Abstract
Neuroticism is a heritable personality trait that is comprised of distinct sub-factors, or facets. Sub-factors of neuroticism are linked to different emotional states or psychiatric symptoms and studying the genetic variants associated with these facets may help reveal the biological mechanisms underlying psychiatric disorders. In the present study, a meta-analysis of genome-wide association studies for six facets of neuroticism was performed in 5584 participants from three cohorts. Additionally, a Gene Set Enrichment Analysis was conducted to find biological pathways associated with each facet. Six neuroticism facets (N1: anxiety, N2: angry hostility, N3: depression, N4: self-consciousness, N5: impulsivity and N6: vulnerability) were assessed using the Korean version of the Revised NEO Personality Inventory. In the single-nucleotide polymorphism-based analysis, results showed genome-wide significance for N2 within the MIR548H3 gene (rs1360001, P=4.14 × 10−9). Notable genes with suggestive associations (P<1.0 × 10−6) were ITPR1 for N1, WNT7A for N2, FGF10 and FHIT for N3, DDR1 for N4, VGLL4 for N5 and PTPRD for N6. In the pathway-based analysis, the axon guidance pathway was identified to be associated with multiple facets of neuroticism (N2, N4 and N6). The focal adhesion and extracellular matrix receptor interaction pathways were significantly associated with N2 and N3. Our findings revealed genetic influences and biological pathways that are associated with facets of neuroticism.
Introduction
Certain personality traits have been associated with risk factors of psychological illnesses. One example is neuroticism, which is a personality trait characterized by a tendency toward various negative emotions such as sadness, fear and anger.1 A number of studies examining the association between personality and psychiatric illness have shown that neuroticism is strongly linked with anxiety, mood and substance use disorders.2 Additionally, neuroticism is a well-known predictive factor of the onset and prognosis of depressive symptoms.3 The five factor model of personality (FFM), which is the most widely used measure of personality, defines neuroticism as a trait that encompasses a broad spectrum of heterogeneous traits, hierarchically organized into lower-level traits called facets. The six facets of neuroticism are anxiety, angry hostility, depression, self-consciousness, impulsivity and vulnerability.4 Each facet of neuroticism may be specifically associated with emotional and behavioral aspects of psychiatric symptoms or mental illness. For example, neuroticism that is characterized by high anxiety and depression is highly correlated with symptoms of anxiety and depressive disorders.
Most studies on the heritability of personality have confirmed that genetic factors substantially influence neuroticism and its facets. Neuroticism shares the genetic and biological components of psychiatric symptoms that are related to mood disorders.2 Genetic association studies have identified suggestive genes linked to neuroticism such as PDE4D,5 SNAP25,6 NKAIN2,7 OR1A2,8 and more specifically the depression facet of neuroticism such as RORA.9 Recently, large GWAS have found genetic loci associated with neuroticism in genome-wide significant levels,10, 11, 12 and genome-wide SNPs explained about 15% of the variance in neuroticism.10, 12 However, there have been few consistent findings of genetic loci linked to neuroticism across the genome-wide association studies, and further studies may be useful to confirm the candidate genes reported so far.
The facet-level traits share common components summarized by a broad dimension of personality as well as reflect specific and narrow phenotypes than the broad factor to which the facet is related.13 Neuroticism score is produced by a summation of the multiple facets, which are variable across individuals, and the broad dimension seems not to reflect specific characteristics of each facet. Genetic association studies of facet-level personality traits may have the advantage of improving statistical power by reducing noise due to the phenotypic variability.9 It was suggested that the heritability of personality traits reflect the heritability of common variance among facets as well as specific heritable components underlying each facet.13 Thus, studying the genetic variants of facets may aid in understanding the biological mechanisms underlying the associations between specific and common components of neuroticism and phenotypically related psychiatric disorders.
Genome-wide associations (GWA) typically evaluate the effect of single-nucleotide polymorphisms (SNP), but such studies have only limited power to detect loci with small to moderate effects on complex traits.14 Personality traits are polygenic phenotypes. Therefore, a large number of genetic variants may be involved, and each may make small contributions and interact with each other. The associations between phenotypes and variants within functionally related gene groups can be examined using pathway-based approaches, which integrate the results of GWA studies with genes or gene sets of certain biological pathways.15 Moreover, such pathway analysis investigates biological pathways that are functionally important for cellular and molecular processes and may provide information on the underlying biological mechanisms of complex traits. This method has the advantages of increased effect size and statistical power and can be used to identify the biological pathways associated with personality16 and neuropsychiatric phenotypes including schizophrenia.17
In the present study, we performed a meta-analysis of GWA results to identify genes that influence the facet-level traits of neuroticism in a total of 5584 participants from three cohorts. The aim of this study was to identify genetic variants associated with each facet of neuroticism, and to explore the respective affected biological pathways.
Materials and methods
Subjects
The total sample size for the GWA study was 5584 subjects from three cohorts. The Ansan Ansung cohort was a rural and urban community-based cohort, which included 1167 men and 1257 women, ranging in age from 29 to 79 years.18 The young women cohort in Korea included 1120 Korean women, aged 18–40 years.19 The Kangbuk Samsung cohort included 1143 men and 897 women ranging in age from 19 to 69 years, and these individuals were recruited from the Health Screening and Examination cohort of the Kangbuk Samsung Hospital in Korea.20 All cohorts were reviewed and approved by local institutional review boards, and written informed consent was obtained. All applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during our research.
Assessment
Neuroticism was assessed using the Korean version of the Revised NEO personality inventory (NEO PI-R; PSI consulting Corp., Seoul, Korea). The NEO PI-R consists of 30 facets, six for each dimension of the five personality traits. The facets of neuroticism are anxiety (N1), angry hostility (N2), depression (N3), self-consciousness (N4), impulsivity (N5) and vulnerability (N6). Raw scores were standardized into T-scores using Korean combined-sex norms (N=7418; mean=50; s.d.=10) to confirm similarities with the Korean normative sample data (PSI Consulting Corp.). The broad dimensions of neuroticism were calculated by summation of raw scores of six facets and converted to T-scores (mean=50, s.d.=1). The Cronbach’s alpha coefficients for neuroticism and six facets were 0.908, 0.657, 0.544, 0.742, 0.494, 0.604 and 0.577, respectively. The Korean version of the NEO PI-R has been demonstrated to provide good reliability and validity.21
Genotyping
SNP genotyping was conducted using Illumina or Affymetrix microarray chips, as displayed in Table 1. Quality control (QC) procedures were conducted to remove SNPs with missing genotyping rates >5%, minor allele frequency (MAF) <0.01, or Hardy–Weinberg equilibrium <1 × 10−6. Sample QC procedures were performed using PLINK version 1.90 to eliminate ineligible subjects with minor allele frequency <0.05, Hardy–Weinberg Equilibrium P-value <10−6, genotyping rate <0.95. The subjects with sex inconsistency and relatedness were also removed. After QC, autosomal SNPs (344 547 for the Ansan Ansung cohort, 636 744 for the Young women cohort, and 226, 706 for the Kangbuk Samsung cohort) were used for imputation. Genotype imputation increases statistical power and the possibility of detecting a causal SNP.22 Imputation was conducted using IMPUTE2 with the 1000 Genomes Phase 3 reference panel containing all populations.
Table 1. Genotype information for the three cohorts in the meta-analysis.
| Cohort | Total (N) | Male (N) | Female (N) | Mean age in years (s.d.) | Genotype platform | Pre-imputed SNPs | Imputed SNPs after QC |
|---|---|---|---|---|---|---|---|
| Ansan ansung | 2424 | 1167 | 1257 | 58.90 (7.94) | Affymetrix | 344 547 | 4 594 951 |
| Young women | 1120 | — | 1120 | 26.14 (4.66) | Illumina | 636 744 | 6 427 507 |
| Kangbuk samsung | 2040 | 1143 | 897 | 39.31 (8.86) | Illumina | 226 706 | 5 927 272 |
SNPs with an imputation quality score (R2) greater than 0.8 and an MAF of > 0.01 were used for the association analysis. The final number of SNPs after imputation was 4 594 951 for the Ansan Ansung cohort, 6 427 507 for the Young women cohort, and 5 927 272 for the Kangbuk Samsung cohort. The number of overlapping SNPs among the three cohorts was 4 352 680.
Statistical analysis
The GWA was performed with best guess imputed genotypes using additive linear regression model in PLINK ver. 1.90. T-scores of neuroticism and six facets were used as quantitative variables to examine the association with SNPs. Age and sex were included in the analysis as covariates. A weighted inverse variance method in METAL was conducted to perform a meta-analysis of the GWA results. The threshold value of P<5 × 10−8 was used to determine statistical significance.23 A less stringent criterion of P<1 × 10−6 was used to denote a suggestive association. Genomic inflation factors (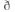 ) ranged from 1.000 to 1.025 for all analyses. Multidimensional scaling (MDS) and principal component analysis (PCA) confirmed that population stratification was not observed in the overall dataset (Supplementary Figure S1).
) ranged from 1.000 to 1.025 for all analyses. Multidimensional scaling (MDS) and principal component analysis (PCA) confirmed that population stratification was not observed in the overall dataset (Supplementary Figure S1).
To identify biological pathways, a Gene Set Enrichment Analysis with Meta-Analysis Gene Set Enrichment of Variant (MAGENTA) algorithms was conducted. MAGENTA tests the enrichment of associations with a disease or trait within gene sets.15 The detailed processes of MAGENTA were described by Segre et al. Briefly, the P-values of the SNPs determined by GWAs were used as input, and gene scores were obtained from the most significant P-value among the SNPs located within each gene. After gene scores were corrected for confounders including the gene size, number of SNPs and linkage disequilibrium-related properties by using step-wise multiple linear regression analysis, gene-set enrichment P-values were obtained for highly ranked gene scores. Gene set enrichment analysis P-values, which were assigned a cut-off threshold set to the 75th percentile, may be suitable to detect weaker signals of associations to polygenic traits or for complex than the 95th threshold.15 Pathway information was obtained from the KEGG (186 pathways) and REACTOME (674 pathways) molecular signature databases (MsigDB) v5.1 (downloaded in April 2014). For meta-analysis of pathway-based association, MAGENTA were applied to all three cohort individually, and the meta P-value were calculated by combining nominal P-values for each pathway from three cohorts using Fisher’s method of ‘metap’ in R. Meta P-values were corrected for multiple testing by a Bonferroni method. Multiple comparison corrections were applied separately in each pathway database (KEGG: 0.05/186=2.69 × 10−4; REACTOME: 0.05/674=7.42 × 10−5) because gene sets between two databases considerably overlap.15 Pathways showing meta P-values with significance levels were considered to be associated with phenotypes.
The bivariate correlation analysis was performed to examine the associations between facet-level traits of neuroticism by using Pearson correlation analysis. Genetic correlations between facets were calculated with by using a LD score regression method (LDSC v1.0.0, https://github.com/bulik/ldsc).
Results
Meta-analysis of SNP-based associations
The Manhattan and quantile-quantile plots for SNP-wide meta-analysis results for the six facets of neuroticism are displayed in Supplementary Figure S2 and 3. Bivariate correlations and genetic correlations between the facets are shown in Supplementary Table S1 and 2. The top-ranked SNPs obtained from the meta-analysis of GWAs for facets are presented in Table 2. One SNP (rs1360001) was identified at the level of genome-wide significance (P<5 × 10−8), and 41 SNPs were identified to have suggestive associations (P<1 × 10−6) in the meta-analysis for facets. Top-ranked results from SNP-based meta-analysis of a broad dimension of neuroticism are shown in Supplementary Table S3.
Table 2. Top-ranked results from the meta-analysis of SNP-based associations for facet-level traits of neuroticism.
| Facet | SNP | Chr | Position | Effect allele | MAF | Z-score | P-value | Directiona | HetChiSq | HetPval | Nearest Geneb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | rs78773901 | 2 | 188731477 | C | 0.027 | −4.665 | 3.09E-06 | −− | 3.571 | 0.168 | . |
| rs4685767 | 3 | 4613530 | G | 0.361 | 4.506 | 6.59E-06 | +++ | 0.404 | 0.817 | ITPR1 | |
| rs2700687 | 7 | 33666459 | A | 0.362 | −4.493 | 7.03E-06 | −− | 0.819 | 0.664 | BBS9 | |
| rs9316411 | 13 | 49321854 | G | 0.079 | −4.445 | 8.78E-06 | −− | 3.469 | 0.177 | CYSLTR2 | |
| rs10970114 | 9 | 31116845 | A | 0.221 | −4.424 | 9.70E-06 | −− | 0.877 | 0.645 | . | |
| N2 | rs1360001 | 9 | 78259530 | T | 0.168 | 5.878 | 4.14E-09 | +++ | 4.434 | 0.109 | MIR548H3 |
| rs74627388 | 3 | 13888606 | T | 0.113 | 4.841 | 1.29E-06 | +++ | 0.822 | 0.663 | WNT7A | |
| rs75400032 | 2 | 35295043 | A | 0.192 | 4.718 | 2.38E-06 | +++ | 0.325 | 0.850 | . | |
| rs117920056 | 5 | 149128924 | T | 0.024 | 4.683 | 2.83E-06 | +++ | 0.111 | 0.946 | PPARGC1B | |
| rs12187180 | 5 | 20815506 | G | 0.286 | 4.524 | 6.08E-06 | +++ | 0.572 | 0.751 | . | |
| N3 | rs2929855 | 5 | 44390364 | A | 0.133 | −4.591 | 4.41E-06 | −− | 4.919 | 0.085 | FGF10 |
| rs1007107 | 6 | 14463419 | G | 0.389 | 4.515 | 6.32E-06 | +++ | 0.648 | 0.723 | . | |
| rs11242622 | 5 | 106373992 | T | 0.308 | −4.501 | 6.75E-06 | −− | 1.614 | 0.446 | LOC102467213 | |
| rs10151594 | 14 | 51864477 | A | 0.099 | −4.491 | 7.08E-06 | −− | 0.044 | 0.978 | LINC00640 | |
| rs17257269 | 3 | 60025505 | C | 0.025 | 4.485 | 7.29E-06 | +++ | 0.879 | 0.644 | FHIT | |
| N4 | rs308254 | 13 | 93672943 | T | 0.287 | −4.822 | 1.43E-06 | −− | 1.283 | 0.527 | . |
| rs79425212 | 6 | 30813282 | A | 0.203 | 4.811 | 1.51E-06 | +++ | 3.195 | 0.202 | DDR1 | |
| rs145239798 | 2 | 188754537 | A | 0.026 | −4.736 | 2.18E-06 | −− | 2.235 | 0.327 | . | |
| rs11691878 | 2 | 22021330 | C | 0.166 | −4.656 | 3.22E-06 | −− | 0.036 | 0.982 | LOC645949 | |
| rs75769602 | 13 | 61788256 | C | 0.051 | 4.603 | 4.16E-06 | +++ | 1.758 | 0.415 | MIR3169 | |
| N5 | rs114649649 | 1 | 201299361 | A | 0.063 | −4.896 | 9.79E-07 | −− | 1.589 | 0.452 | PKP1 |
| rs12494147 | 3 | 11668983 | T | 0.060 | 4.663 | 3.11E-06 | +++ | 1.567 | 0.457 | VGLL4 | |
| rs7967431 | 12 | 82342828 | C | 0.415 | −4.636 | 3.55E-06 | −− | 7.932 | 0.019 | LOC101928449 | |
| rs4569911 | 5 | 160235125 | T | 0.469 | −4.562 | 5.06E-06 | −− | 0.059 | 0.971 | ATP10B | |
| rs1093307 | 12 | 117925643 | T | 0.190 | 4.498 | 6.86E-06 | +++ | 0.260 | 0.878 | KSR2 | |
| N6 | rs2784611 | 9 | 10551203 | A | 0.442 | 4.521 | 6.17E-06 | +++ | 1.031 | 0.597 | PTPRD |
| rs17495034 | 1 | 222181354 | C | 0.138 | 4.459 | 8.22E-06 | +++ | 2.229 | 0.328 | . | |
| rs12944395 | 17 | 28323499 | G | 0.395 | 4.414 | 1.02E-05 | +++ | 0.498 | 0.780 | EFCAB5 | |
| rs183360621 | 2 | 36464282 | A | 0.015 | 4.412 | 1.03E-05 | +++ | 0.151 | 0.928 | . | |
| rs58450504 | 3 | 72555840 | G | 0.174 | −4.379 | 1.19E-05 | −− | 0.752 | 0.687 | RYBP |
Abbreviations: Chr, chromosome; HetChiSq, heterogeneity chi-square statistic; HetPval, heterogeneity P-value; MAF, minor allele frequency; N1, anxiety; N2, angry hostility; N3, depression; N4, self-consciousness; N5, impulsivity; N6, vulnerability; SNP, single-nucleotide polymorphism.
Direction is described the SNP beta-coefficient from Ansan Ansung, Kangbuk Samsung and Young Women cohort in sequence.
Nearest genes are defined as the closest genes to the SNP within signal boundary or the closest genes within a 100 kb window.
For anxiety (N1), the strongest association was found to be with rs78773901 (Z=4.665, P=3.09 × 10−6), which is located in inter-genic regions. Another top-ranked SNP was rs4685767 (Z=−4.506, P=6.59 × 10−6), which is located within the inositol 1,4,5-trisphosphate receptor, type 1 (ITPR1) gene.
For hostility (N2), the most significant association at the genome-wide level (Z=5.878, P=4.14 × 10−9) was with rs1360001, which is located within the microRNA 548h-3 gene (MIR548H3). Additionally, the SNP rs74627388 had a suggestive association (Z=4.841, P=1.29 × 10−6) and is located within the Wnt family member 7A gene (WNT7A).
For depression (N3), the top-ranking SNP was rs2929855 (Z=−4.591, P=4.41 × 10−6), which is located near the fibroblast growth factor 10 gene (FGF10). A suggestive association was found between N3 and rs17257269 (Z=−4.485, P=7.29 × 10−6), which is located within the fragile histidine triad gene (FHIT).
The SNP found to have the highest association with self-consciousness (N4) was rs308254 (Z=4.822, P=1.43 × 10−6), which is located in inter-genic regions. Among the top-ranked SNPs was rs79425212 (Z=4.811, P=1.51 × 10−6), which is located close to the discoidin domain receptor 1 (DDR1) gene.
The strongest association with impulsivity (N5) was identified for rs114649649, which is located within Plakophilin 1 (PKP1) gene (Z=−4.896, P=9.79 × 10−7). The rs12494147 SNP, located in the vestigial like family member 4 gene (VGLL4), was identified to have the suggestive association with N5 (Z=4.663, P=3.11 × 10−6).
The top-ranking SNP associated with vulnerability to stress (N6) was identified as rs2784611 (Z=4.521, P=6.17 × 10−6), which is located within the protein tyrosine phosphatase receptor type D (PTPRD) gene.
Meta-analysis of pathway-based associations
To explore the biological pathways underlying the facet-level traits of neuroticism, we performed a MAGENTA analysis on the results of three GWAs. A combined P-value for each pathway, across the three cohorts, was estimated. The significant pathways associated with the facets of neuroticism are described in Table 3. Top-ranked results from pathway-based meta-analysis of a broad dimension of neuroticism are shown in Supplementary Table S4.
Table 3. Significant results from meta-analysis of pathway-based associations for facet-level traits of neuroticism.
| Facet | DB | Pathway | Meta-pa |
Nominal p |
||
|---|---|---|---|---|---|---|
| AS | KS | YW | ||||
| N1 | REACTOME | Platelet activation signaling and aggregation | 4.50E-06 | 2.80E-03 | 1.00E-04 | 2.36E-02 |
| REACTOME | Developmental biology | 9.19E-06 | 7.40E-03 | 8.10E-05 | 1.10E-03 | |
| REACTOME | Signaling by rho GTPases | 2.05E-05 | 5.66E-02 | 2.50E-05 | 4.00E-02 | |
| KEGG | Focal adhesion | 9.77E-05 | 7.64E-02 | 1.00E-04 | 4.70E-03 | |
| KEGG | Glioma | 1.17E-04 | 9.26E-02 | 1.00E-04 | 2.08E-01 | |
| N2 | KEGG | Focal adhesion | 1.45E-07 | 4.00E-05 | 1.84E-04 | 2.99E-02 |
| REACTOME | Collagen formation | 1.01E-06 | 3.00E-03 | 1.90E-05 | 3.90E-03 | |
| REACTOME | Axon guidance | 2.10E-06 | 1.13E-04 | 1.10E-03 | 1.00E-04 | |
| REACTOME | NCAM signaling for neurite outgrowth | 7.03E-06 | 3.00E-04 | 1.50E-03 | 3.00E-04 | |
| REACTOME | Developmental biology | 1.13E-05 | 2.50E-03 | 3.00E-04 | 1.10E-03 | |
| KEGG | ECM receptor interaction | 5.91E-05 | 4.98E-04 | 8.90E-03 | 4.01E-02 | |
| N3 | KEGG | Focal adhesion | 7.21E-07 | 4.00E-04 | 1.00E-04 | 6.50E-05 |
| REACTOME | Integrin cell surface interactions | 1.37E-06 | 7.89E-02 | 1.00E-06 | 5.45E-02 | |
| KEGG | ECM receptor interaction | 5.88E-06 | 7.30E-05 | 5.10E-03 | 7.80E-03 | |
| REACTOME | Collagen formation | 4.75E-05 | 1.00E-03 | 3.50E-03 | 7.22E-02 | |
| N4 | REACTOME | Axon guidance | 1.71E-06 | 5.00E-04 | 2.00E-04 | 9.90E-07 |
| REACTOME | CA dependent events | 5.84E-05 | 2.19E-02 | 2.00E-04 | 8.66E-01 | |
| REACTOME | DAG and IP3 signaling | 6.72E-05 | 1.02E-02 | 5.00E-04 | 9.02E-01 | |
| N5 | KEGG | Renal cell carcinoma | 2.04E-04 | 1.30E-03 | 1.31E-02 | 2.18E-01 |
| N6 | REACTOME | Axon guidance | 4.31E-05 | 1.00E-04 | 3.15E-02 | 6.40E-03 |
| REACTOME | SLC-mediated transmembrane transport | 5.01E-05 | 2.21E-02 | 1.68E-04 | 3.09E-02 | |
| REACTOME | CRMPS in SEMA3A signaling | 6.50E-05 | 3.30E-05 | 1.49E-01 | 5.52E-02 | |
Abbreviations: AS, Ansan Ansung cohort; KS, Kangbuk Samsung cohort; N1, anxiety, N2, angry hostility, N3, depression, N4, self-consciousness, N5, impulsivity, N6, vulnerability; YW, Young women cohort.
aCombined nominal P-value of three cohorts were calculated by Fisher’s method.
Significance thresholds for pathway-based meta-analysis were determined separately for each database (KEGG: 0.05/186=2.69 × 10−4; REACTOME: 0.05/674=7.42 × 10−5).
Following the pathway analysis for N1, three pathways from the REACTOME dataset and 2 pathways from KEGG were identified as significant. The strongest association was found to be with platelet activation signaling and aggregation from REACTOME (P=4.50 × 10−6). Focal adhesion was a top-ranked pathway from KEGG for N1 (P=9.77 × 10−5).
Among 4 pathways identified in the REACTOME dataset and two pathways identified from the KEGG database to be significantly associated with N2, focal adhesion was the top-ranked pathway (P=1.45 × 10−7). Axon guidance (P=2.10 × 10−6), neural cell adhesion molecule (NCAM) signaling of neurite outgrowth (P=7.03 × 10−6) and the extracellular matrix (ECM) receptor interaction (P=5.91 × 10−5) were also significantly associated with N2.
Focal adhesion gene sets from KEGG comprised the top-ranked pathway associated with N3 (P=7.21 × 10−7). The ECM receptor interaction pathway was observed for N3 (P=5.88 × 10−6).
For N4, Axon guidance which was identified from the REACTOME datasets (P=1.71 × 10−5), was the most significant among the top-ranked pathways. The calcium-dependent event pathway (P=5.84 × 10−5) and diacylglycerol (DAG) and inositol triphosphate (IP3) signaling (P=6.72 × 10−5) pathways were also observed in association with N4.
One significant pathway associated with N5 was renal cell carcinoma from KEGG (P=2.04 × 10−4). For N6, axon guidance was the top-ranked pathway (P=4.31 × 10−5). Significant pathways associated with N6 included solute carrier (SLC)-mediated transmembrane transport (P=5.01 × 10−5) and collapsin response mediator proteins (CRMPs) in the semaphorin 3A (SEMA3A) signaling pathway (P=6.50 × 10−5).
Discussion
Our study reports the results from a meta-analysis of both SNP- and pathway-based data and reveals genetic influences on the six facets of neuroticism. Identifying these genetic factors may provide information critical for understanding the biological basis of psychiatric disorders that share the behavioral and emotional aspects of specific facets.9 We identified several interesting candidate genes, including ITPR1 (which affects the N1 facet), MIR548H3 and WNT7A (which affects N2), FGF10 and FHIT (which affect N3), DDR1 (which affects N4), VGLL4 (which affects N5) and PTPRD (which affects N6), and these genes have previously been associated with other behavioral or psychiatric phenotypes. While these candidate genes did not reach a significant GWA with each facet, we identified significant biological pathways through a meta-analysis of three studies using MAGENTA. Moreover, we found that certain biological pathways were associated with a specific facet, such as calcium-dependent events (N4) and CRMPs in the SEMA3A signaling (N6), while other pathways, such as axon guidance (N2, N4 and N6), focal adhesion and ECM receptor interaction (N2 and N3) were associated with multiple facets.
With respect to anxiety, ITPR1 was the most biologically plausible gene among the top-ranking SNPs. ITPR1 encodes the inositol 1,4,5-trisphosphate receptor, which modulates the concentration and release of Ca2+ by regulating intracellular Ca2+ channels.24 Fluctuations in Ca2+ caused by ITPR1 receptors has been implicated in neural activity and synaptic plasticity.25 Inositol 1,4,5-trisphosphate receptors are enriched in the amygdala, and expression of the receptor subtypes has crucial effects on amygdala functions, such as regulating fear and anxiety.26 Animal studies have shown that the activity of inositol 1,4,5-trisphosphate receptors is dependent on dopaminergic neural transmission and is suggested to underlie one of the mechanisms of drug dependency.27 Anxiety and substance use disorders are highly comorbid, and anxiety disorders are correlated with the occurrence and the severity of alcohol use disorders.28 It is possible that ITPR1 plays a biological role in the clinical and behavioral links between anxiety and drug abuse.
The most significant association at the genome-wide level was found between hostility and rs1360001 located within microRNA 548h-3 (MIR548H3). Emerging evidence suggests that microRNA may play an important role in development of central nervous system and neuropsychiatric diseases.29, 30 Although the function of microRNA in personality has been unknown, our finding was interesting given the fact that neuroticism is strongly associated with symptoms of psychiatric disorders.2 Hostility facet with suggestive association was found in WNT7A gene. This gene encodes a Wnt protein family member that plays a role in cell signaling.31 The WNT7A gene was also identified in a previous GWA study for bipolar disorder.32 The Wnt signaling pathway is important for neural growth and plasticity, which have been suggested as possible mechanisms of depressive disorder.33
With respect to the depression facet, FGF10 and FHIT genes were found to have a suggestive association. FGF10 gene is a component of the fibroblast growth factor system, which has previously been reported as the one of the mechanisms of major depressive disorder.34 Several GWA studies have reported an association between FHIT and depressive disorder.35, 36 This gene is also associated with symptoms of anxiety and psychological distress.37 Our finding suggests that the FHIT gene is involved in the depression facet of neuroticism and is a good candidate for involvement in psychiatric disorders that are phenotypically related to this trait.
Self-consciousness trait showed a possible association with the variant located close to the discoidin domain receptor 1 (DDR1) gene. Previous studies have found that expression of DDR1 increases in parallel with neural myelination during mice brain development.38 This gene has also been suggested to be a susceptibility gene for schizophrenia.39
The suggestive associations with impulsivity were identified in PKP1 and VGLL4 genes. PKP1 gene was identified in a GWA study of panic disorder.40 The function of VGLL4 in personality is unknown; however, this gene is associated with comorbid depression and alcohol dependence.36
Trait vulnerability showed suggestive associations with the PTPRD gene. This gene encodes one of the members of the protein tyrosine phosphatase family, which regulates various cellular processes, including cell growth and differentiation.41 PTPRD was previously associated with other personality traits such as persistence from Cloninger’s Temperament Scales42 and psychiatric symptoms such as anxiety and depression.37, 43 PTPRD is also one of the candidate genes for obsessive compulsive disorder,44 autism,45 and attention-deficit hyperactivity disorder.46 These neuropsychiatric disorders originate from the interplay between environmental and genetic factors. It is possible that PTPRD influences individual differences in the vulnerability to environmental stress, which plays a critical role in the risk for neuropsychiatric disorders. Neural dysfunction, caused by changes in such neurodevelopmental processes as axonal guidance and synapse formation, may explain the associations between PTPRD and neuropsychiatric conditions.47
Meta-analysis using pathway-based approaches revealed several biological pathways that overlapped among the facets and pathways that were specific to each facet. Axon guidance was significantly associated with N2, N4 and N6, which were also correlated with each other. Axon guidance has also previously reported to be linked to a broad dimension of neuroticism.16 Axon guidance is associated with neural developmental processes and hierarchically belongs to a developmental biology pathway in the REACTOME database. Our findings suggest that neural development processes pathway may be a shared genetic component of lower-level traits of neuroticism.
Another notable finding is that focal adhesion and ECM receptor interaction are related to hostility and depression. Focal adhesions are large protein complexes that contain integrin and connect the cytoskeleton of cells to the ECM.48 It has previously been reported that focal adhesion plays an essential role in cell migration during brain development and is involved in the abnormal neurodevelopment underlying schizophrenia.49 The hostility trait is linked to aggressive and violent behaviors and is also closely related to depressive symptoms.50 Our findings suggest that the focal adhesion and ECM receptor pathways are good candidates for future studies that will focus on the biological mechanisms linked to hostility and depression.
Interestingly, we identified an association between calcium-dependent events and self-consciousness. Calcium ions are important for neural functions, such as excitability, synaptic plasticity and transmitter release.51 Calcium signaling in neurons regulates the release of neurotransmitters, such as serotonin and norepinephrine,52 which are functionally important for behaviors and emotions. Self-consciousness is known to be closely linked to shame, social anxiety, and the fear and anxiety emotions.53 It is possible that calcium-related pathways are related to the neural functions involved in self-regulating negative emotions and psychological distress as well as the susceptibility to anxiety and mood disorders.
Several limitations in the present study should be noted. Facet-level traits of neuroticism scored by FFM showed lower reliability of measurements. The present finding may be promising for further genetic studies for neuroticism related phenotypes using higher reliability of measurements. In the SNP-based meta-analysis, we identified one SNP that reached significance at genome-wide levels, but we did not find any SNPs previously associated with neuroticism in other GWAs5, 6, 7, 11, 54 or our previous studies.8 One possible reason stems from differences in the population used in this study compared to that used in previous research, which was mainly performed in Caucasian subjects. Another possible explanation entails differences in the reference panel for imputation, since we used the 1000 Genomes in this study and HapMap in our previous studies.8 Nevertheless, the facet-level pathway analysis herein replicated the neuroticism-associated pathways identified in our previous pathway-based association study on the NEO personality inventory.16 These findings suggest that detection of the genetic influences of complex traits at the single SNP level is difficult and that the interactions and cooperative effects of the genes comprising biological pathways may play more important roles in complex traits, especially in behavioral phenotypes such as personality traits.
In conclusion, the candidate genes identified in this study to influence the facets of neuroticism have been previously implicated in other psychiatric disorders and warrant further study. The pathway-based meta-analysis implicated meaningful biological pathways in the facets of neuroticism; moreover, these pathways, such as axon guidance, have previously been associated with a broad dimension of neuroticism,16 as well as with psychiatric disorders, such as focal adhesion with schizophrenia.49 Additionally, the pathway analysis suggests that pathways underlying neurodevelopmental processes are associated with multiple facets of neuroticism. Taken together, these results contribute to the understanding of the genetic influences and biological mechanisms underlying facet-level traits of neuroticism and phenotypically related disorders.
Acknowledgments
This research was supported by the National Research Foundation of Korea, funded by the Ministry of Education (NRF-2016R1A6A3A11932719) and Ministry of Science, ICT & Future Planning (NRF-2014R1A2A2A04006291) and a grant of Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C0072). The genotype data of Ansan Ansung cohort were gratefully made available by the Center for Genome Science, Korea National Institute of Health, Korea Centers for Disease Control and Prevention. It was also supported with the computing resources by Global Science experimental Data hub Center (GSDC) Project and Korea Research Environment Open NETwork (KREONET) in Korea Institute of Science and Technology Information (KISTI). SNP genotyping data is publicly available upon proper requesting process at the Clinical Omics Data Archive (CODA, http://coda.nih.go.kr).
Footnotes
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Reynaud, E., El Khoury-Malhame, M., Rossier, J., Blin, O. & Khalfa, S. Neuroticism modifies psychophysiological responses to fearful films. PLoS ONE 7, e32413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel, J., Jeronimus, B. F., Kotov, R., Riese, H., Bos, E. H., Hankin, B. et al. Neuroticism and common mental disorders: meaning and utility of a complex relationship. Clin. Psychol. Rev. 33, 686–697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, T. A. & Rosellini, A. J. The direct and interactive effects of neuroticism and life stress on the severity and longitudinal course of depressive symptoms. J. Abnormal. Psychol. 120, 844–856 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, P. & McCrae, R. Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor Inventory (NEO-FFI) Professional Manual, Psychological Assessment Resources, Odessa, FL, USA, (1992). [Google Scholar]
- Shifman, S., Bhomra, A., Smiley, S., Wray, N. R., James, M. R., Martin, N. G. et al. A whole genome association study of neuroticism using DNA pooling. Mol. Psychiatry 13, 302–312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano, A., Sanna, S., Uda, M., Deiana, B., Usala, G., Busonero, F. et al. Genome-wide association scan for five major dimensions of personality. Mol. Psychiatry 15, 647–656 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calboli, F. C., Tozzi, F., Galwey, N. W., Antoniades, A., Mooser, V., Preisig, M. et al. A genome-wide association study of neuroticism in a population-based sample. PLoS ONE 5, e11504 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. N., Roh, S. J., Sung, Y. A., Chung, H. W., Lee, J. Y., Cho, J. et al. Genome-wide association study of the five-factor model of personality in young Korean women. J. Hum. Genet. 58, 667–674 (2013). [DOI] [PubMed] [Google Scholar]
- Terracciano, A., Tanaka, T., Sutin, A. R., Sanna, S., Deiana, B., Lai, S. et al. Genome-wide association scan of trait depression. Biol. Psychiatry 68, 811–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. J., Escott-Price, V., Davies, G., Bailey, M. E., Colodro-Conde, L., Ward, J. et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol. Psychiatry 21, 1644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay, A., Baselmans, B. M., De Neve, J. E., Turley, P., Nivard, M. G., Fontana, M. A. et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor, M. H., van den Berg, S. M., Verweij, K. J., Krueger, R. F., Luciano, M., Arias Vasquez, A. et al. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry 72, 642–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, K. L., McCrae, R. R., Angleitner, A., Riemann, R. & Livesley, W. J. Heritability of facet-level traits in a cross-cultural twin sample: support for a hierarchical model of personality. J. Personal. Soc. Psychol. 74, 1556–1565 (1998). [DOI] [PubMed] [Google Scholar]
- Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005). [DOI] [PubMed] [Google Scholar]
- Segre, A. V., Groop, L., Mootha, V. K., Daly, M. J. & Altshuler, D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 6, e1001058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. N., Kim, B. H., Cho, J., Ryu, S., Shin, H., Sung, J. et al. Pathway analysis of genome-wide association datasets of personality traits. Genes Brain Behav 14, 345–356 (2015). [DOI] [PubMed] [Google Scholar]
- Duncan, L. E., Holmans, P. A., Lee, P. H., O'Dushlaine, C. T., Kirby, A. W., Smoller, J. W. et al. Pathway analyses implicate glial cells in schizophrenia. PLoS ONE 9, e89441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. S., Go, M. J., Kim, Y. J., Heo, J. Y., Oh, J. H., Ban, H. J. et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 41, 527–534 (2009). [DOI] [PubMed] [Google Scholar]
- Kim, B. H., Kim, H. N., Roh, S. J., Lee, M. K., Yang, S., Lee, S. K. et al. GWA meta-analysis of personality in Korean cohorts. J. Hum. Genet. 60, 455–460 (2015). [DOI] [PubMed] [Google Scholar]
- Kim, C. W., Yun, K. E., Jung, H. S., Chang, Y., Choi, E. S., Kwon, M. J. et al. Sleep duration and quality in relation to non-alcoholic fatty liver disease in middle-aged workers and their spouses. J. Hepatol 59, 351–357 (2013). [DOI] [PubMed] [Google Scholar]
- Ahn, C. & Chae, J. Standardization of the Korean version of the Revised NEO Personality Inventory. Korean J. Counsel. Psychother. 9, 443–473 (1997). [Google Scholar]
- Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- Dudbridge, F. & Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch, E. A., Turner, T. J. & Goldin, S. M. Calcium as a coagonist of inositol 1,4,5-trisphosphate-induced calcium release. Science 252, 443–446 (1991). [DOI] [PubMed] [Google Scholar]
- Goto, J. & Mikoshiba, K. Inositol 1,4,5-trisphosphate receptor-mediated calcium release in Purkinje cells: from molecular mechanism to behavior. Cerebellum 10, 820–833 (2011). [DOI] [PubMed] [Google Scholar]
- Chung, S., Kim, I. H., Lee, D., Park, K., Kim, J. Y., Lee, Y. K. et al. The role of inositol 1,4,5-trisphosphate 3-kinase A in regulating emotional behavior and amygdala function. Sci. Rep. 6, 23757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa, K., Mizuno, K. & Ohkuma, S. Possible involvement of type 1 inositol 1,4,5-trisphosphate receptors up-regulated by dopamine D1 and D2 receptors in mouse nucleus accumbens neurons in the development of methamphetamine-induced place preference. Neuroscience 227, 22–29 (2012). [DOI] [PubMed] [Google Scholar]
- Pasche, S. Exploring the comorbidity of anxiety and substance use disorders. Curr. Psychiatry Rep. 14, 176–181 (2012). [DOI] [PubMed] [Google Scholar]
- Mellios, N. & Sur, M. The Emerging Role of microRNAs in Schizophrenia and Autism Spectrum Disorders. Front. Psychiatry 3, 39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo, V., Ciolfi, A., Macri, S. & Pizzuti, A. The emerging role of MicroRNA in schizophrenia. CNS Neurol. Disord. Drug Targets 14, 208–221 (2015). [DOI] [PubMed] [Google Scholar]
- Nusse, R. Wnt signaling in disease and in development. Cell Res. 15, 28–32 (2005). [DOI] [PubMed] [Google Scholar]
- Zandi, P. P., Belmonte, P. L., Willour, V. L., Goes, F. S., Badner, J. A., Simpson, S. G. et al. Association study of Wnt signaling pathway genes in bipolar disorder. Arch. Gen. Psychiatry 65, 785–793 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inkster, B., Nichols, T. E., Saemann, P. G., Auer, D. P., Holsboer, F., Muglia, P. et al. Pathway-based approaches to imaging genetics association studies: Wnt signaling, GSK3beta substrates and major depression. NeuroImage 53, 908–917 (2010). [DOI] [PubMed] [Google Scholar]
- Evans, S. J., Choudary, P. V., Neal, C. R., Li, J. Z., Vawter, M. P., Tomita, H. et al. Dysregulation of the fibroblast growth factor system in major depression. Proc. Natl Acad. Sci. USA 101, 15506–15511 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, E. M., Heath, A. C., Madden, P. A., Pergadia, M. L., Hickie, I. B., Montgomery, G. W. et al. Testing the role of circadian genes in conferring risk for psychiatric disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 165B, 254–260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, A. C., Aliev, F., Bierut, L. J., Bucholz, K. K., Edenberg, H., Hesselbrock, V. et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr. Genet. 22, 31–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano, M., Huffman, J. E., Arias-Vasquez, A., Vinkhuyzen, A. A., Middeldorp, C. M., Giegling, I. et al. Genome-wide association uncovers shared genetic effects among personality traits and mood states. Am J. Med. Genet. B Neuropsychiatr. Genet. 159B, 684–695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Pons, N., Virgos, C., Vogel, W. F., Urena, J. M., Soriano, E., del Rio, J. A. et al. Expression of discoidin domain receptor 1 during mouse brain development follows the progress of myelination. Neuroscience 140, 463–475 (2006). [DOI] [PubMed] [Google Scholar]
- Roig, B., Virgos, C., Franco, N., Martorell, L., Valero, J., Costas, J. et al. The discoidin domain receptor 1 as a novel susceptibility gene for schizophrenia. Mol. Psychiatry 12, 833–841 (2007). [DOI] [PubMed] [Google Scholar]
- Otowa, T., Yoshida, E., Sugaya, N., Yasuda, S., Nishimura, Y., Inoue, K. et al. Genome-wide association study of panic disorder in the Japanese population. J. Hum. Genet 54, 122–126 (2009). [DOI] [PubMed] [Google Scholar]
- van der Geer, P., Hunter, T. & Lindberg, R. A. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol. 10, 251–337 (1994). [DOI] [PubMed] [Google Scholar]
- Service, S. K., Verweij, K. J., Lahti, J., Congdon, E., Ekelund, J., Hintsanen, M. et al. A genome-wide meta-analysis of association studies of Cloninger's Temperament Scales. Transl. Psychiatry 2, e116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosser, A., Butler, A. W., Uher, R., Ng, M. Y., Cohen-Woods, S., Craddock, N. et al. Genome-wide association study of co-occurring anxiety in major depression. World J. Biol. Psychiatry 14, 611–621 (2013). [DOI] [PubMed] [Google Scholar]
- Mattheisen, M., Samuels, J. F., Wang, Y., Greenberg, B. D., Fyer, A. J., McCracken, J. T. et al. Genome-wide association study in obsessive-compulsive disorder: results from the OCGAS. Mol. Psychiatry 20, 337–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Shimada, T., Otowa, T., Wu, Y. Y., Kawamura, Y., Tochigi, M. et al. Genome-wide association study of autism spectrum disorder in the east asian populations. Autism Res. 9, 340–349 (2016). [DOI] [PubMed] [Google Scholar]
- Elia, J., Gai, X., Xie, H. M., Perin, J. C., Geiger, E., Glessner, J. T. et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 15, 637–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensslen-Craig, S. E. & Brady-Kalnay, S. M. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev. Biol 275, 12–22 (2004). [DOI] [PubMed] [Google Scholar]
- Parsons, J. T., Horwitz, A. R. & Schwartz, M. A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., Abrahamsen, G., Mills, R., Calderon, C. C., Tee, J. Y., Leyton, L. et al. Focal adhesion dynamics are altered in schizophrenia. Biol. Psychiatry 74, 418–426 (2013). [DOI] [PubMed] [Google Scholar]
- Terasaki, D. J., Gelaye, B., Berhane, Y. & Williams, M. A. Anger expression, violent behavior, and symptoms of depression among male college students in Ethiopia. BMC Public Health 9, 13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge, M. J. Neuronal calcium signaling. Neuron 21, 13–26 (1998). [DOI] [PubMed] [Google Scholar]
- Lane, J. D. & Aprison, M. H. Calcium-dependent release of endogenous serotonin, dopamine and norepinephrine from nerve endings. Life Sci 20, 665–671 (1977). [DOI] [PubMed] [Google Scholar]
- Stein, D. J. Social anxiety disorder and the psychobiology of self-consciousness. Front. Hum. Neurosci. 9, 489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. J., Escott-Price, V., Davies, G., Bailey, M. E., Colodro-Conde, L., Ward, J. et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol. Psychiatry 21, 749–757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


