity is a measurement used to quantify the compositional differences between two diverse communities; it therefore clusters samples with similar community structures. The resulting dendrogram was divided into 14 clusters (termed urotypes) that were named on the basis of the predominant organism. When a cluster had no predominant organism, it was called a “diverse” urotype.
Table 1. Culture protocols.
| Protocol | Volume | Media | Atmospheric conditions | Time |
|---|---|---|---|---|
| Standard urine culture a | 1 µL | Blood agar MacConkey agar |
Aerobic | 24 h 35 deg |
| Enhanced quantitative urine culture b | 100 µL | Blood agar Chocolate agar CNA agar Anaerobic blood agar |
Aerobic CO 2 Anaerobic |
48 h 35 deg |
Abbreviation: CNA, colistin–naladixic acid.
For standard urine culture, we struck 0.001 mL of urine onto 5% sheep bloodagar plate (BAP) and MacConkey agars (BD BBL prepared plated media), which were incubated aerobically at 35°C for 24 hours. Each separate morphological colony type was counted and identified in any amount. The detection level was 1,000 CFU/mL, represented by one colony of growth on either plate. If no growth was observed, the culture was reported as “no growth,” indicating no growth of bacteria at the lowest dilution, that is, 1:1,000.
For enhanced quantitative urine culture, we struck 0.1 mL of urine onto BAP, chocolate, and CNA agars (BD BBL prepared plated media), which were incubated in 5% CO 2 at 35°C for 48 hours. A second BAP and CDC anaerobe 5% sheep BAP (BD BBL prepared plated media) were also inoculated with 0.1 mL of urine and incubated under anaerobic conditions at 35°C for 48 hours. The detection level was 10 CFU/mL, represented by one colony of growth on any of the plates. Each morphologically distinct colony type was isolated on a different plate of the same medium to prepare a pure culture for identification.
16S rRNA Gene Sequencing
DNA isolation protocols and 16S rRNA gene sequence analysis on the Illumina MiSeq (Illumina, San Diego, CA) have been described previously. 15 Briefly, total DNA was extracted from 1 mL of catheterized urine under a laminar flow hood, using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). The fourth variable region was amplified using Illumina adapted universal primers (515F and 806R). Extraction negative controls (no urine) and polymerase chain reaction (PCR) negative controls (no template) were included to assess contribution of extraneous DNA from reagents. Final PCR products were purified from unincorporated nucleotides and primers using Agencourt AMPure XP-PCR magnetic beads (Beckman Coulter). Purified samples were normalized to equal DNA concentration, as determined by the Bioanalyzer (Agilent, Santa Clara, CA). The sample library and PhiX sequencing control library (Illumina) were denatured and added to the MiSeq 2 × 250 bp sequencing reagent cartridge, according to manufacturer's instructions (Illumina).
Sequence Processing
Each specimen was sequenced in duplicate. MiSeq sequence reads were processed following mothur's MiSeq SOP at http://www.mothur.org/wiki/MiSeq_SOP, 19 with minor modifications. Mothur software (version 1.34.4) 20 was used to process raw reads and, using default mothur parameters, to remove low quality and chimeric sequences. Taxonomic classification from phylum to genus level of sequence reads was performed by the RDP Classifier (version 2.5), 21 using the default 0.8 confidence threshold. To determine if each sample is accurate and reliable, all samples must have more than 2,000 reads, be distinct from the corresponding extraction control, and be comparable to its replicate.
Using genus-level classification, from a single replica, α diversities were measured, the Bray–Curtis distance calculated, 17 and the complete method was used for hierarchical clustering using RStudio. 18 The resulting dendrogram was divided into six clusters, termed urotypes, named on the basis of the predominant organism. When a cluster had no predominant organism, it was termed the “diverse” urotype.
Statistical Analysis
For cultured organisms, abundance was measured using total CFU/mL. Continuous variables are reported as means and standard deviations or medians and interquartile ranges. Post hoc pairwise comparisons were made using Wilcoxon's rank-sum tests for continuous variables with significant overall p -values using statistical software, SAS software v9.4 (SAS Institute, Cary, NC).
Results
All 51 recruited participants were included in the study analysis. Table 2 displays the cohort's demographic characteristics. The average age was 30 years, and most (68%) participants were obese. The average gestational age at sample collection was 36 weeks, majority (74%) were enrolled within 2 days of delivery, and 17 (33%) subjects delivered prior to 37 weeks.
Table 2. Demographics.
| N = 51 | |
|---|---|
| Mean maternal age (y) | 30 (19–41) |
| Nulliparity (%) | 49.0% |
| Race/ethnicity | N (%) |
| Caucasian | 16 (31.3%) |
| African American | 17 (33.3%) |
| Hispanic | 14 (27.5%) |
| Asian | 4 (7.8%) |
| BMI (kg/m 2 ) | |
| Normal (18.5–24.9) | 5 (9.8%) |
| Overweight (25–29.9) | 11 (21.6%) |
| Obese (30–34.5) | 19 (37.3%) |
| Morbidly obese (>35) | 16 (31.3%) |
| Mean BMI (range) | 34.3 (22.0–58.4) |
| Education | |
| High school | 11 (21.6%) |
| Some college College |
19 (37.3%) 11 (21.6%) |
| Graduate | 10 (19.6%) |
| Gestational age at delivery (wk and d) Mdn (IQR) | 38 2/7 (2 5/7 ) |
| Gestational age at collection (wk and d) Mdn (IQR) | 37 6/7 (3 5/7 ) |
| GU infection/colonization | |
| GBS positive | 6 (11.8%) |
| UTI during this pregnancy | 4 (7.8%) |
| History of any UTI | 12 (23.5%) |
| Medical comorbidities | |
| Gestational diabetes mellitus | 3 (5.9%) |
| Diabetes mellitus | 2 (3.9%) |
| Gestational hypertension | 2 (3.9%) |
| Chronic hypertension | 6 (11.8%) |
| Renal disease | 2 (3.9%) |
Abbreviations: GBS, group B streptococcus; GU, genitourinary; IQR, interquartile ranges; Mdn, median; UTI, urinary tract infection.
Sequencing Results
A bladder microbiome was detected in 68.6% (35/51) of participants by 16S rRNA sequencing, which resulted in a total of 3,605,762 reads from 7 phyla, 14 classes, 18 orders, 28 families, and 31 genera. Fig. 1 displays the maternal bladder microbiome profile for each participant grouped by predominant organism. The majority of sequenced individuals were of the Lactobacillus -predominant urotype (60%, 21/35), followed by Gardnerella (25.7%, 9/35) ( Table 3 ). A small number of individuals were dominated by the genus Staphylococcus ( N = 1), the order Bacillales (which includes the genus Enterococcus , N = 2), or the family Enterobacteriaceae (which contains the species E. coli , N = 1). Only one patient had no predominant organism, thereby that individual's urotype was considered to be “diverse.”
Fig. 1.
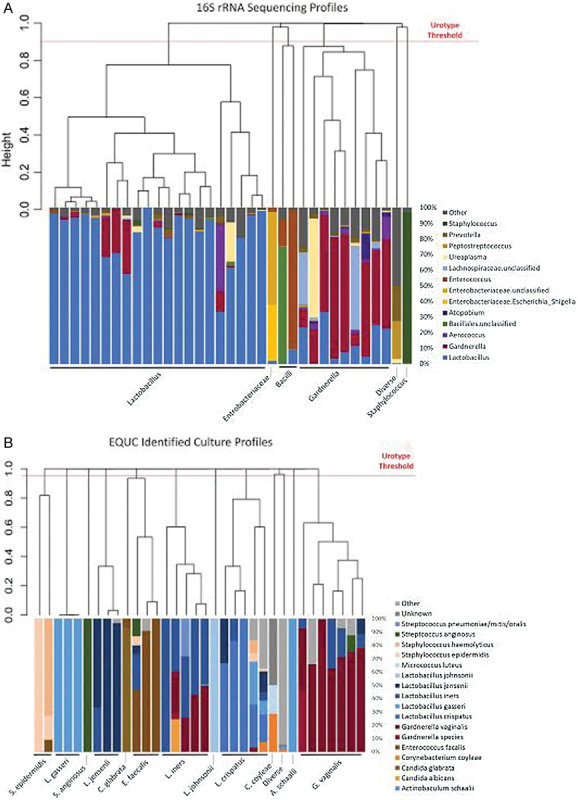
Sequencing and culture profiles of the maternal urinary microbiome. The sequencing profiles ( A ) and culture profiles ( B ) are shown. Each bar represents a urine sample from one patient. On the y -axis is the relative abundance or percentage of classified sequencing reads (A) or colony forming units per milliliter ( B ) per individual. Each bar or taxon is color coded by family or genus ( A ) or species ( B ). All samples were phylogenetically compared with each other using Bray–Curtis similarity, resulting in a phylogenetic tree or dendrograms. The length of each branch of the dendrograms represents the similarity between the samples. We divided the samples into clades by identifying a profile threshold. The clades group together by dominant organism; these groups are termed “sequence profiles” ( A ) or “culture profiles” ( B ), which are listed underneath. The sequencing dataset ( A ) divided into six profiles: Lactobacillus , Enterobacteriaceae, Bacilli, Gardnerella , Staphylococcus , and a mixed profile termed “diverse.” The culture profile ( B ) divided into 13 profiles: S. epidermidis , L. gasseri , S. anginosus , L. jensenii , C. glabrata , E. faecalis , L. iners , L. johnsonii , C. coyleae , A. schaalii , G. vaginalis , and a mixed profile termed “diverse.”
Table 3. Frequency of profiles using either sequence or enhanced quantitative urine culture data.
| Frequency of sequencing profiles | Frequency of culture profiles | ||
|---|---|---|---|
| BDT | 16/51 (31.4%) | BDT | 17/51 (33.3%) |
| Sequenced | 35/51 (68.6%) | Cultured | 34/51 (66.7%) |
| Lactobacillus | 21/35 (60%) | G. vaginalis | 7/34 (22.6%) |
| Gardnerella | 9/35 (25.7%) | L. iners | 5/34 (14.3%) |
| Bacilli/Enterococcus | 2/35 (5.7%) | L. crispatus | 5/34 (14.3%) |
| Enterobacteriaceae | 1/35 (2.9%) | E. faecalis | 3/34 (8.8%) |
| Staphylococcus | 1/35 (2.9%) | L. gasseri | 3/34 (8.8%) |
| L. jensenii | 3/34 (2.9%) | ||
| Diverse a | 1/35 (2.9%) | S. epidermidis | 2/34 (5.9%) |
| A. schaalii | 1/34 (2.9%) | ||
| C. coyleae | 1/34 (2.9%) | ||
| C. glabrata | 1/34 (2.9%) | ||
| Diverse | 1/34 (2.9%) | ||
| L. johnsonii | 1/34 (2.9%) | ||
| S. anginosus | 1/34 (2.9%) | ||
Abbreviation: BDT, below detection threshold.
A diverse profile indicates a diverse community lacking a single predominant organism.
The microbiome of participants with a Lactobacillus urotype was significantly less diverse than Gardnerella urotype subjects as determined by the Shannon diversity index and inverse Simpson index, which measure both richness and evenness ( Table 4 ). The microbiomes of participants with a Gardnerella urotype were more even, as measured by the Pielou's index. This is visible in the Gardnerella urotype profiles, which show a greater presence of other genera, including Lactobacillus , Ureaplasma , Aerococcus , Atopobium , and/or Lachnospiraceae ( Fig. 1A ).
Table 4. Differences in α diversity measurements between the Lactobacillus and Gardnerella sequencing .
| Gardnerella urotype ( N = 9) | Lactobacillus urotype ( N = 21) | p -Value | |
|---|---|---|---|
| Shannon diversity index, Mdn (IQR) | 1.28 (1.13–1.53) | 0.54 (0.35–0.91) | 0.0003 |
| Inverse Simpson index, Mdn (IQR) | 2.5 (2.02–3.10) | 1.8 (1.10–1.53) | 0.0005 |
| Total number of species, Mdn (IQR) | 83 (63–94) | 77 (74–87) | 0.8562 |
| Pielou index, Mdn (IQR) | 0.3 (0.28–0.34) | 0.13 (0.08–0.20) | 0.0003 |
Abbreviation: IQR, interquartile range.
Four different α diversity measurements were calculated from the sequencing data: Shannon diversity (a measurement of richness and evenness), inverse Simpson (richness and evenness), total number of species (richness only), and Pielou (evenness only). p -Values were generated using a Wilcoxon rank-sum test.
Enhanced Culture Results
Enhanced quantitative urine culture detected live microbes in the majority of participants (66.7%, 34/51), similar to that proportion of samples that were positive by sequencing ( Table 3 , Fig. 1B ) . At the genus level, the most common culture profile was predominated by Lactobacillus (50%, 17/34), similar to that observed by sequencing. This genus level Lactobacillus profile was composed of the following species: L. gasseri , L. jensenii , L. iners , L. johnsonii , and L. crispatus. Thus, with respect to the major dominant profiles, sequencing and enhanced quantitative urine culture obtained similar results.
The combination of 16S sequencing and enhanced quantitative urine culture detected bacteria in 80% (41/51) of participants and reached the same conclusion regarding urotype in 64% (33/51) of all the participants. Considering only participants with positive results for both methods, these two methods reached the same conclusion in 82% (23/28) of participants ( Fig. 2 ).
Fig. 2.
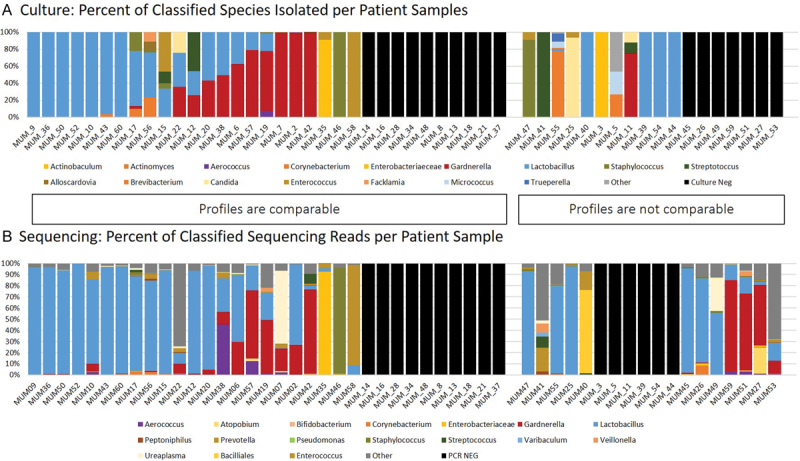
Comparison of culture and sequencing for all 51 individuals. To compare each individual's sequencing results and culture results, we lined up the relative abundance graphs for each. The relative abundance (or total classified CFU/mL per individual) culture profiles ( A ) are compared with the relative abundance (or total classified reads per individual) of the sequencing profiles ( B ). The culture classification is given at the genus level to better compare with sequencing results. Each genus is represented as a color-coded bar for each individual, where each color represents a family or genus.
Standard Urine Culture Results versus Enhanced Quantitative Urine Culture
Standard urine culture results were available for all participants. The standard urine culture was negative for all samples. In contrast, enhanced quantitative urine culture detected bacteria in 34/51 subjects. Thus, the standard urine culture had a 100% false-negative rate. In 15 participants, the standard protocol also did not detect several known or emerging urinary pathogens, including Actinobaculum schaalii ( N = 1), Alloscardovia omnicolens ( N = 1), Candida albicans ( N = 1), Candida parapsilosis ( N = 1), Corynebacterium urealyticum ( N = 1), Enterococcus faecalis ( N = 7), E. coli ( N = 1), Staphylococcus aureus ( N = 1), S. agalactiae (also known as GBS, N = 1), and Streptococcus anginosus ( N = 5) ( Table 5 ) . Five patients grew two or more of these uropathogens. Some of these organisms ( A. schaalii , A. omnicolens , and C. urealyticum ) would not normally grow under standard urine culture conditions. Many of these patients contained organisms at thresholds below the detectable level of standard urine culture (10–100 CFU/mL). However, four individuals contained one of these organisms at more than 1,000 CFU/mL; thus, in theory, these bacteria could have been detected by standard culture, but they were not.
Table 5. Uropathogens detected by enhanced quantitative urine culture in the urine samples of 15 patients.
| Uropathogen (CFU/mL) | Gestational age at collection (wk) | Gestational age at delivery (wk) | Indication for delivery | Other complications (including infection) | |
|---|---|---|---|---|---|
| MUM_03 | Actinobaculum schaalii (10) | 22 3/7 | 36 4/7 | Preeclampsia | Chronic hypertension, gestational diabetes |
| MUM_07 | Staphylococcus aureus (10) | 22 0/7 | 25 4/7 | Preterm labor | Aortic stenosis |
| MUM_11 |
Candida parapsiosis
(10)
Streptococcus anginosus (10) |
31 0/7 | 32 4/7 | pPROM | Chorioamnionitis |
| MUM_15 |
Enterococcus faecalis
(70)
Streptococcus anginosus (20) |
36 6/7 | 36 6/7 | Labor | Post-partum hemorrhage |
| MUM_19 |
Enterococcus faecalis
(10)
Streptococcus anginosus (10) |
39 1/7 | 39 2/7 | Induction for diabetes | Gestational diabetes |
| MUM_22 | Candida albicans (310) | 30 5/7 | 39 3/7 | Labor | N/A |
| MUM_25 | Enterococcus faecalis (700) | 39 0/7 | 39 1/7 | Labor | N/A |
| MUM_35 |
Enterococcus faecalis
(10,000)
Escherichia coli (1,000) |
39 2/7 | 39 3/7 | Oligohydramnios | Chorioamnionitis |
| MUM_41 | Streptococcus anginosus (1,100) | 40 4/7 | 40 4/7 | Labor | Chorioamnionitis |
| MUM_42 |
Enterococcus faecalis
(10)
Streptococcus agalactiae (30) Streptococcus anginosus (60) |
39 4/7 | 39 4/7 | Labor | N/A |
| MUM_47 | Enterococcus faecalis (10) | 37 1/7 | 38 2/7 | Labor | N/A |
| MUM_55 | Corynebacterium urealyticum (100) | 32 1/7 | 32 6/7 | Preeclampsia | Chronic hypertension, sleep apnea, gestational diabetes, |
| MUM_56 | Alloscardovia omnicolens (100) | 36 4/7 | 36 4/7 | Prior classical cesarean delivery | N/A |
| MUM_58 | Enterococcus faecalis (5,000) | 40 0/7 | 40 0/7 | Labor | N/A |
Abbreviation: pPROM, preterm premature rupture of the membrane.
History of UTIs during the Current Pregnancy
Uropathogens have been known to recur and it is possible that some of these patients had a prior infection. To determine if the patients with detectable uropathogens had previously been treated for UTIs, we looked at the medical records for prior UTI treatment during the current pregnancy. Four patients reported UTI-like symptoms during their current pregnancy. One PTL/preterm premature rupture of the membrane patient had an Enterococcus UTI in the first trimester and was treated with cephalexin. An indicated preterm patient was treated in the second trimester with cephalexin for a UTI caused by an unknown organism. Two full-term patients documented a single UTI during this pregnancy: one was treated early in their pregnancy but their records are unavailable; the second patient was treated with ceftriaxone for gonorrhea in the first trimester and nitrofurantoin in the second trimester for symptoms with a negative urine culture. At the time of sample collection, none of these four patients had detectable uropathogens by either enhanced or standard culture.
Discussion
In this prospective observational pilot study of pregnant women admitted to labor and delivery and undergoing a clinically indicated bladder catheterization, a large majority of pregnant women had evidence of bladder microbes by 16S RNA sequencing or enhanced quantitative urine culture or both. Most importantly, enhanced quantitative urine culture demonstrates the existence of live microbes in catheterized urine samples obtained from women late in gestation. This high rate of microbe detection stands in stark contrast to the low rate of detection by the standard urine culture method, which had a 100% false-negative rate for identification of bladder microbes compared with enhanced quantitative urine culture. Critically, standard urine culture did not identify several known or emerging uropathogens.
The evidence of live microbes in the bladders of most pregnant women questions the notion that the urine of pregnant women is sterile. In addition, these findings challenge the utility of the term ASB; if the bladders of most pregnant women contain bacteria, then what does the term ASB mean? We believe these data support the findings of studies from nonpregnant women, namely, that the majority of women have an identifiable urinary microbiome. As with microbiota of other human microbial niches, the maternal bladder microbiota likely vary in response to nearby microbial niches and/or system influences, such as antibiotic use, systemic disease, or perturbations in the immune system. As current standard of care suggests screening and treating ASB during pregnancy, an expanded understanding of the maternal urinary microbiome is needed using new approaches that refine our working definition of what are problematic urinary microbes during pregnancy. Further research is needed to understand how the microbiota might also play preventative or protective roles and to determine the dysbiotic effects of antibiotics.
The strengths of this investigation are the prospective design, the use of catheterized urine specimens, and the use of the robust combination of enhanced quantitative urine culture and 16S rRNA sequencing techniques. 14 15 22 Together, these techniques illustrate the limited view provided by the standard urine culture, which was specifically designed to detect fast growing bacteria with limited nutrient requirements and no aversion to oxygen (especially E. coli , the most common cause of UTI). In contrast, enhanced quantitative urine culture utilizes larger urine volumes, varied media and incubation states, and longer incubation times to also detect anaerobes, slow growing bacteria, and those that require special nutrients 14 ( Table 1 ). As enhanced quantitative urine culture can classify bacteria to the species level, the total number of culture profiles was much greater than the number of profiles detected by sequencing, which typically classifies only to the genus level. However, 16S rRNA gene sequencing complements enhanced quantitative urine culture by detecting bacteria independent of culture. 23 24
We acknowledge that our study has limitations. First, the small sample size reduces our ability to statistically compare important subgroups, such as preterm versus term deliveries. In addition, we were unable to account for potential confounding factors, such as demographics or medical comorbidities. Further studies are also needed to characterize the bladder microbiome throughout each trimester of pregnancy. Second, samples were collected over a wide gestational age range and from women with both normal and complicated pregnancies. Finally, while all samples were obtained as part of routine care, the clinical indication for catheter placement was not recorded as part of our study.
A theoretical concern exists that the detected bacteria may represent urethral or vaginal contamination. However, as described previously, a prior investigation comparing the bacterial profiles of vaginal swabs and urine obtained by transurethral catheter, suprapubic aspirate, and the mid-stream voided method concluded that bacteria detected in urine obtained by suprapubic aspirate or transurethral catheter represent true residents of the bladder. As the samples in this study were obtained by transurethral catheterization, we conclude that our findings are not the result of contamination and instead document the presence of live bladder microbiota in pregnant women.
We also recognize that enhanced quantitative urine culture and 16S sequencing did not universally agree regarding specimen profile, potentially due to differences in sample storage prior to processing. The culture tubes contain a preservative that prevents bacterial growth at room temperature, but the sequencing tubes did not. Therefore, bacterial growth in the sequencing tubes could account for the majority of the discrepancies between the data ( Fig. 2 ). This is an issue that should be considered in the design of future studies. Even given these discrepancies with sequencing data, however, the culture data clearly shows the presence of more than 10 3 CFUs of many different organisms, including known and emerging uropathogens.
As this study also documents the poor performance of the standard urine culture, these findings offer us the opportunity to improve our understanding of the maternal bladder microbiota, especially as it relates to clinically relevant definitions of ASB. Of particular concern was the finding of bacteria by our enhanced quantitative urine culture technique that had gone undetected by the standard urine culture method. Of the bacteria identified by enhanced quantitative urine culture, several are known or emerging urinary pathogens, including E. faecalis , S. anginosus , and A. schaalii (recently renamed as Actinotignum schaalii 25 ). These results require confirmation in a larger study.
Currently, we do not recommend a change in clinical practice because we do not yet know the impact of the previously undetected microbes. Some of the newly detected microbes are likely beneficial and antibiotic treatment could preclude their ability to protect. Future study into the roles of the detected microbes is indicated.
While the majority of the pregnant women in this study possessed detectable and identifiable bladder microbiota, the microbiota of several women was below the detection threshold, similar to previous reports of perimenopausal women. 1 2 3 26 27 We use the term below the detection threshold instead of “negative” or “sterile” to emphasize that these urines likely have a low biomass that cannot be detected using the current methods. Since we are unable to prove sterility, we believe this is a more accurate scientific approach. Indeed, a recent study showed that the use of larger volumes of urine could result in detection of bacteria by DNA sequencing in all samples. 28
In conclusion, we believe this study documents the presence of live microbiota in the bladders of pregnant women, an observation that warrants further study. Future investigations should clarify the normal bladder microbiota in pregnancy and identify changes over the course of pregnancy. This would allow for a deeper understanding of the normal maternal bladder microbiota and how they can become altered in ways that lead to pathologic conditions. For example, this new knowledge would clarify the definition and risk factors associated with ASB during pregnancy. Other factors should be investigated, especially the relationship between the maternal bladder microbiota and those of other pelvic floor niches, especially the vagina.
Acknowledgments
We kindly thank Mary Tulke, RN (Department of Ob/Gyn, Loyola University Medical Center, no financial disclosures) for her assistance with project coordination and set-up; Travis Price, MS (Department of Microbiology and Immunology, Stritch School of Medicine, Loyola University Chicago, no financial disclosures) for assistance with laboratory set-up, team training, and sample processing; James Grayczk, BS (Department of Microbiology and Immunology, Stritch School of Medicine, Loyola University Chicago, no financial disclosures) for his help in sample processing and DNA sequencing; Linda Brubaker, MD, MS, for critical editing of the article (Department of Obstetrics and Gynecology, Loyola University Chicago and Loyola University Medical Center, no financial disclosures), and Jean Goodman, MD (Department of Ob/Gyn, Loyola University Medical Center, no financial disclosures) for her support of this project. We would also like to thank the Loyola University Chicago Health Sciences Division's Office of Informatics and Systems Development for their expertise and for the computational resources utilized in support of this research. This research was supported by grants from the NIH to AJW (1R56DK104718–01 and R01DK104718).
Footnotes
Conflict of Interest Dr. Wolfe reports grants from Astellas Scientific and Medical Affairs, outside the submitted work. All the other authors report no conflict of interest.
References
- 1.Stein G, Fünfstück R. Asymptomatic bacteriuria [in German] Med Klin (Munich) 2000;95(04):195–200. doi: 10.1007/pl00002106. [DOI] [PubMed] [Google Scholar]
- 2.Whalley P. Bacteriuria of pregnancy. Am J Obstet Gynecol. 1967;97(05):723–738. doi: 10.1016/0002-9378(67)90458-9. [DOI] [PubMed] [Google Scholar]
- 3.Hill J B, Sheffield J S, McIntire D D, Wendel G D., Jr Acute pyelonephritis in pregnancy. Obstet Gynecol. 2005;105(01):18–23. doi: 10.1097/01.AOG.0000149154.96285.a0. [DOI] [PubMed] [Google Scholar]
- 4.Fisher M. Baltimore, Maryland: Williams & Wilkins; 1989. Guide to Clinical Preventive Services. [Google Scholar]
- 5.American College of Obstetricians and Gynecologists.ACOG educational bulletin. Antimicrobial therapy for obstetric patients. Number 245, March 1998 (replaces no. 117, June 1988) Int J Gynaecol Obstet 19986103299–308. [PubMed] [Google Scholar]
- 6.Levinson W J. New York: Lange Medical Books/McGraw-Hill; 2000. Ernest Medical Microbiology and Immunology. [Google Scholar]
- 7.Madigan M T, Brock T D. San Francisco, CA: Pearson Education; 2009. Brock Biology of Microorganisms. [Google Scholar]
- 8.Brubaker L, Wolfe A J. The new world of the urinary microbiota in women. Am J Obstet Gynecol. 2015;213(05):644–649. doi: 10.1016/j.ajog.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakeman M M, Roovers J P. Urinary tract infections in women with urogynaecological symptoms. Curr Opin Infect Dis. 2016;29(01):92–97. doi: 10.1097/QCO.0000000000000229. [DOI] [PubMed] [Google Scholar]
- 10.Whiteside S A, Razvi H, Dave S, Reid G, Burton J P. The microbiome of the urinary tract--a role beyond infection. Nat Rev Urol. 2015;12(02):81–90. doi: 10.1038/nrurol.2014.361. [DOI] [PubMed] [Google Scholar]
- 11.Wolfe A J, Brubaker L. “Sterile urine” and the presence of bacteria. Eur Urol. 2015;68(02):173–174. doi: 10.1016/j.eururo.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas-White K, Brady M, Wolfe A J, Mueller E R. The bladder is not sterile: history and current discoveries on the urinary microbiome. Curr Bladder Dysfunct Rep. 2016;11(01):18–24. doi: 10.1007/s11884-016-0345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe A J, Toh E, Shibata N et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 2012;50(04):1376–1383. doi: 10.1128/JCM.05852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilt E E, McKinley K, Pearce M M et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52(03):871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearce M M, Hilt E E, Rosenfeld A B et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. 2014;5(04):e01283–e14. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price T K, Dune T, Hilt E E et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54(05):1216–1222. doi: 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Development Core team. R: A Language and Environment for Statistical ComputingR Foundation for Statistical Computing. 2014
- 18.RS Team.RStudio: Integrated Development for RBoston, MA. RStudio Inc. Available at:http://www.rstudio.com/.2015
- 19.Kozich J J, Westcott S L, Baxter N T, Highlander S K, Schloss P D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss P D, Westcott S L, Ryabin T et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity G M, Tiedje J M, Cole J R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas-White K J, Hilt E E, Fok Cet al. Incontinence medication response relates to the female urinary microbiotaInt Urogynecol J Pelvic Floor Dysfunct2015 [DOI] [PMC free article] [PubMed]
- 23.Dubnau D, Smith I, Morell P, Marmur J. Gene conservation in Bacillus species. I. Conserved genetic and nucleic acid base sequence homologies . Proc Natl Acad Sci U S A. 1965;54(02):491–498. doi: 10.1073/pnas.54.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woese C R, Olsen G J, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64(01):202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lotte R, Lotte L, Ruimy R.Actinotignum schaalii (formerly Actinobaculum schaalii ): a newly recognized pathogen-review of the literature Clin Microbiol Infect2015 [DOI] [PubMed]
- 26.Romero R, Hassan S S, Gajer P et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schieve L A, Handler A, Hershow R, Persky V, Davis F. Urinary tract infection during pregnancy: its association with maternal morbidity and perinatal outcome. Am J Public Health. 1994;84(03):405–410. doi: 10.2105/ajph.84.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karstens L, Asquith M, Davin S et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol. 2016;6:78. doi: 10.3389/fcimb.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]


