Abstract
Plant-specific NAC proteins comprise one of the largest transcription factor families in plants and play important roles in plant development and the stress response. Gossypium hirsutum L. is a major source of fiber, but its growth and productivity are limited by many biotic and abiotic stresses. In this study, the NAC domain gene GhNAC79 was functionally characterized in detail, and according to information about the cotton genome sequences, it was located on scaffold42.1, containing three exons and two introns. Promoter analysis indicated that the GhNAC79 promoter contained both basic and stress-related elements, and it was especially expressed in the cotyledon of Arabidopsis. A transactivation assay in yeast demonstrated that GhNAC79 was a transcription activator, and its activation domain was located at its C-terminus. The results of qRT-PCR proved that GhNAC79 was preferentially expressed at later stages of cotyledon and fiber development, and it showed high sensitivity to ethylene and meJA treatments. Overexpression of GhNAC79 resulted in an early flowering phenotype in Arabidopsis, and it also improved drought tolerance in both Arabidopsis and cotton. Furthermore, VIGS-induced silencing of GhNAC79 in cotton led to a drought-sensitive phenotype. In summary, GhNAC79 positively regulates drought stress, and it also responds to ethylene and meJA treatments, making it a candidate gene for stress studies in cotton.
Keywords: GhNAC79, cotton, stress, drought, development
Introduction
As a major source of fiber, cotton (Gossypium hirsutum L.) shows greater tolerance to drought and salt than wheat and rice, but with the changing of global climate and increasing level of pollution, abiotic stress is becoming a major limiting factor for cotton growth and productivity (Ahuja et al., 2010). Among these factors, high salinity and drought are the main stresses (Wang et al., 2001; Rabbani et al., 2003); currently, more than 10% of arable land is experiencing water shortages, leading to a 50% average reduction in the yield of major crops (Bartels and Sunkar, 2005). Senescence, which consists of a variety of molecular and physiological events, is the last stage of leaf development, and it positively impacts plant reproduction. However, premature senescence negative affects cotton yield and quality. Therefore, improving stress tolerance and delaying leaf senescence in cotton through genetic engineering is a promising production strategy for which candidate genes need to be identified.
NAC proteins are plant-specific transcription factors characterized by a conserved NAC domain (Christianson et al., 2009), whose name was originally derived from the names of proteins containing NAM (no apical meristem), ATAF1/2 and CUC2 (cup-shaped cotyledon). NAM is required for SAM formation during embryogenesis (Souer et al., 1996); ATAF1/2 has dual functions in both abiotic and biotic stress responses (Lu et al., 2006; Jensen et al., 2008); and CUC2 is involved in organ separation (Aida et al., 1997). Recent studies have reported that NAC transcription factors play diverse roles in a variety of stress responses (Fujita et al., 2004; He et al., 2005; Seo et al., 2010) and developmental processes (Xie et al., 2000; Kim et al., 2008), and some are important in the development of roots and floral organs (Ishida et al., 2000; Xie et al., 2000; He et al., 2005), the development of secondary cell walls and lignin (Chai et al., 2015; Xu et al., 2015), fruit ripening and carotenoid accumulation (Xu et al., 2015; Zhou et al., 2015), leaf senescence (Wu et al., 2012; Zhao et al., 2015), and cell death (Wang et al., 2015). Furthermore, some NAC members are involved in the responses to abiotic and biotic stresses, such as drought, salinity, and cold (Li et al., 2014; Wang et al., 2014; Yu X. et al., 2014; Yang et al., 2015), as well as pathogen attack and wounds (Wu et al., 2009; Seo et al., 2010).
Since the publication of the cotton genome sequences (Paterson et al., 2012; Yu J. et al., 2014; Li F. et al., 2015; Zhang et al., 2015), more NACs (GhNACs) have been studied in detail. There are more than 100 NAC members in cotton, and they have been shown to have a variety of functions (Balazadeh et al., 2010; Kim et al., 2011; Matallana-Ramirez et al., 2013; Qiu et al., 2015) such as in development and in responses to biotic and abiotic stresses (Shah et al., 2013, 2014). In cotton, an NAC gene, GhXND1, negatively regulates xylem development (Li et al., 2014); GhNAC12 promotes leaf senescence (Zhao et al., 2016); and SNAC1 improves drought and salt resistance (Liu et al., 2014). Therefore, the potential functions of GhNACs in cotton development and stress responses should be thoroughly studied.
Our study mainly describes an NAC domain gene, GhNAC79, that is predominantly expressed in cotyledons and fibers. It promotes flowering in Arabidopsis and enhances drought tolerance in both Arabidopsis and cotton, and it also responds to drought and different plant hormones, such as ethylene and ABA. This work not only complements previous functional studies of GhNACs in cotton but also provides important material for cotton breeding for drought resistance.
Materials and Methods
Plant Materials, Growth Conditions and Stress Treatments
The cotton cultivar CCRI10 (premature senescence) was used for gene cloning and subjected to different treatments. Healthy and uniform seeds were selected and cultivated in a culture room at 25°C with a 16-h light/8-h dark cycle. Arabidopsis thaliana ecotype Col-0 was used as the parent strain for Arabidopsis transformation. Surface-sterilized seeds were sown in 1/2 MS medium and incubated at 4°C for 3 days in dark conditions to break dormancy, and the plants were then cultivated at 22°C with a 16-h light/8-h dark cycle and a relative humidity of approximately 80%.
Cotton was subjected to drought and salinity treatments in three ways. First, detached true leaves were submerged in water for 1 day and then transferred into a 20% PEG6000 or 200 mM NaCl solution, and controls were submerged in water. Samples were collected at 0, 2, 4, 6, 8, 12, and 24 h. Second, roots were submerged in a 20% PEG6000 or 200 mM NaCl solution, and the same amount of water was used for the control. The leaves were collected at 0 Day post anthesis (DPA), 5, 10, 15, and 20 Days. Third, healthy seeds were germinated in sealed glass jars containing either 200 mM mannitol or 200 mM NaCl, and the leaves and roots were collected separately once the seedlings showed an obvious phenotype. The control consisted of only the medium.
Two plant hormone treatments were carried out. First, seedlings at the three-leaf stage were sprayed with 50 μM ABA, 200 μM meJA or 200 μM ethephon (ethephon releases ethylene when dissolved in water), and controls were sprayed with water. The leaf samples were collected at 0, 4, 12, 24, and 48 h after treatment. Second, 25 μM ABA, 100 μM meJA and 100 μM ethephon were individually applied to the sealed glass jars, and then the leaf and root samples were collected separately. The control consisted of only MS medium.
The cotyledons, true leaves, stems, roots and flowers of cotton seedlings were collected for gene expression analyses. Fibers at 0, 5, 10, 15, 20, and 25 DPA were obtained as described previously (Singh et al., 2009). To evaluate the function of GhNAC79 during cotyledon development, two short-season cotton cultivars, CCRI10 (premature senescence) and Liao4086 (no premature senescence), were selected and cultivated under normal water and nitrogen management. Cotyledon samples were collected every 7 days in each of eight developmental stages from the non-senescent stage to the completely senescent stage. There were three biological replicates for each sample.
Malondialdehyde (MDA) and Soluble Protein Measurements
The enzymatic solution was prepared as follows: 0.5 g samples were quickly ground in a cold mortar with 8 μl of cold extraction medium (0.05 mol/L, pH = 7.8, Na2HPO4-NaH2PO4), and all the liquid was then transferred into a 25 ml centrifuge tube and centrifuged at 20,000 rpm for 30 min at 2°C. The supernatants were collected for further use as the enzymatic solution.
The MDA content was determined according to the method described by Jingqing Zhao et al. (2012) with slight modification. A mixture of 1.5 ml of buffer (0.05 mol/L, pH = 7.8, Na2HPO4-NaH2PO4) and 1.5 ml of enzymatic solution was placed in a 10 ml centrifuge tube (the control consisted of 1.5 ml of buffer and 1.5 ml of H2O), and 2.5 ml of 0.5% thiobarbituric acid (TBA) was then added to each tube. Finally, the reaction mixture was mixed well, incubated at 100°C for 20 min, and then quickly put on ice. After centrifugation at 1800 g for 10 min, the absorbances of the supernatant at 450, 532, and 663 nm were determined with a spectrophotometer. The soluble protein was determined according to the method described by Bradford (1976).
RNA Extraction, cDNA Synthesis and DNA Preparation
Total RNA was extracted with an RNAprep Pure kit (Tiangen, China), which was suitable for plants rich in polyphenols and amylase. The quality and concentration of the RNA were confirmed by 1% agarose gel electrophoresis and a spectrophotometer, and DNaseI was used to remove the genomic DNA. cDNA synthesis was performed by strictly following the manufacturer’s protocol for ReverTra Ace qPCR RT Master Mix (TOYOBO, Japan). Total genomic DNA was extracted from the cotton leaf tissues by the CTAB method (Song et al., 1998).
Gene Cloning and Sequence Analysis
The primers used for gene cloning, qRT-PCR, VIGS and promoter cloning were designed with OLIGO7, as shown in Table 1. The conserved domain of GhNAC79 was searched for in NCBI1. Using the G. hirsutum genome database (Zhang et al., 2015), GhNAC79 was pitched at scaffold42.1, and the intron and exon structure was analyzed by comparing the genomic and coding sequences. At the same time, an unrooted phylogenetic tree was constructed with 126 AtNACs by MEGA 5.1 (Tamura et al., 2011).
Table 1.
Primers for gene and promoter cloning, qRT-PCR and VIGS.
| Name | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) | PCR product size (bp) |
|---|---|---|---|
| GhNAC79 | AGACTGGCATGAATAAACAAG | AGCTTCTCCATCTACACATCA | 759 |
| qRT-PCR-GhNAC79 | AATCACATTAATCGGTTTACT | CTAAAGATTCCAAAACCCATC | 117 |
| VIGS-GhNAC79 | CTACACCTCCTATCCTAAGAC | AGCTTCTCCATCTACACAT | 272 |
| PGhNAC79 | GTGATGTGTAAAACAGTCTAT | TTCTTCAGTAGGATAGAACCG | 1031 |
The GhNAC79 primer was used for gene cloning; the qRT-PCR-GhNAC79 primer was used for fluorescent qRT-PCR; the VIGS-GhNAC79 primer was used for VIGS; and the PGhNAC79 primer was used for promoter cloning.
Transcriptional Activation Activity Analysis
According to the domain features of GhNAC79, the entire open reading frame (ORF) was divided into four parts, which were inserted into the pGBKT7 vector with EcoRI and BamHI. After sequencing with the T7 universal primer, all the constructed plasmids were transferred into a yeast strain (Y2H) by strictly following the Clontech method2. The positive yeast strain was selected using SD/-Trp/X-α-gal/25 mM 3-AT medium, and PCR was used to verify the results. The primers were designed based on the sequences of the four fragments and are shown in Table 2.
Table 2.
Primers for the transactivation assay in yeast.
| Name | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) | PCR product size (bp) |
|---|---|---|---|
| ADfra-1 | CATGAATAAACAAGTATAGGC | CATGGTCTTGAATAGCTTTAT | 462 |
| ADfra-2 | CATGAATAAACAAGTATAGGC | TTCTCCCATCTTTAATATCCC | 590 |
| ADfra-3 | GACCATGTTTCTTTAGCTAAT | CATCTACACATCATCTCTAAA | 289 |
| ADfra-4 | CATGAATAAACAAGTATAGGC | CATCTACACATCATCTCTAAA | 744 |
Promoter Cloning and Analysis
For promoter cloning, approximately 1.0 kb sequences of the 5′ UTR were isolated based on the information from the genome database. Using XbaI and ScaI cutting sites, the promoter was inserted into the pBI121::GUS expression vector and transferred into Arabidopsis through Agrobacterium tumefaciens strain LBA4404 with GUS as the reporter. At the same time, PlantCARE3 was used to predict the important elements in the promoter.
Histochemical staining for GUS activity was conducted by incubating fresh Arabidopsis tissues in the GUS staining solution (Jefferson et al., 1987) with the wild type as the control. After one night of incubation at 37°C, the stained tissues were bleached with ethanol ranging from a high concentration to a low concentration until the wild type became white. Tissues were then photographed directly under a stereomicroscope.
Quantitative RT-PCR (qRT-PCR) Assays
The primer was designed with OLIGO7 according to the C-terminus of the sequence, and the specificity of the primer was ensured by the melting peaks and dissociation curves. The reactions were performed with Go Taq qPCR Master Mix (Promega, United States) on an ABI7500 instrument (Applied Biosystems, United States) with GhHis3 as the endo-reference gene (Tu et al., 2007). A total reaction volume of 20 μl was used for qRT-PCR as follows: 2 μl of diluted cDNA, 1.0 μl of both the forward and reserve primers, 6 μl of sterile H2O, and 10 μl of Go Taq qPCR Master Mix. The running procedure was strictly established as described in the manual, and each collected sample contained three technical replicates. The entire operation was performed under low-light conditions, and the results were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001), where ΔCt1 = Ct(GhNAC79)-Ct(GhHis3) and ΔCt2 = Ct1(GhNAC79)-Ct1(GhNAC79of control). Statistical significance was calculated by analysis of variance (ANOVA) or t-tests using SAS software (Knapp et al., 1985).
Transformation of Arabidopsis and Cotton
The 35S::GhNAC79 plasmid was constructed by inserting the coding region into a binary vector, pBI121, with XbaI and SacI cutting sites. After sequencing, the 35S::GhNAC79 plasmid was transformed into A. tumefaciens strain LBA4404 by the heat shock method (Bhuiyan et al., 2011), and Arabidopsis was transfected with an improved inflorescence-dip method (Clough and Bent, 1998). The T0 transgenic plants were germinated on 1/2 MS medium with kanamycin. After 14 days, the green plants were transferred to nutritive soil in a culture room, and PCR was used to analyze the positive plants.
The transformation of cotton was completed at the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences (CAAS). Seeds of CCRI24 (an upland cotton cultivar) were sterilized and germinated on 1/2 MS medium at 28°C in a culture room, and hypocotyls cut from sterile seedlings served as the transformation receptors. The construction of 35S::GhNAC79 was transferred into cotton via Agrobacterium (Zhang, 2008). The positive cotton lines were verified with PCR and kanamycin, and the T1 generations of the GhANC79-overexpression lines were used for further analysis. The primer used for PCR was designed based on the 35S promoter and the GhNAC79 sequence, as shown in Supplementary Table S1.
Modified Virus-Induced GhNAC79 Silencing in Cotton
To improve the robustness of the virus-induced cotton GhNAC79 silencing results, we used two silencing systems: pCLCrVA-pCLCrVB and pYL156-pYL192. Approximately 200 bp of the GhNAC79 sequence was inserted into the pCLCrVA vector with SpeI and AscI cutting sites and pCLCrVB as the helper vector (Gu et al., 2014). pCLCrVA::GhNAC79 and pCLCrVB were transferred into cotton cotyledons through Agrobacterium as previously described (Kumagai et al., 1995) with pLCrVA::PDS as the indicator and pLCrVA-infected plants as the negative control. At the same time, 200 bp of the GhNAC79 sequence was inserted into the pYL156 vector with EcoRI and BamHI with pYL192 as the helper vector, pYL156::PDS as the indicator, and pYL156-infected plants as the negative control (Unver and Budak, 2009). The cotton used for infection was cultivated at 22°C until the cotyledons flattened, and the infected cotton seedlings were grown at 22°C under low humidity.
Results
Cloning and Characterization of GhNAC79
GhNAC79 (KU963586, unpublished) was cloned from a senescent cotyledon of G. hirsutum. According to the genome information, GhNAC79 was located on scaffold42.1 and contained 3 exons and 2 introns. It further contained a conserved NAM domain and encoded a protein consisting of 231 amino acids with a molecular weight of 26.972 kDa.
The GhNAC79 sequence was divided into four fragments. The first fragment was from 23 to 485 bp and contained the conserved NAM domain. The second fragment was slightly longer than the first fragment, from 23 to 613 bp, which was selected to verify the definite transcriptional activation domain. The third fragment was from 479 to 767 bp, which avoided the conserved domain. Finally, the fourth fragment contained the complete ORF. The constructed plasmids pGBKT7-fra1, pGBKT7-fra2, pGBKT7-fra3, and pGBKT7-fra4 were transformed into Y2H, and the results are shown in Figure 1. The positive control stains (pGBKT7-p53 + pGADT7-largeT) appeared blue in color on X-α-Gal substrate, whereas the negative control (pGBKT7-laminC + pGADT7-largeT) exhibited no change in color. The GhNAC79 fragments, except pGBKT7-fra1, appeared blue in color, which indicated that GhNAC79 functioned as a transcriptional activator and that its transactivation domain was located at the C-terminus. To ensure the accuracy of the results, all of the stains were affirmed by PCR.
FIGURE 1.
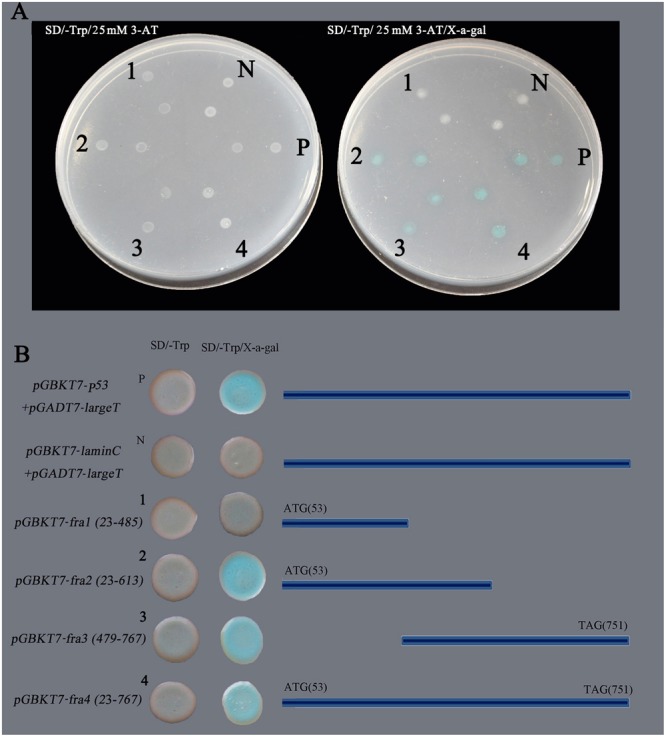
Transcriptional activation activity analysis of GhNAC79. The GhNAC79 sequence was divided into four fragments, which are marked with 1–4 respectively. (A) The phenotype of yeast strains growing on SD/-Trp/25 mM 3-AT or SD/-Trp/25 mM 3-AT/x-a-gal media. (B) The sequence information for different fragments. N/pGBKT7-LaminC + pGADT7-LargeT: negative control; P/pGBKT7-53 + pGADT7-LargeT: positive control.
The Expression Patterns of GhNAC79 in Special Tissues
To detect the special expression patterns of GhNAC79 in different tissues, the roots, cotyledons, stems, true leaves, flowers and fiber at different stages were collected. As shown in Figure 2A, GhNAC79 was mainly expressed in the cotyledon and later stages of fiber development, such as 20 and 25 DPA. In contrast, the expression level was relatively low in the stem and the initial fiber developmental stage.
FIGURE 2.
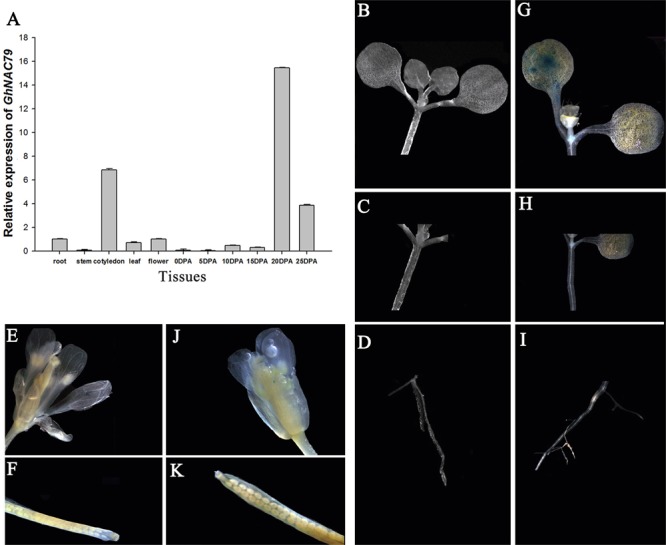
The expression patterns of GhNAC79 in special tissues. (A) Samples of roots, stems, cotyledons, true leaves, flowers, and different developmental fibers were collected, and qRT-PCR was conducted to explore the expression patterns of GhNAC79. Data are shown as the mean ± SD (n = 3). GhHIS3 was used as the reference gene. (B–K) The GhNAC79 promoter was transformed into Arabidopsis with GUS as the indicator, and photographs were taken under a stereomicroscope. (B–F) Different tissues of wild type Arabidopsis. (G–K) Different tissues of transgenic Arabidopsis.
The GhNAC79 promoter was cloned, and the sequenced promoter contained parts of the ORF sequences, which demonstrated consistency from the ORF to 5′ UTR. Based on the PlantCARE analysis, the 1.0 kb sequences contained all basal elements, such as the TATA box, CAAT box and 5 UTR Py-rich stretch. Additionally, the promoter contained some stress- and regulation-related cis-elements, such as those related to light, circadian rhythm, drought and ethylene, as shown in Table 3. The reconstructed vector promoterGhNAC79::GUS was transferred into Arabidopsis, and histochemical staining was conducted to detect the GUS activity. As shown in Figures 2B–F, there was no GUS activity in the wild type, while GUS activity was preferentially detected in the cotyledons and cotyledon bases in transgenic Arabidopsis, which is presented in Figures 2G–K.
Table 3.
Stress- and regulation-related cis-elements identified in the GhNAC79 promoter.
| Site name | Species | Position | Sequence | Function |
|---|---|---|---|---|
| AT1-motif | Solanum tuberosum | +42 | ATTAATTTTACA | Part of a light-responsive module |
| Box 4 | Petroselinum crispum | +42/-648/-782/-893 | ATTAAT | Part of a conserved DNA module involved in light responsiveness |
| HSE | Brassica oleracea | +125/+126/+495 | AAAAAATTTC | cis-acting element involved in heat stress responsiveness |
| Skn-1_motif | Oryza sativa | –172/+262/-453 | GTCAT | cis-acting regulatory element required for endosperm expression |
| MBS | Arabidopsis thaliana | –232/+615/+916 | TAACTG/ CAACTG | MYB-binding site involved in the induction of drought stress |
| ACE | Petroselinum crispum | +465 | AAAACGTTTA | cis-acting element involved in light responsiveness |
| GT1-motif | Avena sativa | –514 | GGTTAAT | Light-responsive element |
| GT1-motif | Arabidopsis thaliana | –515 | GGTTAA | Light-responsive element |
| GARE-motif | Brassica oleracea | +626 | AAACAGA | Gibberellin-responsive element |
| circadian | Lycopersicon esculentum | +665 | CAANNNNATC | cis-acting regulatory element involved in circadian control |
| TCT-motif | Arabidopsis thaliana | –701/+840 | TCTTAC | Part of a light-responsive element |
| TC-rich repeats | Nicotiana tabacum | +836 | GTTTTCTTAC | cis-acting element involved in defense and stress responsiveness |
| GT1-motif | Solanum tuberosum | +867 | AATCCACA | Light-responsive element |
| CCAAT-box | Hordeum vulgare | –877 | CAACGG | MYBHv1-binding site |
| Sp1 | Zea mays | +887 | CC(G/A)CCC | Light-responsive element |
| LTR | Hordeum vulgare | –1007 | CCGAAA | cis-acting element involved in low-temperature responsiveness |
The promoter of GhNAC79-induced GUS was especially expressed in the cotyledon, which was consistent with the expression patterns of GhNAC79 in cotton tissues. The high expression level of GhNAC79 in the cotyledon and fiber indicated its special function in these tissues.
GhNAC79 Was Also Significantly Upregulated in Later Cotyledon Development Stages
During development, the color of the cotyledon became yellow in both cultivars (Figure 3A), and the MDA content sharply increased in CCRI10 but was relatively steady in Liao4086, as shown in Figure 3B. As a product of membrane lipid peroxidation, MDA is an important indicator of cell damage (Kramer et al., 1991), and the high level of MDA in CCRI10 indicated premature senility. In terms of soluble protein, the two cultivars did not differ greatly, and the soluble protein contents of both decreased during leaf senescence, as shown in Figure 3C.
FIGURE 3.
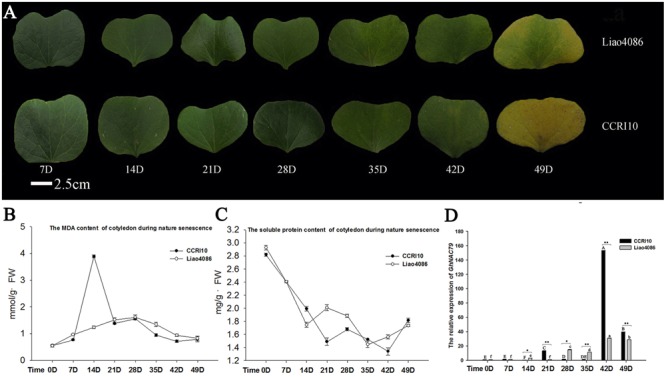
Expression patterns of GhNAC79 during cotyledon development. (A) Seven cotyledon developmental stages. Bar = 2.5 cm. (B,C) Changes in MDA and soluble protein contents during cotyledon development. D indicates the number of days after the cotyledon had spread out. (D) Expression patterns of GhNAC79 during cotyledon development. One-way ANOVA was based on varying the times for the two varieties. Different letters indicate a significant difference between two values (p < 0.01); capital letters are used for CCRI10 and lowercase for Liao4086. A t-test was conducted between two varieties at the same time point. ∗Values between two varieties are significantly different at the 0.05 confidence level; ∗∗Values between two varieties are significantly different at the 0.01 confidence level. Data are presented as the mean ± SD (n = 3). GhHIS3 was used as the reference gene.
Figure 3D shows that GhNAC79 was very highly expressed in CCRI10 and Liao4086 in later cotyledon development stages, and the highest changes in expression level were approximately 170-fold in CCRI10 and 40-fold in Liao4086. The expression of GhNAC79 was significantly higher in CCRI10 than in Liao4086. As CCRI10 was more sensitive to senescence than Liao4086, the higher expression of GhNAC79 in CCRI10 indicated that it might function in cotyledon senescence.
GhNAC79 Was Induced by Abiotic Stresses
To explore the role of GhNAC79 in response to different stresses, cotton seeds were sown in sealed glass jars containing MS medium subjected to different treatments. As shown in Figures 4A,B, GhNAC79 was induced in leaves by the meJA and drought treatments, but its expression in roots was repressed in all treatments, especially the meJA, ethylene and ABA treatments, by approximately 5∼10 fold. These results indicated that GhNAC79 might be involved in responses to abiotic stresses.
FIGURE 4.
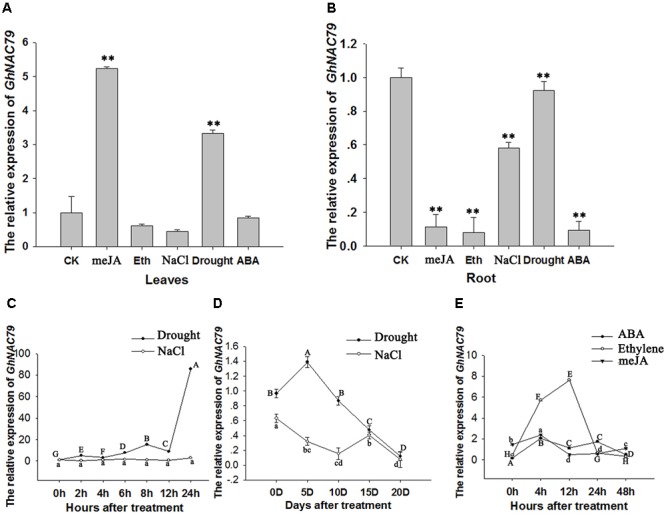
The expression patterns of GhNAC79 in response to stresses and plant hormones. (A,B) Cotton seedlings were cultivated in sealed glass bottles containing MS medium with different treatments. After an obvious phenotype appeared, root and leaf samples were collected separately. (A) Expression patterns of GhNAC79 in response to different stresses and plant hormones in leaves. (B) Expression patterns of GhNAC79 in response to different stresses and plant hormones in roots. (C,D) Expression patterns of GhNAC79 in response to drought and salt treatments. (C) Detached leaves were submerged in 20% PEG6000 or 200 mM NaCl, and samples were collected at different time points with h indicating the number of hours after treatment. (D) Roots of cotton seedlings were submerged in 20% PEG6000 or 200 mM NaCl, and samples were collected at different time points with D indicating the number of days after treatment. (E) Expression patterns of GhNAC79 in response to ABA, meJA and ethylene treatments with h indicating the number of hours after treatment. Data are presented as the mean ± SD (n = 3). GhHIS3 was used as the reference gene.
To verify the response of GhNAC79 to drought and salt, detached cotton leaves were submerged in 20% PEG6000 or 200 mM NaCl. Figure 4C shows that GhNAC79 was sharply induced at 24 h after drought treatment, whereas GhNAC79 remained constant under salt treatment. At the same time, after submerging the roots of cotton seedlings in 20% PEG6000 or 200 mM NaCl, GhNAC79 exhibited an expression pattern indicating sensitivity to drought treatment, as shown in Figure 4D. In terms of the response to different plant hormones, GhNAC79 was very sensitive to ethylene, but its response to ABA and meJA remained relatively steady, as illustrated in Figure 4E.
Overexpression of GhNAC79 in Arabidopsis Resulted in an Early Flowering Phenotype
35S::GhNAC79 was structured (Figure 5A) and transformed into Arabidopsis with selective 1/2 MS medium, and the results were verified by PCR. The positive plants were selected and grown in a culture room, and the T4-generation transgenic plants were used for phenotype analysis. At the transcriptional level, the expression of GhNAC79 in the four lines was higher than that of the wild type by approximately 5∼80 fold, as shown in Figure 5B. Transgenic Arabidopsis also exhibited an early flowering phenotype, which is shown in Figure 5C, and the flowering time was approximately 5 days earlier than the wild type, as presented in Table 4.
FIGURE 5.
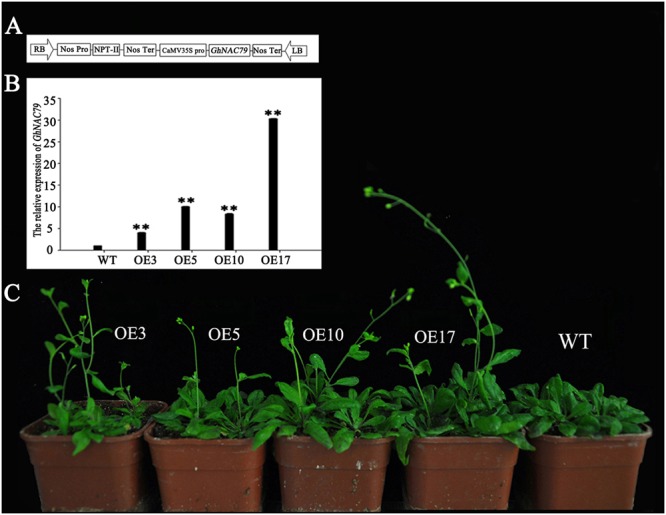
Phenotypes of transgenic Arabidopsis. (A) Construction of 35S::GhNAC79 vector. (B) Expression level of GhNAC79 in transgenic Arabidopsis and wild type. Data are presented as the mean ± SD (n = 3) with GhHIS3 as the reference gene. ∗∗Values significantly different from wild type at the 0.01 confidence level. (C) An early flowering phenotype of transgenic Arabidopsis compared with wild type; Line 3, Line 5, Line 10, and Line 17 were the four lines of transgenic plants.
Table 4.
Comparison of the flowering time for GhNAC79-transgenic Arabidopsis.
| Genotypea | Anthesis (DAS)b | N |
|---|---|---|
| Wild type | 33.48 ± 1.52 | 40 |
| OE3 | 28.35 ± 1.35∗∗ | 40 |
| OE5 | 29.73 ± 1.73∗∗ | 40 |
| OE10 | 29.20 ± 1.08∗∗ | 40 |
| OE17 | 28.78 ± 1.22∗∗ | 40 |
Plants were grown under a 16-h light/8-h dark cycle. ∗∗Values significantly different from Col-0 at the 0.01 confidence level. aGenetic backgrounds: wild type, and OE3, OE5, OE10, and OE17, which were the four lines of transgenic Arabidopsis.bIndicators of anthesis (days after sowing) and data are shown as the mean ± standard deviation (SD). N represents the number of plants used for analysis.
Overexpression of GhNAC79 in Arabidopsis Enhanced Drought Tolerance
To explore the function of GhNAC79 during drought stress, transgenic and wild type Arabidopsis were treated with 20% PEG6000 after 20 days of sowing. Under normal water management, all plants reached the vegetable developmental stage, and these results are shown in Figure 6A. After drought treatment, transgenic Arabidopsis began bolting, but the wild type was still at the vegetable stage, as shown in Figure 6B. Additionally, transgenic Arabidopsis and wild type were treated with 20% PEG6000 after 14 days of sowing. After 3 days of drought stress, wild type exhibited obvious wilting, while the transgenic Arabidopsis remained fresh. Furthermore, the stomatal aperture of transgenic Arabidopsis was smaller than that of the wild type, as shown in Figures 6C,D.
FIGURE 6.
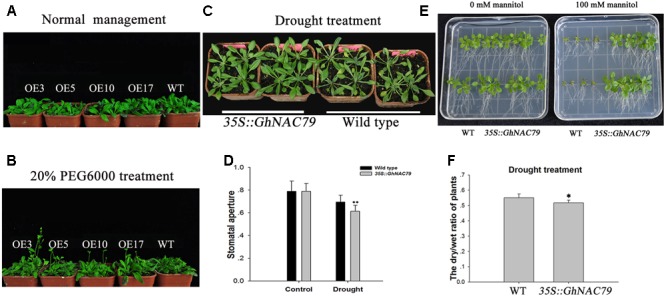
Overexpression of GhNAC79 in Arabidopsis enhanced drought tolerance. (A,B) Plants were treated with 20% PEG6000 at 20 days after sowing; wild type represents the wild type, and OE3, OE5, OE10, and OE17 represent the four lines of transgenic Arabidopsis. (A) Phenotypes of transgenic Arabidopsis and wild type under normal management. (B) Phenotypes of all plants after drought treatment. (C,D) Transgenic plants and wild type were treated with 20% PEG6000 at 14 days after sowing. (C) Phenotypes of transgenic plants and wild type. (D) Stomatal aperture of transgenic Arabidopsis and wild type. (E) Phenotypes of transgenic Arabidopsis and wild type after treatment with 100 mM mannitol in 1/2 MS medium. (F) Differences in dry/wet ratios between transgenic plants and the wild type after mannitol treatment. ∗Values significantly different from wild type at the 0.05 confidence level. ∗∗Values significantly different from wild type at the 0.01 confidence level.
Transgenic Arabidopsis and wild type were germinated in 1/2 MS medium containing 100 mM mannitol, and after approximately 15 days, the transgenic plants displayed an obvious phenotype. As shown in Figure 6E, the transgenic plants were stronger than the wild type, and their roots were longer. The dry/wet ratio of the transgenic plants was significantly lower than that of the wild type, as visualized in Figure 6F, and the high dry/wet ratio in the wild type implied relatively high water loss. The enhanced drought tolerance in Arabidopsis indicated the positive role of GhNAC79 in drought response.
GhNAC79 Played a Positive Role in Drought Stress in Cotton
To study the function of GhNAC79 in cotton during drought stress, two virus-induced gene silencing (VIGS) systems were used (pCLCrVA-pCLCrVB and pYL156-pYL192). After injection, cotton seedlings were covered with black boxes for one night and then grown in a culture room. As the leaves of the indicator plants exhibited an albino phenotype, the expression level of GhNAC79 was detected by qRT-PCR. In the pCLCrVA-pCLCrVB system, the expression level of GhNAC79 was 0.2∼0.6% lower in the infected plants compared with the control (pCLCrVA vector-infected plants), as shown in Figure 7A. After treatment with 20% PEG6000, the infected plants showed severe wilting, and all leaves were soft and drooping. However, the young leaves of the control plants remained fresh, as shown in Figure 7B. For the pYL156-pYL192 system, the expression of GhNAC79 decreased by 0.2∼0.9% relative to the control plants (pYL156 vector-infected plants), and after 20% PEG6000 treatment, the infected plants showed more wilting than the control, as shown in Figures 7C,D.
FIGURE 7.
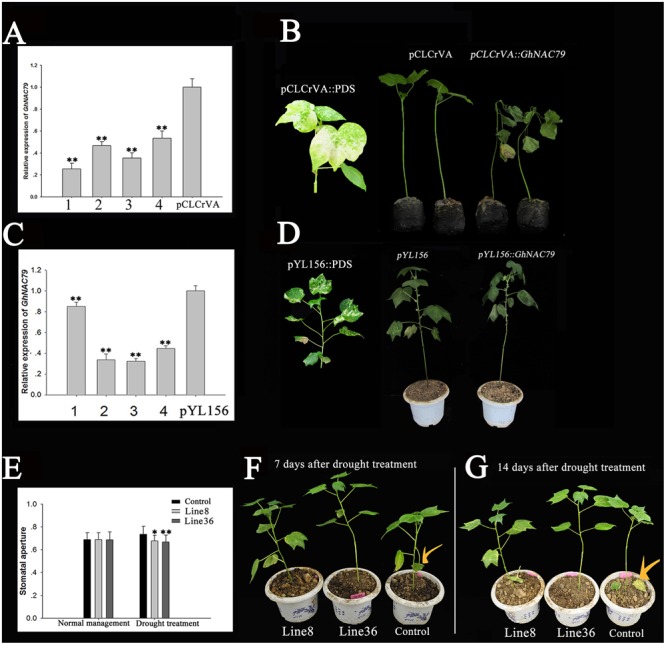
Overexpression of GhNAC79 enhanced drought tolerance in cotton. (A,B) pCLCrVA-pCLCrVB system. (A) The expression of GhNAC79 in virus-infected plants and the control (pCLCrVA). All virus-infected plants were divided into four groups and marked 1–4. (B) After drought treatment, the phenotypes of virus-infected cotton seedling and the control with pCLCrVA::PDS as the indicator, pCLCrVA as the control, and pCLCrVA::GhNAC79 representing the virus-infected cotton seedlings. (C,D) pYL156-pYL192 system. (C) The expression level of GhNAC79 in virus-infected plants and the control (pYL156). All virus-infected plants were divided into four groups and marked 1–4. (D) After drought treatment, the phenotypes of virus-infected cotton seedlings and the control with pYL156::PDS as the indicator, pYL156 as the control, and pYL156::GhNAC79 representing the virus-infected cotton seedlings. (E–G) The phenotype of cotton overexpressing GhNAC79 at the T1 stage. Line 8 and Line 36 were two lines of transgenic cotton, and the control was CCRI24 (a cotton cultivar used as a transgenic recipient). (E) Stomatal movement of transgenic cotton and control. (F,G) After drought treatment, transgenic cotton showed higher drought resistance compared with the control. (F) Seven days after drought treatment; (G) 14 days after drought treatment. ∗Values significantly different from the control at the 0.05 confidence level. ∗∗Values significantly different from the control at the 0.01 confidence level. Data are presented as the mean ± SD (n = 3). GhHIS3 was the reference gene.
To analyze the overexpression of GhNAC79 in different cotton lines, the roots of two transgenic cotton lines and a wild type (CCRI24) were submerged in 20% PEG6000, and the results are shown in Figures 7E–G. After 7 days of treatment, the leaves of the wild type appeared wilted or had dropped, while the transgenic cotton showed normal growth. After 14 days, the wild type had dropped three true leaves, but the two transgenic lines had dropped one leaf and one cotyledon and had another unhealthy cotyledon. Furthermore, there was no significant difference in stoma number and the size of the stomatal aperture between the transgenic lines and the wild type under normal water management, but after drought treatment, the stomatal apertures were smaller in the two transgenic lines compared to the wild type.
Two VIGS-induced GhNAC79 silencing systems resulted in the cotton being more sensitive to drought, while overexpression of GhNAC79 increased the drought tolerance of cotton, which demonstrated the positive role of GhNAC79 in drought tolerance in cotton.
Discussion
A phylogenetic tree was built with 126 AtNACs and GhNAC79 protein sequences, as shown in Supplementary Figure S1. GhNAC79 belongs to the NAM subfamily, many members of which are involved in plant development; for example, AT2G17040.1/ANAC036 are associated with a dwarf phenotype and distorted leaves in Arabidopsis (Kato et al., 2010). Overexpression of GhNAC79 in Arabidopsis promotes flowering, which enriches the function of NACs in cotton development. Four homologous genes were obtained with three CDs genomes in cotton (A, D and AD), and a multiple alignment was created by DNAMAN software, as shown in Supplementary Figure S2. GhNAC79 differs from CotAD_11909 by only a single base (a gene from G. hirsutum may have originated from G. arboreum), and this single-base difference may have been caused by a sequencing error or cultivar variation.
Overexpression of GhNAC79 in Arabidopsis leads to an early flowering phenotype, indicating that GhNAC79 promotes flowering. GhNAC79 also exhibits special expression patterns in three differently maturing cotton varieties, as shown in Supplementary Figure S3, CCRI74 (early maturing) > Shan70 (middle-maturing) > Bo1 (late-maturing), which further indicate the potential function of GhNAC79 in flowering. The fiber of G. arboreum is spinnable (Xu et al., 2010), but that of G. raimondii is not. GhNAC79 is predominantly expressed in later fiber development stages and comes from the A-subgenome, which implies a potential role of GhNAC79 in fiber development. The high sensitivity to ethylene exhibited by GhNAC79 indicates that ethylene plays important roles in fiber elongation (Shi et al., 2006; Qin et al., 2007), so we compare the fiber length between CCRI24 and 35S-GhNAC79 plants at T2 stage. As shown in Supplementary Figure S4, the length of 35S-GhNAC79 plants is longer than CCRI24, but not significant, but there need more data in different years and areas to support this result.
Additionally, we collected half-yellow rosette leaves from transgenic and wild type Arabidopsis to assess the expression of some senescence-related genes with qRT-PCR, and the results are shown in Figure 8. ORE1 coordinates with EIN3 to regulate leaf senescence through ethylene (Qiu et al., 2015), and the expressions levels of ORE1 and EIN3 were higher in the four transgenic lines than in wild type. Because GhNAC79 in cotton is very sensitive to ethylene treatment, its promoter contains an ethylene-related element, and its overexpression in Arabidopsis resulted in an early bolting phenotype under ethylene treatment (Supplementary Figure S5). Therefore, GhNAC79 may play some roles in the ethylene pathway. ABA plays a very important role in drought tolerance (Belimov et al., 2014; Osakabe et al., 2014; Shinohara and Leskovar, 2014) and regulates growth after germination through ABI5 (Lopez-Molina et al., 2002; Albertos et al., 2015). The expression of ABI5 in transgenic lines is lower than in the wild type, and overexpression of GhNAC79 enhanced the tolerance of Arabidopsis to ABA treatment, which illustrates the vital role of GhNAC79 in the ABA pathway. LhCB1 functions in state transitions during light harvesting in Arabidopsis photosynthesis (Pietrzykowska et al., 2014), and photosynthetic ability is an important indicator of leaf senescence. In contrast, SAG12 is a negative marker gene for leaf senescence (Chang et al., 2003; Merewitz et al., 2011). LhCB1 had a relatively high expression level in the transgenic lines, while the expression level of SAG12 was low, which indicated that senility was delayed in the transgenic lines. SEN4 has a positive influence on plant survival as its homologous gene is related to DNA damage (Takeda et al., 2004), and SEN4 expression was relatively high in transgenic plants, which indicated the role of GhNAC79 in plant development. The expression levels of ABI5, LhCB1, SAG12, and SEN4 implied that GhNAC79 may negatively regulate leaf senescence in Arabidopsis. However, high levels of ORE1 and EIN3 promoted leaf senescence, and this contradiction may be due to the complexity of leaf senescence.
FIGURE 8.
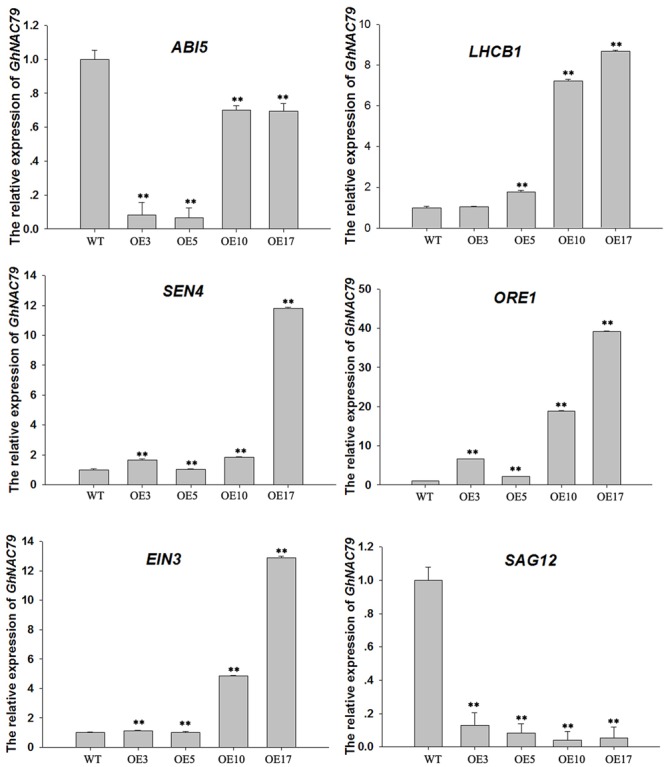
Expression levels of 6 senescence-related genes in transgenic and wild type Arabidopsis. ∗∗Values significantly different from wild type at the 0.01 confidence level. Data are presented as the mean ± SD (n = 3). GhHIS3 was used as the reference gene.
It is interesting that GhNAC79 was induced in leaves by meJA and drought treatments, but the expression of GhNAC79 in roots was repressed in all treatments. For this result, it may caused by the experiment method we used. As all treatments applied in medium, when phenotype appearing in leaf, the condition of roots is not good. Bad condition of roots may be related with low level of GhNAC79, so GhNAC79 may a positive regulator in stress responses. In the same time, drought, Eth and ABA treatments all results in an early bolting phenotype in transgenic Arabidopsis, and we think it caused by stress reaction of plant. For plants, they will try their best to accomplish reproduction, when stress coming, through earlier blooming, they get seeds, which make their life continuing.
Under open and sealed conditions, drought stress sharply induced GhNAC79 in leaves. Overexpression of GhNAC79 enhanced drought tolerance in Arabidopsis and cotton, and GhNAC79 repression made cotton more sensitive to drought, which demonstrates its potential function in drought stress. Drought is always associated with ABA (Li C. et al., 2015; Tombesi et al., 2015; Yin et al., 2015), so we explored the relationship between GhNAC79 and ABA. Supplementary Figure S6A shows that transgenic Arabidopsis exhibited an early bolting phenotype after 15 days of ABA treatment, and wild type growth was obviously inhibited. ABA regulates drought stress through stomatal movements (Cai et al., 2015; Cohen et al., 2015), so the number and aperture size of the stomas were measured after 3 days of ABA treatment. As shown in Supplementary Figure S6B, the stomatal aperture decreased after ABA treatment, but the average stomatal aperture was smaller in transgenic Arabidopsis. Therefore, we conclude that GhNAC79 may regulate drought stress in an ABA-dependent manner in Arabidopsis. Upland cotton is allotetraploid, and the internal mechanism underlying drought signal transmittance is complex. The expression of GhNAC79 in leaves was not strongly affected by ABA treatment, but under sealed conditions, the GhNAC79 in the roots was greatly altered by ABA treatment. Therefore, the repression of GhNAC79 makes cotton more sensitive to drought stress, whereas overexpression enhances drought tolerance. Furthermore, GhNAC79 is highly expressed in drought-resistant cotton varieties after drought treatment (Supplementary Figure S7). The promoter of GhNAC79 contains a drought-related cis-element, so we consider GhNAC79 to be a drought-response gene.
Conclusion
This study proves that GhNAC79 promotes flowering in Arabidopsis, and it also acts as a positive regulator during drought stress through stomatal movement and may be involved in an ABA signal-related pathway. At the same time, GhNAC79 appears to be involved in the ethylene signal pathway, and because its promoter contains an ethylene-related cis-element, its expression is highly induced by ethylene, and its overexpression makes Arabidopsis more sensitive to ethylene. Interestingly, GhNAC79 comes from the A-genome, which is preferentially expressed in later fiber development, and it shows high sensitivity to ethylene treatment, indicating the preferential role of GhNAC79 in fiber elongation. However, confirming this role requires additional evidence.
Author Contributions
SY, SF, HlW, and CP designed the experiments. LD and LG collected the sequences, YG performed the experiments and wrote the manuscript, HtW, XJ, JS, and QM revised the language. All the authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. We thank The National Key Research and Development Program of China (grant no. 2016YFD0101006) for the financial support provided to this project.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01657/full#supplementary-material
Phylogenetic relationships among GhNAC79 and 126 AtNACs. Multiple alignments of GhNAC79 and AtNACs were executed using Clustal X, and the phylogenetic tree was constructed using MEGA 5.1 and the neighbor-joining (NJ) method.
Sequence alignments of GhNAC79 and its predicted homologous genes in A, D and AD genomes.
Expression patterns of GhNAC79 in the apical bud of three varieties. CCRI74: an early maturing variety; Shan70: a middle-maturity variety; Bo1: a late-maturing variety. Three-leaf to 6-leaf represent the different developmental stages of cotton.
The comparison of fiber length between CCRI24 (wild type) and 35S-GhNAC79 (transgenic cotton).
Phenotypes of transgenic and wild type Arabidopsis after different treatments.
Phenotypes of transgenic and wild type Arabidopsis after ABA treatment. (A) Plants were treated with ABA. (B) Stomatal aperture of transgenic and wild type Arabidopsis after ABA treatment. Control: transgenic and wild type plants were treated with water.
The expression levels of GhNAC79 after drought treatment in different drought-resistant cotton varieties. ZhongjianD228 and ZhongH177: two drought-resistant cotton varieties. ZhongS9612 and Zhongjian9648: two drought-sensitive cotton varieties. Data are presented as the mean ± SD (n = 3). ∗: Values significantly different from wild type at the 0.05 confidence level, ∗∗: Values significantly different from wild type at the 0.01 confidence level. GhHIS3 was the reference gene.
Primer used for selection of transgenic Arabidopsis and cotton.
References
- Ahuja I., de Vos R. C., Bones A. M., Hall R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15 664–674. 10.1016/j.tplants.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertos P., Romero-Puertas M. C., Tatematsu K., Mateos I., Sánchez-Vicente I., Nambara E., et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6 8669 10.1038/ncomms9669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S., Siddiqui H., Allu A. D., Matallana-Ramirez L. P., Caldana C., Mehrnia M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62 250–264. 10.1111/j.1365-313X.2010.04151.x [DOI] [PubMed] [Google Scholar]
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24 23–58. 10.1080/07352680590910410 [DOI] [Google Scholar]
- Belimov A. A., Dodd I. C., Safronova V. I., Dumova V. A., Shaposhnikov A. I., Ladatko A. G., et al. (2014). Abscisic acid metabolizing rhizobacteria decrease ABA concentrations in planta and alter plant growth. Plant Physiol. Biochem. 74 84–91. 10.1016/j.plaphy.2013.10.032 [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S. U., Min S. R., Jeong W. J., Sultana S., Choi K. S., Lim Y. P., et al. (2011). An improved method for Agrobacterium-mediated genetic transformation from cotyledon explants of Brassica juncea. Plant Biotechnol. 28 17–23. 10.5511/plantbiotechnology.10.0921a [DOI] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Cai S., Jiang G., Ye N., Chu Z., Xu X., Zhang J., et al. (2015). A key ABA catabolic gene, OsABA8ox3, is involved in drought stress resistance in rice. PLOS ONE 10:e0116646 10.1371/journal.pone.0116646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai M., Bellizzi M., Wan C., Cui Z., Li Y., Wang G. (2015). The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 58 44–51. 10.1007/s12374-014-0400-y [DOI] [Google Scholar]
- Chang H., Jones M. L., Banowetz G. M., Clark D. G. (2003). Overproduction of cytokinins in petunia flowers transformed with P(SAG12)-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol. 132 2174–2183. 10.1104/pp.103.023945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J. A., Wilson I. W., Llewellyn D. J., Dennis E. S. (2009). The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 149 1724–1738. 10.1104/pp.108.131912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cohen A. C., Bottini R., Pontin M., Berli F. J., Moreno D., Boccanlandro H., et al. (2015). Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 153 79–90. 10.1111/ppl.12221 [DOI] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39 863–876. 10.1111/j.1365-313X.2004.02171.x [DOI] [PubMed] [Google Scholar]
- Gu Z., Huang C., Li F., Zhou X. (2014). A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12 638–649. 10.1111/pbi.12169 [DOI] [PubMed] [Google Scholar]
- He X. J., Mu R. L., Cao W. H., Zhang Z. G., Zhang J. S., Chen S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44 903–916. 10.1111/j.1365-313X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- Ishida T., Aida M., Takada S., Tasaka M. (2000). Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 41 60–67. 10.1093/pcp/41.1.60 [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. K., Hagedorn P. H., de Torres-Zabala M., Grant M. R., Rung J. H., Collinge D. B., et al. (2008). Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. Sp. hordei in Arabidopsis. Plant J. 56 867–880. 10.1111/j.1365-313X.2008.03646.x [DOI] [PubMed] [Google Scholar]
- Kato H., Motomura T., Komeda Y., Saito T., Kato A. (2010). Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167 571–577. 10.1016/j.jplph.2009.11.004 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Chung K. M., Woo H. R. (2011). Three positive regulators of leaf senescence in Arabidopsis Ore 1, ORE3 and ORE9, play roles in crosstalk among multiple hormone-mediated senescence pathways. Genes Genom. 33 373–381. 10.1007/s13258-011-0044-y [DOI] [Google Scholar]
- Kim S. G., Lee A. K., Yoon H. K., Park C. M. (2008). A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 55 77–88. 10.1111/j.1365-313X.2008.03493.x [DOI] [PubMed] [Google Scholar]
- Knapp S. J., Stroup W. W., Ross W. M. (1985). Exact confidence intervals for heritability on a progeny mean basis. Crop Sci. 25 192–194. 10.2135/cropsci1985.0011183X002500010046x [DOI] [Google Scholar]
- Kramer G. F., Norman H. A., Krizek D. T., Mirecki R. M. (1991). Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry 30 2101–2108. 10.1016/0031-9422(91)83595-C [DOI] [Google Scholar]
- Kumagai M. H., Donson J., della-Cioppa G., Harvey D., Hanley K., Grill L. K. (1995). Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. U.S.A. 92 1679–1683. 10.1073/pnas.92.5.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Shen H., Wang T., Wang X. (2015). ABA regulates subcellular redistribution of OsABI-LIKE2, a negative regulator in ABA signaling, to control root architecture and drought resistance in Oryza sativa. Plant Cell Physiol. 56 2396–2408. 10.1093/pcp/pcv154 [DOI] [PubMed] [Google Scholar]
- Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R. J., et al. (2015). Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33 524–530. 10.1038/nbt.3208 [DOI] [PubMed] [Google Scholar]
- Li W., Huang G. Q., Zhou W., Xia X. C., Li D. D., Li X. B. (2014). A cotton (Gossypium hirsutum) gene encoding a NAC transcription factor is involved in negative regulation of plant xylem development. Plant Physiol. Biochem. 83 134–141. 10.1016/j.plaphy.2014.07.022 [DOI] [PubMed] [Google Scholar]
- Liu G., Li X., Jin S., Liu X., Zhu L., Nie Y., et al. (2014). Overexpression of Rice NAC Gene SNAC1 improves drought and Salt tolerance by enhancing root development and reducing transpiration Rate in transgenic cotton. PLOS ONE 9:e86895 10.1371/journal.pone.0086895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D. T., Chait B. T., Chua N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32 317–328. 10.1046/j.1365-313X.2002.01430.x [DOI] [PubMed] [Google Scholar]
- Lu P.-L., Chen N.-Z., An R., Su Z., Qi B.-S., Ren F., et al. (2006). A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 63 289–305. 10.1007/s11103-006-9089-8 [DOI] [PubMed] [Google Scholar]
- Matallana-Ramirez L. P., Rauf M., Farage-Barhom S., Dortay H., Xue G. P., Dröge-Laser W., et al. (2013). NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 6 1438–1452. 10.1093/mp/sst012 [DOI] [PubMed] [Google Scholar]
- Merewitz E. B., Gianfagna T., Huang B. (2011). Photosynthesis, water use, and root viability under water stress as affected by expression of SAG12-ipt controlling cytokinin synthesis in Agrostis stolonifera. J. Exp. Bot. 62 383–395. 10.1093/jxb/erq285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L. S. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202 35–49. 10.1111/nph.12613 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J., Jin D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- Pietrzykowska M., Suorsa M., Semchonok D. A., Tikkanen M., Boekema E. J., Aro E. M., et al. (2014). The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 26 3646–3660. 10.1105/tpc.114.127373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y. M., Hu C. Y., Pang Y., Kastaniotis A. J., Hiltunen J. K., Zhu Y. X. (2007). Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19 3692–3704. 10.1105/tpc.107.054437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu K., Li Z., Yang Z., Chen J., Wu S., Zhu X., et al. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLOS Genet. 11:e1005399 10.1371/journal.pgen.1005399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani M. A., Maruyama K., Abe H., Khan M. A., Katsura K., Ito Y., et al. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133 1755–1767. 10.1104/pp.103.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Kim M. J., Park J. Y., Kim S. Y., Jeon J., Lee Y. H., et al. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61 661–671. 10.1111/j.1365-313X.2009.04091.x [DOI] [PubMed] [Google Scholar]
- Shah S. T., Pang C., Fan S., Song M., Arain S., Yu S. (2013). Isolation and expression profiling of GhNAC transcription factor genes in cotton (Gossypium hirsutum L.) during leaf senescence and in response to stresses. Gene 531 220–234. 10.1016/j.gene.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Shah S. T., Pang C., Hussain A., Fan S., Song M., Zamir R., et al. (2014). Molecular cloning and functional analysis of NAC family genes associated with leaf senescence and stresses in Gossypium hirsutum L. Plant Cell Tiss. Organ Cult. 117 167–186. 10.1007/s11240-014-0430-7 [DOI] [Google Scholar]
- Shi Y. H., Zhu S. W., Mao X. Z., Feng J. X., Qin Y. M., Zhang L., et al. (2006). Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18 651–664. 10.1105/tpc.105.040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T., Leskovar D. I. (2014). Effects of ABA, antitranspirants, heat and drought stress on plant growth, physiology and water status of artichoke transplants. Sci. Hortic. 165 225–234. 10.1016/j.scienta.2013.10.045 [DOI] [Google Scholar]
- Singh B., Avci U., Inwood S. E. E., Grimson M. J., Landgraf J., Mohnen D., et al. (2009). A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles. Plant Physiol. 150 684–699. 10.1104/pp.109.135459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G. L., Cui R. X., Wang K. B., Guo L. P., Li S. H., Wang C. Y., et al. (1998). A rapid improved CTAB method for extraction of cotton genomic DNA. Acta Gossypii Sin. 10 273–275. [Google Scholar]
- Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. (1996). The No apical meristem gene of Petunia is required for Pattern Formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85 159–170. 10.1016/S0092-8674(00)81093-4 [DOI] [PubMed] [Google Scholar]
- Takeda S., Tadele Z., Hofmann I., Probst A. V., Angelis K. J., Kaya H., et al. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18 782–793. 10.1101/gad.295404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombesi S., Nardini A., Frioni T., Soccolini M., Zadra C., Farinelli D., et al. (2015). Stomatal closure is induced by hydraulic signals and maintained by ABA in drought-stressed grapevine. Sci. Rep. 5:12449 10.1038/srep12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L., Zhang X., Liu D., Jin S., Cao J., Zhu L., et al. (2007). Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chin. Sci. Bull. 52 3110–3117. 10.1007/s11434-007-0461-0 [DOI] [Google Scholar]
- Unver T., Budak H. (2009). Virus-induced gene silencing, a post transcriptional gene silencing method. Int. J. Plant Genomics 2009:198680 10.1155/2009/198680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Guo X., Wang C., Ma J., Niu F., Zhang H., et al. (2015). Identification and characterization of plant-specific NAC gene family in canola (Brassica napus L.) reveal novel members involved in cell death. Plant Mol. Biol. 87 395–411. 10.1007/s11103-015-0286-1 [DOI] [PubMed] [Google Scholar]
- Wang B., Lüttge U., Ratajczak R. (2001). Effects of salt treatment and osmotic stress on V-ATPase and V-PPase in leaves of the halophyte Suaeda salsa. J. Exp. Bot. 52 2355–2365. 10.1093/jexbot/52.365.2355 [DOI] [PubMed] [Google Scholar]
- Wang Z., Rashotte A. M., Moss A. G., Dane F. (2014). Two NAC transcription factors from Citrullus colocynthis, CcNAC1, CcNAC2 implicated in multiple stress responses. Acta Physiol. Plant. 36 621–634. 10.1007/s11738-013-1440-5 [DOI] [Google Scholar]
- Wu A., Allu A. D., Garapati P., Siddiqui H., Dortay H., Zanor M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24 482–506. 10.1105/tpc.111.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Deng Z., Lai J., Zhang Y., Yang C., Yin B., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19 1279–1290. 10.1038/cr.2009.108 [DOI] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 3024–3036. 10.1101/gad.852200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Wang W., Zeng J., Zhang J., Grierson D., Li X., et al. (2015). A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Technol. 102 25–31. 10.1016/j.postharvbio.2015.02.002 [DOI] [Google Scholar]
- Xu Z., Yu J. Z., Cho J., Yu J., Kohel R. J., Percy R. G. (2010). Polyploidization altered Gene functions in cotton (Gossypium spp.). PLOS ONE 5:e14351 10.1371/journal.pone.0014351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wang X., Ji L., Yi Z., Fu C., Ran J., et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 34 943–958. 10.1007/s00299-015-1756-2 [DOI] [PubMed] [Google Scholar]
- Yin X., Huang L., Zhang X., Wang M., Xu G., Xia X. (2015). Expression of rice gene OsMSR4 confers decreased ABA sensitivity and improved drought tolerance in Arabidopsis thaliana. Plant Growth Regul. 75 549–556. 10.1007/s10725-014-0020-z [DOI] [Google Scholar]
- Yu J., Jung S., Cheng C. H., Ficklin S. P., Lee T., Zheng P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42 D1229–D1236. 10.1093/nar/gkt1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Peng H., Liu Y., Zhang Y., Shu Y., Chen Q., et al. (2014). CarNAC2, a novel NAC transcription factor in chickpea (Cicer arietinum L.), is associated with drought-response and various developmental processes in transgenic arabidopsis. J. Plant Biol. 57 55–66. 10.1007/s12374-013-0457-z [DOI] [Google Scholar]
- Zhang C. (2008). Establishment of High Frequency Regeneration System and Genetic Analysis for Mature Leaf Petioles in Upland Cotton (G. hirsutum L.). Beijing: Chinese Academy of Agricultural Sciences. [Google Scholar]
- Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33 531–537. 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
- Zhao D., Derkx A. P., Liu D. C., Buchner P., Hawkesford M. J. (2015). Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 17 904–913. 10.1111/plb.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Ma J., Li L., Fan S., Guo Y., Song M., et al. (2016). GhNAC12, a neutral candidate gene, leads to early aging in cotton (Gossypium hirsutum L). Gene 576 268–274. 10.1016/j.gene.2015.10.042 [DOI] [PubMed] [Google Scholar]
- Zhao J., Li S., Jiang T., Liu Z., Zhang W., Jian G., et al. (2012). Chilling stress–the key predisposing factor for causing Alternaria alternata infection and leading to cotton (Gossypium hirsutum L.) leaf senescence. PLOS ONE 7:e36126 10.1371/journal.pone.0036126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Lin-Wang K., Wang H., Gu C., Dare A. P., Espley R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82 105–121. 10.1111/tpj.12792 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationships among GhNAC79 and 126 AtNACs. Multiple alignments of GhNAC79 and AtNACs were executed using Clustal X, and the phylogenetic tree was constructed using MEGA 5.1 and the neighbor-joining (NJ) method.
Sequence alignments of GhNAC79 and its predicted homologous genes in A, D and AD genomes.
Expression patterns of GhNAC79 in the apical bud of three varieties. CCRI74: an early maturing variety; Shan70: a middle-maturity variety; Bo1: a late-maturing variety. Three-leaf to 6-leaf represent the different developmental stages of cotton.
The comparison of fiber length between CCRI24 (wild type) and 35S-GhNAC79 (transgenic cotton).
Phenotypes of transgenic and wild type Arabidopsis after different treatments.
Phenotypes of transgenic and wild type Arabidopsis after ABA treatment. (A) Plants were treated with ABA. (B) Stomatal aperture of transgenic and wild type Arabidopsis after ABA treatment. Control: transgenic and wild type plants were treated with water.
The expression levels of GhNAC79 after drought treatment in different drought-resistant cotton varieties. ZhongjianD228 and ZhongH177: two drought-resistant cotton varieties. ZhongS9612 and Zhongjian9648: two drought-sensitive cotton varieties. Data are presented as the mean ± SD (n = 3). ∗: Values significantly different from wild type at the 0.05 confidence level, ∗∗: Values significantly different from wild type at the 0.01 confidence level. GhHIS3 was the reference gene.
Primer used for selection of transgenic Arabidopsis and cotton.


