Abstract
In this study, we confirmed that HOXC13 might be a potential oncogene in lung adenocarcinoma through an analysis of The Cancer Genome Atlas (TCGA) datasets. Further analysis revealed that the expression of HOXC13 was significantly higher in lung adenocarcinoma tissues than in adjacent normal tissues; importantly, its expression correlated with poor clinical characteristics and worse prognosis. In vitro experiments showed that HOXC13 expression generally increased in lung adenocarcinoma cell lines. Moreover, knockdown of HOXC13 inhibited lung adenocarcinoma cell proliferation, and induced G1-phase arrest via downregulation of CCND1 and CCNE1. Conversely, HOXC13 overexpression promoted lung adenocarcinoma cell proliferation, and decreased the percentage of cells in G1-phase via upregulation of CCND1 and CCNE1. We also found that miR-141 downregulated HOXC13, by directly targeting its 3’UTR, and inhibited proliferation of lung adenocarcinoma cells. Taken together, our results suggest that HOXC13, which is directly targeted by miR-141, is highly expressed in lung adenocarcinoma, and promotes proliferation of lung adenocarcinoma by modulating the expression of CCND1 and CCNE1.
Keywords: HOXC13, lung adenocarcinoma, CCND1, CCNE1, miR-141
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide [1,2]; the most common type is non-small-cell lung cancer (NSCLC) [3], which accounts for ~85% of all lung cancer deaths [4]. Despite recent advances in multi-modality therapy, the 5-year lung cancer survival rate is 4-17%, depending on stage and regional differences [5]. Adenocarcinoma is currently the predominant histological subtype of NSCLC [6,7]; therefore, screening for new functional genes and biomarkers in lung adenocarcinoma development may yield alternative approaches for managing lung cancer.
By analyzing the TCGA datasets, we obtained three lung adenocarcinoma-related candidate genes (PITX2, DMBX1, and HOXC13) that were expressed at significantly higher levels in lung adenocarcinoma tissues than in adjacent normal tissues. The literature reports that HOXC13 is highly expressed in ameloblastoma [8], odontogenic tumors [9], metastatic melanoma [10], and liposarcoma [11]; moreover, knockdown of HOXC13 affected cell growth, and resulted in cell cycle arrest, in colon cancer [12]. However, expression and function of HOXC13 in lung cancer remains to be investigated, and, therefore, HOXC13 was chosen as the subject of our study. HOXC13 is one of several homeobox HOXC genes located in a cluster on chromosome 12, and it has been reported to play a role in the development of hair, nail, and filiform papilla [13-15]. Identification of the role of HOXC13 in lung adenocarcinoma development is expected to provide new biomarkers and therapeutic targets.
MicroRNAs (miRNAs) are a class of non-coding RNAs that bind to the 3’ untranslated region (UTR) of messenger RNA (mRNA), and either inhibit protein translation or destabilize target mRNA [16]. MicroRNAs play important roles in physiological processes, including developmental timing, cell death and proliferation, hematopoiesis, and patterning of the nervous system [17]. Studies are increasingly implicating miRNAs in the pathogenesis of many types of cancer, including lung cancer [18-22]. Herein, we show that miR-141 targets the 3’UTR of HOXC13. This miRNA has been widely studied in oncology, and is involved in the inhibition of tumor growth or migration in colorectal cancer [23], esophageal squamous cell carcinoma [24], breast cancer [25], and gastric cancer [26]. Further studies on the influence of miR-141 on HOXC13, and its role in lung adenocarcinoma, are meaningful and necessary.
Materials and methods
Data sources and bioinformatics
Two TCGA datasets, named TCGA_LUNG_exp_HiseqV2-2015-02-24 and TCGA_LUAD_exp_HiSeqV2-2015-02-24, were downloaded using the UCSC Cancer Browser (https://genome-cancer.ucsc.edu/) [27]. All normalized gene expression values can be obtained from “genomicMatrix” files. Using the co-expression tool in cBioPortal (http://www.cbioportal.org/) [28], we obtained a list of 313 genes with high co-expression correlation (Pearson score >0.3) with HOXC13. These genes were submitted to DAVID Bioinformatics Resources 6.8 (http://david.abcc.ncifcrf.gov/) [29] for KEGG pathway (GO) enrichment analysis, and to the Reactome Pathway Database (http://www.reactome.org/), [30] for reactome pathway enrichment analysis.
Tissue samples, cell culture, shRNA, plasmid DNA, microRNA mimics, and transfection
In total, 60 lung adenocarcinoma tissue samples were obtained from patients who had undergone curative surgical resection at Nanjing Chest Hospital, from 2013 to 2016. None of the patients had received preoperative chemotherapy or radiotherapy. A549 and H1299 cells were cultured in RPMI1640 media (KeyGEN, Nanjing, China) supplemented with 10% fetal bovine serum and penicillin/streptomycin (KeyGEN, Nanjing, China), and cultured at 37°C in a humidified incubator containing 5% CO2. Transfection of small hairpin RNA (shRNA), plasmid DNA, and miR-141 mimics were performed according to the Lipofectamine 3000 reagent (Invitrogen, USA) protocol; nonsense shRNA (sh-nc), empty plasmid (pcDNA), and negative control mimic (miR-NC) were used as the respective controls. Transfection efficiency was evaluated by quantitative real-time RT-PCR (qRT-PCR) and western blot.
The sequences used were as follows: shRNA-1 for HOXC13, 5’-GAGCCTTATGTACGTCTATGATTCAAGAGATCATAGACGTACATAAGGCTCTTTTTT-3’; shRNA-2 for HOXC13, 5’-GCAAATCGAAAGCGCCTCATCTTCAAGAGAGATGAGGCGCTTTCGATTTGCTTTTTT-3’; sh-nc, 5’-GCACCCAGTCCGCCCTGAGCAAATTCAAGAGATTTGCTCAGGGCGGACTGGGTGCTTTTT-3’; miR-141 mimics, 5’-UAACACUGUCUGGUAAAGAUGG-3’, and miR-NC mimics, 5’-UUCUCCGAACGUGUCACGUTT-3’. Overexpression of HOXC13, CCND1, and CCNE1 was achieved using plasmids containing the respective full-length human genes purchased from Vigene Biosciences, Shandong, China.
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was extracted from tissue samples or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. Reverse transcription was performed with 1000 ng total RNA in a final volume of 20 μL, using a Reverse Transcription Kit (Takara, cat: RR036A, KeyGEN, Nanjing, China). For qRT-PCR, SYBR Select Master Mix (Applied Biosystems, Cat: 4472908. KeyGEN, Nanjing, China) was used, and the reaction was performed in a QuantStudioTM 6 Flex Real-Time PCR System as follows: initial denaturation step at 95°C for 10 min, followed by 40 cycles at 92°C for 15 s and 60°C for 1 min. Primers are shown in Table 2 and ACTB was used as housekeeping gene. The comparative CT method (ΔΔCT) was used to measure relative gene expression.
Table 2.
Sequences of qRT-PCR primers
| Gene | Sense | Antisense |
|---|---|---|
| HOXC13 | AAGCTTACGACTTCGCTGCTCCTGC | GGATCCTCAGGTGGAGTGGAGATGAGGC |
| CDKN1B (p27) | TGGAGAAGCACTGCAGAGAC | GCGTGTCCTCAGAGTTAGCC |
| CDKN1A (p21) | GCAGACCAGCATGACAGATTT | GGATTAGGGCTTCCTCTTGGA |
| CCND1 | GCGCTTCCAACCCACCCTCCATG | GCGCCGCAGGCTTGACTCCAGAA |
| CCNE1 | TTCTTGAGCAACACCCTCTTCTGCAGCC | TCGCCATATACCGGTCAAAGAAATCTTGTGCC |
| ACTB | GAAATCGTGCGTGACATTAA | AAGGAAGGCTGGAAGAGTG |
Protein extracts and western blot analysis
Cells were harvested and treated with lysis buffer (RIPA, KeyGEN, Nanjing, China) on ice, and protein concentration was determined using a BCA kit (KeyGEN, Nanjing, China). Comparable amounts of extracts were loaded on SDS-PAGE gels and subjected to electrophoresis. After separation on the gel, proteins were transferred to a PVDF membrane. Membranes were blocked in 2% BSA in TBS-T for 1 h, and subsequently incubated overnight, at 4°C, with antibodies against HOXC13 (Santa Cruz, sc514377; 1:1,000), p21 (Santa Cruz, sc-397; 1:500), p27 (Santa Cruz, sc-528; 1:500), cyclin D1 (CST, #2978; 1:1,000), cyclin E1 (Abcam, ab7959; 1:1,000), or β-actin (Cell Signaling, 8H10D10; 1:1,000). After washing in TBS-T, membranes were incubated with goat anti-rabbit or goat anti-mouse HRP-conjugated secondary antibodies (both from Abcam; 1:10,000), for 2 h at room temperature. Blots were visualized using ECL detection (Thermo Scientific). All experiments were repeated at least three times, independently.
Cell counting kit-8 (CCK-8), colony formation, 5-ethynyl-2’-deoxyuridine (EDU), and xCELLigence system assays
The assays were performed 24 h after transfection. For CCK-8 assay, cells were plated in 96-well plates at a density of 2,000 cells/100 µL, and the absorbance was measured at 450 nm in an ELx-800 Universal Microplate Reader. For colony formation assay, a total of 200 cells were placed in a fresh 6-well plate and cultured in medium containing 10% FBS, with medium replacement every 3-4 d. After two weeks, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Visible colonies were manually counted. For EDU assay, 10,000 cells/100 µL were plated in 96-well plates. After incubation with EDU solution for 2 h, cells were fixed with 4% paraformaldehyde for nucleus and DNA staining. Images were obtained from fluorescence microscope for further calculation of proliferation rates. For the xCELLigence system, 8,000 cells/100 µL were seeded in E-plates, and the plates were locked into the RTCA DP device in the incubator. The proliferative ability in each well was automatically monitored by the xCELLigence system, and expressed as a “cell index” value. All experiments were repeated at least three times.
Cell cycle analysis
Flow-cytometry analysis was performed to detect cell cycle distribution. Cells were fixed, for at least 2 h at 4°C, in centrifuge tubes containing 4.5 mL 70% ethanol. Next, cells were centrifuged for 5 min at 300×g. The pellet was resuspended in 5 mL of PBS, for ~30 s, and centrifuged again at 300×g for 5 min. Finally, the pellet was resuspended in 1 mL of PI staining solution, and kept in the dark at 37°C, for 10 min. Samples were analyzed using a FACSCalibur flow cytometer. The percentage of cells in G1, S, and G2-M phases were counted and compared. Each experimental group was analyzed at least three times.
Luciferase reporter assay
To construct HOXC13 3’-UTR plasmid, a wild type (WT) 3’-UTR fragment of HOXC13 containing the putative miR-141 binding sequence was amplified by PCR and cloned into XbaI and SacI sites of the pmirGLO dual-luciferase miRNA target expression vector (Promega, Madison, WI, USA). This plasmid was named as WT-HOXC13 3’-UTR. The mutant variant of HOXC13 3’-UTR (named Mut-HOXC13 3’-UTR) was generated based on WT-HOXC13 3’-UTR, by mutating six nucleotides that potentially bind to miR-141. For the luciferase assay, A549 cells were seeded in 24-well plates, at 2×105 cells per well, and co-transfected with WT-HOXC13 3’-UTR or Mut-HOXC13 3’-UTR, and miR-141 mimics or miR-NC mimics, using the Lipofectamine 2000 reagent (Invitrogen). The relative Firefly luciferase activity, normalized with that of Renilla luciferase, was measured 48 h after transfection by using the Dual-Light luminescent reporter gene assay (Promega, Madison, WI, USA). All experiments were repeated at least three times.
Statistical analyses
All results are presented as the mean ± standard deviation (SD). Student’s t test, chi-square test, Cox regression analysis, Pearson test, one-way ANOVA analysis, and Kaplan-Meier survival analysis were used to analyze the data using SPSS Statistics software (version 20.0, Chicago, Ill, USA). P<0.05 was considered statistically significant. Graphs were made using the GraphPad Prism 6.0 software package.
Results
Bioinformatics and tissue sample analysis indicates that HOXC13 is highly expressed in lung adenocarcinoma tissues, correlating with poorer prognosis and more aggressive clinical characteristics
To screen for differentially expressed genes between lung adenocarcinoma tissues and adjacent normal tissues, we analyzed two datasets: TCGA_LUNG_exp_HiseqV2-2015-02-24 and TCGA_LUAD_exp_HiSeqV2-2015-02-24. The screening criteria firstly required the fold change in average gene expression, between tumor tissues and adjacent normal tissues, to be >10. Secondly, our criteria required the average gene expression in tumor tissues to be >3. This screen yielded three genes: PITX2, DMBX1, and HOXC13 (Figure 1A). For the reasons detailed in the introduction, HOXC13 was chosen as our research object. Further analysis of TCGA_LUAD_exp_HiSeqV2-2015-02-24 revealed that expression of HOXC13 is significantly higher in lung adenocarcinoma tissues (513 cases) than in normal tissues (58 cases) (Figure 1B). For the 511 cases of lung adenocarcinoma tissues with clinical prognosis and TNM stage information, we interpreted cases in the upper quartile of HOXC13 expression as having “high HOXC13 expression”, and cases in the lower quartile as having “low HOXC13 expression”. Survival curve analysis demonstrated that patients with low expression of HOXC13 presented a higher percentage of overall and disease-free survival than patients with high expression of HOXC13 (for median overall survival, 1,725 d versus 1,258 d, with P=0.0247; for disease-free survival, P=0.0202; Figure 1C and 1D). Additionally, correlation analysis between HOXC13 expression and clinical characteristics indicated that expression of HOXC13 is closely linked to lymph node metastasis (P=0.000005) and TNM stage (P=0.000001) (Table 1). By measuring the mRNA expression of HOXC13 in lung adenocarcinoma tissues from patients at the Nanjing Chest Hospital, we found that HOXC13 was upregulated in 83.3% of 60 lung adenocarcinoma tissues (Figure 1E), and overexpression of HOXC13 was associated with greater T stage (P<0.0001), N stage (P=0.0025), and TNM stage (P=0.0006) (Figure 1F).
Figure 1.

HOXC13 is generally highly expressed in lung adenocarcinoma, and correlates with poorer prognosis. A. Venn Diagram for gene screening: TCGA dataset genes with fold change >10 and tumor expression >3 are included. Following this screen, three genes were obtained. B. TCGA datasets show that expression of HOXC13 is significantly higher in lung adenocarcinoma tissues than in normal tissues (P<0.0001). C. Lung adenocarcinoma patients with low expression of HOXC13 show a higher percentage of overall survival than patients with high expression of HOXC13 (median survival 1,725 d versus 1,258 d; P=0.0247). D. Lung adenocarcinoma patients with low expression of HOXC13 show a higher percentage of disease-free survival than patients with high expression of HOXC13 (P=0.0202). E. Quantitative RT-PCR analysis showed that HOXC13 was upregulated in 83.3% of 60 lung adenocarcinoma tissues (normalized to adjacent normal tissues). F. Overexpression of HOXC13 is associated with greater T stage (P<0.0001), N stage (P=0.0025), and TNM stage (P=0.0006). G and H. HOXC13 mRNA and protein are highly expressed in lung adenocarcinoma cell lines.
Table 1.
Correlation between HOXC13 expression and clinical characteristics (n=243)
| Characteristics | Low level of HOXC13 expression number | High level of HOXC13 expression number | p-Value |
|---|---|---|---|
| Age (years | 0.840837 | ||
| ≤65 | 53 | 55 | |
| >65 | 68 | 67> | |
| Sex | 0.484293 | ||
| Male | 60 | 54 | |
| Female | 68 | 73 | |
| Metastasis | 0.352308 | ||
| M0 | 88 | 94 | |
| M1-MX | 40 | 33 | |
| Lymph node | 0.000005* | ||
| N0 | 98 | 85 | |
| N1 | 29 | 18 | |
| N2-NX | 1 | 24 | |
| Primary Tumor | 0.363068 | ||
| T1 | 43 | 39 | |
| T2 | 72 | 72 | |
| T3 | 13 | 13 | |
| T4 | 0 | 3 | |
| TNM stage | 0.000001* | ||
| I | 85 | 73 | |
| II | 43 | 27 | |
| III | 0 | 22 | |
| IV | 0 | 5 |
Significant correlation.
Knockdown of HOXC13 inhibits in vitro lung adenocarcinoma proliferation and induces G1-phase arrest, while overexpression of HOXC13 yields the opposite outcomes
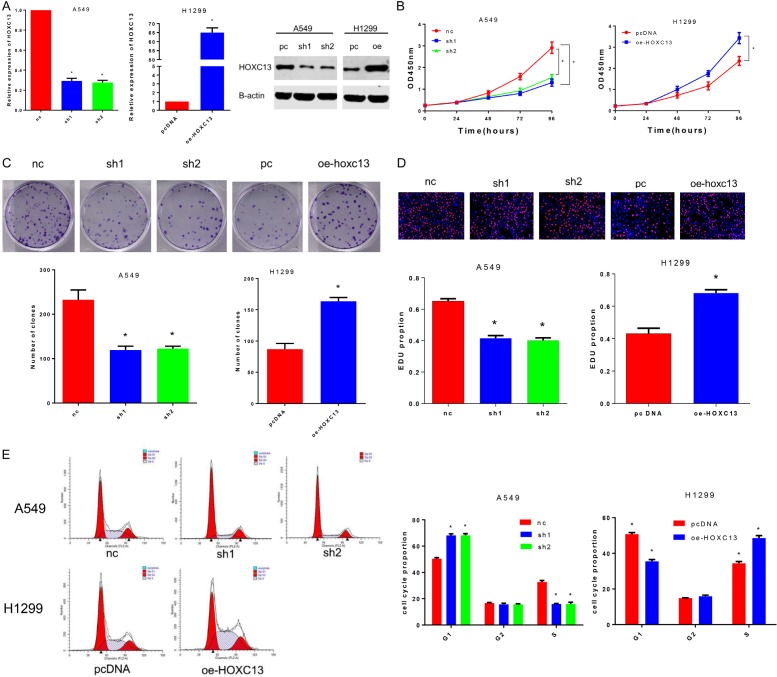
Confirmed by qRT-PCR and western blot, HOXC13 mRNA level and protein expression were generally higher in lung adenocarcinoma cell lines compared with normal human bronchial epithelial (HBE) cells, with the A549 cell line showing the highest expression values and H1299 the lowest (Figure 1G and 1H). For in vitro investigation of the biological function of HOXC13, we constructed two shRNA (sh1 and sh2) to knockdown HOXC13 in A549 cells and a plasmid DNA (oe-HOXC13) to overexpress HOXC13 in H1299 cells. Both shRNAs and the plasmid DNA were able to effectively decrease or increase, respectively, HOXC13 mRNA and protein expression (Figure 2A).
Figure 2.
Knockdown of HOXC13 inhibits A549 cell proliferation and induces G1-phase arrest, while overexpression of HOXC13 promotes H1299 cell proliferation and decreases the percentage of G1-phase cells. A. Two specific shRNA (sh1 and sh2) and an overexpression plasmid (oe-HOXC13) of HOXC13 were designed and synthesized, and the transfection efficiency in A549 and H1299 cell lines was measured by qRT-PCR and western blot. B. Knockdown of HOXC13 inhibited proliferation of A549 cells (sh1, P<0.0001; sh2, P<0.0001), and overexpression of HOXC13 promoted proliferation of H1299 cells (P<0.0001). C. Colony numbers of A549 cells transfected with shRNA-HOXC13 were lower than those of A549 cells transfected with shRNA-nc (sh1, P=0.0012; sh2, P=0.0012), while colony numbers of H1299 cells transfected with oe-HOXC13 were greater than those for H1299 cells transfected with pcDNA (P=0.0003). D. The proliferation ratios of A549 cells transfected with shRNA-HOXC13 were lower than those of A549 cells transfected with shRNA-nc (sh1, P=0.0061; sh2, P=0.0052), while H1299 cells transfected with oe-HOXC13 proliferated more than H1299 cells transfected with pcDNA (P=0.0122). E. Transfection of A549 cell line with shRNA-HOXC13 yielded more cells in G1-phase than did transfection of A549 cells with shRNA-nc (sh1, P=0.0036; sh2, P=0.0037), while overexpression of HOXC13 in H1299 cells (oe-HOXC13) caused a decrease in the number of cells in G1 phase, compared with this measure for cells transfected with the control plasmid (pcDNA, P=0.0004).
Cell counting kit-8 (CCK-8) assays revealed that knockdown of HOXC13 inhibited proliferation of A549 cells, and overexpression of HOXC13 promoted proliferation of H1299 cells (Figure 2B). In the colony formation test, colony numbers of A549 cells transfected with shRNA-HOXC13 were lower than those transfected with the control shRNA-nc, while H1299 cells transfected with oe-HOXC13 were more numerous than those transfected with the control pcDNA (Figure 2C). Similar results were obtained for the proliferation ratio calculated by the EDU assay (Figure 2D). Finally, the effect of HOXC13 on cell cycle distribution was evaluated by flow cytometry analysis. Knockdown of HOXC13 caused a G1-phase arrest in A549 cells, while overexpression of HOXC13 in H1299 cells caused a decrease in the number of cells in G1 phase and an increase in the number of cells in S phase (Figure 2E).
Knockdown of HOXC13 downregulates CCND1 and CCNE1, and overexpression of CCND1 and CCNE1 partially rescues HOXC13 knockdown effect on proliferation and cell cycle, in vitro
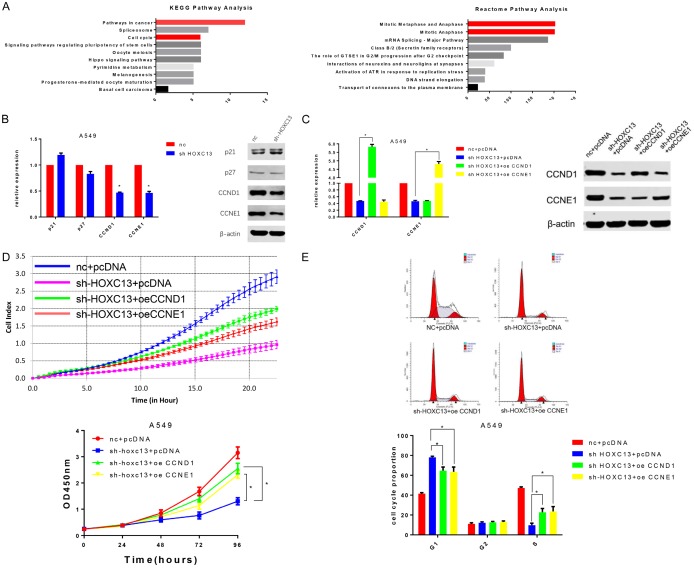
To further investigate HOXC13 biological function, a list of 313 genes with high co-expression correlation (Pearson score >0.3) with HOXC13 was obtained from lung adenocarcinoma TCGA datasets. KEGG and Reactome pathway enrichment analysis of the 313 genes revealed that HOXC13 is closely related to cancer and the cell cycle (Figure 3A); this finding is consistent with our present experimental results.
Figure 3.
HOXC13 promotes cell cycle progression of lung adenocarcinoma cells by influencing CCND1 and CCNE1. A. KEGG and Reactome pathway enrichment analysis showed that genes co-expressed with HOXC13 were enriched in the “pathway in cancer”, “cell cycle”, and “mitotic metaphase and anaphase”. B. Quantitative RT-PCR and western blot indicated that CCND1 (P<0.0001) and CCNE1 (P<0.0001) expressions were downregulated in A549 cells transfected with shRNA-HOXC13 compared with cells transfected with shRNA-nc. C. Transfection of A549 cell line with CCND1 or CCNE1 overexpression plasmids (oe-CCND1 and oe-CCNE1) increased the expression of CCND1 and CCNE1 (both P<0.0001), respectively. D. Overexpression of CCND1 or of CCNE1 partially reversed the inhibitory effect of shRNA-HOXC13 on the proliferation of A549 cells (both P<0.0001). E. Overexpression of CCND1 (P=0.014) or of CCNE1 (P=0.019) partially reversed shRNA-HOXC13-mediated G1-phase arrest.
As knockdown of HOXC13 induced G1-phase arrest, we investigated the influence of HOXC13 on cell cycle-related genes that play vital roles in G1 phase. Using qRT-PCR and western blot analysis, we found that the expression of cyclins D1 and E1 (CCND1 and CCNE1) was significantly downregulated in A549 cells transfected with shRNA-HOXC13, compared with cells transfected with the control shRNA-nc; however, CDKN1A (p21) and CDKN1B (p27), two G1-checkpoint cyclin dependent kinase inhibitors, had no significant change (Figure 3B). To clarify whether the effect of HOXC13 on proliferation and cell cycle relied on CCND1 and CCNE1, CCND1- or CCNE1-overexpression plasmids (oe-CCND1 and oe-CCNE1) were introduced into sh-HOXC13-treated A549 cells. The transfection efficiency was determined by qRT-PCR and western blot (Figure 3C). Both oe-CCND1 and oe-CCNE1 could partially reverse the shRNA-HOXC13 inhibitory effect on the proliferation of A549 cells (Figure 3D), as well as the G1-phase arrest (Figure 3E).
miR-141 inhibits proliferation and induces G1-phase arrest in A549 cells via downregulation of HOXC13 by directly targeting its 3’UTR
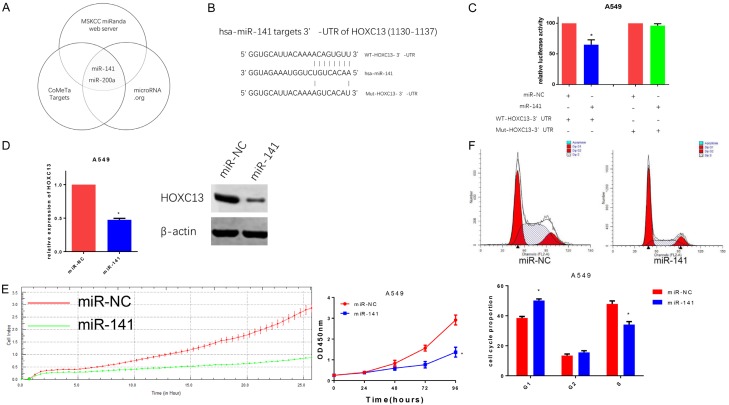
MicroRNAs have been implicated in the pathogenesis of many cancer types, including lung cancer. To identify potential miRNAs that target HOXC13, three prediction software packages (MSKCC, http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl?type=miRanda [31]; CoMeTa targets, http://cometa.tigem.it/ [32], and microRNA.org, http://www.microrna.org/microrna/home.do [33]) were used. Two miRNAs, miR-141 and miR-200a, were identified (Figure 4A). Using DIANA TOOLS [34], which predicts the binding score of miRNA and genes, miR-141 was shown to present the highest score (0.987935 vs 0.982702) in the interaction with the 3’UTR of HOXC13. Luciferase reporter assays demonstrated that miR-141 inhibited luciferase activity when the reporter plasmid carried the wild type version of the HOXC13 3’-UTR (WT-HOXC13 3’-UTR), but not the mutant 3’-UTR (Mut-HOXC13 3’-UTR) (Figure 4C). Quantitative RT-PCR and western blot also revealed that miR-141 was able to inhibit the expression of HOXC13 mRNA and protein (Figure 4D). Further investigation showed that miR-141 inhibited proliferation (Figure 4E), and induced G1-phase arrest (Figure 4F) in A549 cells.
Figure 4.
miR-141 directly targets HOXC13 and inhibits proliferation of A549 cells. A. Venn Diagram for HOXC13-targeted miRNA prediction: three prediction software packages showed that miR-141 and miR-200a might target HOXC13. B. Sequence in HOXC13 3’-UTR that is predicted to be targeted by miR141 (nucleotides 1130 to 1137) and mutated sequence constructed to prevent HOXC13 3’-UTR targeting by miR-141. C. Compared with miR-NC, miR-141 decreased the luciferase activity of the reporter plasmid carrying the wild type HOXC13 3’-UTR (P=0.0085) but not its mutated version, in A549 cells. D. Quantitative RT-PCR and western blot showed that miR-141 downregulated the expression of HOXC13 (P=0.0002). E. miR-141 inhibited the proliferation of A549 cells (P<0.0001). F. miR-141 induced G1-phase arrest in A549 cells (P=0.0048).
Overexpression of HOXC13 partially reverses miR-141 effect on in vitro proliferation and cell cycle
To further investigate the relationship between miR-141 and HOXC13, we performed a rescue experiment in A549 cells. Western blot analysis showed that miR-141 downregulated the expression of HOXC13, CCND1, and CCNE1, while overexpression of HOXC13 could reverse the downregulation of HOXC13, CCND1, and CCNE1 (Figure 5A). The miR141-mediated inhibition of cell proliferation was also partially rescued after overexpression of HOXC13 (Figure 5B and 5C). Flow-cytometry analysis showed that G1-phase arrest induced by miR-141 was reverted after overexpression of HOXC13 (Figure 5D).
Figure 5.
miR-141 inhibits proliferation and induces G1-phase arrest of A549 cells by suppressing the expression of HOXC13. A. miR-141 downregulated the expression of HOXC13, CCND1, and CCNE1, while oe-HOXC13 reversed these effects. B and C. Compared with pcDNA, oe-HOXC13 partially reversed the miR-141-mediated inhibition of A549 cell proliferation (P<0.0001). D. Overexpression of HOXC13 (oe-HOXC13) partially reversed the miR-141-mediated G1-phase arrest (P<0.0001).
Discussion
Lung adenocarcinoma is the most common subtype of lung cancer, and it has a high mortality rate [35]. The poor prognosis of lung adenocarcinoma is largely due to late diagnosis and lack of effective therapeutic targets [36,37]; therefore, there is a compelling need to screen for new early detection markers and pharmacological targets. Through an analysis of TCGA datasets, we found that expression of the HOXC13 gene is significantly elevated in lung adenocarcinomas. A member of the homeobox HOXC gene family, HOXC13 correlates with the development of hair, nail, and filiform papilla [13-15]. Recent studies show that HOXC13 is overexpressed in human liposarcomas [11], squamous cell carcinoma [38], skin tumors [39], and prostate cancer [40]. BMI-1, which is a regulator of HOXC13, and is overexpressed in breast and other cancers, promotes self-renewal of cancer stem-like cells [41]; knockdown of BMI-1 causes cell-cycle arrest, and derepresses p16INK4a in HeLa cells [42]. As a transcription factor, HOXC13 synergistically regulates the expression of Zfp521, which has been identified as a B-cell proto-oncogene causing leukemia [43]. It was also reported that knockdown of HOXC13 affected cell growth, and resulted in cell cycle arrest, in colon cancer [12]. In accordance with these studies, we have uncovered an oncogenic role for HOXC13 in lung adenocarcinoma, via upregulation of CCND1 and CCNE1 expression.
A well-balanced cell cycle progression is necessary for cell proliferation, while dysregulation of the cell cycle components may lead to tumor formation [44]. In our study, we confirmed that HOXC13 was inextricably linked to lung adenocarcinoma cell cycle G1/S transition. Cyclins, as fundamental regulators of the cell cycle, play an important role in tumorigenesis [45]. Cyclin D1, encoded by CCND1, is a well-documented regulator of the G1/S transition through the activation of CDK4/6 kinase and subsequent phosphorylation of Rb [46]. Additionally, the interaction between cyclin E1 and CDK2 plays an essential role in the G1/S phase transition via phosphorylation of p27 [47].
CCND1 and CCNE1 are well-recognized oncogenes, as well as poor prognostic indicators in lung cancer [48-50]. These studies suggest that CCND1 and CCNE1 and/or their upstream regulators can affect proliferation of tumor cells. Further verification of the influence of HOXC13 on cell cycle showed that CCND1 and CCNE1 were significantly downregulated after knockdown of HOXC13. The rescue experiment demonstrated that overexpression of CCND1 or of CCNE1 could partially reverse the inhibition of cell proliferation and G1-phase arrest caused by downregulation of HOXC13. We thus conclude that HOXC13 can regulate cell proliferation by influencing the expression of CCND1 and CCNE1, during G1 phase.
MicroRNAs regulate post-transcriptional gene expression by base pairing with complementary sequences in the 3’-UTRs of target mRNAs, and subsequently inducing mRNA degradation or translational repression [51]. In general, one miRNA appears to be able to regulate several hundreds of genes; thus, miRNAs are involved in a variety of physiological and pathological processes, including the occurrence and development of tumors [52,53]. Identification of a particular miRNA that regulates HOXC13 may lead to promising therapeutic targets and prognostic biomarkers in lung adenocarcinoma diagnosis and treatment. The present study confirmed that miR-141 directly targets the 3’UTR of HOXC13, downregulating the expression of HOXC13, CCND1, and CCNE1, while overexpression of HOXC13 partially rescues the effect of miR-141 on proliferation and cell cycle. These data indicated that a miR-141-HOXC13-CCND1/CCNE1 axis participates in the regulation of lung adenocarcinoma proliferation.
In conclusion, our study showed that HOXC13 expression was significantly elevated in lung adenocarcinoma, and correlated with worse clinical characteristics and poorer prognosis. HOXC13 promoted proliferation and cell cycle progression via upregulation of CCND1 and CCNE1. Moreover, miR-141 was able to inhibit lung adenocarcinoma cell proliferation by suppressing the expression of HOXC13, CCND1, and CCNE1 (Figure 6).
Figure 6.

Mechanism of miR-141-targeted HOXC13 promoting proliferation and cell cycle progression via modulation of CCND1 and CCNE1 expression.
Acknowledgements
We thank professor Lin Xu (Jiangsu Cancer Hospital) for providing the facilities to perform the experiments in this study.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, Matthews N, Rowan A, Chambers T, Moore DA, Turajlic S, Xu H, Lee SM, Forster MD, Ahmad T, Hiley CT, Abbosh C, Falzon M, Borg E, Marafioti T, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Shah R, Joseph L, Quinn AM, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Dentro S, Taniere P, O’Sullivan B, Lowe HL, Hartley JA, Iles N, Bell H, Ngai Y, Shaw JA, Herrero J, Szallasi Z, Schwarz RF, Stewart A, Quezada SA, Le Quesne J, Van Loo P, Dive C, Hackshaw A, Swanton C TRACERx Consortium. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 4.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 6.Fry WA, Phillips JL, Menck HR. Ten-year survey of lung cancer treatment and survival in hospitals in the United States: a national cancer data base report. Cancer. 1999;86:1867–1876. doi: 10.1002/(sici)1097-0142(19991101)86:9<1867::aid-cncr31>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Kaisermann MC, Trajman A, Madi K. Evolving features of lung adenocarcinoma in Rio de Janeiro, Brazil. Oncol Rep. 2001;8:189–192. doi: 10.3892/or.8.1.189. [DOI] [PubMed] [Google Scholar]
- 8.Zhong M, Wang J, Gong YB, Li JC, Zhang B, Hou L. [Expression of HOXC13 in ameloblastoma] . Zhonghua Kou Qiang Yi Xue Za Zhi. 2007;42:43–46. [PubMed] [Google Scholar]
- 9.Hong YS, Wang J, Liu J, Zhang B, Hou L, Zhong M. [Expression of HOX C13 in odontogenic tumors] . Shanghai Kou Qiang Yi Xue. 2007;16:587–591. [PubMed] [Google Scholar]
- 10.Cantile M, Scognamiglio G, Anniciello A, Farina M, Gentilcore G, Santonastaso C, Fulciniti F, Cillo C, Franco R, Ascierto PA, Botti G. Increased HOX C13 expression in metastatic melanoma progression. J Transl Med. 2012;10:91. doi: 10.1186/1479-5876-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantile M, Galletta F, Franco R, Aquino G, Scognamiglio G, Marra L, Cerrone M, Malzone G, Manna A, Apice G, Fazioli F, Botti G, De Chiara A. Hyperexpression of HOXC13, located in the 12q13 chromosomal region, in welldifferentiated and dedifferentiated human liposarcomas. Oncol Rep. 2013;30:2579–2586. doi: 10.3892/or.2013.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasiri S, Ansari KI, Hussain I, Bhan A, Mandal SS. Antisense oligonucleotide mediated knockdown of HOXC13 affects cell growth and induces apoptosis in tumor cells and over expression of HOXC13 induces 3D-colony formation. RSC Adv. 2013;3:3260–3269. doi: 10.1039/C2RA22006G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulessa H, Turk G, Hogan BL. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. EMBO J. 2000;19:6664–6674. doi: 10.1093/emboj/19.24.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkatchenko AV, Visconti RP, Shang L, Papenbrock T, Pruett ND, Ito T, Ogawa M, Awgulewitsch A. Overexpression of Hoxc13 in differentiating keratinocytes results in downregulation of a novel hair keratin gene cluster and alopecia. Development. 2001;128:1547–1558. doi: 10.1242/dev.128.9.1547. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 18.Hu C, Lv L, Peng J, Liu D, Wang X, Zhou Y, Huo J. MicroRNA-375 suppresses esophageal cancer cell growth and invasion by repressing metadherin expression. Oncol Lett. 2017;13:4769–4775. doi: 10.3892/ol.2017.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Wu C, Zhao Y. miRNA-34a enhances the sensitivity of gastric cancer cells to treatment with paclitaxel by targeting E2F5. Oncol Lett. 2017;13:4837–4842. doi: 10.3892/ol.2017.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou R, Wang D, Lu J. MicroRNA-10b inhibits proliferation, migration and invasion in cervical cancer cells via direct targeting of insulinlike growth factor-1 receptor. Oncol Lett. 2017;13:5009–5015. doi: 10.3892/ol.2017.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Jin X, Meng H, Hu B, Zhang T, Yu J, Chen S, Guo X, Wang W, Jiang W, Wang J. Morin promotes prostate cancer cells chemosensitivity to paclitaxel through miR-155/GATA3 axis. Oncotarget. 2017;8:47849–47860. doi: 10.18632/oncotarget.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, Ju S, Yuan F, Chen G, Shu Y, Li C, Xu Y, Luo J, Xia L. microRNA-664 enhances proliferation, migration and invasion of lung cancer cells. Exp Ther Med. 2017;13:3555–3562. doi: 10.3892/etm.2017.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye J, Wang Z, Zhao J, Chen W, Wu D, Wu P, Huang J. MicroRNA-141 inhibits tumor growth and minimizes therapy resistance in colorectal cancer. Mol Med Rep. 2017;15:1037–1042. doi: 10.3892/mmr.2017.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan H, Zhu Y, Zhang J, Peng L, Ji T. miR141 expression is downregulated and negatively correlated with STAT5 expression in esophageal squamous cell carcinoma. Exp Ther Med. 2016;11:1803–1808. doi: 10.3892/etm.2016.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Lu H, Mukherjee D, Lahiri SK, Shen C, Yu L, Zhao J. Identification of epidermal growth factor receptor and its inhibitory microRNA141 as novel targets of Kruppel-like factor 8 in breast cancer. Oncotarget. 2015;6:21428–21442. doi: 10.18632/oncotarget.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du Y, Wang L, Wu H, Zhang Y, Wang K, Wu D. MicroRNA-141 inhibits migration of gastric cancer by targeting zinc finger E-box-binding homeobox 2. Mol Med Rep. 2015;12:3416–3422. doi: 10.3892/mmr.2015.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, Haussler D, Zhu J. The UCSC cancer genomics browser: update 2015. Nucleic Acids Res. 2015;43:D812–817. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vastrik I, D’Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Gennarino VA, D’Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, Sardiello M, Banfi S. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163–1172. doi: 10.1101/gr.130435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 36.Neri S, Yoshida J, Ishii G, Matsumura Y, Aokage K, Hishida T, Nagai K. Prognostic impact of microscopic vessel invasion and visceral pleural invasion in non-small cell lung cancer: a retrospective analysis of 2657 patients. Ann Surg. 2014;260:383–388. doi: 10.1097/SLA.0000000000000617. [DOI] [PubMed] [Google Scholar]
- 37.Wanders R, Steevens J, Botterweck A, Dingemans AM, Reymen B, Baardwijk A, Borger J, Bootsma G, Pitz C, Lunde R, Geraedts W, Lambin P, De Ruysscher D. Treatment with curative intent of stage III non-small cell lung cancer patients of 75 years: a prospective population-based study. Eur J Cancer. 2011;47:2691–2697. doi: 10.1016/j.ejca.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 38.Marcinkiewicz KM, Gudas LJ. Altered histone mark deposition and DNA methylation at homeobox genes in human oral squamous cell carcinoma. J Cell Physiol. 2014;229:1405–1416. doi: 10.1002/jcp.24577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battistella M, Carlson JA, Osio A, Langbein L, Cribier B. Skin tumors with matrical differentiation: lessons from hair keratins, betacatenin and PHLDA-1 expression. J Cutan Pathol. 2014;41:427–436. doi: 10.1111/cup.12313. [DOI] [PubMed] [Google Scholar]
- 40.Komisarof J, McCall M, Newman L, Bshara W, Mohler JL, Morrison C, Land H. A four gene signature predictive of recurrent prostate cancer. Oncotarget. 2017;8:3430–3440. doi: 10.18632/oncotarget.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiraki M, Maeda T, Bouillez A, Alam M, Tagde A, Hinohara K, Suzuki Y, Markert T, Miyo M, Komura K, Ahmad R, Rajabi H, Kufe D. MUC1-C activates BMI1 in human cancer cells. Oncogene. 2017;36:2791–2801. doi: 10.1038/onc.2016.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen F, Li Y, Wang L, Hu L. Knockdown of BMI-1 causes cell-cycle arrest and derepresses p16INK4a, HOXA9 and HOXC13 mRNA expression in HeLa cells. Med Oncol. 2011;28:1201–1209. doi: 10.1007/s12032-010-9634-9. [DOI] [PubMed] [Google Scholar]
- 43.Yu M, Al-Dallal S, Al-Haj L, Panjwani S, McCartney AS, Edwards SM, Manjunath P, Walker C, Awgulewitsch A, Hentges KE. Transcriptional regulation of the proto-oncogene Zfp521 by SPI1 (PU.1) and HOXC13. Genesis. 2016;54:519–533. doi: 10.1002/dvg.22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Peng Z, Zhao Y, Chen L. microRNA-25 inhibits cell apoptosis of human gastric adenocarcinoma cell line AGS via regulating CCNE1 and MYC. Med Sci Monit. 2016;22:1415–1420. doi: 10.12659/MSM.896118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musgrove EA, Lee CS, Buckley MF, Sutherland RL. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci U S A. 1994;91:8022–8026. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donnellan R, Chetty R. Cyclin E in human cancers. FASEB J. 1999;13:773–780. doi: 10.1096/fasebj.13.8.773. [DOI] [PubMed] [Google Scholar]
- 48.Caputi M, Groeger AM, Esposito V, Dean C, De Luca A, Pacilio C, Muller MR, Giordano GG, Baldi F, Wolner E, Giordano A. Prognostic role of cyclin D1 in lung cancer. Relationship to proliferating cell nuclear antigen. Am J Respir Cell Mol Biol. 1999;20:746–750. doi: 10.1165/ajrcmb.20.4.3366. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Sun Q, Chen C, Yin R, Huang X, Wang X, Shi R, Xu L, Ren B. ZYG11A serves as an oncogene in non-small cell lung cancer and influences CCNE1 expression. Oncotarget. 2016;7:8029–8042. doi: 10.18632/oncotarget.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi R, Sun J, Sun Q, Zhang Q, Xia W, Dong G, Wang A, Jiang F, Xu L. Upregulation of FAM83D promotes malignant phenotypes of lung adenocarcinoma by regulating cell cycle. Am J Cancer Res. 2016;6:2587–2598. [PMC free article] [PubMed] [Google Scholar]
- 51.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3:563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 53.Singh R, Ramasubramanian B, Kanji S, Chakraborty AR, Haque SJ, Chakravarti A. Circulating microRNAs in cancer: hope or hype? Cancer Lett. 2016;381:113–121. doi: 10.1016/j.canlet.2016.07.002. [DOI] [PubMed] [Google Scholar]