Abstract
Background: Mutations in KRAS are negative predictors of the response to anti-EGFR therapies in the treatment of metastatic colorectal cancer. Yet, the ideal tissue to test for KRAS mutation-primary or metastatic-remains unknown, as is the validity of testing only 1 area of the primary tumor. The aim of this study was to determine the heterogeneity of KRAS mutational status between areas of the primary lesion and between paired primary CRC and the corresponding lymph node (LN), liver, and lung metastasis with a high-sensitivity sequencing method. Design: DNA from 2 or 3 areas from the primary tumor and 1 area of metastatic tissue was obtained from formalin-fixed paraffin-embedded specimens from 102 metastatic CRC patients. Mutations in KRAS codons 12, 13, and 61 were analyzed by pyrosequencing. Results: Ninety-one cases had DNA extracted from more than 1 area of the primary tumor. Only 1 patient showed intratumor heterogeneity, which involved KRAS mutation type, not KRAS mutational status. We examined KRAS mutations in 97 primaries and matched metastatic samples, recording 2 discordant cases, representing 2.1% of our cohort of matched samples. Conclusion: KRAS status is highly homogeneous throughout primary CRC tumor areas and consistent between the primary tumor and metastatic tissue in the same patient. Our data suggest that testing KRAS mutations in only 1 area of the primary or metastatic tissue is suitable for predicting the response to anti-EGFR treatment and guiding clinical decisions.
Keywords: Genetic heterogeneity, precision medicine, colorectal neoplasms, molecular pathology, RAS protein, molecular sequence data
Introduction
In the early 2000s, the use of epidermal growth factor receptor (EGFR) inhibitors became a new and beneficial treatment strategy for metastatic colorectal cancer (CRC) patients. Although the advantages of this approach were well documented, it was soon recognized that not all patients with metastatic CRC respond to it. This subgroup of patients was shown to have alterations in the EGFR pathway, involving downstream markers [1].
It is now well established that mutations in KRAS lead to chronic activation of EGFR signaling [2]; thus, the patients who respond to anti-EGFR are those whose tumors harbor no KRAS mutation. KRAS mutation has become a negative predictor of the response to anti-EGFR, allowing unqualified patients to avoid the unnecessary toxicity and costs that are associated with this treatment [3,4]. Today, KRAS mutation testing is mandatory before anti-EGFR therapy is begun [2,5,6].
Recently, NRAS mutations have been shown to be associated with poor responses to anti-EGFR as well [6]. NRAS mutations are seen in approximately 5% of CRC cases and usually involve codons 12, 13, and 61. Simultaneous mutations in KRAS and NRAS are not observed frequently in CRC tumors [7,8].
Tumor heterogeneity is a significant concern in CRC. There are no definitive data on the ideal tissue-the primary tumor or the metastatic lesion-to test for KRAS mutations in metastatic CRC [2]. Further, there are no findings regarding the validity, representation, and reproducibility of obtaining a single sample of the primary tumor for KRAS mutation testing. Several reviews have reported comparative analyses of KRAS status between primary tumors and their respective metastases. The percentage of discordance in KRAS status varies significantly between studies, ranging from 0% to 30% [9,10], as do the detection methods that are used and the selection of the area of interest [11-13].
Thus, to examine this crucial issue in molecular diagnostics, we analyzed KRAS mutational heterogeneity between several areas of the primary tumor and the concordance of KRAS mutational status between primary and matched metastatic tissues by pyrosequencing, a sensitive method of detecting mutations, and performed a rigorous pathological assessment of tumor area selection.
Material and methods
Case selection and clinical-pathological data
The study cohort consisted of male and female patients from any age group with a confirmed pathological diagnosis of metastatic colorectal adenocarcinoma who had it operated in A. C. Camargo Cancer Center or who had their resections performed by outside clinics but were followed at this institution. Patients with adenocarcinomas from rectum or either right or left colon were included, independent of any previous treatment status. Formalin-fixed paraffin-embedded (FFPE) tumor blocks were retrieved from the A. C. Camargo Cancer Center pathology files. Clinical-pathological data were obtained from institutional electronic charts.
Selection of tumor area for DNA extraction
Tumor samples were drawn from the clinical diagnostic FFPE material. Hematoxylin and eosin slides from the primary and metastatic colorectal adenocarcinomas were reviewed by an experienced pathologist (MPM) to confirm the diagnosis and select the best representative area of the tumor for DNA extraction. In each slide, a circular area of approximately 1.0 cm2 was chosen, containing at least 30% neoplastic nuclei and avoiding stromal cells and inflammatory infiltrates.
DNA extraction from FFPE blocks
Five 5.0-um-thick unstained sections were cut from the previously selected paraffin blocks for each case. The slides were deparaffinized (5 min with xylene 3 times and 2 min with absolute alcohol 2 times), and the neoplastic tissue sample was obtained by scraping the tumor area from the slide using a scalpel (macrodissection) and transferring it to an Eppendorf tube. DNA was extracted using a commercial kit (QIAamp DNA FFPE Tissue Kit®) per the manufacturer’s instructions. The DNA concentration was measured on a Nandrop 2000®, and the minimum DNA concentration for the experiments was set to 10 ng/ul.
KRAS mutation testing
Pyrosequencing (PyroMarkTM Q24 Qiagen) of KRAS mutations in codons 12, 13, and 61 was performed in primary and metastatic tumor samples per the manufacturer’s instructions. DNA from cell lines with previously known mutations were used as positive controls (LS174T for codon 12 c.35G>A mutation and HCT116 for codon 13 c.38G>A mutation). Commercial genomic DNA without KRAS mutation was used as a negative control. The results were categorized as KRAS mutated or KRAS wild-type, including characterization of the mutation and the percentage of mutated alleles. Heterogeneity was determined with regard to KRAS status (wild-type versus mutated) and mutation type (specific type of mutation). All cases that showed intratumoral heterogeneity in the primary tumor or were discordant between primary and metastatic tissues regarding both KRAS status and mutation type had their findings confirmed by repeat DNA extraction and sequencing reactions.
Study design to assess intratumoral heterogeneity of KRAS mutation profile and concordance between primary and matched metastatic tissues
To examine intratumoral heterogeneity regarding KRAS mutational profile in single primary CRC lesions we tested 2 or 3 regions of a primary tumor, based on the availability of tissue from the pathology files. The regions were defined as distinct areas in routine macroscopic tumor blocks that were representative of the primary lesion (Figure 1). No morphological aspects of the neoplasia or depth of invasion was used as inclusion criteria in the selection of these areas. To determine the concordance of KRAS mutational status of primary and matched metastatic tissues, we also tested 1 area of the paired metastatic tissue, including the regional lymph node, liver, and lung, based on the availability of tissue in the pathology files.
Figure 1.
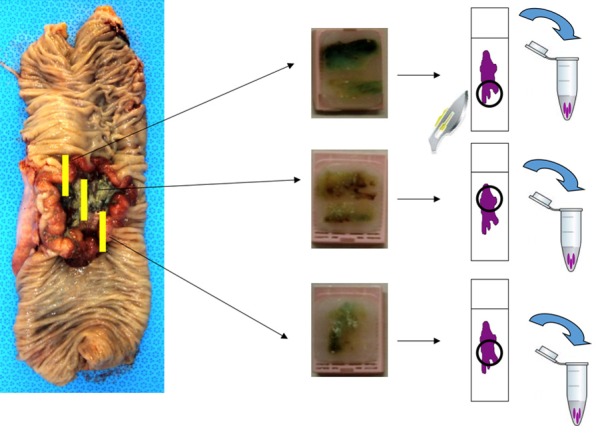
Representative scheme of routine macroscopic sampling of primary tumors for assessing intratumoral heterogeneity between 2 or 3 areas of the primary tumor, depending on tissue availability.
Ethics committee review
This study is part of a scientific project that was approved by the local ethics committee (AC Camargo Cancer Center) (number 1543/11, dated April 12, 2011).
Results
Clinical pathological and mutational data of cases
One hundred two cases of metastatic CRC were selected: 61 (59.8%) were from male patients and 41 were from (40.2%) females. The mean age (± standard deviation [interval]) was 57.13 (±12.49 [26-80]) years. The distribution of cases regarding tumor localization was as follows: 14 cases (13.7%) with right-sided tumors, 55 (53.9%) with left-sided tumors, and 33 (32.4%) rectal. The distribution of tumors concerning pathological tumor stage (pTNM) [14] was as follows: 15 patients (14.7%) with stage pT4, 79 (77.5%) with stage pT3, 7 patients (7%) with stage 2, and 1 patient (1%) with pT1.
KRAS mutation was found in 41 of 102 patients (40.1%)-75% of mutated cases were in codon 12, compared with 23% in codon 13 and 2% in codon 61. Of the mutations in codon 12, 48% had the KRAS c.35G>A (p.G12D) mutation, followed by 36% with c.35G>T (p.G12V), 6% with c.34G>A (p.G12S), 6% with c.34G>T (p.G12C), and 4% with c.35G>C (p.G12A). In codon 13, 100% of mutations were c.38G>A (p.G13D). The only case with a mutation in codon 61 was c.183A>C (p.Q61K).
Intratumoral KRAS mutational heterogeneity
Of the 102 cases, we obtained DNA from more than 1 area of the primary tumor from 91 patients-3 representative areas from 71 patients and 2 areas from 20 patients. A total of 253 primary tumor samples from 91 patients were examined with regard to intratumoral heterogeneity of KRAS mutations in the primary tumor.
We noted 100% concordance regarding KRAS mutational status (wild-type versus mutated) in the various areas of the primary tumor.
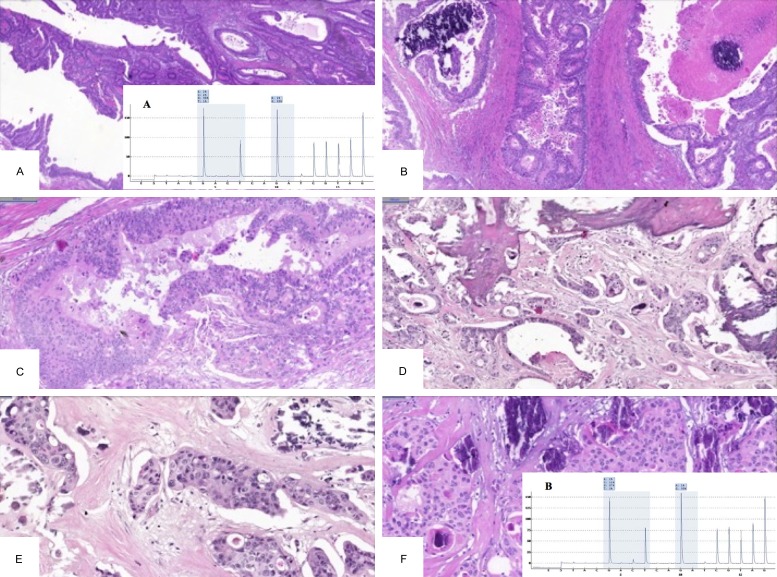
One case (1%) showed heterogeneity of KRAS mutation between areas of a primary tumor (case A)-a 59-year-old male with an 8.5-cm right-sided colorectal adenocarcinoma with LN and liver metastasis. All 3 areas in the primary tumor harbored a KRAS mutation, although the mutations differed-1 with the c.35G>T mutation, with 24% mutated alleles, and the other 2 areas with the c.35G>A mutant allele (48% and 57%). The liver and LN metastases had the c.35G>T mutation. In this case, the mutation type correlated with the morphology between areas. Both areas from the primary tumor with the c.35G>A mutation had a villous papillary histology with elongated structures. The area of the tumor with the c.35G>T mutation formed small tubular glands, which were also observed in the LN and liver metastases that had the same mutation pattern (Figure 2).
Figure 2.
Morphological characteristics and mutational data of the case (case A) with intratumoral heterogeneity between areas of the primary tumor. (A and B) Elongated villous papillary morphology of 2 distinct primary regions with the c.35G>A KRAS mutation, contrasting the tubular morphology (C) of the primary tumor region with the c.35G>T KRAS mutation. Lymph node (D) and liver (E) harbor the c.35G>T mutation and have a similar tubular morphology as the primary tumor region with the same mutation type.
Concordance of KRAS mutational status between primary and matched metastatic lesion
The concordance of KRAS mutational status between primary and metastatic tissues in the same patients was analyzed in 97 matched samples. Ninety samples had already been shown to be homogeneous for KRAS status between areas of the primary lesion; 7 cases had only 1 area of the primary tissue tested and thus were not evaluated as part of the primary intratumoral subgroup. We examined 144 metastatic samples from 97 patients, including LN, lung, and liver. The metastatic sites, their corresponding primary tumors and number of tested samples are listed in Figure 3.
Figure 3.
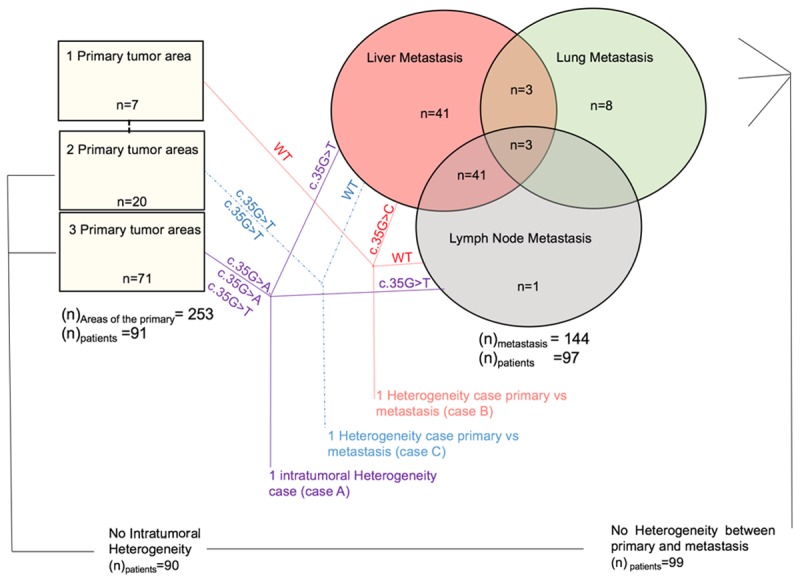
Schematic of metastatic sites analyzed for intratumoral and intertumoral heterogeneity between primary and matched metastastatic lesions regarding KRAS mutation in CRC, and the results for KRAS mutation.
We observed concordance of KRAS mutational status in 95 of the 97 (98%) patients between the primary tissue and corresponding metastatic samples. No case was discordant regarding KRAS mutation type.
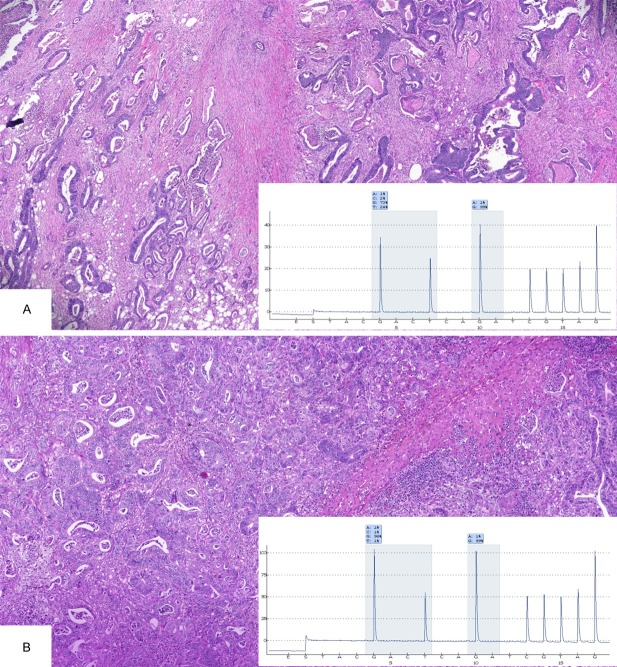
One of the heterogeneous cases (case B)-a 34-year-old female with a left-sided CRC-had wild-type KRAS in the 1 area of primary tumor that was analyzed and in the LN metastasis. The liver metastasis harbored the c.35G>C KRAS mutation, with 11% of alleles mutated. The tumor infiltrated the serosa and presented with perineural infiltration. She developed LN metastasis in 4 of the 27 pericolic LNs that were studied. The liver metastasis that was tested for KRAS mutation was resected 1 year and 4 months after the primary lesion resection. The patient underwent 2 rounds of chemotherapy after the primary lesion resection-FOLFOX (12 cycles) and FOLFIRI (6 cycles) as adjuvant treatment and conversion chemotherapy for resection of the liver metastasis but was not treated with monoclonal antibodies. The tumor presented with an unusual morphology for CRC, with papillary and cribriform areas, foci of necrosis and calcification, and large eosinophilic tumor cells in the primary tumor and both metastatic sites (Figure 4).
Figure 4.
Case with heterogeneity of KRAS mutation comparing primary and metastatic lesions of a CRC patient (case B). Morphological characteristics of the primary lesion with wild-type KRAS showing areas of papillary (A) and cribiform architecture (B), with dystrophic calcification, necrosis, and squamous component (C). Same aspects as observed in lymph node metastasis with wild-type KRAS (D and E) and liver metastasis with KRAS c.35G>C (F).
The second heterogeneous case (case C) had only 2 areas of the primary tumor that were tested, both of which harbored the c.35G>T KRAS mutation, whereas the liver metastasis was wild-type for KRAS. This 57-year-old patient had a left-sided CRC and synchronous liver metastasis that was resected in 2008. The tumor infiltrated the subserous soft tissue and presented with lymphatic and perineural invasion and metastasis in 2 of the 14 pericolic LNs. Morphologically, both areas of the primary tumor and the liver metastasis assumed a tubular formation (Figure 5). It was not possible to obtain representative DNA from the LN metastasis due to the limited amount of tumor tissue. The patient died 4 years after the initial diagnosis. Figure 3 shows a schematic representation of tested samples and heterogeneous cases with mutation findings.
Figure 5.
Morphological characteristics of a case with heterogeneity of KRAS mutation comparing primary and metastatic lesions of a CRC patient (case C). One picture representing the same tubular morphological characteristics shared by 2 representative areas of the primary lesions with c.35G>T KRAS mutation (A) and similar morphology in the liver metastasis with wild-type KRAS (B).
Discussion
In this study, by a sensitive sequencing method, the concordance of KRAS mutations was high between areas of primary tumors and between primary and paired metastatic samples in a large cohort of CRC patients.
The cohort had a KRAS mutation frequency of 40%, similar to what has been reported for CRC. The distribution of mutations among codons 12, 13, and 61 and the most frequent nucleotide changes in each codon were also similar to the the literature, in which codon 12 is the most commonly mutated codon in CRC and c.35G>A is the most frequent mutation [3,4,15-18]. These data show that our study population and sequencing technique are representative of global findings.
KRAS mutation heterogeneity was noted in 3 of 102 patients (3%). Intratumoral heterogeneity in the primary tumor was seen in 1 case (1%), regarding only the type of mutation, not the status of the gene (WT versus mutated). We also observed high concordance in KRAS mutational status between the primary and matched metastatic tumors. Discordance was seen in only 2 cases (2%), both of which concerned mutational status; in 1 case, the primary tumor was mutated and the metastatic tissue was wild-type, and the other case had the opposite pattern.
KRAS mutations vary widely among primary and metastatic tumor tissue. A large review of 18 articles on comparative KRAS mutational analysis between primary and metastatic lesions [9] noted that discordance rates ranged from 0% to 31%, but most of the articles were based on a small number of patients. Only 2 articles from this review had a similar number of patients as in our study, reporting 5% discordance in a cohort of 93 patients [19] and 4% discordance in 99 patients, both by direct sequencing [20]. KNIJN et al. also reported a comparative analysis of KRAS mutational status between 1 area of the primary tumor and liver metastasis from 305 patients by direct sequencing and found 11 cases to be heterogeneous (3.6%) [9]. DÓCS et al. compared the mutation status of KRAS in 18 metastatic samples at various time points and found discordance in 6 cases [21].
More recent publications have trended toward a smaller percentage of heterogeneity. MIGLIO et al. studied 45 patients with metastatic CRC, including LN, lung, and liver, and did not find any heterogeneity in KRAS mutations status in any case [22]. This group performed rigorous selection of the tumor area and used a high-sensitivity sequencing method. VAKIANI et al. studied 84 paired samples of primary and metastatic CRC and 31 patients with pairs of metastases and recorded only 2 cases with divergent KRAS status [23], whereas another group noted 90% concordance in a cohort of 31 paired samples [24].
Next-generation sequencing with larger gene mutation testing panels has been applied in certain studies to determine concordance rates for KRAS mutations in primary and metastatic lesions. One study found 100% concordance only for KRAS mutations among 69 paired samples from primary and metastatic CRC samples, whereas if other genes were considered, like those in The Cancer Genomic Atlas network publication for Colorectal Cancer [25], the concordance rate was 93% [26]. Another group reported 80% concordance for gene mutations in a 16-patient cohort using a 50-gene panel but 93% concordance when only KRAS mutations were analyzed [27].
One author analyzed KRAS mutational status between areas of the primary tumor in 75 cases of CRC, describing 50% heterogeneity between the center and periphery of the tumor by pyrosequencing [28]. This rate is higher than in our study regarding intratumoral heterogeneity. The tumor periphery contains more inflammatory infiltrate, and the contamination of tumor DNA with nontumor sources in the tumor stroma interface must be considered.
In our study, although 3 of 102 cases were heterogeneous, only 2-those with differences in KRAS status between primary and metastatic tumors-have clinical significance and represent a shift in clinical decision-making. Current clinical recommendations argue against anti-EGFR for patients with KRAS mutations, regardless of mutation type [6,29].
Reports and ongoing studies trying to determine the impact of the various types of mutations on the response to anti-EGFR. Some data suggest that patients who harbor a mutation in codon 13 will benefit from EGFR inhibitors, but this approach remains absent from official treatment guidelines [30]. Thus, patients with intratumoral heterogeneity between areas of the primary tumor regarding the type of KRAS mutation would not represent a clinical problem. Our analysis is the first study to actively examine several areas of the primary tumor with metastatic sites, including lung, and enriched tumor areas using a sensitive sequencing method.
None of the heterogeneous cases in our cohort involved lung metastasis. The number of lung metastases in this cohort was limited compared with those of the liver.
Our patient who presented with intratumoral heterogeneity in the primary tumor regarding mutation type had an 8.5-cm mass in the right colon with various morphologies throughout the specimen. Although it was considered a single lesion macroscopically, it was not possible to exclude the possibility of this tumor being the aggregate of 2 synchronous primary lesions, due to the right-sided location, the large size of the lesion, and the range of morphologies. Synchronous primary CRC lesions have a high frequency of heterogeneity in KRAS mutations [31-33].
The patient who had a WT primary tumor and mutated metastasis underwent chemotherapy between resection of the primary and metastatic tissue. Although the frequency of somatic mutations has been shown to increase after chemotherapy [34], the data specifically on KRAS mutation demonstrate concordant KRAS mutational status in biopsies before and after neodadjuvant treatment [35-37]. We did not analyze the concordant cases regarding their chemotherapeutic treatment between the resection of the primary and metastatic tumors. One explanation for the presence of a mutated metastatic focus and a WT KRAS primary tumor is the occurrence of small subpopulations of mutated cells in the primary tumor that expanded during treatment and tumor progression, becoming detectable in the metastatic site.
The heterogeneous cases in our cohort that showed 2 areas with the primary tumor mutated and WT liver metastasis had both specimens resected simultaneously, but we do not have information regarding their treatment. Another possible source of discordance in KRAS mutational status between the primary and metastatic samples is DNA degradation [38].
Although most neoplasias have significant genetic heterogeneity [39] -specifically because CRC is related to many molecular pathways-when we searched for genetic alterations in CRC solely in the spectrum of KRAS mutations, we noted that most tumors that harbored a KRAS mutation expressed it in the primary lesion and maintained it in the metastatic lesion, regardless of site. These findings corroborate the colorectal carcinogenic model [40] in which KRAS mutations develop early during carcinogenesis, because the precursor lesions in CRC are considered driver mutations (not passenger) in this cancer, consistent with its high homogeneity throughout various regions of the tumor and with the concordance between the primary tumor and metastasis sites.
Recent studies have shown that mutations in KRAS and NRAS are negative predictors of the response to anti-EGFR treatment. NRAS mutations are seen in approximately 5% of CRC cases [7,8]. In our cohort, we did not test for NRAS mutations. The frequency of NRAS mutations in CRC is much lower compared with KRAS mutations in codons 12 and 13, and although studies on heterogeneity in these sites might be beneficial, a larger cohort would need to be tested to determine the existence of this heterogeneity.
We conclude that intratumoral genetic heterogeneity in CRC is minor and that primary and metastatic tumors have high concordance regarding KRAS mutational status. Thus, in clinical decision-making, we suggest testing only 1 area of the primary tumor or metastasis for the presence of KRAS mutations to select patients who would benefit from anti-EGFR treatment, prioritizing the tissue in which viable tumor is most highly represented and with the least contamination with nontumor cells.
Acknowledgements
We acknowledge Fundação de Amparo à Pesquisa do Estado de São Paulo FAPESP, grant 2011/08510-2 for financial support and Fundação Antônio Prudente-AC Camargo Cancer Center, São Paulo, SP, Brazil for institutional support. We acknowledge Severino da Silva Ferreira and Carlos Carlos Ferreira Nascimento for support in preparing all of the slides for the experiments.
Disclosure of conflict of interest
None.
References
- 1.Soulieres D, Greer W, Magliocco AM, Huntsman D, Young S, Tsao MS, Kamel-Reid S. KRAS mutation testing in the treatment of metastatic colorectal cancer with anti-EGFR therapies. Curr Oncol. 2010;17(Suppl 1):S31–40. doi: 10.3747/co.v17is1.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Krieken JH, Jung A, Kirchner T, Carneiro F, Seruca R, Bosman FT, Quirke P, Flejou JF, Plato Hansen T, de Hertogh G, Jares P, Langner C, Hoefler G, Ligtenberg M, Tiniakos D, Tejpar S, Bevilacqua G, Ensari A. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453:417–431. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 3.Plesec TP, Hunt JL. KRAS mutation testing in colorectal cancer. Adv Anat Pathol. 2009;16:196–203. doi: 10.1097/PAP.0b013e3181a9d4ed. [DOI] [PubMed] [Google Scholar]
- 4.Wang HL, Lopategui J, Amin MB, Patterson SD. KRAS mutation testing in human cancers: The pathologist’s role in the era of personalized medicine. Adv Anat Pathol. 2010;17:23–32. doi: 10.1097/PAP.0b013e3181c6962f. [DOI] [PubMed] [Google Scholar]
- 5.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J. Clin. Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 6.Allegra CJ, Rumble RB, Hamilton SR, Mangu PB, Roach N, Hantel A, Schilsky RL. Extended RAS Gene Mutation Testing in Metastatic Colorectal Carcinoma to Predict Response to Anti-Epidermal Growth Factor Receptor Monoclonal Antibody Therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J. Clin. Oncol. 2016;34:179–185. doi: 10.1200/JCO.2015.63.9674. [DOI] [PubMed] [Google Scholar]
- 7.Irahara N, Baba Y, Nosho K, Shima K, Yan L, Dias-Santagata D, Iafrate AJ, Fuchs CS, Haigis KM, Ogino S. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19:157–163. doi: 10.1097/PDM.0b013e3181c93fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocakova I, Ruff P, Blasinska-Morawiec M, Smakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 9.Knijn N, Mekenkamp LJ, Klomp M, Vink-Borger ME, Tol J, Teerenstra S, Meijer JW, Tebar M, Riemersma S, van Krieken JH, Punt CJ, Nagtegaal ID. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011;104:1020–1026. doi: 10.1038/bjc.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han CB, Li F, Ma JT, Zou HW. Concordant KRAS mutations in primary and metastatic colorectal cancer tissue specimens: a metaanalysis and systematic review. Cancer Invest. 2012;30:741–747. doi: 10.3109/07357907.2012.732159. [DOI] [PubMed] [Google Scholar]
- 11.Italiano A, Hostein I, Soubeyran I, Fabas T, Benchimol D, Evrard S, Gugenheim J, Becouarn Y, Brunet R, Fonck M, Francois E, Saint-Paul MC, Pedeutour F. KRAS and BRAF mutational status in primary colorectal tumors and related metastatic sites: biological and clinical implications. Ann Surg Oncol. 2010;17:1429–1434. doi: 10.1245/s10434-009-0864-z. [DOI] [PubMed] [Google Scholar]
- 12.Richman SD, Chambers P, Seymour MT, Daly C, Grant S, Hemmings G, Quirke P. Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost-effectiveness of multiple sample testing. Anal Cell Pathol (Amst) 2011;34:61–66. doi: 10.3233/ACP-2011-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oltedal S, Aasprong OG, Moller JH, Korner H, Gilje B, Tjensvoll K, Birkemeyer EM, Heikkila R, Smaaland R, Nordgard O. Heterogeneous distribution of K-ras mutations in primary colon carcinomas: implications for EGFR-directed therapy. Int J Colorectal Dis. 2011;26:1271–1277. doi: 10.1007/s00384-011-1233-5. [DOI] [PubMed] [Google Scholar]
- 14.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 15.Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruine AP, Goldbohm RA, van den Brandt PA. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703–710. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- 16.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 17.Seth R, Crook S, Ibrahem S, Fadhil W, Jackson D, Ilyas M. Concomitant mutations and splice variants in KRAS and BRAF demonstrate complex perturbation of the Ras/Raf signalling pathway in advanced colorectal cancer. Gut. 2009;58:1234–1241. doi: 10.1136/gut.2008.159137. [DOI] [PubMed] [Google Scholar]
- 18.de Macedo MP, de Melo FM, Lisboa BC, Andrade LD, de Souza Begnami MD, Junior SA, Ribeiro HS, Soares FA, Carraro DM, da Cunha IW. KRAS gene mutation in a series of unselected colorectal carcinoma patients with prognostic morphological correlations: a pyrosequencing method improved by nested PCR. Exp Mol Pathol. 2015;98:563–567. doi: 10.1016/j.yexmp.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 19.Cejas P, Lopez-Gomez M, Aguayo C, Madero R, de Castro Carpeno J, Belda-Iniesta C, Barriuso J, Moreno Garcia V, Larrauri J, Lopez R, Casado E, Gonzalez-Baron M, Feliu J. KRAS mutations in primary colorectal cancer tumors and related metastases: a potential role in prediction of lung metastasis. PLoS One. 2009;4:e8199. doi: 10.1371/journal.pone.0008199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santini D, Loupakis F, Vincenzi B, Floriani I, Stasi I, Canestrari E, Rulli E, Maltese PE, Andreoni F, Masi G, Graziano F, Baldi GG, Salvatore L, Russo A, Perrone G, Tommasino MR, Magnani M, Falcone A, Tonini G, Ruzzo A. High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist. 2008;13:1270–1275. doi: 10.1634/theoncologist.2008-0181. [DOI] [PubMed] [Google Scholar]
- 21.Docs O, Fazakas F, Horvath NL, Toth L, Andras C, Horvath Z, Mehes G. Changes of KRAS Exon 2 Codon 12/13 Mutation Status in Recurrent Colorectal Cancer. Pathol Oncol Res. 2015;21:399–404. doi: 10.1007/s12253-014-9834-2. [DOI] [PubMed] [Google Scholar]
- 22.Miglio U, Mezzapelle R, Paganotti A, Allegrini S, Veggiani C, Antona J, Gentilli S, Monga G, Alabiso O, Boldorini R. Mutation analysis of KRAS in primary colorectal cancer and matched metastases by means of highly sensitivity molecular assay. Pathol Res Pract. 2013;209:233–236. doi: 10.1016/j.prp.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Vakiani E, Janakiraman M, Shen R, Sinha R, Zeng Z, Shia J, Cercek A, Kemeny N, D’Angelica M, Viale A, Heguy A, Paty P, Chan TA, Saltz LB, Weiser M, Solit DB. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J. Clin. Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paliogiannis P, Cossu A, Tanda F, Palmieri G, Palomba G. mutational concordance between primary and metastatic colorectal adenocarcinoma. Oncol Lett. 2014;8:1422–1426. doi: 10.3892/ol.2014.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, Kania K, Viale A, Oschwald DM, Vacic V, Emde AK, Cercek A, Yaeger R, Kemeny NE, Saltz LB, Shia J, D’Angelica MI, Weiser MR, Solit DB, Berger MF. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crumley SM, Pepper KL, Phan AT, Olsen RJ, Schwartz MR, Portier BP. Next-Generation Sequencing of Matched Primary and Metastatic Rectal Adenocarcinomas Demonstrates Minimal Mutation Gain and Concordance to Colonic Adenocarcinomas. Arch Pathol Lab Med. 2016;140:529–35. doi: 10.5858/arpa.2015-0261-SA. [DOI] [PubMed] [Google Scholar]
- 28.Kosmidou V, Oikonomou E, Vlassi M, Avlonitis S, Katseli A, Tsipras I, Mourtzoukou D, Kontogeorgos G, Zografos G, Pintzas A. Tumor heterogeneity revealed by KRAS, BRAF, and PIK3CA pyrosequencing: KRAS and PIK3CA intratumor mutation profile differences and their therapeutic implications. Hum Mutat. 2014;35:329–340. doi: 10.1002/humu.22496. [DOI] [PubMed] [Google Scholar]
- 29.Murphy JE, Ryan DP. American Society of Clinical Oncology 2010 colorectal update. Expert Rev Anticancer Ther. 2010;10:1371–1373. doi: 10.1586/era.10.123. [DOI] [PubMed] [Google Scholar]
- 30.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J. Clin. Oncol. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 31.de Macedo MP, de Melo FM, Ribeiro Jda S, de Mello CA, de Souza Begnami MD, Soares FA, Carraro DM, da Cunha IW. RAS mutations vary between lesions in synchronous primary colorectal cancer: testing only one lesion is not sufficient to guide anti-EGFR treatment decisions. Oncoscience. 2015;2:125–130. doi: 10.18632/oncoscience.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balschun K, Haag J, Wenke AK, von Schonfels W, Schwarz NT, Rocken C. KRAS, NRAS, PIK3CA exon 20, and BRAF genotypes in synchronous and metachronous primary colorectal cancers diagnostic and therapeutic implications. J Mol Diagn. 2011;13:436–445. doi: 10.1016/j.jmoldx.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogino S, Brahmandam M, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. Epigenetic profiling of synchronous colorectal neoplasias by quantitative DNA methylation analysis. Mod Pathol. 2006;19:1083–1090. doi: 10.1038/modpathol.3800618. [DOI] [PubMed] [Google Scholar]
- 34.Kubota M, Lin YW, Hamahata K, Sawada M, Koishi S, Hirota H, Wakazono Y. Cancer chemotherapy and somatic cell mutation. Mutat Res. 2000;470:93–102. doi: 10.1016/s1383-5742(00)00043-0. [DOI] [PubMed] [Google Scholar]
- 35.Kawamoto Y, Tsuchihara K, Yoshino T, Ogasawara N, Kojima M, Takahashi M, Ochiai A, Bando H, Fuse N, Tahara M, Doi T, Esumi H, Komatsu Y, Ohtsu A. KRAS mutations in primary tumours and post-FOLFOX metastatic lesions in cases of colorectal cancer. Br J Cancer. 2012;107:340–344. doi: 10.1038/bjc.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ondrejka SL, Schaeffer DF, Jakubowski MA, Owen DA, Bronner MP. Does neoadjuvant therapy alter KRAS and/or MSI results in rectal adenocarcinoma testing? Am J Surg Pathol. 2011;35:1327–1330. doi: 10.1097/PAS.0b013e3182253800. [DOI] [PubMed] [Google Scholar]
- 37.Boissiere-Michot F, Lopez-Crapez E, Frugier H, Berthe ML, Ho-Pun-Cheung A, Assenat E, Maudelonde T, Lamy PJ, Bibeau F. KRAS genotyping in rectal adenocarcinoma specimens with low tumor cellularity after neoadjuvant treatment. Mod Pathol. 2012;25:731–739. doi: 10.1038/modpathol.2011.210. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko Y, Kuramochi H, Nakajima G, Inoue Y, Yamamoto M. Degraded DNA may induce discordance of KRAS status between primary colorectal cancer and corresponding liver metastases. Int J Clin Oncol. 2014;19:113–120. doi: 10.1007/s10147-012-0507-4. [DOI] [PubMed] [Google Scholar]
- 39.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 40.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]