Abstract
It is still a controversy whether the role of Sirtuin 7 (SIRT7) is an oncogene or a tumor suppressor gene in cancer as SIRT7 may have different functions in different types of cancer. Particularly, the specific roles of SIRT7 in the progression of osteosarcoma remain undiscovered. The main aim of this study is to identify the expression of SIRT7 in osteosarcoma and explore the biological functions of SIRT7 in regulating cellular processes of osteosarcoma cells. Here, we show that SIRT7 expression was significantly higher in osteosarcoma tissues and osteosarcoma cell lines than in non-tumor tissues and an immortalized normal cell line, respectively. Moreover, elevated SIRT7 levels in clinical samples indicate a poor prognosis of osteosarcoma patients. SIRT7 knockdown reduces proliferation, migration, invasion, tumor formation, and metastasis of osteosarcoma cells, while SIRT7 overexpression has the opposite effects. Mechanistically, SIRT7 down regulates H3K18ac expression and decreases H3K18ac binding to the promoter region of CDC4, leading to the inhibition of CDC4 transcription. Furthermore, the silencing of CDC4 partially rescued SIRT7 knockdown-mediated inhibitory effects on proliferation, migration, and invasion of osteosarcoma cells. In summary, our results show that SIRT7 promotes proliferation, migration, and invasion of osteosarcoma cells through targeting CDC4, suggesting a potential therapeutic target for SIRT7 based therapy for osteosarcoma.
Keywords: SIRT7, CDC4, osteosarcoma, H3K18ac
Introduction
Osteosarcoma is a common bone tumor in adolescents [1]. The development of treatment for high-grade osteosarcoma, which combines surgery with multiagent chemotherapy, has contributed to significantly improved survival rates [2]. However, osteosarcoma has a high metastatic potential. Several molecular pathways contributing to osteosarcoma development and progression have recently been discovered, but the detailed mechanisms of osteosarcoma metastasis remain unclear [3].
Sirtuin 7 (SIRT7) is one of the least understood human sirtuins, which is concentrated in the nucleoli and activates transcription of rRNA genes [4]. Under certain stress, SIRT7 can translocate from nucleoli to nucleoplasm resulting in hyperacetylation of PAF53, which inhibits Pol I transcription [5]. By deacetylating histone H3 Lys18 (H3K18), SIRT7 also suppresses mRNA transcription mediated by the Pol II machinery [6]. Moreover, SIRT7 regulates the transcription of nucleus-encoded mitochondrial biogenesis genes by deacetylating and activating a transcription factor [7]. A recent study discovered a specific target of SIRT7 and identified a crucial role for SIRT7 in the maintenance of cancer phenotype and transformation [8]. SIRT7 specifically targets a single histone marker, H3K18Ac, which directly links to the control of its expression [9,10]. SIRT7 is overexpressed in several cancer types, including leukemia, bladder, prostate, breast and gastric cancer [11-13]. However, the role of SIRT7 in osteosarcoma remains largely unknown.
This study aims to investigate the possible roles of SIRT7 in the regulation of osteosarcoma formation and to further explore the potential signaling pathways and molecular mechanisms in osteosarcoma.
Materials and methods
Cells and antibodies
This research used osteosarcoma cell lines (hFOB 1.19, SJSA-1, Hs755, MG-63, and D-17) obtained from American Type Culture Collection (ATCC, Manassas, VA). All lines were cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin at 37°C, 5% CO2. Rabbit monoclonal SIRT7, CDC4, E-cadherin, N-cadherin, α-catenin, vimentin, and snail antibodies purchased from Abcam. Mouse monoclonal aurora-A, cyclin E, c-Myc, EZH2, and ENO1 antibodies from Cell Signaling Technology (CST, Danvers, MA). Mouse monoclonal β-actin antibody was the product of Santa Cruz Biotech (Santa Cruz, CA).
Osteosarcoma samples
Osteosarcoma tissue samples were obtained from Cancer Hospital of China Medical University. All investigations described in this study were carried out after obtaining informed consent and in accordance with an Institutional Review Board protocol approved by the Partners Human Research Committee at the China Medical University.
qRT-PCR
RNA was extracted from tissues and cells, then reverse transcription was performed by using the Thermoscript RT System (Invitrogen). Hotstart PCR conditions were set as follows: 45 s at 94°C, 30 s at 55°C, and 1 min at 72°C for 45 cycles. The relative expression of SIRT7 and CDC4, normalized to GAPDH was quantified using the 2-ΔΔCt. The following primers were used in this study: SIRT7: sense 5’-ACTTGGTCGTCTACACAGGC-3’ and antisense 5’-AAATGGGATCCTGCCACCTG-3’; CDC4: sense 5’-TCATCGAAACTCGTCCACGG-3’ and antisense 5’-ATCACTGCGTTCAGGAGTGG-3’; GAPDH: sense 5’-TGCCTCCTGCACCACCAACT-3’ and antisense 5’-CCCGTTCAGCTCAGGGATGA-3’.
Western blot
Proteins from cells were transferred to polyvinylidene difluoride membranes. The membranes were first probed with a primary antibody and then with secondary antibody. The bound antibody was detected by enhanced chemiluminescence detection reagents. The band intensity was quantitated.
Plasmids and generation of stable cell lines
Human cDNA of SIRT7 was cloned as previously reported [14]. The full-length cDNAs were subcloned into the multiple cloning sites of the vector plasmid, forming the pBabe-SIRT7 expression plasmids. Short hair-pin RNA (shRNA) targeting SIRT7, CDC4, and their control shRNA were purchased from Sigma. D-17 cell line was transfected with the vector or pBabe-SIRT7 plasmid using the Lipofectamine 2000. SJSA-1 cells were transfected with the control or shSIRT7/shCDC4 plasmid using the Lipofectamine 2000. Stable transfectants were obtained after selection by puromycin.
Cell proliferation, colony formation, migration and invasion assays
Osteosarcoma cell lines were treated with knockdown or overexpression SIRT7. Cell proliferation, colony formation, migration, and invasion assays were measured using procedures described previously [15].
Sphere formation assay
Sphere formation assay was performed as described previously [16]. Single-cell suspensions were plated in an ultralow attachment 96-well. Cells were grown in a serum-free mammary epithelial growth medium, supplemented with 1:50 B27, 20 ng/ml EGF, 20 ng/ml basic fibroblast growth factor (bFGF), and 10 μg/ml heparin. The numbers of spheroids were counted after 14 days.
Immunohistochemistry staining
Paraffin-embedded sections were deparaffinized, blocked, and incubated with antibody at 4°C overnight. Secondary antibody was then added, and incubated for 1 hour at room temperature. The sections were developed using a 3,3’-diaminobenzidine tetrahydrochloride (DAB) substrate kit at room temperature for 5 minutes and then counterstained with hematoxylin.
Xenografted tumor formation and metastasis assay
This experiment was conducted according to the international regulations of the usage and welfare of laboratory animals, and approved by the Institutional Animal Care and Use Committee of China Medical University (#: 2016082). The cell suspension (0.1 mL; 5 × 106 cells) was subcutaneously injected into the right flank of 5- to 6-week-old female BALB/c-nu/nu mice (n = 5/group). Tumor volume (mm3) was measured every 3 days in two dimensions using a caliper and was calculated as 0.4 × (short length)2 × long length. Mice were humanely euthanized when they became moribund or when the subcutaneous tumors reached 15 mm in diameter. For experimental metastasis assays, age-matched nude mice were injected with 1 × 106 cells (resuspended in PBS) via the tail vein. Liver metastases were detected by hematoxylin and eosin staining.
TCGA genetic alteration analysis
Analysis of genetic alterations in osteosaroma TCGA data was performed on cBioPortal (http://www.cbioportal.org). A Fisher’s exact test was performed on the confusion matrix of co-occurrence of alterations in SIRT7 and CDC4. The supplemental excel file lists the osteosaroma samples and their GISTIC 2.0 copy-number alterations, whole exome sequencing mutation data, and RNASeqV2 RSEM expression data used in the study. P-values were calculated using a pairwise t-test in R, performing pairwise comparisons between group levels with a Holm correction for multiple testing.
TCGA RNASeqV2 expression correlation and outcome analysis
The Kaplan-Meier overall survival plot and the log-rank p-value for TCGA osteosarcoma were generated using R with osteosarcoma samples (TCGA Network, 2015) that had RNASeqV2 data and clinical information with at least 6 months of follow-up information or a death event. The samples were split according to whether the expression measured by RNASeqV2 fell into the lower 1/3 quantile or the upper 2/3 quantile of expression.
Microarray-based gene expression analysis
RNA for the D-17 with the vector and SIRT7 were hybridized onto Affymetrix HuGene1v1 microarrays. Raw values from the CEL files were processed and normalized using RMA in the R Bioconductor package. Probe sets on the HuGene1v1 microarray were mapped to genes using the chip file HuGene1v1.chip in the molecular signature database. P-values were calculated using a hypergeometric test in R.
Statistical analysis
Experimental data are shown as mean ± standard deviation (S.D.). The results from different treatment groups were compared using a two-tailed Student’s t-test. Differences were considered statistically significant at a value of P less than 0.05. Statistical analysis was done with SPSS/Win11.0 software (SPSS, Inc., Chicago, Illinois, USA).
Results
Higher SIRT7 expression correlates with poor prognosis in osteosarcoma
In order to discover the role of SIRT7 in the development of osteosarcoma, we first used the TCGA data for bioinformatic analyses. We found a negative correlation between overall patient survival and SIRT7 expression using the Kaplan-Meier plotter assessment tool. The results show that high expression of SIRT7 is correlated with poor overall survival in osteosarcoma (Figure 1A and 1B). Analysis of genomic copy-number data from TCGA provided a genetic mechanism for high-level SIRT7 expression in osteosarcoma. A substantial proportion of these tumors (24%) exhibit genomic amplification or mRNA upregulation in the SIRT7 gene (Figure 1C). In addition, we analyzed the public microarray data from the GEO repository and found that SIRT7 expression increased significantly in osteosarcoma tissues compared with adjacent normal tissues (Figure 1D). Consistently, the qRT-PCR analysis of normal and osteosarcoma tissues (from our own Cancer Hospital of China Medical University) confirmed that SIRT7 mRNA expression was significantly higher in osteosarcoma tissues than in the adjacent non-tumor tissues (Figure 1E). Also, the Western blot results further confirmed those changes (Figure 1F). Additionally, the expression of SIRT7 in osteosarcoma cell lines was measured by qRT-PCR and Western blots. SIRT7 expression level was markedly increased in four osteosarcoma cell lines (SJSA-1, Hs755, MG-63, and D-17) compared with that in hFOB 1.19 (an immortalized human fetal osteoblastic cell line) (Figure 1G-I). Taken together, these results indicate that SIRT7 is overexpressed in osteosarcoma and correlate with worse osteosarcoma prognosis.
Figure 1.
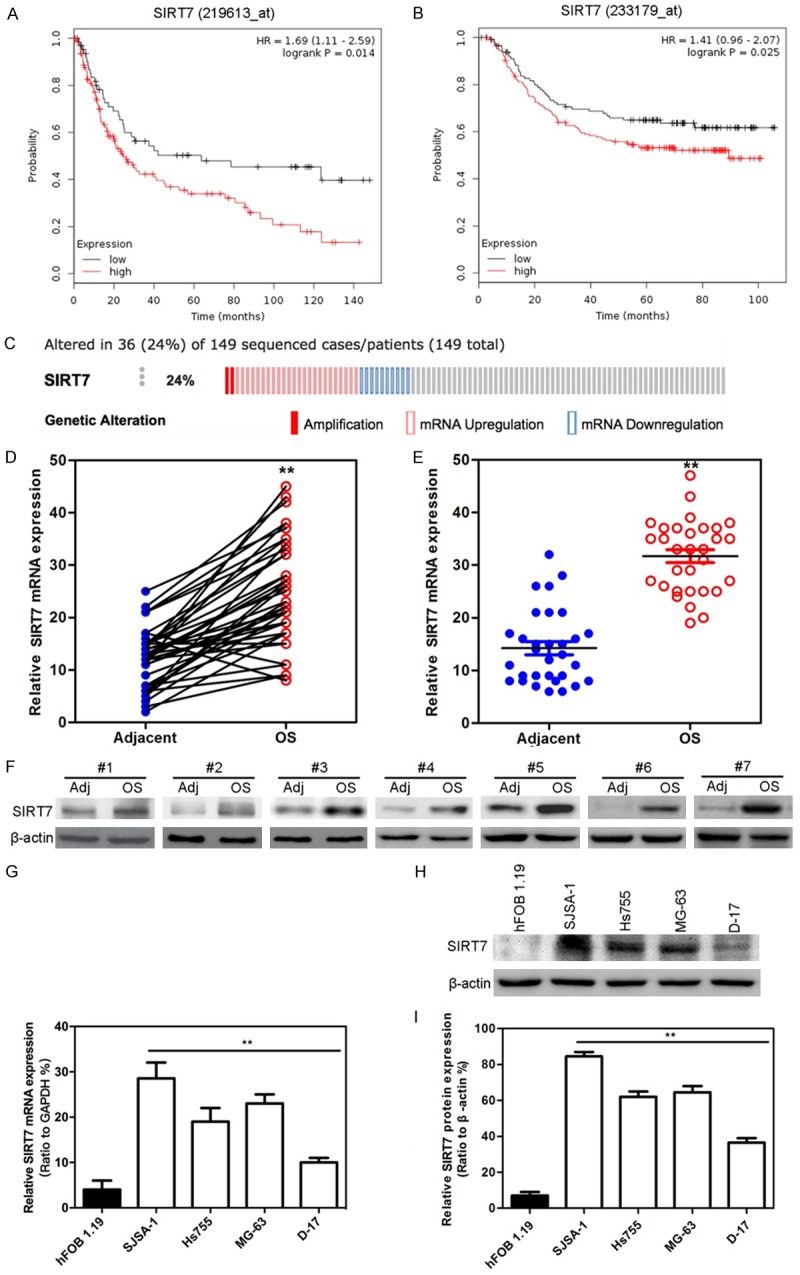
Higher SIRT7 expression correlates with poor prognosis in osteosarcoma. (A and B) Kaplan-Meier analysis of two probes of SIRT7 (A: 219613_at; B: 233179_at) in survival of osteosarcoma patients (log-rank test; n = 149). The data were obtained from TCGA database. (C) SIRT7 genomic amplification or mRNA expression was analyzed by TCGA database (n = 149). (D) SIRT7 mRNA expression in 58 pairs of osteosarcoma and adjacent normal tissues. The data were obtained from the GEO database. (E) SIRT7 mRNA expression in 32 pairs of osteosarcoma and adjacent normal tissues obtained from Cancer Hospital of China Medical University. (F) The expression of SIRT7 protein were measured by Western blot pairs of osteosarcoma and adjacent normal tissues obtained from Cancer Hospital of China Medical University. (G) The expression of SIRT7 mRNA in osteosarcoma cells were measured by qRT-PCR. (H) The expression of SIRT7 protein in osteosarcoma cells were measured by Western blot. (I) The relative quantitative description of (H). **P < 0.01 (in D, E, G, and I) is based on Student’s t-test. Error bars indicate standard deviation.
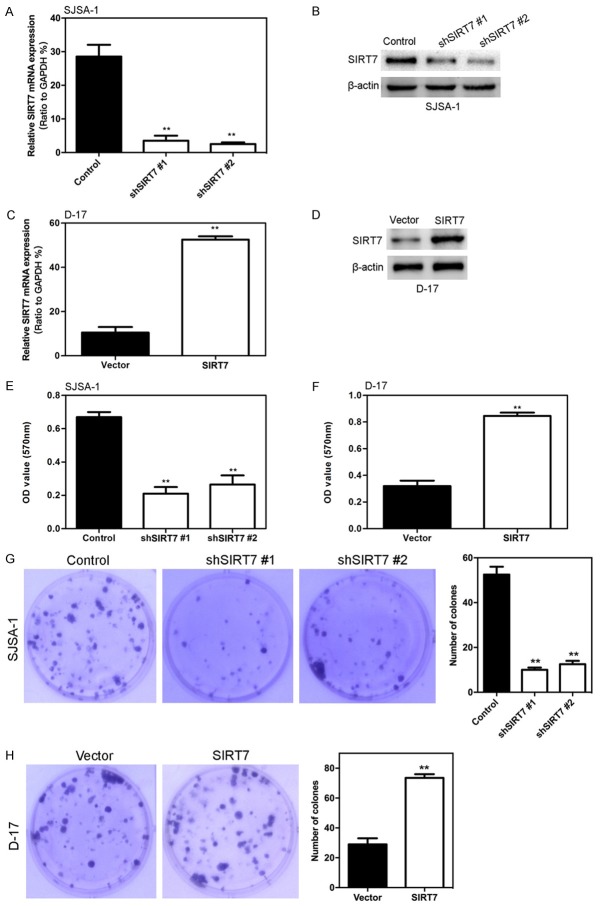
Stable SIRT7 transfected osteosarcoma cell line formation
To investigate the biological function of SIRT7 in osteosarcoma formation, we first chose the SJSA-1 cells to stably transfect SIRT7 shRNA. After being selected with G418, the expression of SIRT7 was greatly suppressed in shRNA group cells (Figure 2A and 2B). In addition, we ectopically expressed SIRT7 in the D-17 cell line and showed that the expression of SIRT7 was significantly up-regulated in D-17-SIRT7 cells compared with D-17-vector only cells (Figure 2C and 2D).
Figure 2.
SIRT7 induced osteosarcoma cell proliferation in vitro. (A) SIRT7 protein expression was examined by Western blot assay in SIRT7 knockdown SJSA-1 cells. (B) SIRT7 mRNA expression was examined by qRT-PCR assay in SIRT7 knockdown SJSA-1 cells. (C) SIRT7 protein expression was examined by Western blot assay in SIRT7 ectopic expression D-17 cells. (D) SIRT7 mRNA expression was examined by qRT-PCR assay in SIRT7 ectopic expression D-17 cells. (E) Knockdown of SIRT7 inhibited the ability of SJSA-1 cell proliferation measured by MTT. (F) Overexpression of SIRT7 promotes the ability of D-17 cell proliferation measured by MTT. (G) Knockdown of SIRT7 inhibited the ability of SJSA-1 cell proliferation measured by clone formation. (H) Overexpression of SIRT7 promotes the ability of D-17 cell proliferation measured by clone formation. **P < 0.01 (in A, C, E-H) is based on Student’s t-test. Error bars indicate standard deviation.
SIRT7 induced osteosarcoma cell proliferation, migration and invasion in vitro
The ability of SIRT7 to modulate the proliferation of osteosarcoma cells was measured by the MTT assay. As shown in Figure 2E, silencing of SIRT7 in SJSA-1 cells significantly decreased cell proliferation. In contrast, ectopic expression of SIRT7 in D-17 cells significantly increases cell proliferation (Figure 2F). Consistently, the clone formation assay results also show that knocking down SIRT7 reduced the clone number of SJSA-1 cells (Figure 2G), while overexpression of SIRT7 robustly increased the clone number of D-17 cells (Figure 2H). In addition, the effects of SRIT7 on cell migration was assessed by the transwell migration assay. Silencing SIRT7 dramatically decreased the number of cells through the chamber compared to its control cells (Figure 3A). Moreover, silencing SIRT7 also decreased the degree of invasion through matrigel (Figure 3A). Similarly, ectopic expression of SIRT7 in D-17 cells dramatically increased their migration and invasion capacity (Figure 3B).
Figure 3.
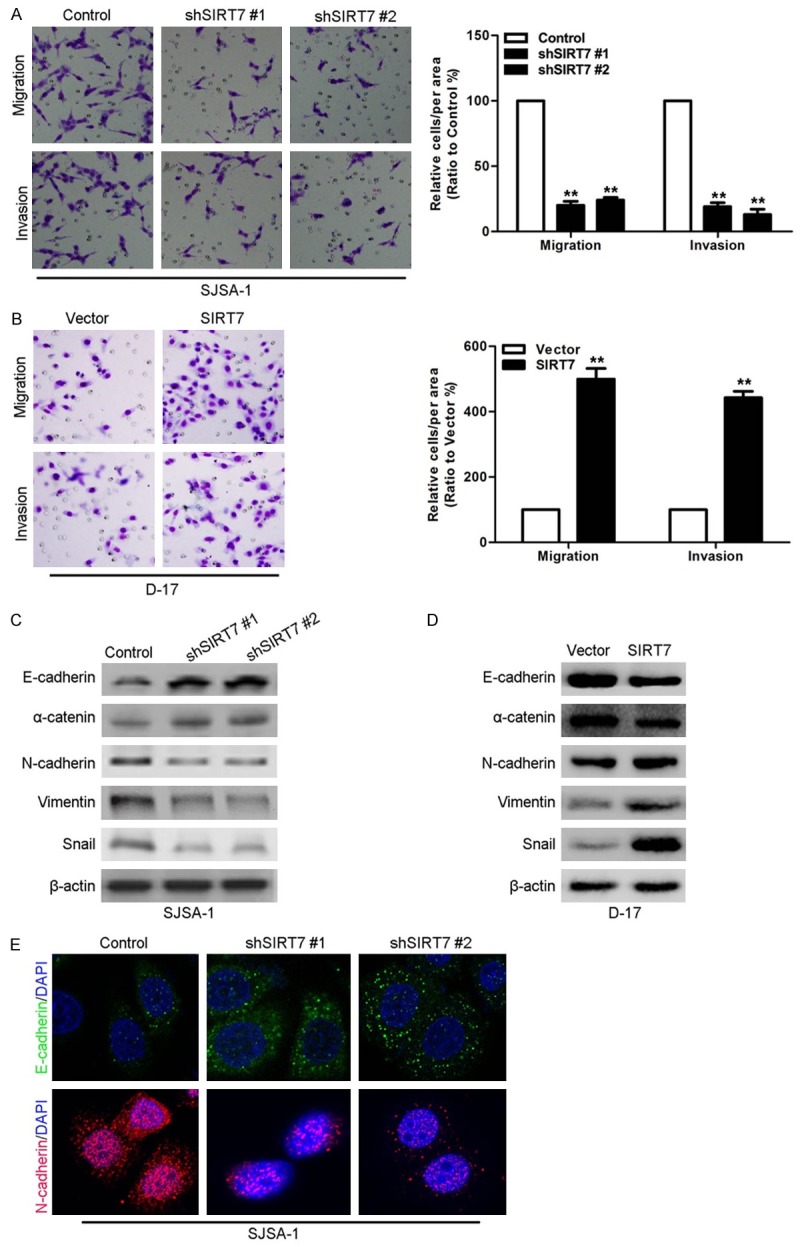
SIRT7 induced osteosarcoma cell migration and invasion in vitro. (A) SIRT7 knockdown and its control SJSA-1 cells were subjected to transwell migration (top) and matrigel invasion (bottom) assays. (B) SIRT7 overexpression and its control D-17 cells were subjected to transwell migration (top) and matrigel invasion (bottom) assays. (C) The expression of EMT markers were measured by Western blot in SIRT7 knockdown and its control SJSA-1 cells. (D) The expression of EMT markers were measured by Western blot in SIRT7 overexpression and its control D-17 cells. (E) The expression of EMT markers were measured by IF in SIRT7 knockdown and its control SJSA-1 cells. **P < 0.01 (in A and B) is based on Student’s t-test. Error bars indicate standard deviation.
To further investigate the cause of increased migration and invasion behavior due to the modulation of SIRT7, epithelial-mesenchymal transition (EMT) markers were measured by Western blotting as EMT is known to alter the ability of cells to migrate and invade. Silencing SIRT7 in SJSA-1 cells caused significant upregulation of epithelial markers (i.e., E-cadherin and α-catenin) and downregulation of mesenchymal markers (i.e., N-cadherin, vimentin, and snail) (Figure 3C), while ectopic expression of SIRT7 in D-17 cells had the reversed effect (Figure 3D). In addition, the IF assays also shown that silencing SIRT7 induced the E-cadherin expression and inhibited the Ncadherin expression in D-17 cells (Figure 3E). All these results demonstrate that SIRT7 promotes proliferation, migration, and invasion behaviors in osteosarcoma cells in vitro.
SIRT7 induced osteosarcoma cell sphere formation
Sphere formation is a rigorous assay for tumor malignancy, in which cells are grown in an ultra-low attachment and serum-free environment. The ability to form spheres under these conditions correlates with expression of stem cell markers and enhanced tumorigenesis in vivo. The results showed that SIRT7 knockdown effectively suppressed clonogenic capacity in the sphere formation assay in SJSA-1 cells (Figure 4A-C), and the overexpression of SIRT7 in D-17 significantly promoted this capacity (Figure 4D-F). Taken as a whole, these finding suggest an important role for SIRT7 in the maintenance of oncogenicity in osteosarcoma cells.
Figure 4.
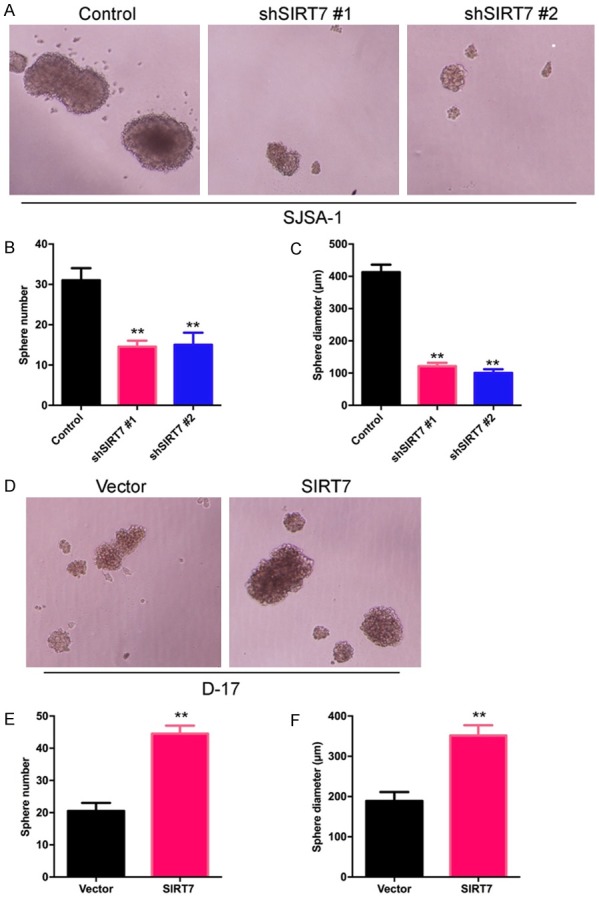
SIRT7 induced osteosarcoma cell sphere formation. (A) Representative phase-contrast images of sphere formation of SIRT7 silenced and its control SJSA-1 cells. (B and C) Quantification of sphere diameter (B) and number (C) of SIRT7 silenced and its control SJSA-1 cells. (D) Representative phase-contrast images of sphere formation of SIRT7 over-expression and its control D-17 cells. (E and F) Quantification of sphere diameter (E) and number (F) of SIRT7 over-expression and its control D-17 cells. **P < 0.01 (in B, C, E, and F) is based on Student’s t-test. Error bars indicate standard deviation.
SIRT7 promotes osteosarcoma tumor formation in vivo
To extend our in vitro observations, we investigated whether SIRT7 could regulate tumorigenic capacity of osteosarcoma cells in vivo. SJSA-1-shSIRT7, D-17-SIRT7, and their corresponding control cells were subcutaneously injected into nude mice. Tumor size was measured up to 28 days after injection. As expected, the tumors from SJSA-1-shSIRT7 cells grew less rapidly at the implantation site than the control cells (Figure 5A-C), and the ectopic expression of SIRT7 in the D-17 cells led to a dramatic increase in tumor volume and weight (Figure 5D-F). Increased cell proliferation in derived tumors was further confirmed by Ki67 level (Figure 5G and 5H). These results suggest that SIRT7 is an important regulator of proliferation in osteosarcoma cells in vivo.
Figure 5.
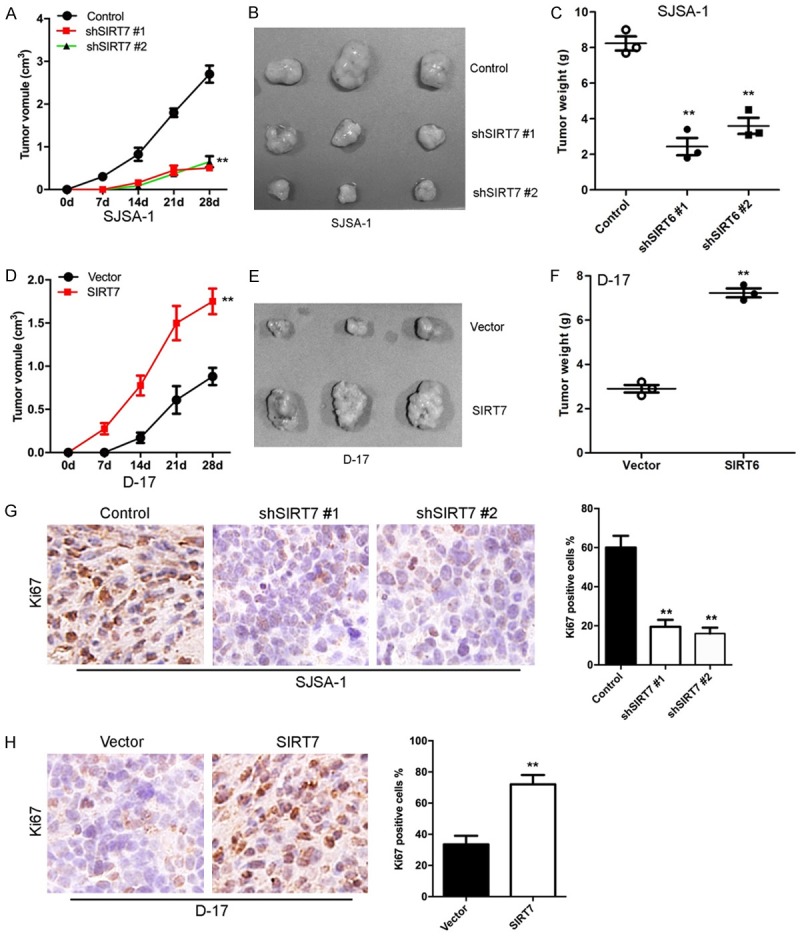
SIRT7 promotes osteosarcoma tumor formation in vivo. (A) Growth curve of tumors formed by SIRT7 silenced and its control SJSA-1 cells. (B) Representative images of tumor from SIRT7 silenced and its control SJSA-1 cells. (C) The weight of tumors formed by SIRT7 silenced and its control SJSA-1 cells. (D) Growth curve of tumors formed by SIRT7 overexpression and its control D-17 cells. (E) Representative images of tumor from SIRT7 overexpression and its control D-17 cells. (F) The weight of tumors formed by SIRT7 overexpression and its control D-17 cells. (G) Ki67 expression was measured by immunohistochemistry in tumors from SIRT7 silencing and its control cell. (H) Ki67 expression was measured by immunohistochemistry in tumors from SIRT7 overexpression and its control cell. **P < 0.01 (in C, F, G, and H) is based on Student’s t-test. Error bars indicate standard deviation.
SIRT7 promotes osteosarcoma cell metastasis in vivo
We then investigated the functional relevance of SIRT7 for metastasis in vivo. SJSA-1-shSIRT7, D-17-SIRT7, and their corresponding control cells were injected into nude mice through the tail vein. We observed that silencing SIRT7 significantly decreased the number of metastatic tumors in the liver of each mouse (Figure 6A-C); whereas, the ectopic expression of SIRT7 in D-17 cells increased the number of metastatic tumors in the liver of each mouse (Figure 6D-F). Therefore, the in vivo results further demonstrate the critical role of SIRT7 in osteosarcoma metastasis.
Figure 6.
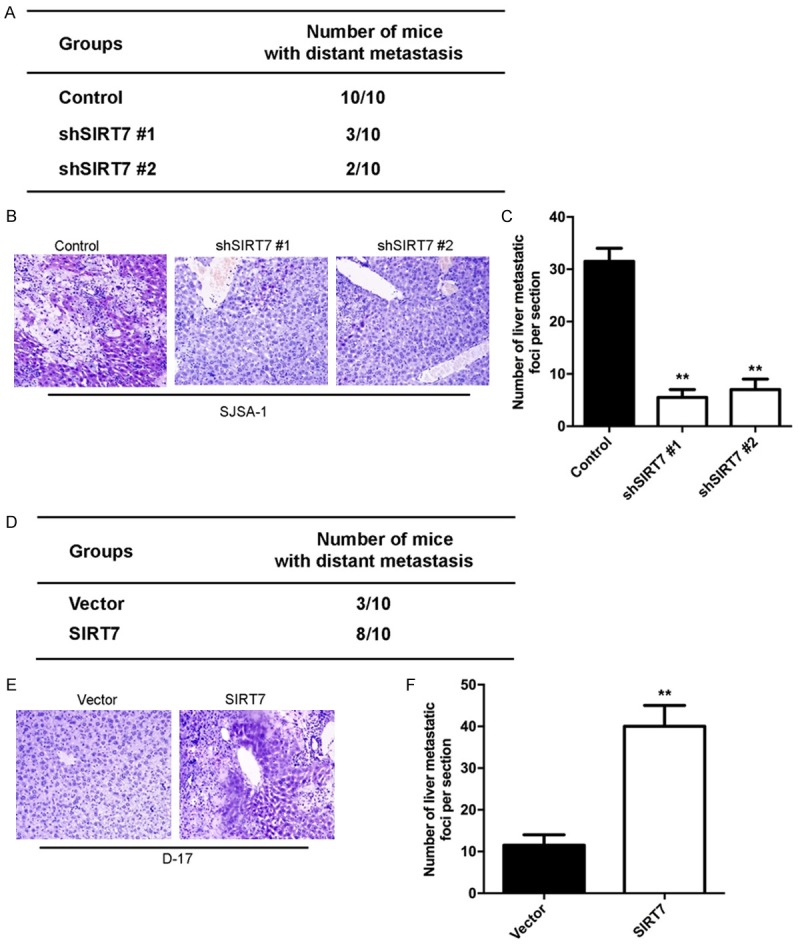
SIRT7 promotes osteosarcoma cell metastasis in vivo. (A) The total numbers of mice with liver distant metastasis at 30 days after injection of SJSA-1-shSIRT7 or its control cells into tail vein. (B) Representative images of liver tissues measured by H&E stain with injection of SJSA-1-shSIRT7 or its control cells into tail vein. (C) The numbers of metastatic foci per section in liver of individual mouse with injection of SJSA-1-shSIRT7 or its control cells into tail vein. (D) The total numbers of mice with liver distant metastasis at 30 days after injection of D-17-SIRT7 or its control cells into tail vein. (E) Representative images of liver tissues measured by H&E stain with injection of D-17-SIRT7 or its control cells into tail vein. (F) The numbers of metastatic foci per section in liver of individual mouse with injection of D-17-SIRT7 or its control cells into tail vein. **P < 0.01 (in C and F) is based on Student’s t-test. Error bars indicate standard deviation.
SIRT7 inhibits the expression of CDC4
To better understand the mechanisms by which SIRT7 is engaged in proliferation, migration, and invasion functionality, we performed gene expression profiling on D-17-SIRT7 and its control cells. Microarray analyses identified a list of significantly differentially expressed genes after SIRT7 overexpression, including the down-regulation of CDC4 (Figure 7A and 7B). The data from the TCGA showed that the expression of SIRT7 negatively correlated with CDC4 expression (Figure 7C). The same correlation was also observed in our osteosarcoma tissues by measuring the mRNA expressions of SIRT7 and CDC4 using qRT-PCR (Figure 7D). Together with previous literature linking CDC4 with tumorigenesis and metastasis in carcinomas [17], we examined whether CDC4 is a downstream target of SIRT7. Expression of CDC4 in the cells with altered SIRT7 expression was evaluated by Western blot and qRT-PCR. Silencing SIRT7 in SJSA-1 cells dramatically increased CDC4 expression at protein and mRNA levels (Figure 8A and 8B). Moreover, overexpression SIRT7 in D-17 cells greatly decreased CDC4 expression at protein and mRNA levels (Figure 8C and 8D). It has been demonstrated that CDC4 targets many proteins for degradation, such as aurora-A, cyclin E, c-Myc, EZH2, and ENO1. Thus, we measured the level of these proteins in SIRT7 altered cells. The results showed that silencing SIRT7 significantly inhibited the expression of these proteins (Figure 8E), while ectopic expression of SIRT7 drastically upregulated these proteins (Figure 8F). All of these results indicate that CDC4 is the downstream target of SIRT7.
Figure 7.
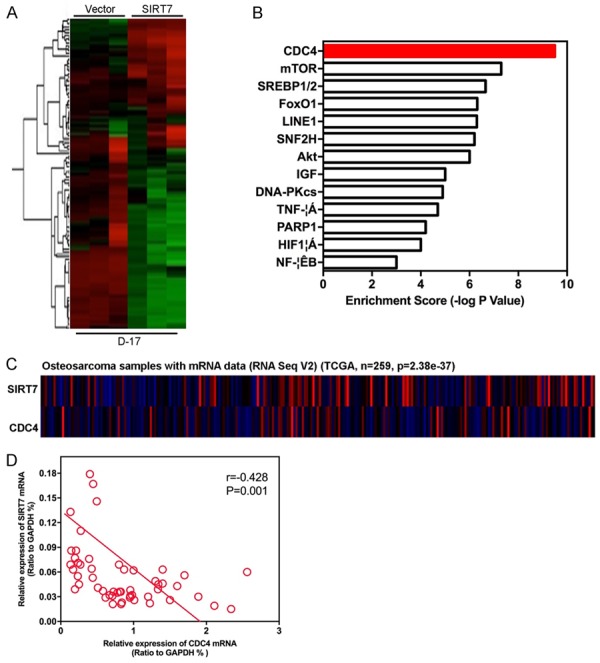
SIRT7 expression is relevant to CDC4. A: Gene expression profiling on D-17-SIRT7 and its control cells were assayed by microarray analyses. B: Gene set enrichment analysis was carried out using ConceptGen. C: The data from the TCGA showed that the expression of SIRT7 negatively correlated with CDC4 expression in osteosarcoma. D: CDC4 mRNA expression was negatively correlated with SIRT7 mRNA in osteosarcoma tissues.
Figure 8.
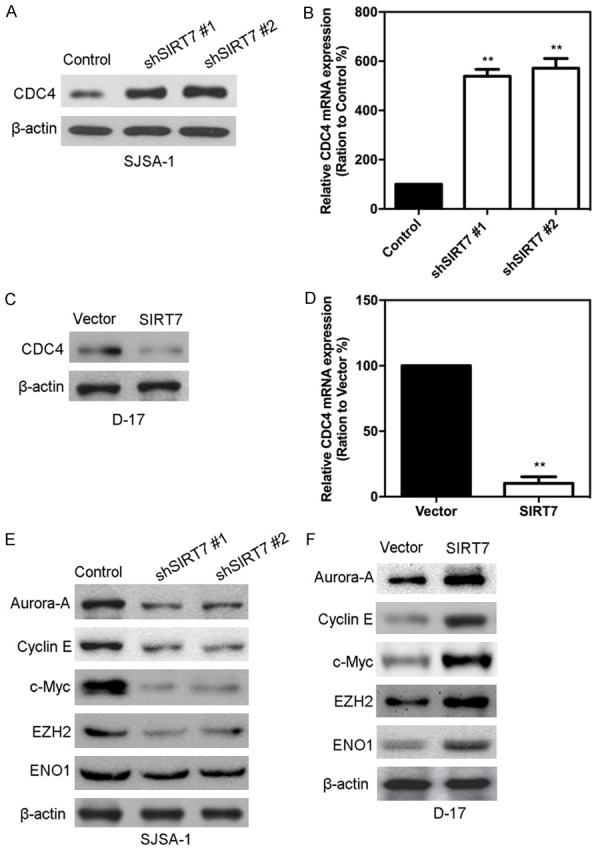
SIRT7 inhibits the expression of CDC4. (A) Expression of CDC4 protein in SJSA-1-shSIRT7 and its control cells were measured by Western blot. (B) Expression of CDC4 mRNA in SJSA-1-shSIRT7 and its control cells were measured by qRT-PCR. (C) Expression of CDC4 protein in D-17-SIRT7 and its control cells were measured by Western blot. (D) Expression of CDC4 mRNA in D-17-SIRT7 and its control cells were measured by qRT-PCR. (E) CDC4 target proteins were measured by Western blot in SJSA-1-shSIRT7 and its control cells. (F) CDC4 target proteins were measured by Western blot in D-17-SIRT7 and its control cells. **P < 0.01 (in B and D) is based on Student’s t-test. Error bars indicate standard deviation.
SIRT7 down regulates H3K18ac expression, and decrease H3K18ac binding to the promoter region of CDC4
We then explored the mechanism of how SIRT7 regulates CDC4 expression. Histone deacetylases are frequently involved in chromatin regulation and histone modifications that play important roles in tumor progression. To determine whether SIRT7 activity was associated with specific histone modifications in osteosarcoma cells, histone modification patterns were measured after modulation of SIRT7 expression. We found that H3K18ac protein expression was affected by changes in SIRT7 expression, in that the silencing of SIRT7 increased the protein levels of H3K18ac (Figure 9A), while ectopic expression of SIRT7 decreased this modification (Figure 9B). Because H3K18ac is associated with active transcription, we tested whether SIRT7 expression correlated with the H3K18ac modification at the CDC4 gene promoter in osteosarcoma cells. ChIP assay was performed in SJSA-1-shSIRT7, D-17-SIRT7, and their control cells. Antibodies against H3K18ac and IgG were used to pull down the chromatin complex, and three pairs of primers against the CDC4 gene promoter region were used to assess the occupancy of the CDC4 gene promoter (Figure 9C). Silencing SIRT7 expression was associated with increased H3K18ac levels at #2 and #3 of the CDC4 gene promoter region (Figure 9D). Less occupancy of those CDC4 gene promoter regions by H3K18ac was detected in D-17-SIRT7 cells (Figure 9E). These results clearly indicate that SIRT7 induces transcriptional inactivation of CDC4 expression through regulating H3K18ac and decreased H3K18ac binding to the CDC4 gene promoter in osteosarcoma cells.
Figure 9.
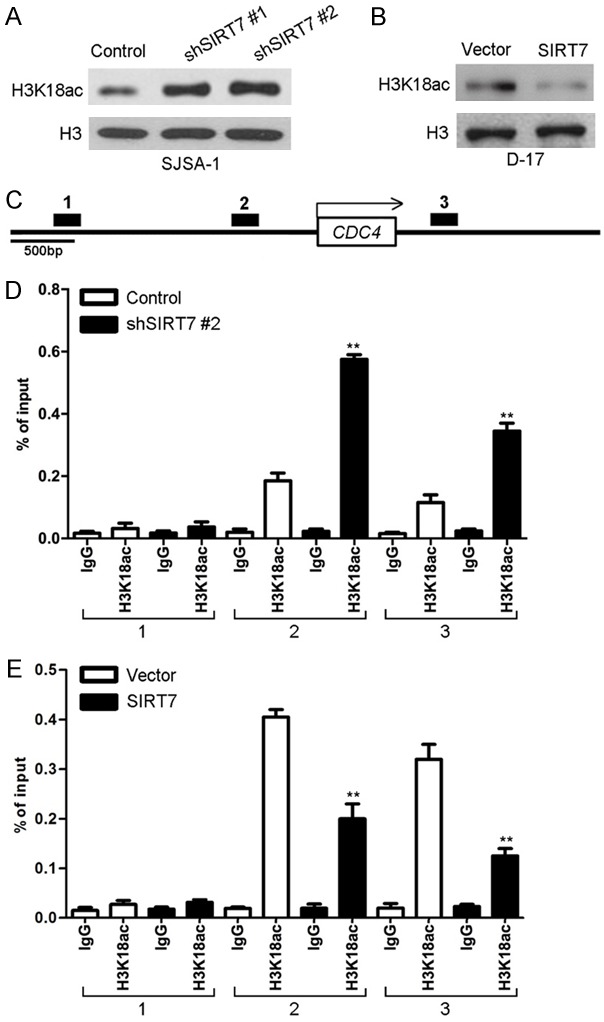
SIRT7 down regulates H3K18ac expression, and decreases H3K18ac binding to the promoter region of CDC4. (A) The expression of H3K18ac was assayed by Western blot in SJSA-1-shSIRT7 and its control cells. (B) The expression of H3K18ac was assayed by Western blot in D-17-SIRT7 and its control cells. (C) Schematic presentation of three regions relative to the CDC4 transcriptional start site used as primers to test H3K18ac occupied abundance. (D) qChIP was performed to assess H3K18ac occupancy to CDC4 transcriptional start site in SJSA-1-shSIRT7 and its control cells. IgG was used as negative control. (E) qChIP was performed to assess H3K18ac occupancy to CDC4 transcriptional start site in D-17-SIRT7 and its control cells. IgG was used as negative control. **P < 0.01 (in D and E) is based on Student’s t-test. Error bars indicate standard deviation.
CDC4 is a mediator of SIRT7-induced malignant behaviors in osteosarcoma cells
To test whether SIRT7-induced malignant behaviors in osteosarcoma cells were mediated by CDC4, shRNA was used to silence the CDC4 gene expression in SJSA-1-shSIRT7 cells (Figure 10A). The MTT result showed that the knockdown of CDC4 in SJSA-1-shSIRT7 cells resulted in increased proliferation (Figure 10B). To verify whether CDC4 mediates SIRT7 induced migration and invasion in vitro, transwell and matrigel assays were performed. As shown in Figure 10C, knockdown of CDC4 in SJSA-1-shSIRT7 cells significantly decreases their migratory and invasive capacities. In addition, knocking down CDC4 rescued the sphere formation ability caused by silencing SIRT7 in SJSA-1 cells (Figure 10D-F). Taken together, these results show that CDC4 mediates SIRT7-induced malignant behaviors in osteosarcoma cells.
Figure 10.
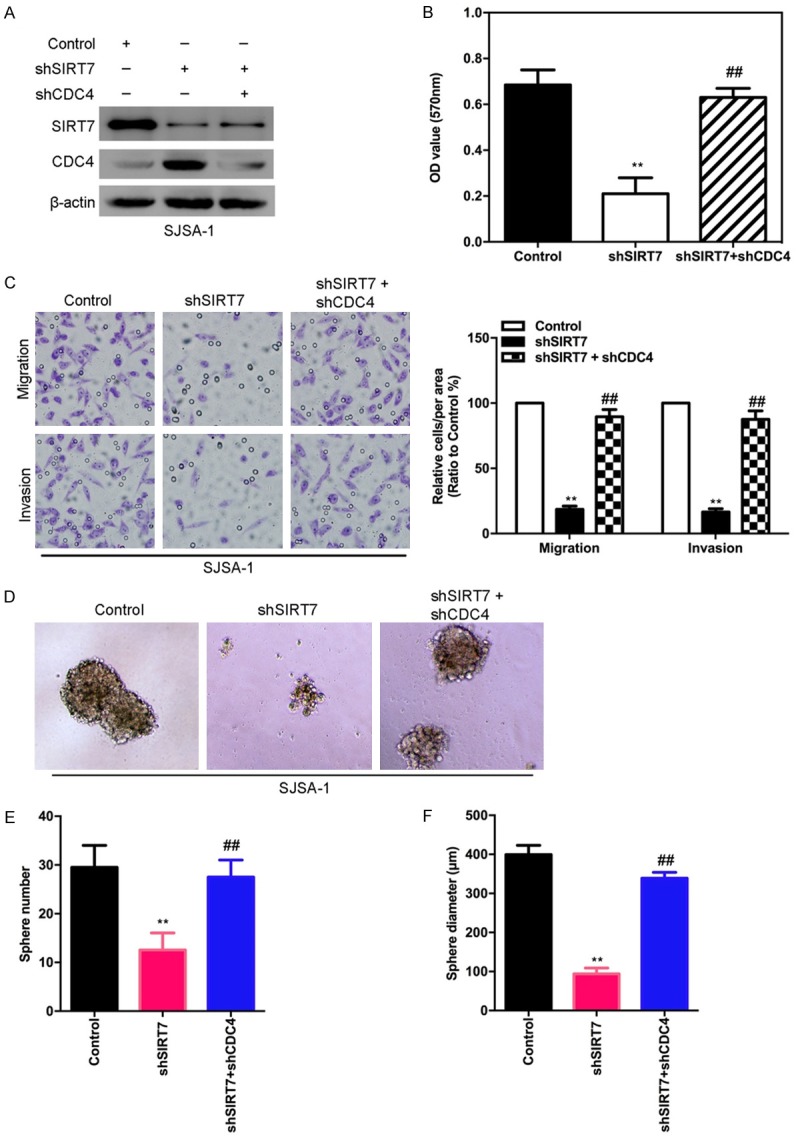
CDC4 is a mediator of SIRT7-induced malignant behaviors in osteosarcoma cells. (A) The silencing of CDC4 was measured by Western blot in SJSA-1-shSIRT-7 cells. (B) The proliferation was measured by MTT in knockdown of CDC4 in SJSA-1-shSIRT7 cells. (C) CDC4 knockdown and its control cells were subjected to transwell migration (top) and matrigel invasion (bottom) assays. (D) Representative phase-contrast images of sphere formation of CDC4 silenced and its control SJSA-1-shSIRT7 cells. (E and F) Quantification of sphere diameter (E) and number (F) of CDC4 silenced and its control SJSA-1-shSIRT7 cells. **,##P < 0.01 (in B, C, E, and F) is based on Student’s t-test. Error bars indicate standard deviation.
Discussion
Many researchers have demonstrated that histone modifications play a crucial function in the tumorigenesis and development [18]. Histone H3K18Ac deacetylase SIRT7 plays a pivotal role in regulating histone modification, which participates in the activation of RNA polymerase I transcription and plays an important part in rRNA expression [10]. Recent studies show that SIRT7 is upregulated in ovarian and colorectal cancer [6,19], yet no study has assessed SIRT7’s exact activity in osteosarcoma. Here, we aim to detect the role of SIRT7 in osteosarcoma development and its potential molecular mechanism.
In this study, we find for the first time the clinical significance of SIRT7 in osteosarcoma, and the mechanistic role of SIRT7 in regulating osteosarcoma cells’ proliferation and metastasis. To our knowledge, this is the first study to show that SIRT7 plays a crucial functional role in osteosarcoma cells’ proliferation and metastasis. Overexpression of SIRT7 in osteosarcoma cells induces proliferation, migration, and invasion traits in vitro and enhanced metastatic capacity in vivo; and silencing SIRT7 reverses these events in invasive osteosarcoma cells. We also show that a mechanistic link exists between SIRT7 and CDC4 through SIRT7-mediated regulation of H3K18ac, which subsequently leads to transcriptional downregulation of CDC4 expression. Knockdown of CDC4 attenuates silencing SIRT7-induced effects. These results lead us to propose a model for SIRT7 regulation of proliferation and metastasis through transcriptional regulation of CDC4 in osteosarcoma cells.
The putative role of SIRT7 as an oncogene in cancer development is supported by the observations that SIRT7 is highly expressed in colorectal cancers, breast cancers, and other tumors [19,20]. Our study points to a novel function of SIRT7 in osteosarcoma metastasis through promoting two essential characteristics of metastatic disease in osteosarcoma: proliferation and invasion. All of these characteristics induced by SIRT7 in vitro culminate in an increased number of distant metastases in vivo. These empirical findings provide a mechanistic framework to explain the clinical observations that osteosarcoma patients with high levels of SIRT7 in tissue samples have more chances of distant metastasis and a significantly shorter overall and disease-free survival.
In our effort to discover the molecular mechanism through which SIRT7 modulates proliferation and metastasis in osteosarcoma cells, we focus on CDC4 as a crucial mediator of SIRT7-induced malignancy property. CDC4 functions as a substrate recognition subunit of the SCF E3 ubiquitin ligase complex to regulate a network of proteins with central roles in cell division, growth, and differentiation and has been characterized as a general tumor suppressor [21]. CDC4 mutation or deletion is often observed in multiple human cancers and loss of CDC4 function results in tumorigenesis [22,23]. The mechanistic connection between SIRT7 and CDC4 has been previously unknown. In this study, we found that SIRT7 induces transcriptional inactivation of CDC4 expression through regulating H3K18ac and decreasing the binding of H3K18ac to the CDC4 gene promoter in osteosarcoma cells. Thus, we conclude that SIRT7 transcriptionally inactivates CDC4 expression and consequently promotes migration in vitro and metastasis in vivo.
Metastasis and proliferation properties are essential for osteosarcoma cells to disseminate from adjacent tissues and seed new tumors in distant sites. Our results demonstrate that SIRT7 regulates these two essential characteristics of metastatic disease, providing us an optimal therapeutic option to manipulate SIRT7 levels in clinical osteosarcoma practice.
Disclosure of conflict of interest
None.
References
- 1.Wen ZQ, Li XG, Zhang YJ, Ling ZH, Lin XJ. Osteosarcoma cell-intrinsic colony stimulating factor-1 receptor functions to promote tumor cell metastasis through JAG1 signaling. Am J Cancer Res. 2017;7:801–815. [PMC free article] [PubMed] [Google Scholar]
- 2.Liu MH, Cui YH, Guo QN, Zhou Y. Elevated ASCL2 expression is associated with metastasis of osteosarcoma and predicts poor prognosis of the patients. Am J Cancer Res. 2016;6:1431–1440. [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar RM, Arlt MJ, Kuzmanov A, Born W, Fuchs B. Sunitinib malate (SU-11248) reduces tumour burden and lung metastasis in an intratibial human xenograft osteosarcoma mouse model. Am J Cancer Res. 2015;5:2156–2168. [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Redondo P, Santos-Barriopedro I, Vaquero A. A big step for SIRT7, one giant leap for Sirtuins… in cancer. Cancer Cell. 2012;21:719–721. doi: 10.1016/j.ccr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 5.Paredes S, Villanova L, Chua KF. Molecular pathways: emerging roles of mammalian sirtuin SIRT7 in cancer. Clin Cancer Res. 2014;20:1741–1746. doi: 10.1158/1078-0432.CCR-13-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X, Zhou Y, Wang H, Pan C, Huang W. Overexpression of sirt7 exhibits oncogenic property and serves as a prognostic factor in colorectal cancer. Clin Cancer Res. 2014;20:3434–3445. doi: 10.1158/1078-0432.CCR-13-2952. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez BN, Thackray JK, Simonet NG, Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, Bunting S, Vaquero A, Tischfield JA, Serrano L. SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 2016;35:1488–1503. doi: 10.15252/embj.201593499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L, Liang J, Cheng Z, Shi L, Shang Y, Yu W. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonet NG, Vaquero A. Raising the list of SirT7 targets to a new level. Proteomics. 2017:17. doi: 10.1002/pmic.201700137. [DOI] [PubMed] [Google Scholar]
- 10.Zhang PY, Li G, Deng ZJ, Liu LY, Chen L, Tang JZ, Wang YQ, Cao ST, Fang YX, Wen F, Xu Y, Chen X, Shi KQ, Li WF, Xie C, Tang KF. Dicer interacts with SIRT7 and regulates H3K18 deacetylation in response to DNA damaging agents. Nucleic Acids Res. 2016;44:3629–3642. doi: 10.1093/nar/gkv1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu ML, Hsu CM, Yang MY. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 2013;34:1847–1854. doi: 10.1007/s13277-013-0726-y. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, Hu Y, Cai T. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5:9787. doi: 10.1038/srep09787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Lee HS, Jung W, Lee E, Chang H, Choi JH, Kim HG, Kim A, Kim BH. SIRT7, H3K18ac, and ELK4 immunohistochemical expression in hepatocellular carcinoma. J Pathol Transl Med. 2016;50:337–344. doi: 10.4132/jptm.2016.05.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbi ME, Hu H, Kshitiz , Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J Biol Chem. 2013;288:20768–20775. doi: 10.1074/jbc.M113.476903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang P, He X, Wang Q, Huang Y, Jen KY, LaBarge MA, You L, Kogan SC, Gray JW, Mao JH, Wei G. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014;74:520–531. doi: 10.1158/0008-5472.CAN-13-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Wang Y, Fan C, Gao P, Wang X, Wei G, Wei J. Estrogen promotes stemness and invasiveness of ER-positive breast cancer cells through Gli1 activation. Mol Cancer. 2014;13:137. doi: 10.1186/1476-4598-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhang P, Wang Y, Zhan P, Liu C, Mao JH, Wei G. Distinct interactions of EBP1 isoforms with FBXW7 elicits different functions in cancer. Cancer Res. 2017;77:1983–1996. doi: 10.1158/0008-5472.CAN-16-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kofman AE, Huszar JM, Payne CJ. Transcriptional analysis of histone deacetylase family members reveal similarities between differentiating and aging spermatogonial stem cells. Stem Cell Rev. 2013;9:59–64. doi: 10.1007/s12015-012-9392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HL, Lu RQ, Xie SH, Zheng H, Wen XM, Gao X, Guo L. SIRT7 exhibits oncogenic potential in human ovarian cancer cells. Asian Pac J Cancer Prev. 2015;16:3573–3577. doi: 10.7314/apjcp.2015.16.8.3573. [DOI] [PubMed] [Google Scholar]
- 20.Liu GF, Lu JY, Zhang YJ, Zhang LX, Lu GD, Xie ZJ, Cheng MB, Shen YF, Zhang Y. C/EBPalpha negatively regulates SIRT7 expression via recruiting HDAC3 to the upstream-promoter of hepatocellular carcinoma cells. Biochim Biophys Acta. 2016;1859:348–354. doi: 10.1016/j.bbagrm.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Tan Y, Sangfelt O, Spruck C. The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett. 2008;271:1–12. doi: 10.1016/j.canlet.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 22.Chiang CH, Chu PY, Hou MF, Hung WC. MiR-182 promotes proliferation and invasion and elevates the HIF-1alpha-VEGF-A axis in breast cancer cells by targeting FBXW7. Am J Cancer Res. 2016;6:1785–1798. [PMC free article] [PubMed] [Google Scholar]
- 23.Grim JE. Fbxw7 hotspot mutations and human colon cancer: mechanistic insights from new mouse models. Gut. 2014;63:707–709. doi: 10.1136/gutjnl-2013-305144. [DOI] [PubMed] [Google Scholar]