Abstract
The heat-labile toxins (LT) produced by enterotoxigenic Escherichia coli display adjuvant effects to coadministered antigens, leading to enhanced production of serum antibodies. Despite extensive knowledge of the adjuvant properties of LT derivatives, including in vitro-generated non-toxic mutant forms, little is known about the capacity of these adjuvants to modulate the epitope specificity of antibodies directed against antigens. This study characterizes the role of LT and its non-toxic B subunit (LTB) in the modulation of antibody responses to a coadministered antigen, the dengue virus (DENV) envelope glycoprotein domain III (EDIII), which binds to surface receptors and mediates virus entry into host cells. In contrast to non-adjuvanted or alum-adjuvanted formulations, antibodies induced in mice immunized with LT or LTB showed enhanced virus-neutralization effects that were not ascribed to a subclass shift or antigen affinity. Nonetheless, immunosignature analyses revealed that purified LT-adjuvanted EDIII-specific antibodies display distinct epitope-binding patterns with regard to antibodies raised in mice immunized with EDIII or the alum-adjuvanted vaccine. Notably, the analyses led to the identification of a specific EDIII epitope located in the EF to FG loop, which is involved in the entry of DENV into eukaryotic cells. The present results demonstrate that LT and LTB modulate the epitope specificity of antibodies generated after immunization with coadministered antigens that, in the case of EDIII, was associated with the induction of neutralizing antibody responses. These results open perspectives for the more rational development of vaccines with enhanced protective effects against DENV infections.
Keywords: heat-labile toxins, labile toxins, adjuvants, dengue virus, envelope protein, vaccines, antibodies, immunosignature
Introduction
Adjuvants are essential components of vaccines, particularly those composed of purified antigens, which naturally have lower immunogenicity than live attenuated microorganisms. Adjuvants are known for their ability to enhance the magnitude of adaptive immune responses, particularly antibody responses, to coadministered antigens; they may impact the longevity, antigen avidity, and modulation of isotype and IgG subclass switches (1, 2). To date, only a limited number of adjuvants have been incorporated into human vaccines, but several different substances, including bacterial toxins, are known for their strong and rather versatile adjuvant effects in various mammalian hosts (3, 4). We are investigating the changes in epitope recognition elicited by adjuvants.
Heat-labile toxins (LT) naturally produced by some enterotoxigenic Escherichia coli strains are enterotoxins endowed with potent adjuvant effects when coadministered with purified soluble or particulate antigens after delivery via different parenteral or mucosal routes (5–9). LTs, similarly to cholera toxin, are composed of one toxic A subunit with enzymatic ADP-ribosylation activity and a B subunit pentamer that binds to host cell receptors (10, 11). LT adjuvant effects, as well as those associated with in vitro-generated non-toxic LT derivatives, including purified B subunits and mutated LT forms, have primarily been ascribed to the activation of dendritic cells and B and T lymphocytes, leading to enhanced mucosal and serum antigen-specific antibody responses (6, 12–19). In addition, previous evidence indicated that LT derivative may modulate the epitope specificity of antibodies raised against coadministered antigen (12). Indeed, adjuvant-mediated modulation of epitope specificity of antibodies is still a poorly understood issue, particularly in situations involving the design of vaccines capable of inducing protective immunity to various infectious diseases.
Dengue fever is caused by an arbovirus transmitted by Aedes aegypti mosquitoes. Fifty to one hundred million people are affected by the disease in tropical and subtropical regions annually, and thousands are potentially at risk to develop more serious side effects, which may lead to death caused by the illness (20, 21). The dengue virus (DENV) has an envelope composed of a single protein (E) and encodes two other structural (prM and C proteins) and seven non-structural proteins (22–24). The glycoprotein E is composed of three structural and functional domains that mediate the initial steps of the DENV infectious cycle. Receptor binding is mainly triggered by domain III [envelope glycoprotein domain III (EDIII)], which leads to the subsequent entry and replication of virus into susceptible host cells (22, 24, 25). EDIII-specific antibodies can confer protection to DENV infection and have been considered protection correlates for anti-DENV vaccines (26–31). Nonetheless, depending on the specificity, concentration and affinity of antibodies targeting the E protein, antibodies may either block virus infection or promote enhancement of infection (the antibody-dependent enhancement effect—ADE) via cell entry mediated by Fc-γ receptors (32–35).
Proteomics platforms such as microarrays have been exploited as promising high-throughput assays to measure humoral immune responses, assisting our understanding of the subtle characteristics of successful immunization, a key challenge in the rational development of effective vaccines (36–38). The immunosignature technology is a microarray-based platform that uses unnatural randomized-sequence peptides, spotted in high density, as a universal and robust assay to identify antibody-binding patterns of natural or induced responses in both humans and mice. Due to its successful applicability for both infectious and non-infectious diseases, this platform can be used as a powerful tool for systematic studies of various vaccine approaches (39–46).
In this study, we compared the adjuvant effects of alum, LT and LTB in vaccine formulations containing a recombinant form of the DENV EDIII antigen. The results demonstrate that, in contrast to mice immunized with the alum-adjuvanted formulation, mice parenterally immunized with EDIII admixed with LT or LTB mounted a more efficient antibody response capable of inhibiting DENV infection. Immunosignature analysis of the humoral immune response elicited in vaccinated mice showed that LT differentially modulates the epitope specificity of the EDIII-specific IgG antibodies. In addition, the results led to the identification of an EDIII-derived peptide specifically involved in the infectivity of the virus into host cells. Together, this study emphasizes the role of adjuvants in the modulation of the epitope specificity of antibodies induced upon vaccination and, thus, may impact the development of anti-DENV and other subunit vaccines.
Materials and Methods
Cloning of Adjuvant LTB
LTB was obtained after cloning the nucleotide sequence of the eltB gene (encoding the B subunit of the LT from H10407 strain) into the pET22b vector. The eltB sequence (450 bp) was amplified by PCR using the forward primer 5′-CAGAAGGCGGAATGACATATGAATAA-3′ and reverse primer 5′-TTAAAAGCATGTCTCTCGAGAGAACC-3′ (the underlined sequences indicate the NdeI and XhoI restriction sites, respectively). The insert generated by the PCR reaction was digested and ligated into the corresponding NdeI and XhoI restriction sites of the pET22b expression vector (Novagen-Merck Bioscience, MA, USA), generating the recombinant plasmid pET22bLTB, which was subsequently used to transform chemically competent E. coli DH5α. Transformants were analyzed by digestion with the enzymes NdeI and XhoI. The recombinant DNA fragment was sequenced and compared to data reported for the LT sequence (GenBank database GI: 408994). A chemically competent E. coli BL21 (DE3) strain was transformed with the pET22bLTB plasmid and denominated as E. coli BL-LTB.
Expression and Purification of Adjuvants
The LT and LTB adjuvants were obtained from E. coli strains harboring the plasmid pML19 (47, 48) or pET22bLTB, respectively. The purification of LT was based on a previously described method (47). The recombinant E. coli strains were cultivated in Luria-Bertani (LB) medium or Terrific Broth (TB) supplemented with 100 µg/ml of ampicillin at 37°C in an orbital shaker (200 rpm). While LTB expression was obtained after 0.75 mM IPTG induction of the E. coli BL-LTB strain cultures for 6 h, LT production was achieved by constitutive expression of the eltAB gene in overnight cultures of a pML19-harboring E. coli strain. The cell pellets were suspended, and the bacterial cells were disrupted by high pressure in a homogenizer (APLAB-10 model, ARTEPEÇAS, Brazil). The LT and LTB contained in the bacterial cell homogenates were purified by affinity chromatography on immobilized d-galactose columns (Pierce, Waltham, MA, USA) previously equilibrated with TEAN buffer (50 mM Tris, 1 mM EDTA, 3 mM NaN3, and 0.2 M NaCl, pH 7.4) in an Akta FPLC (Amersham Pharmacia Biotech, Buckinghamshire, UK). The elution step was performed with TEAN supplemented with 0.3 M d-galactose. The proteins were monitored by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The concentrations of the purified proteins were determined by absorbance measurements in a spectrophotometer (Gene Quant spectrophotometer GE Amersham Biosciences, Buckinghamshire, UK) as previously described (49).
Cloning, Expression, and Purification of Antigen
The pE1D2 plasmid (50), harboring gene encoding the ectodomain of the envelope glycoprotein from the DENV2 New Guinea C (NGC) strain, was kindly provided by Dr. Ada Alves and used as template for PCR reactions. The sequences of the forward and reverse primers used in PCR reactions were 5′-ACATGCGAGGATCCGGAATGTCATACTCTAT-3′ (the underlined sequence indicates the BamHI restriction site) and 5′-GCCTTCTACTCGAGTTACGATAGAACTTCCTTTCTTA-3′ (the underlined sequence indicates the XhoI restriction site), respectively. The PCR product was introduced into pET28a(+) expression vector (Novagen, Germany), generating the recombinant plasmid pDEDIII. The E. coli BL21 (DE3) strain harboring the plasmid pDEDIII was cultivated in LB medium containing 50 µg/ml kanamycin. The culture at an OD600nm of 0.5 was induced by IPTG to a final concentration of 0.5 mM and incubated for 4 h. The bacterial pellet was suspended in buffer A (0.1 M Na2PO4, 0.5 M NaCl, pH 6.8) and lysed in the APLAB-10 homogenizer (ARTEPEÇAS, Brazil). The inclusion bodies were solubilized in buffer A supplemented with 8 M urea, and the recombinant protein was refolded in buffer A. The soluble protein was submitted to nickel affinity chromatography using a HisTrap™ HP column (GE Healthcare Life Sciences, Buckinghamshire, UK). The purified EDIII was evaluated by 15% SDS-PAGE and measured by spectrophotometry.
Endotoxin Removal from Purified Protein Preparations
The endotoxin was removed using a Detoxi-gel Endotoxin Removing Gel (Pierce, Waltham, MA, USA), and any residual endotoxin was detected by the Limulus Amebocyte Lysate (LAL) QCL 1000 kit (Lonza, Basel, Switzerland). Following treatment for LPS removal, the residual endotoxin contents in the LT, LTB, and EDIII samples were less than 0.1 EU/μg of protein, corresponding to the levels allowed for preclinical research (51).
Biological Characterization of LT and LTB Adjuvants
The cytotonic activity of the LT derivatives on the adrenal Y1 cells was evaluated as previously described (6). The Y1 cells were seeded in 96-well plates (5 × 104 cells/well) and exposed to 1 µg of LT or LTB, or to phosphate-buffered saline (PBS) as negative control, diluted in Dulbecco’s modified Eagle’s medium (DMEM, Vitrocell Embriolife, Brazil) supplemented with 2% fetal bovine serum (FBS, Vitrocell Embriolife, Brazil). After 8 h of incubation, the cells were observed for the cytotonic effect. The GM1-ELISA was performed as previously described (52) to evaluate LT binding to the cognate ganglioside GM1. Microtiter plates (Polysorp, Thermo Fisher-Nunc, Roskilde, Denmark) were coated with GM1 ganglioside (1 µg/ml) (Sigma-Aldrich, St. Louis, MO, USA) diluted in PBS (pH 7.4) and incubated overnight at room temperature in a humid chamber. Plates were blocked with PBS containing 0.1% BSA. Then, serially twofold diluted LT samples were applied in duplicate wells. To detect LT derivatives, the plates were sequentially incubated with anti-LT serum (titer equal to 105) and horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibodies (Sigma-Aldrich) diluted to 1:1,000 and 1:3,000, respectively, in PBS containing 0.05% Tween-20 and 0.1% BSA. The chromogenic reactions developed with ortho-phenylenediamine dihydrochloride (Sigma-Aldrich) and H2O2 were stopped with 1 M H2SO4 and measured at A492nm.
Analysis of the Biological Activity of Recombinant EDIII in Vero Cells
This assay was performed as previously described (53). African Green Monkey Kidney Epithelial Cells (Vero Line), which express receptors for E glycoprotein, were cultured in minimum essential medium (MEM) supplemented with 2% FBS and incubated at 37°C in an atmosphere containing 5% CO2 for 24 h. Cells were incubated with 100 µg/ml of EDIII protein in MEM at 37°C in 5% CO2 for 30 min. After washing with PBS with 2% FBS, Vero cells were fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 10 min, treated with anti-EDIII sera harvested from mice immunized with EDIII plus Freund’s adjuvant for 60 min and subsequently with goat anti-mouse IgG conjugated to FITC (Invitrogen, Paisley, UK) for further 60 min. The FITC-stained Vero cells were detected by BD LSRFortessa flow cytometry (BD Bioscience, San Jose, CA, USA), and the data were analyzed using the FlowJo v10 program.
Animal Immunizations
All experiments involving mice were approved by the Committee on the Ethical Use of Laboratory Animals of the Institute of Biomedical Sciences (CEUA 198) at the University of São Paulo (USP), in accordance with the guidelines for the care and use of laboratory animals adopted by the National Council of Animal Experimentation (CONCEA). The mice were obtained from the Isogenic Mouse Breeding Facility of Department of Parasitology, Institute of Biomedical Sciences—USP. Female BALB/c mice (6–8 weeks old) were immunized via the subcutaneous (s.c.) route at days 0, 14, and 28 of the immunization protocol with the vaccine formulations. Groups of five BALB/c mice were treated with 100 µl of PBS alone as a negative control or PBS containing 10 µg of EDIII with or without one of the following adjuvants: 1.0 µg of LT, 3.2 µg of LTB, or 12.5 µg of Al(OH)3 (Rehydragel, Reheis, NJ, USA). Serum samples were harvested 2 weeks after each vaccine dose and stored at −20°C until immunological analyses were conducted.
ELISA
ELISA for anti-EDIII antibodies was performed as previously described (53). Microtiter plates (Maxisorp Thermo Fisher-Nunc, Roskilde, Denmark) were coated with EDIII (1 µg/ml) diluted in PBS pH 7.4 and incubated overnight at 4°C. For ELISA using denaturing conditions, the EDIII protein was submitted to heat treatment at 100°C for 15 min and used to coat the plates. Plates were blocked with PBS containing 3% gelatin for 2 h at 37°C. The serum samples harvested from mice immunized with EDIII alone or coadministered with each adjuvant were applied to duplicate wells and serially twofold diluted. The serum IgGs were measured with HRP-conjugated anti-mouse IgG (1:3,000), IgG1 (1:10,000), or IgG2a (1:3,000) antibodies (Sigma-Aldrich, St. Louis, MO, USA). Absorbance at 492 nm was obtained as previously described and used to calculate the antibody titers, defined as the highest sample dilution able to generate an A492nm of 0.2 above the preimmune sera.
Purification of Serum IgG Fractions
Anti-EDIII IgG antibody fractions were obtained from serum samples harvested from mice submitted to EDIII-based immunization regimens. First, the recombinant 6×-His-tagged EDIII was immobilized in a column filled with a nickel-coupled resin. After serum delipidation, as previously described (54), the sera were submitted to affinity chromatography on EDIII-immobilized resin using 0.05 M PBS, pH 7.4. The anti-EDIII antibodies were eluted in 0.1 M glycine buffer (pH 4), neutralized with 1 M Tris–HCl (pH 9.0), and dialyzed in PBS (pH 7.4). Subsequently, the anti-EDIII antibodies were subjected to affinity chromatography to protein G sepharose according to the manufacturer’s instructions (GE Healthcare, Buckinghamshire, UK), and the IgG fraction was obtained following the elution step described above. The purified EDIII-specific IgG antibodies were measured by BCA assay (Pierce, Waltham, MA, USA) and monitored by 12.5% SDS-PAGE.
Affinity Assay for Anti-EDIII IgG Antibodies
The antibody affinity index was measured using a modified ELISA protocol with ammonium thiocyanate (47). Briefly, the plates were coated with EDIII as described above. The sera collected from mice from different immunization groups were tested at dilutions corresponding to an OD492nm of 0.8, while purified anti-EDIII IgG antibodies were evaluated at an OD492nm of 0.5. After incubation with sera or purified antibodies, different concentrations of sodium thiocyanate were added to wells and incubated for 15 min. The plates were washed and incubated with HRP-conjugated anti-mouse IgG antibody. The percentage of antibodies bound to EDIII was determined: OD492nm in the presence of ammonium thiocyanate × 100/OD492nm in the absence of ammonium thiocyanate. The values obtained with the serum samples represent the antibody avidity, which under our conditions depends on the affinity and valence of the immunoglobulins since the parameter of structural arrangement was standardized by the use of the antigen immobilized on the solid support. The analyses of the purified IgG antibodies showing constant Ig valence generated average affinity values for each immunization group.
Plaque Reduction Neutralization Test (PRNT)
To perform the PRNT, the DENV2 NGC strain, kindly provided by Laura Helen Vera Gonzales Gil (Fiocruz, Recife, Brazil), was purified, and the Vero cells were cultivated as previously described (35). Briefly, the NGC strain was propagated in the Aedes albopictus cell line C6/36 cultured in Leibovitz medium containing 5% FBS and grown at 28°C for 7 days. The supernatant of C6/36 infected with the NGC strain was collected and titrated for the determination of PFU/ml. For the neutralization test, Vero cells were maintained in DMEM supplemented with 10% FBS and seeded in 24-well plates (1 × 105 cells/well) 24 h before infection. Serum samples from immunized mice were inactivated for complement proteins for 30 min at 56°C and serially twofold diluted. Purified EDIII-specific IgGs were tested at different concentrations. C6/36 cell supernatants containing 500 PFU of DENV/ml were incubated with each dilution of anti-EDIII sera or purified IgG antibodies for 1 h at 37°C. Vero cells were washed with serum-free DMEM and infected in duplicate wells with 200 µl of the neutralization mixture for 1 h at 37°C. The viral suspension was removed, and the cells were overlaid with 1 ml of complete DMEM [2.5% FBS and 1% carboxymethylcellulose (Synth, São Paulo, Brazil)]. Following incubation of the plates at 37°C for 7 days, the cells were fixed for 15 min with 4% paraformaldehyde and stained with crystal violet for 10 min. The percentage of plaque reduction compared to the positive control (DENV not exposed to antibodies) was calculated. Neutralizing antibody titers were expressed as the serum dilution, or IgG concentration, yielding 50% plaque number reduction (PRNT50).
Immunosignature Assay
For the peptide microarray assay, slides were manufactured using in situ synthesis of 330,000 (CIM330K) sequences of non-natural randomized peptides as previously described (40). Statistical analyses of microarray data were performed using JMP 12 software (Statistical Discovery Software from SAS) by importing data analysis from Excel and image-processed data from GenePix Pro-6.0 (Molecular Devices, Sunnyvale, CA, USA). Raw intensities were normalized to each slide by dividing all values per array by the median of that array (median normalization). Poor quality spots were excluded from analysis by flagging them as “absent” upon visual inspection. Values less than 0.01 were set to 0.01, and values from duplicate arrays were averaged and used in the analysis. Important peptides were determined by Student’s two-tailed t-test, with Benjamini and Hochberg multiple test correction applied to a p value < 0.05. For principal component analysis, we used significant peptides for each vaccine approach and an analysis using JMP 12 software (39). The prediction of recognized epitopes was based on lists of informative peptides compared to the EDIII sequence from the DENV2 strain NGC (AHG97599.1) using GuiTope (55). The amino acid substitution matrix was provided by the peptide library and protein sequence. An inversion weight of 1 was selected. Library subtracted scores were returned using the mean of 10 random samplings of the peptide library and a minimum score cutoff of 8.0. The results were graphed using GraphPad Prism v5 using a moving average of 15.
Competition Assay for DENV2 Using Peptides
This assay was adapted from a protocol previously described (56, 57). Vero cells (1 × 105 cells/well, on 96-well plates) were cultured in MEM supplemented with 3% FBS and incubated at 37°C in an atmosphere containing 5% CO2 for 18 h. Intact or heat-denatured EDIII protein, as well as peptide 47 (369-AEPPFGDSYIIIGVEPGQLK-388), custom-made by GeneScript (Piscataway, NJ, USA), at different molar concentrations, was incubated with the DENV2 strain NGC (multiplicity of infection—equal to 1.0). The mixtures were added to Vero cells at 37°C in 5% CO2 for 1 h. The mixtures were removed, and MEM supplemented with 2% FBS was added to each well. Following 18 h of incubation, the culture plates were washed with PBS and treated with 50 µl/well of trypsin (2.5 mg/ml) plus EDTA at 37°C in 5% CO2 for 10 min to remove the cells. Vero cells were resuspended with PBS containing 10% FBS and fixed and permeabilized with Cytofix/Citoperm kit (BD Bioscience, San Jose, CA, USA) for 10 min. The cells were incubated for 1 h with mAb 4G2, which recognizes the fusion loop at the extremity of the domain II of the envelope protein (ATCC-HB112), and subsequently labeled with anti-mouse IgG conjugated with Alexa 488 (BD Biosciences) for 30 min. The stained cells were measured by flow cytometry using a BD LSRFortessa (BD Bioscience, San Jose, CA, USA) instrument, and the data were analyzed with FlowJo v10 software to determine the amount of DENV-positive Vero cells. Viral replication analysis was carried out at 24 h post-infection based on a protocol previously described (58). RNA was extracted from 100 µl of infected Vero cell, using a guanidine isothiocyanate phenol method (Trizol LS, Invitrogen), according to the manufacturer’s instructions. The quantitative real-time polymerase-chain-reaction was carried out with a set of primers and probes with FAM as dye reporter for the probe. Primers/probes used in assay for DENV were previously described (58). The assay was performed using the AgPath-IDTM One-Step RT-PCR reagents (Applied Biosystems). Succinctly, we used 5 µl of the extracted RNA in 1 µl of the mix from primers/probe (10 pM/μl) and 19 µl of the reagent mix from AgPath-IDTM One-Step RT-PCR kit following the manufacturer’s instructions.
Structural Analyses
The amino acid sequence composing the main predicted epitope was indicated in a previously published structural model of the quaternary DENV2 envelope glycoprotein (PDB accession number 1OKE) using PyMol v1.8.4.0.
Statistical Analyses
The data are represented as arithmetic means ± SD and analyzed for variance (ANOVA) with a subsequent Bonferroni’s multiple comparison test using GraphPad Prism v5 software. For the data generated by the PRNT and affinity assay, the statistical analyses were calculated from two independent experiments, while all other results were analyzed from three independent experiments.
Results
LT and LTB Enhance EDIII-Specific Antibody Responses
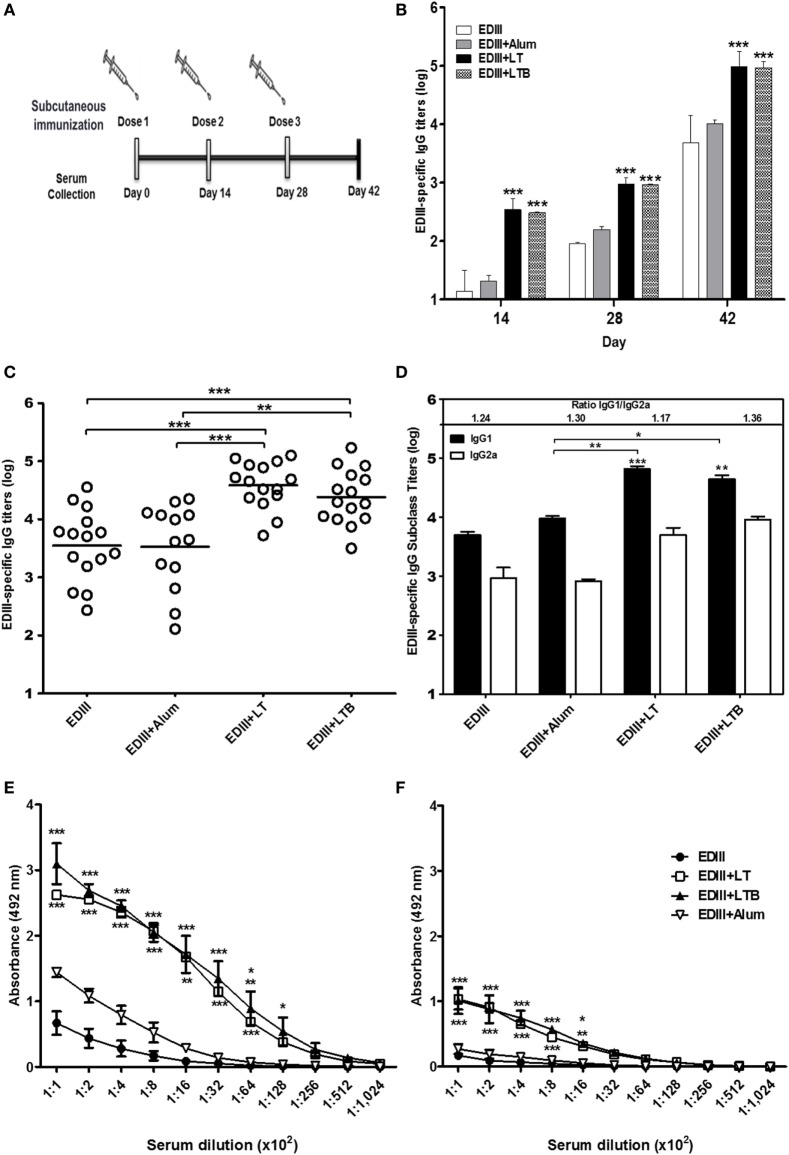
The recombinant forms of LT, LTB and type 2 DENV EDIII were purified by affinity chromatography (Figures S1 and S2 in Supplementary Material). Both LT and LTB bind to host cell receptors, but as expected, only LT had cytotonic effects to Y1 cells (Figures S1C,D in Supplementary Material). The recombinant EDIII also preserved the receptor-binding function of the native viral protein (Figure S2B in Supplementary Material). Mouse groups were immunized with purified EDIII and vaccine formulations containing EDIII admixed with alum, LT, or LTB. Mice received three s.c. doses at intervals of 2 weeks and were bled one day before each dose and 2 weeks after the last vaccine dose (Figure 1A). Significant increases in the EDIII-specific IgG antibody levels were detected in serum samples of mice immunized with vaccines containing LT and LTB, but not with alum compared to EDIII alone (Figures 1B,C). The antigen-specific IgG1/IgG2a ratios in vaccinated mice were similar, indicating that the adjuvants were not able to modulate the IgG subclass responses induced by the antigen alone. Although anti-EDIII IgG1 and IgG2a titers detected in mice immunized with LT and LTB enhanced in comparison with the non-adjuvanted EDIII, significant differences were only found regarding to IgG1 levels (Figure 1D). In addition, EDIII-specific antibodies in serum samples collected from vaccinated mice were capable of recognizing both conformational and linear epitopes of the antigen, as demonstrated by reactivity with both intact and heat-denatured antigen. Serum samples from mice immunized with LT or LTB showed significantly higher reactivity with both conformational and linear epitopes than those collected from mice immunized with non-adjuvanted EDIII or EDIII admixed with alum (Figures 1E,F).
Figure 1.
Analysis of envelope glycoprotein domain III (EDIII)-specific antibody responses induced in mice immunized with different vaccine formulations. (A) Schematic representation of the vaccine regimen tested in this study. BALB/c female mice were s.c. immunized with three doses of EDIII alone (10 µg) or EDIII coadministered with one of the tested adjuvants [LT1 (1 µg), LT1-B (3.2 µg), or alum (12.5 µg)]. (B) Analyses of the antigen-specific IgG responses performed in blood samples collected 2 weeks after each vaccine dose. (C,D) Evaluation of the anti-EDIII total IgG (C) or IgG subclass (D) responses was carried out 2 weeks after the third dose. (E,F) Reactivity of serum antibodies carried out by ELISA with the native purified EDIII antigen (E) or the same antigen submitted to a heat denaturation treatment (F) used as solid phase bound antigens. Anti-EDIII titers were determined in three independent experiments with at least five animals per immunization group. Values represent means ± SD of the IgG titers. ***p < 0.001, **p < 0.01, and *p < 0.05, comparing adjuvanted EDIII-immunized mice to non-adjuvanted EDIII-immunized mice or alum-adjuvanted mice (ANOVA with Bonferroni post hoc test).
Antigen Reactivity and DENV-Neutralization Properties of Purified EDIII-Specific Antibodies
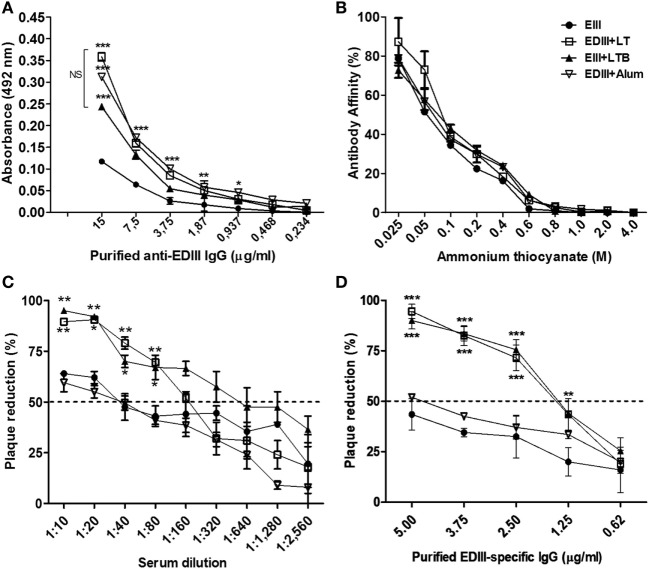
To evaluate the role of adjuvants in the modulation of the effector activities of antigen-directed antibodies induced by subunit vaccines, EDIII-specific antibodies raised in different mouse groups were purified by affinity chromatography (Figure S3 in Supplementary Material). The purified anti-EDIII antibodies reacted with the native antigen but did not recognize the heat-denatured antigen, suggesting that the purified antibodies are specifically directed toward conformational epitopes (Figure 2A; data not shown). No difference in antigen affinity was detected in EDIII-specific antibodies raised in the different immunization groups (Figure 2B). Anti-EDIII antibodies, either in serum or purified fractions from vaccinated mice showed virus-neutralization activity in vitro; however, anti-EDIII antibodies elicited with LT or LTB were more efficient than those raised with antigen alone or admixed to alum (Figures 2C,D; Table S1 in Supplementary Material). The mice immunized with LTB coadministered to EDIII elicited antigen-specific sera with twofold higher neutralizing capability than LT-adjuvanted vaccination (Figure 2C; Table S1 in Supplementary Material). The purified anti-EDIII IgG from both LT and LTB-adjuvanted mice yielded neutralization titers (PRNT50) of 1.6 ng/ml and thereby revealed strong virus-neutralizing effects (Figure 2D).
Figure 2.
Characterization of purified envelope glycoprotein domain III (EDIII)-specific IgG antibodies raised in mice immunized with different adjuvants. (A) Reactivity of antibodies with the purified antigen following ELISA. (B) Affinity of purified antibodies to the purified EDIII antigen. Values are expressed by percentage of antibodies that remain bound to the solid phase-adsorbed antigen under presence of ammonium thiocyanate in regard to the reaction performed without ammonium thiocyanate. Immunization groups represented by different symbols as indicated in the insert. (C,D) Plaque reduction neutralization test of a DENV2 strain (New Guinea C) carried out with serially diluted serum samples (C) or various amounts of purified EDIII-specific IgG antibodies (D) derived from mice immunized with the different vaccine formulations, as indicated in panel (B). Results are based on two independently performed experiments carried out with duplicate samples. Values represent means ± SD. ***p < 0.001, **p < 0.01, and *p < 0.05, significant differences with regard to mice immunized with non-adjuvanted EDIII (two-way ANOVA with Bonferroni post hoc test). NS: values without statistically significant differences.
Immunosignatures of Anti-EDIII Antibodies
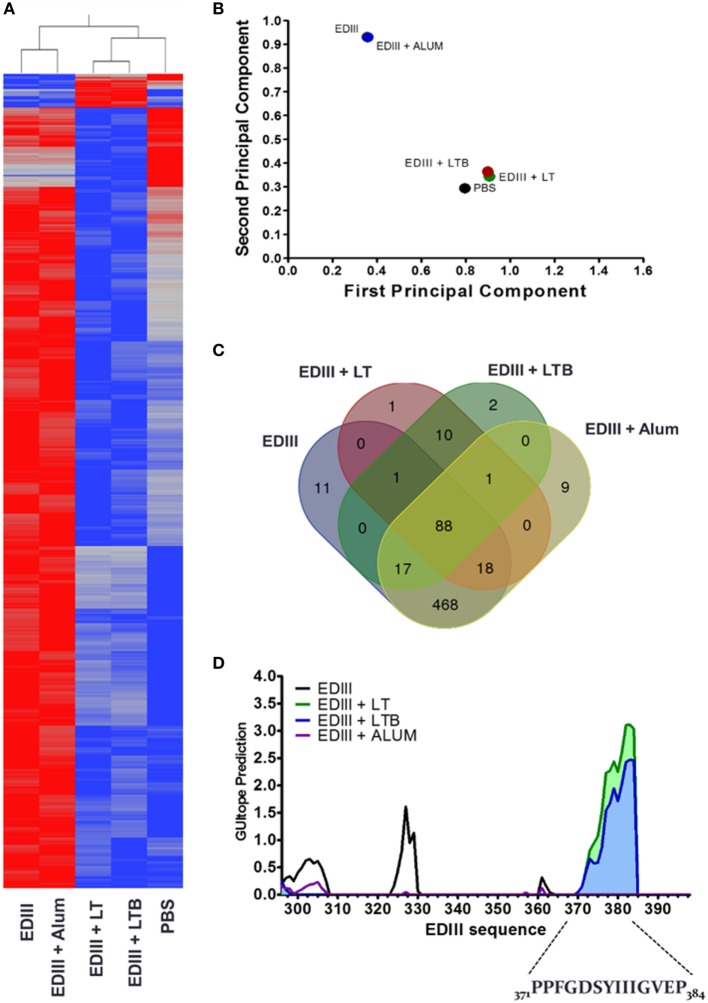
To gain a better understanding of the enhanced virus-neutralization activity of the anti-EDIII antibodies raised in mice immunized with different vaccine formulations, we determined the immunosignatures of purified EDIII-specific antibodies raised in different immunization groups (Figure 3). By profiling the antibodies with random linear peptide sequences spotted on microarray slides, we observed that antibodies from mice immunized with EDIII and LT or LTB had distinct immunological signatures for those raised in mice immunized without adjuvant or with alum (Figures 3A,B). In addition, fewer peptides were recognized with high binding intensity by EDIII-specific IgGs adjuvanted with LT or LTB, suggesting an increased specificity of these antibodies (Figures 3A,C). Analysis of peptide-binding profiles indicated that antibodies raised in mice immunized with LT and LTB reacted with 119 peptides sequences each, while antibodies raised in mice immunized only with antigen, or with alum admixed with the antigen, reacted with 605 and 603 peptides, respectively. Among these peptides, only 10 were solely recognized by antibodies raised in mice immunized with LT or LTB and 88 peptides among all groups (Figure 3C).
Figure 3.
Immunosignature analyses of purified envelope glycoprotein domain III (EDIII)-specific IgG antibodies. (A) Heat map demonstrating the immunosignatures detected with EDIII-specific IgG antibodies purified from serum samples collected from mice submitted to the different immunization regimens and a mock mouse group (control). A Student’s t-test (p < 3.33 × 10−6) between vaccine groups was used to select the informative peptides. Hierarchical clustering using EUCLIDEAN distance was used as a measure of similarity to cluster the selected peptides (x axis) and vaccine groups (y axis). Peptides’ intensity is colored where blue corresponding to low intensity and red to high intensity. (B) Variance of immune responses among different vaccine groups. Variation among vaccine groups [pooled sera per group (n = 5)] for all significant peptides sequences in the principal component analysis plot, where the first two principal components are plotted and individuals are colored according to the vaccination groups. (C) Venn diagram between vaccine groups. Results summarize the overlapping peptides that are significantly different above 1.3-fold change of the mock group versus each vaccine group. (D) Antigenic epitope prediction for vaccine groups using random peptide arrays. Significant peptides sequences recognized by antibodies of each vaccine group were submitted to epitope prediction with the GuiTope software and using the EDIII protein sequence from DENV2 New Guinea C (AHG97599.1) as reference. Each line graph represents the GuiTope prediction score for each vaccine group.
Epitopes Predicted by Immunosignatures Are Critical for DENV Infection
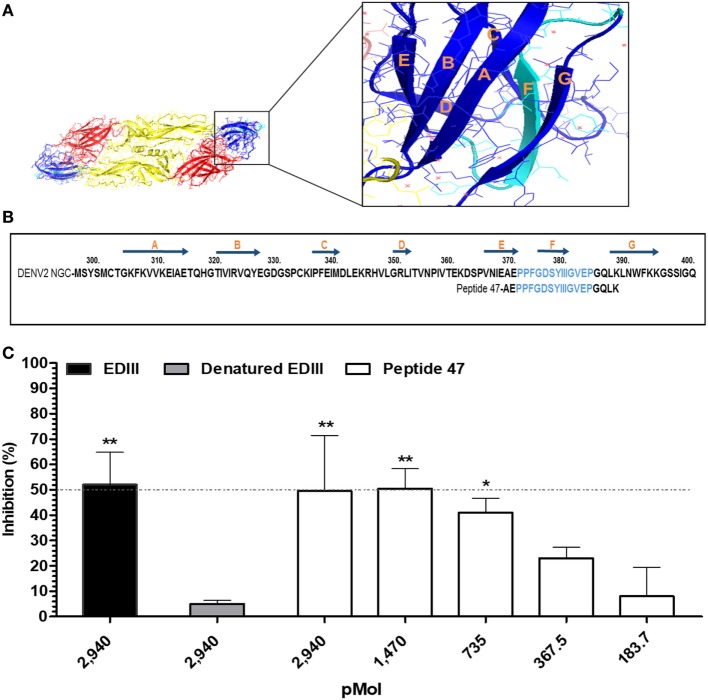
Prediction of epitopes present in the EDIII antigen and recognized by the different set of purified IgG antibodies showed that mice immunized with both LT derivatives reacted with a specific sequence comprising the EF loop, F beta-sheet, and FG loop between amino acid residues 371–384 (Figures 3D and 4A,B). To further investigate the biological relevance of this finding, we tested a 21-mer synthetic peptide encompassing the complete predicted epitope of the EDIII antigen (Figure 4B). As shown in Figure 4C and Figure S4 in Supplementary Material, the peptide (EDIII369–388) efficiently blocked the infectivity of DENV2 NGC strain in Vero cells. A similar inhibition of DENV infectious activity was achieved with the purified EDIII antigen, whereas the heat-denatured antigen lost the virus-infection inhibitory effect.
Figure 4.
Identification of a linear envelope glycoprotein domain III (EDIII)-derived epitope involved in binding of DENV2 to host cells. (A) 3D representation of DENV E protein dimer. Domains I (yellow), II (red), and III (blue) are highlighted. The insert displays EDIII structural features. The amino acid sequence marked in light blue encompasses the epitope recognized by antibodies raised in mice immunized with labile toxins (LT)-adjuvanted vaccines. (B) Amino acid sequence of DENV2 EDIII (GenBank accession number: AF038403.1). The positions of the EDIII β strands are indicated above the sequence by dark blue arrows, while the sequences of the synthetic peptide used in the competition assays are shown below. The epitope predicted by the immunosignature is marked in light blue. (C) Inhibition of DENV2 infection. Different molar concentrations of synthetic peptide 47 were mixed with DENV2 New Guinea C and subsequently added to cells. Purified EDIII and heat-denatured EDIII were used as positive and negative controls in the inhibition of virus infectivity assay, as indicated in the figure. Cells stained with primary mAb 4G2 and subsequently with Alexa 488-conjugated anti-mouse IgG were measured by cytometry, and the number of DENV-positive infected Vero cells was determined for each group. Values express the reduction in the percentage of the number of virus-infected cells compared to the control group denatured EDIII- (untreated DENV2). Dashed line indicates 50% inhibition of virus infection. Bars represent the mean values and SEM from three independent experiments. ***p < 0.001, **p < 0.001, and *p < 0.05 represent significant differences with regard to denatured EDIII-treated and untreated DENV2 (two-way ANOVA with Bonferroni post hoc test).
Discussion
A considerable amount of information regarding the use of bacterial toxins such as LT as vaccine adjuvants has accumulated during the last four decades. Nonetheless, perspectives on the clinical use of non-toxic LT derivatives have been overshadowed by the induction of undesirable side effects (transient facial paralysis) observed in patients submitted to intranasal immunization trials (59). However, recent observations describing the successful use of LT derivatives as parenterally administered adjuvants rekindled interest in these adjuvants under both experimental and clinical conditions (8, 9, 60–62). In this study, we evaluated the adjuvant properties of LT and a non-toxic derivative composed of the B subunit pentamer as parenterally delivered adjuvants admixed with a recombinant form of the DENV2 EDIII. Our results demonstrated that, unlike alum, LT and LTB enhanced antibody responses to EDIII and induced the generation of antibodies with increased capacity to neutralize DENV2 particles. In addition, we demonstrated that both LT and LTB drastically changed the epitope-binding profiles of EDIII-specific IgG antibodies. Immunosignature analyses performed with peptide microarrays led us to identify an epitope located between the EDIII EF and FG loops, which is specifically recognized by antibodies raised in mice immunized with LT or LTB but not by antibodies raised in mice immunized with alum-adjuvanted EDIII or non-adjuvanted antigen. Remarkably, a synthetic peptide encompassing the epitope identified by the immunosignature analyses interfered with the infectivity of DENV2 in mammalian cells, opening perspectives for therapeutic and preventive interventions. Collectively, this study demonstrated that LT derivatives can modulate the epitope specificity of antibodies after parenteral immunization with purified proteins and thus may contribute to the rational design of subunit vaccines for DENV and other infectious agents.
The evidence presented here demonstrates that LT used as a parenteral adjuvant promotes strong neutralizing antibody responses to DENV infection. Antibody neutralization titers achieved in mice immunized with EDIII and LT or LTB were significantly higher than those elicited in mice immunized with alum. In accordance with these findings, the EDIII-specific IgG fraction purified from mice immunized with LT or LTB showed greater virus-neutralizing activity than antibodies purified from mice immunized with alum. Furthermore, purified IgG collected from mice immunized with LT or LTB revealed a strong virus-neutralizing effect compared to the previously described anti-EDIII mAbs, such as 2H12, 9D12, and 1A1D-2 that block DENV infection under in vitro conditions with different intensity (26, 32, 63, 64). The results emphasize the fact that parenterally administered LT represents a promising adjuvant alternative comparable to the most potent adjuvants presently in use under both experimental and clinical conditions. In addition, the observation that purified LT B subunits exert similar adjuvant effects as LT indicated that the local inflammatory reactions induced by the enzymatically active toxin can be abrogated without reducing the immunomodulatory effects to coadministered antigens.
Our results obtained with purified IgGs demonstrated that the specificity, and not the magnitude, of the antibody response is critical in virus neutralization. In fact, antibody-mediated virus-neutralization activity has been strongly correlated with antibody/antigen interaction force and/or epitope recognition patterns (12, 26, 47). In our case, the distinct neutralization effects of the EDIII-specific polyclonal antibodies were not associated with antigen affinity. In accordance with these results, several studies have reported that the virus-neutralizing activity of anti-EDIII mAbs or sera does not necessarily correlate with antigen affinity (33, 35, 64–67). However, epitope recognition patterns of EDIII-specific mAbs showed a positive correlation with virus-infection blockage activity (26, 32–34, 65). In this context, we demonstrated that the immunosignature patterns of antibodies raised in mice immunized with LT derivatives differed drastically from those raised in mice immunized with non-adjuvanted EDIII or alum-adjuvanted antigen. More importantly, such differences correlated with an enhanced virus-neutralizing activity of the IgG antibodies induced by LT forms. Although corroborative evidence remains to be gathered, this study clearly demonstrated that adjuvant-mediated modulation of epitope recognition patterns by the antibodies raised is a critical aspect of vaccine-induced immune responses, particularly with regard to the induction of immunity to DENV.
In this report, the immunosignature analyses of EDIII-specific polyclonal antibodies disclosed that, in contrast to alum, incorporation of LT or LTB to vaccine formulation resulted in antibodies with increased specificity to the epitopes located at the EDIII EF and FG loops in addition to the F β-strand. In support of this concept, our study demonstrated that a synthetic peptide comprising the predicted epitope interfered with the in vitro infectivity of DENV, as measured with Vero cells. In addition, the lateral FG loop has been implicated in DENV infectivity both in mosquito and mammalian cells (57, 68, 69). More importantly, part of the EDIII epitope unveiled by the immunosignature analyses, comprising the F β-strand and FG loops, has also been identified as a target epitope for anti-DENV and anti-ZIKV mAbs isolated from high responders in clinical studies with ZIKV-infected individuals (70). Together, these findings emphasize the relevance of the EDIII lateral ridge region as a target for antibodies capable of conferring protection from flaviviruses.
The role of LT derivatives, in the context of active immunization practices, on the modulation of epitope recognition profiles of antibodies has been scarcely investigated. Here, and in previous reports, we have demonstrated that LT derivatives shape the epitope recognition profile of antibodies to different pathogens [(12), unpublished data]. Previous studies have demonstrated that LT derivatives improve the expression of co-stimulatory molecules on B lymphocytes and dendritic cells and, consequently, on prime CD4+ T cells, leading to modulation of Th1/Th2/Th17 cytokine production patterns (12, 17, 19, 71–73). Although future studies will have to be performed, the capacity to program B cells using cognate T helper lymphocytes, concomitant with direct activation effects, may represent possible alternatives to explain the action of LT derivatives on the modulation of the epitope recognition pattern of antibodies.
Synthetic peptides have been largely exploited in therapeutic approaches against viral diseases such as hepatitis, acquired immunodeficiency syndrome and influenza (74). However, little effort has investigated the use of peptides for treatment of arboviroses. Anti-dengue peptides under pre-clinical evaluation were designed after in silico analyses and target the stem region, hydrophobic pocket or FG loop of the E glycoprotein (57, 69, 75, 76). These peptides showed half-maximal inhibitory concentration (IC50) for virus infection or binding to host cells at micromolar levels (57, 75, 76). Here, based on a different experimental approach, we report the identification of a peptide that affects DENV infectivity with an IC50 in nanomolar concentration range. In contrast to previous approaches, our results demonstrate that, besides having a direct effect on the host cell/virus interaction, such peptides may be amenable to generated protective antibody responses and therefore find both prophylactic and therapeutic applications.
Epitope mapping based on linear peptides has been a routine technique for the characterization of both monoclonal and specific polyclonal antibodies. The antibody immunosignature approach relies on the use of random peptides microarrays that broader the epitope screening inasmuch not specific to a pathogen or a vaccine, resulting in reduced cost and time. In addition, random peptides can be reactive with antibodies directed against conformational epitopes, which are not possible with conventional peptide microarray (40, 76). In fact, our data showed that even without the specific EDIII sequences on the array it was possible to detect DENV neutralizing peptide demonstrating the applicability of this technique.
In conclusion, our results demonstrated that LT and LTB enhance both the magnitude and the specificity of the antibody responses elicited in mice immunized with an anti-DENV subunit vaccine, promoting changes to the epitope-binding profiles that, in turn, lead to enhanced virus-neutralization effects with regard to alum. Such findings bring relevant insights into the design of vaccine formulations based on purified proteins and the induction of antibodies capable of preventing infections associated with different pathogens.
Ethics Statement
All experiments involving mice were approved by the Committee on the Ethical Use of Laboratory Animals of the Institute of Biomedical Sciences (CEUA 198) at the University of São Paulo (USP), in accordance with the guidelines for the care and use of laboratory animals adopted by the National Council of Animal Experimentation (CONCEA). The mice were obtained from the Isogenic Mouse Breeding Facility of Department of Parasitology, Institute of Biomedical Sciences—USP.
Author Contributions
Conceived and designed the experiments: DM, CM-S, SS, LF, and JR. Performed the experiments: DM, MB, LP, MJ, SP, SS, VS, and DO. Analyzed the data: DM, MB, JA, SJ, LF, and JR. Contributed reagents/materials/analysis tools: JA, SB, EM, SJ, LF, and JR. Wrote the paper: DM, MB, LF, and JR. All the authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer, KF, and handling editor declared their shared affiliation.
Acknowledgments
This study was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grants to LCSF: 2012/51204-2, 2014/17595-0), the Conselho Nacional do Desenvolvimento Científico e Tecnológico (CNPq grants to JR and EF-M, respectively: 485546/2012-4 and 309392/2015-2), and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES grant: 88887.116618/2016-01–AUXPE 0448/2016). At ASU by a grant from the Department of Homeland Security to SJ and the CIM Peptide Array Core (HSHQDC-15-C-B0008). The authors thank Dr. Phillip Stafford for his invaluable assistance, and the authors acknowledge the helpful technical assistance of E. G. Martins, C. Bertelli, Pattie Madjidi, and Penny Gwynne.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.01175/full#supplementary-material.
References
- 1.Pasquale A, Preiss S, Silva F, Garçon N. Vaccine adjuvants: from 1920 to 2015 and beyond. Vaccines (Basel) (2015) 3:320–43. 10.3390/vaccines3020320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity (2010) 33:492–503. 10.1016/j.immuni.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Hagan DT, De Gregorio E. The path to a successful vaccine adjuvant – “The long and winding road”. Drug Discov Today (2009) 14:541–51. 10.1016/j.drudis.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 4.Savelkoul HFJ, Ferro VA, Strioga MM, Schijns VEJC. Choice and design of adjuvants for parenteral and mucosal vaccines. Vaccines (Basel) (2015) 3:148–71. 10.3390/vaccines3010148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Hora VP, Conceição FR, Dellagostin OA, Doolan DL. Non-toxic derivatives of LT as potent adjuvants. Vaccine (2011) 29:1538–44. 10.1016/j.vaccine.2010.11.091 [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues JF, Mathias-Santos C, Sbrogio-Almeida ME, Amorim JH, Cabrera-Crespo J, Balan A, et al. Functional diversity of heat-labile toxins (LT) produced by enterotoxigenic Escherichia coli: differential enzymatic and immunological activities of LT1 (hLT) AND LT4 (pLT). J Biol Chem (2011) 286:5222–33. 10.1074/jbc.M110.173682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luiz WB, Rodrigues JF, Crabb JH, Savarino SJ, Ferreira LCS. Maternal vaccination with a fimbrial tip adhesin and passive protection of neonatal mice against lethal human enterotoxigenic Escherichia coli challenge. Infect Immun (2015) 83:4555–64. 10.1128/IAI.00858-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braga CJM, Rodrigues JF, Medina-Armenteros Y, Farinha-Arcieri LE, Ventura AM, Boscardin SB, et al. Parenteral adjuvant effects of an enterotoxigenic Escherichia coli natural heat-labile toxin variant. Front Immunol (2014) 4:487. 10.3389/fimmu.2013.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu JC, Mathias-Santos C, Greene CJ, King-Lyons ND, Rodrigues JF, Hajishengallis G, et al. Intradermal administration of the type II heat-labile enterotoxins LT-IIb and LT-IIc of enterotoxigenic Escherichia coli enhances humoral and CD8+ T cell immunity to a co-administered antigen. PLoS One (2014) 9:e113978. 10.1371/journal.pone.0113978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev (1998) 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lencer WI, Hirst TR, Holmes RK. Membrane traffic and the cellular uptake of cholera toxin. Biochim Biophys Acta (1999) 1450:177–90. 10.1016/S0167-4889(99)00070-1 [DOI] [PubMed] [Google Scholar]
- 12.Batista MT, Souza RD, Ferreira EL, Robinette R, Crowley PJ, Rodrigues JF, et al. Immunogenicity and in vitro and in vivo protective effects of antibodies targeting a recombinant form of the Streptococcus mutans P1 surface protein. Infect Immun (2014) 82:4978–88. 10.1128/IAI.02074-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Q, Vickers TJ, Fleckenstein JM. Immunogenicity and protective efficacy against enterotoxigenic Escherichia coli colonization following intradermal, sublingual, or oral vaccination with EtpA adhesin. Clin Vaccine Immunol (2016) 23:628–37. 10.1128/CVI.00248-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giuliani MM, Del Giudice G, Giannelli V, Dougan G, Douce G, Rappuoli R, et al. Mucosal adjuvanticity and immunogenicity of LTR72, a novel mutant of Escherichia coli heat-labile enterotoxin with partial knockout of ADP-ribosyltransferase activity. J Exp Med (1998) 187:1123–32. 10.1084/jem.187.7.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Haan L, Verweij W, Agsteribbe E, Wilschut J. The role of ADP-ribosylation and GM1-binding activity in the mucosal immunogenicity and adjuvanticity of the Escherichia coli heat-labile enterotoxin and Vibrio cholerae cholera toxin. Immunol Cell Biol (1998) 76:270–9. 10.1046/j.1440-1711.1998.00745.x [DOI] [PubMed] [Google Scholar]
- 16.Summerton NA, Welch RW, Bondoc L, Yang H-H, Pleune B, Ramachandran N, et al. Toward the development of a stable, freeze-dried formulation of Helicobacter pylori killed whole cell vaccine adjuvanted with a novel mutant of Escherichia coli heat-labile toxin. Vaccine (2010) 28:1404–11. 10.1016/j.vaccine.2009.10.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan EJ, McNeela E, Pizza M, Rappuoli R, O’Neill L, Mills KH. Modulation of innate and acquired immune responses by Escherichia coli heat-labile toxin: distinct pro- and anti-inflammatory effects of the nontoxic AB complex and the enzyme activity. J Immunol (2000) 165:5750–9. 10.4049/jimmunol.165.10.5750 [DOI] [PubMed] [Google Scholar]
- 18.Hajishengallis G, Arce S, Gockel CM, Connell TD, Russell MW. Immunomodulation with enterotoxins for the generation of secretory immunity or tolerance: applications for oral infections. J Dent Res (2005) 84:1104–16. 10.1177/154405910508401205 [DOI] [PubMed] [Google Scholar]
- 19.Brereton CF, Sutton CE, Ross PJ, Iwakura Y, Pizza M, Rappuoli R, et al. Escherichia coli heat-labile enterotoxin promotes protective Th17 responses against infection by driving innate IL-1 and IL-23 production. J Immunol (2011) 186:5896–906. 10.4049/jimmunol.1003789 [DOI] [PubMed] [Google Scholar]
- 20.Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ, et al. The global burden of dengue: an analysis from the Global Burden of Disease Study 2013. Lancet Infect Dis (2016) 16:712–23. 10.1016/S1473-3099(16)00026-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO. Dengue Vaccine: WHO Position Paper. (2016). Available from: http://www.who.int/wer/2016/wer9130.pdf
- 22.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell (2002) 108:717–25. 10.1016/S0092-8674(02)00660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinz FX, Stiasny K. Flaviviruses and their antigenic structure. J Clin Virol (2012) 55:289–95. 10.1016/j.jcv.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann B, Rossmann MG. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect (2011) 13:1–9. 10.1016/j.micinf.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol (2005) 3:13–22. 10.1038/nrmicro1067 [DOI] [PubMed] [Google Scholar]
- 26.Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, et al. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol (2007) 81:12816–26. 10.1128/JVI.00432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NTH, et al. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe (2010) 8:271–83. 10.1016/j.chom.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang C-Y, Hsieh C-H, Chen M-Y, Tsai J-P, Liu H-H, Liu S-J, et al. Recombinant lipidated dengue-4 envelope protein domain III elicits protective immunity. Vaccine (2014) 32:1346–53. 10.1016/j.vaccine.2014.01.041 [DOI] [PubMed] [Google Scholar]
- 29.Chiang C-Y, Huang M-H, Hsieh C-H, Chen M-Y, Liu H-H, Tsai J-P, et al. Dengue-1 envelope protein domain III along with PELC and CpG oligodeoxynucleotides synergistically enhances immune responses. PLoS Negl Trop Dis (2012) 6:e1645. 10.1371/journal.pntd.0001645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etemad B, Batra G, Raut R, Dahiya S, Khanam S, Swaminathan S, et al. An envelope domain III-based chimeric antigen produced in Pichia pastoris elicits neutralizing antibodies against all four dengue virus serotypes. Am J Trop Med Hyg (2008) 79:353–63. [PubMed] [Google Scholar]
- 31.Bernardo L, Pavon A, Hermida L, Gil L, Valdes I, Cabezas S, et al. The two component adjuvant IC31 potentiates the protective immunity induced by a dengue 2 recombinant fusion protein in mice. Vaccine (2011) 29:4256–63. 10.1016/j.vaccine.2011.03.040 [DOI] [PubMed] [Google Scholar]
- 32.Lok S-M, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol (2008) 15:312–7. 10.1038/nsmb.1382 [DOI] [PubMed] [Google Scholar]
- 33.Cockburn JJB, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, et al. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure (2012) 20:303–14. 10.1016/j.str.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 34.Li X-Q, Qiu L-W, Chen Y, Wen K, Cai J-P, Chen J, et al. Dengue virus envelope domain III immunization elicits predominantly cross-reactive, poorly neutralizing antibodies localized to the AB loop: implications for dengue vaccine design. J Gen Virol (2013) 94:2191–201. 10.1099/vir.0.055178-0 [DOI] [PubMed] [Google Scholar]
- 35.Poggianella M, Slon Campos JL, Chan KR, Tan HC, Bestagno M, Ooi EE, et al. Dengue E protein domain III-based DNA immunisation induces strong antibody responses to all four viral serotypes. PLoS Negl Trop Dis (2015) 9:e0003947. 10.1371/journal.pntd.0003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price JV, Jarrell JA, Furman D, Kattah NH, Newell E, Dekker CL, et al. Characterization of influenza vaccine immunogenicity using influenza antigen microarrays. PLoS One (2013) 8:e64555. 10.1371/journal.pone.0064555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentini D, Rao M, Rane L, Rahman S, Axelsson-Robertson R, Heuchel R, et al. Peptide microarray-based characterization of antibody responses to host proteins after bacille Calmette–Guérin vaccination. Int J Infect Dis (2017) 56:140–54. 10.1016/j.ijid.2017.01.027 [DOI] [PubMed] [Google Scholar]
- 38.Wumkes ML, van der Velden AMT, de Bruin E, Meerveld-Eggink A, Koopmans MPG, Rimmelzwaan GF, et al. Microarray profile of the humoral immune response to influenza vaccination in breast cancer patients treated with chemotherapy. Vaccine (2017) 35:1299–305. 10.1016/j.vaccine.2017.01.039 [DOI] [PubMed] [Google Scholar]
- 39.Stafford P, Cichacz Z, Woodbury NW, Johnston SA. Immunosignature system for diagnosis of cancer. Proc Natl Acad Sci U S A (2014) 111:E3072–80. 10.1073/pnas.1409432111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Donnell B, Maurer A, Papandreou-Suppappola A, Stafford P. Time-frequency analysis of peptide microarray data: application to brain cancer immunosignatures. Cancer Inform (2015) 14:219–33. 10.4137/CIn.s17285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legutki JB, Johnston SA. Immunosignatures can predict vaccine efficacy. Proc Natl Acad Sci U S A (2013) 110:18614–9. 10.1073/pnas.1309390110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Legutki JB, Magee DM, Stafford P, Johnston SA. A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine (2010) 28:4529–37. 10.1016/j.vaccine.2010.04.061 [DOI] [PubMed] [Google Scholar]
- 43.Hughes AK, Cichacz Z, Scheck A, Coons SW, Johnston SA, Stafford P. Immunosignaturing can detect products from molecular markers in brain cancer. PLoS One (2012) 7:e40201. 10.1371/journal.pone.0040201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sykes KF, Legutki JB, Stafford P. Immunosignaturing: a critical review. Trends Biotechnol (2013) 31:45–51. 10.1016/j.tibtech.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 45.Singh S, Stafford P, Schlauch KA, Tillett RR, Gollery M, Johnston SA, et al. Humoral immunity profiling of subjects with myalgic encephalomyelitis using a random peptide microarray differentiates cases from controls with high specificity and sensitivity. Mol Neurobiol (2016). 10.1007/s12035-016-0334-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnston SA, Thamm DH, Legutki JB. The immunosignature of canine lymphoma: characterization and diagnostic application. BMC Cancer (2014) 14:657. 10.1186/1471-2407-14-657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lasaro MA, Mathias-Santos C, Rodrigues JF, Ferreira LCS. Functional and immunological characterization of a natural polymorphic variant of a heat-labile toxin (LT-I) produced by enterotoxigenic Escherichia coli (ETEC). FEMS Immunol Med Microbiol (2009) 55:93–9. 10.1111/j.1574-695X.2008.00506.x [DOI] [PubMed] [Google Scholar]
- 48.Lasaro MA, Rodrigues JF, Mathias-Santos C, Guth BEC, Balan A, Sbrogio-Almeida ME, et al. Genetic diversity of heat-labile toxin expressed by enterotoxigenic Escherichia coli strains isolated from humans. J Bacteriol (2008) 190:2400–10. 10.1128/JB.00988-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry (1967) 6:1948–54. 10.1021/bi00859a010 [DOI] [PubMed] [Google Scholar]
- 50.Azevedo AS, Yamamura AMY, Freire MS, Trindade GF, Bonaldo M, Galler R, et al. DNA vaccines against dengue virus type 2 based on truncate envelope protein or its domain III. PLoS One (2011) 6:e20528. 10.1371/journal.pone.0020528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malyala P, Singh M. Endotoxin limits in formulations for preclinical research. J Pharm Sci (2008) 97:2041–4. 10.1002/jps.21152 [DOI] [PubMed] [Google Scholar]
- 52.Lasaro MAS, Rodrigues JF, Mathias-Santos C, Guth BEC, Régua-Mangia A, Piantino Ferreira AJ, et al. Production and release of heat-labile toxin by wild-type human-derived enterotoxigenic Escherichia coli. FEMS Immunol Med Microbiol (2006) 48:123–31. 10.1111/j.1574-695X.2006.00134.x [DOI] [PubMed] [Google Scholar]
- 53.Amorim JH, Diniz MO, Cariri FAMO, Rodrigues JF, Bizerra RSP, Gonçalves AJS, et al. Protective immunity to DENV2 after immunization with a recombinant NS1 protein using a genetically detoxified heat-labile toxin as an adjuvant. Vaccine (2012) 30:837–45. 10.1016/j.vaccine.2011.12.034 [DOI] [PubMed] [Google Scholar]
- 54.Kostner GM, Avogaro P, Bon GB, Cazzolato G, Quinci GB. Determination of high-density lipoproteins: screening methods compared. Clin Chem (1979) 25:939–42. [PubMed] [Google Scholar]
- 55.Halperin RF, Stafford P, Emery JS, Navalkar KA, Johnston SA. GuiTope: an application for mapping random-sequence peptides to protein sequences. BMC Bioinformatics (2012) 13:1. 10.1186/1471-2105-13-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L, Liu Y, Wang S, Sun J, Wang P, Xin Q, et al. Antiviral activity of peptide inhibitors derived from the protein E stem against Japanese encephalitis and Zika viruses. Antiviral Res (2017) 141:140–9. 10.1016/j.antiviral.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 57.Alhoot MA, Rathinam AK, Wang SM, Manikam R, Sekaran SD. Inhibition of dengue virus entry into target cells using synthetic antiviral peptides. Int J Med Sci (2013) 10:719–29. 10.7150/ijms.5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner D, de With K, Huzly D, Hufert F, Weidmann M, Breisinger S, et al. Nosocomial acquisition of dengue. Emerg Infect Dis (2004) 10:1872–3. 10.3201/eid1010.031037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis DJM, Huo Z, Barnett S, Kromann I, Giemza R, Galiza E, et al. Transient facial nerve paralysis (Bell’s palsy) following intranasal delivery of a genetically detoxified mutant of Escherichia coli heat labile toxin. PLoS One (2009) 4:e6999. 10.1371/journal.pone.0006999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mathias-Santos C, Rodrigues JF, Sbrogio-Almeida ME, Connell TD, Ferreira LCS. Distinctive immunomodulatory and inflammatory properties of the Escherichia coli type II heat-labile enterotoxin LT-IIa and its B pentamer following intradermal administration. Clin Vaccine Immunol (2011) 18:1243–51. 10.1128/CVI.00012-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diaz-Montero CM, Chiappori A, Aurisicchio L, Bagchi A, Clark J, Dubey S, et al. Phase 1 studies of the safety and immunogenicity of electroporated HER2/CEA DNA vaccine followed by adenoviral boost immunization in patients with solid tumors. J Transl Med (2013) 11:62. 10.1186/1479-5876-11-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clements JD, Freytag LC. Parenteral vaccination can be an effective means of inducing protective mucosal responses. Clin Vaccine Immunol (2016) 23:438–41. 10.1128/CVI.00214-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to vero cells. J Virol (2001) 75:7769–73. 10.1128/JVI.75.16.7769-7773.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Midgley CM, Flanagan A, Tran HB, Dejnirattisai W, Chawansuntati K, Jumnainsong A, et al. Structural analysis of a dengue cross-reactive antibody complexed with envelope domain III reveals the molecular basis of cross-reactivity. J Immunol (2012) 188:4971–9. 10.4049/jimmunol.1200227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wahala WMPB, Donaldson EF, De Alwis R, Accavitti-Loper MA, Baric RS, De Silva AM. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog (2010) 6:e1000821. 10.1371/journal.ppat.1000821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Costin JM, Zaitseva E, Kahle KM, Nicholson CO, Rowe DK, Graham AS, et al. Mechanistic study of broadly neutralizing human monoclonal antibodies against dengue virus that target the fusion loop. J Virol (2013) 87:52–66. 10.1128/JVI.02273-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodenhuis-Zybert IA, da Silva Voorham JM, Torres S, van de Pol D, Smit JM. Antibodies against immature virions are not a discriminating factor for dengue disease severity. PLoS Negl Trop Dis (2015) 9:e0003564. 10.1371/journal.pntd.0003564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erb SM, Butrapet S, Moss KJ, Luy BE, Childers T, Calvert AE, et al. Domain-III FG loop of the dengue virus type 2 envelope protein is important for infection of mammalian cells and Aedes aegypti mosquitoes. Virology (2010) 406:328–35. 10.1016/j.virol.2010.07.024 [DOI] [PubMed] [Google Scholar]
- 69.Hung J-J, Hsieh M-T, Young M-J, Kao C-L, King C-C, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. J Virol (2004) 78:378–88. 10.1128/JVI.78.1.378-388.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, Gazumyan A, et al. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell (2017) 169:597–609.e11. 10.1016/j.cell.2017.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tritto E, Muzzi A, Pesce I, Monaci E, Nuti S, Galli G, et al. The acquired immune response to the mucosal adjuvant LTK63 imprints the mouse lung with a protective signature. J Immunol (2007) 179:5346–57. 10.4049/jimmunol.179.8.5346 [DOI] [PubMed] [Google Scholar]
- 72.Olafsdottir TA, Hannesdottir SG, Giudice GD, Trannoy E, Jonsdottir I. Effects of LT-K63 and CpG2006 on phenotype and function of murine neonatal lymphoid cells. Scand J Immunol (2007) 66:426–34. 10.1111/j.1365-3083.2007.01970.x [DOI] [PubMed] [Google Scholar]
- 73.Ji J, Griffiths KL, Milburn PJ, Hirst TR, O’Neill HC. The B subunit of Escherichia coli heat-labile toxin alters the development and antigen-presenting capacity of dendritic cells. J Cell Mol Med (2015) 19:2019–31. 10.1111/jcmm.12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skalickova S, Heger Z, Krejcova L, Pekarik V, Bastl K, Janda J, et al. Perspective of use of antiviral peptides against influenza virus. Viruses (2015) 7:5428–42. 10.3390/v7102883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panya A, Bangphoomi K, Choowongkomon K, Yenchitsomanus P. Peptide inhibitors against dengue virus infection. Chem Biol Drug Des (2014) 84:148–57. 10.1111/cbdd.12309 [DOI] [PubMed] [Google Scholar]
- 76.Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog (2010) 6:e1000851. 10.1371/journal.ppat.1000851 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.