Abstract
Increasing studies showed that long non-coding RNAs (lncRNAs) played important roles in the development and progression of tumors. Previous evidences suggested that Tumor suppressor candidate 7 (TUSC7) was involved in several tumors initiation. However, the role of TUSC7 in colorectal cancer is still unknown. In this study, we indicated that the expression of TUSC7 was downregulated in colorectal cancer cell lines and tissues. Moreover, the expression of TUSC7 was lower in the high-grade (Dukes C and D) colorectal cancer patients compared to that in the low-grade colorectal cancer patients (Dukes A and B). Colorectal cancer patients with a lower level of TUSC7 expression had worse overall survival rate. Elevated expression of TUSC7 suppressed SW480 and HT29 cell proliferation and invasion. In addition, we demonstrated that overexpression of TUSC7 inhibited the expression of miR-10a and enhanced the expression of PTEN and EphA8, which were the direct target genes of miR-10a. Furthermore, the expression of miR-10a was upregulated in colorectal cancer cell lines and tissues. TUSC7 suppressed colorectal cancer cell proliferation and invasion partly through targeting miR-10a. These results suggested that TUSC7 played as a tumor suppressor gene in colorectal cancer partly through inhibiting miR-10a expression.
Keywords: Colorectal cancer, LncRNAs, TUSC7, miR-10a
Introduction
Colorectal cancer is a common digestive tract cancer and is the 3rd leading cause of the tumor deaths [1-4]. More than one million new patients are diagnosed annually and this disease is a serious economic and demographic problem worldwide [5-7]. Nevertheless, there is a paucity of the effective therapy for patients with metastatic colorectal cancer [8-11]. Therefore, it is of great importance to understand the molecular mechanism underlying the development of colorectal cancer and identify new markers and effective therapeutic strategies for colorectal cancer patients.
LncRNAs (long non-coding RNAs) are a subgroup of ncRNAs which are less than 200 nucleotides in length with no or limited protein-coding capacity [12-15]. Recent studies have demonstrated that lncRNAs participate in many cellular processes such as cell development, differentiation, proliferation, invasion, apoptosis, stem cell pluripotency and cell migration [16-20]. Increasing evidences suggested that dysfunction of lncRNAs were found in a broad range of tumors such as gastric cancer, ovarian cancer, glioblastoma, renal cell carcinoma, cutaneous squamous cell carcinoma and also colorectal cancer [21-25]. LncRNA tumor suppressor candidate 7 (TUSC7) is located at the 3q13.31 and consists of four exons of about 2 kb in length [26-28]. Recent evidences have demonstrated that TUSC7 expression is downregulated in osteosarcoma, gastric cancer and acute myeloid leukemia [26,27,29]. However, the role of TUSC7 in colorectal cancer was still unknown.
In this study, we investigated the expression and role of TUSC7 in colorectal cancer. We indicated that the expression of TUSC7 was downregulated in colorectal cancer cell lines and tissues. Elevated expression of TUSC7 suppressed SW480 and HT29 cell proliferation and invasion.
Materials and methods
Cell lines and tumor samples
Thirty pairs of human colorectal cancer samples and matched non-cancerous tissues were collected with primary colorectal cancer patients at our department. All patients did not receive any preoperative therapy, such as chemotherapy or radiotherapy. Our study was approved by the Research Ethical Committee of Cang Zhou Central Hospital. Written informed consents were obtained from all patients. Four colorectal cancer cell lines (HT29, SW480, SW620 and DLD-1) and one normal colon epithelium cell line (FHC) were bought from the ATCC (American Type Culture Collection, Manassas, USA) and kept in the Dulbecco’s modified Eagle’s medium (DMEM) supplemented with PBS, penicillin and streptomycin. The TUSC7 vector or control vector and miR-10a mimic or scramble were collected from Shanghai GenePharma (Shanghai, China) and were transfected to SW480 cells by using the Lipofectamine 2000 (Invitrogen, USA) in accordance with manufacturer’s instruction.
Quantitative Real-time-PCR
Total RNAs from cells or frozen tissues were extracted by using the TRIzol Reagent (Invitrogen, USA) n accordance with the manufacturer’s protocol. The expression of TUSC7, ki-67, cyclin D1, PTEN, EphA8 and GAPDH was determined by using SYBR Green qRT-PCR analysis. Specific primers were designed and synthesized from Sangon (Shanghai, China). GAPDH was used as the internal control. The primers were shown in the Table 1.
Table 1.
Primer sequence
| Name | Sequence (5’-3’) |
|---|---|
| Real-time PCR primer sequence | |
| TUSC7 | CACTGCCTATGTGCACGACT |
| AGAGTCCGGCAAGAAGAACA | |
| U6 | CTCGCTTCGGCAGCACATATACT |
| ACGCTTCACGAATTTGCGTGTC | |
| GAPDH | AATGGGCAGCCGTTAGGAAA |
| TGAAGGGGTCATTGATGGCA | |
| cyclin D1 | CTGTGCTGCGAAGTGGAAACCAT |
| TTCATGGCCAGCGGGAAGACCTC | |
| ki-67 | TCCTTTGGTGGGCACCTAAGACCTG |
| TGATGGTTGAGGTCGTTCCTTGATG | |
| PTEN | GGGACGAACTGGTGTAATGA |
| CGCCTCTGACTGGGAATAG | |
| EphA8 | CCACCAGGGTATGTAAATATC |
| TGTGCTTTGAAGACCATTT |
Cell growth, colony formation and invasion assays
Cell growth was measured by using MTT analysis (Sigma, USA). Cells were cultured in the 96-well plate and 20 μl of MTT was put into each hole and continued to culture for additonal 4 hours. The OD (absorbance value) was determined at the 490 nm on the microplate reader. For cell colony formation, cell were plated in the 6-well plate and cultured for 2 weeks. These colonies were stained with the crystal violet and counted. For cell invasion, Matrigel (BD Biosciences) and Matrigel was coated onto the transwell upper chamber of the cell was cultured on the upper chamber. The serum was added on the lower chamber and the invasive cell was fixed with methanol, stained with crystal violet.
Statistical analysis
All data was shown as the mean ± standard deviation (SD). The data was tested by SPSS 17.0 version software (Chicago, IL, USA). The two-tailed Student’s t-test was used to determine the statistical significance differences between groups. P<0.05 was indicated to be statistically significant difference.
Results
TUSC7 expression was downregulated in colorectal cancer cell lines and samples
TUSC7 expression was lower in four colorectal cancer cell lines (HT29, SW480, SW620 and DLD-1) compared to in normal colon epithelium cell line (FHC) (Figure 1A). The expression of TUSC7 was also lower in colorectal cancer samples than in matched non-cancerous tissues (Figure 1B). Of 40 colorectal cancer samples, TUSC7 expression was downregulated in 29 cases (27/40; 67.5%) compared with that in the adjacent non-cancerous tissues (Figure 1C). Moreover, compared to colorectal cancer samples with low-grade (Dukes A and B), high-grade (Dukes C and D) samples expressed lower TUSC7 (Figure 2A). The colorectal cancer patients with lower TUSC7 expression had worse overall survival rate compared with those with high TUSC7 expression (Figure 2B).
Figure 1.
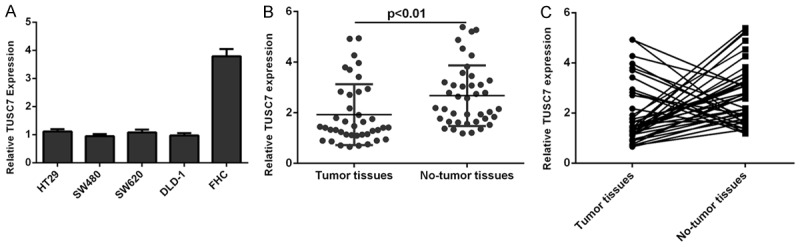
The expression of TUSC7 was downregulated in colorectal cancer cell lines and samples. A: The expression of TUSC7 in four colorectal cancer cell lines (HT29, SW480, SW620 and DLD-1) and normal colon epithelium cell line (FHC) was measured by qRT-PCR. GAPDH was used for TUSC7 normalization. B: TUSC7 expression was lower in colorectal cancer tissues compared to in the matched non-cancerous tissues. The expression of TUSC7 was measured by qRT-PCR. GAPDH was used for TUSC7 normalization. C: The expression of TUSC7 in colorectal cancer tissues and matched non-cancerous tissues was measured by qRT-PCR. Of 40 colorectal cancer samples, TUSC7 expression was downregulated in 29 cases (27/40; 67.5%) compared with that in the adjacent non-cancerous tissues.
Figure 2.
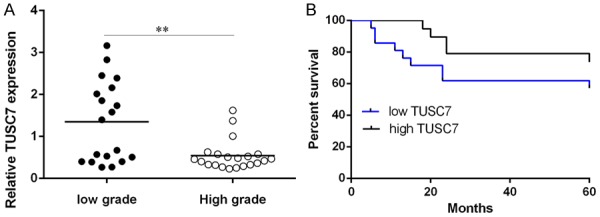
Downexpression of TUSC7 in colorectal cancer confers poor prognosis. A: The expression in the high-grade (Dukes C and D) samples was lower compared to in the low-grade (Dukes A and B). B: Kaplan-Meier OS analysis showed that the colorectal cancer patients with a lower level of TUSC7 expression had worse overall survival rate compared with those with high TUSC7 expression. **P<0.01.
Overexpression of TUSC7 suppressed colorectal cancer cell proliferation and invasion
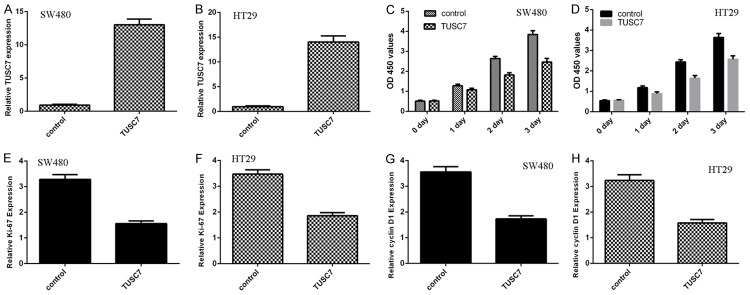
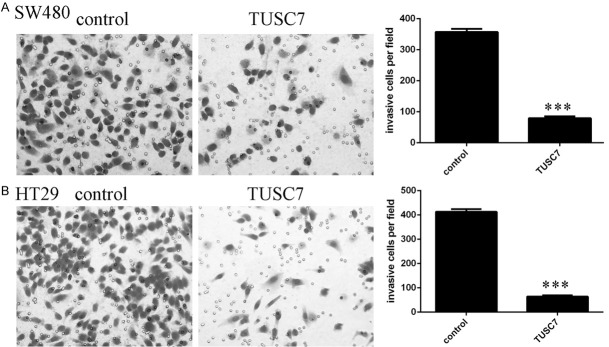
The expression of TUSC7 was significantly upregulated in both colorectal cancer cell line SW480 (Figure 3A) and HT29 (Figure 3B). Overexpression of TUSC7 suppressed cell proliferation in both SW480 and HT29 cell lines (Figure 3C and 3D). In addition, elevated expression of TUSC7 decreased ki-67 expression in SW480 cell (Figure 3E) and HT29 cell (Figure 3F). Moreover, Overexpression of TUSC7 inhibited cyclin D1 expression in SW480 cell (Figure 3G) and HT29 cell (Figure 3H). Ectopic expression of TUSC7 suppressed SW480 cell invasion (Figure 4A). In line with this, overexpression of TUSC7 decreased HT29 cell invasion (Figure 4B).
Figure 3.
Overexpression of TUSC7 suppressed the colorectal cancer cell proliferation and invasion. A: The expression of TUSC7 in the SW480 cell that treated with pcDNA-TUSC7 or control vector was determined by qRT-PCR. B: The expression of TUSC7 in the HT29 cell was determined by qRT-PCR. C: MTT analysis was performed to measure the cell proliferation. Overexpression of TUSC7 suppressed the SW480 cell growth. D: Overexpression of TUSC7 decreased the HT29 cell proliferation by using MTT analysis. E: Elevated expression of TUSC7 suppressed the ki-67 expression in SW480 cell. F: The ki-67 mRNA expression was measured by qRT-PCR. G: Ectopic expression of TUSC7 decreased the cyclin D1 expression in the SW480 cell. H: The cyclin D1 mRNA expression in the HT29 cell was measured by qRT-PCR. Elevated expression of TUSC7 decreased cyclin D1 mRNA expression in the HT29 cell.
Figure 4.
TUSC7 suppressed the colorectal cancer cell invasion. A: Ectopic expression of TUSC7 decreased the SW480 cell invasion. The relative invasive cells were shown in the right. B: Overexpression of TUSC7 suppressed the HT29 cell invasion. The relative invasive cells were shown in the right. ***P<0.001.
Overexpression of TUSC7 inhibited the miR-10b expression
Ecoptic expression of TUSC7 suppressed miR-10b expression in SW480 cell (Figure 5A) and HT29 cell (Figure 5B). Moreover, overexpression of TUSC7 enhanced the expression of PTEN (Figure 5C) and EphA8 (Figure 5D), which were the direct target genes of miR-10a. The expression of miR-10a was upregulated in colorectal cancer cell lines and samples.
Figure 5.
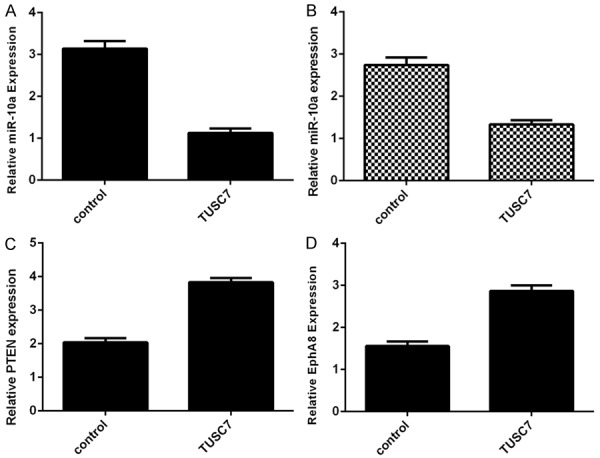
Overexpression of TUSC7 inhibited the miR-10b expression. A: The expression of miR-10b in the SW480 cell was determined by qRT-PCR. B: Ecoptic expression of TUSC7 suppressed miR-10b expression in the HT29 cell. C: The expression of PTEN was determined by qRT-PCR. D: The expression of EphA8 was determined by qRT-PCR.
The expression of miR-10a was higher in four colorectal cancer cell lines (HT29, SW480, SW620 and DLD-1) compared to that in the normal colon epithelium cell line (FHC) (Figure 6A). The expression of miR-10a was also higher in colorectal cancer samples than in the matched non-cancerous tissues (Figure 6B). Of 40 colorectal cancer samples, miR-10a expression was upregulated in 28 cases (28/40; 70%) compared with adjacent non-cancerous tissues (Figure 6C). The expression of TUSC7 was negatively correlated with the expression of miR-10a in colorectal cancer samples (Figure 6D).
Figure 6.
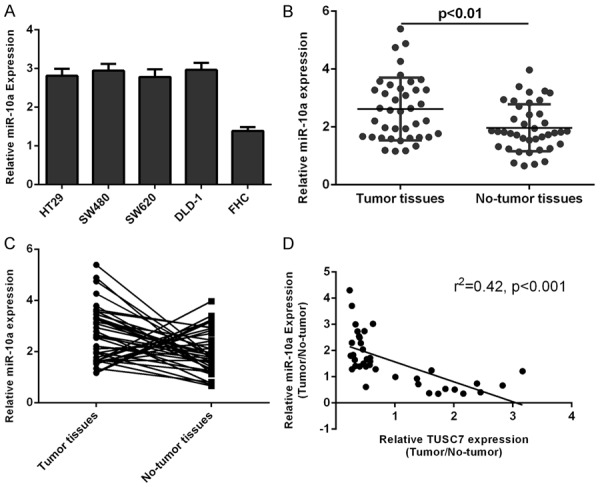
The expression of miR-10a was upregulated in the colorectal cancer cell lines and samples. A: The expression of miR-10a in the four colorectal cancer cell lines (HT29, SW480, SW620 and DLD-1) and one normal colon epithelium cell line (FHC) was determined by qRT-PCR. U6 was performed as the control. B: miR-10a expression was higher in colorectal cancer tissues compared to in the matched non-cancerous tissues. The expression of miR-10a in the four colorectal cancer tissues was determined by qRT-PCR. U6 was performed as the control. C: The expression of miR-10a in colorectal cancer tissues and matched non-cancerous tissues was measured by qRT-PCR. Of 40 colorectal cancer samples, miR-10a expression was upregulated in 28 cases (28/40; 70%) compared with adjacent non-cancerous tissues. D: The expression of TUSC7 was negatively correlated with the expression of miR-10a in the colorectal cancer samples.
TUSC7 suppressed colorectal cancer cell proliferation and invasion through targeting miR-10a
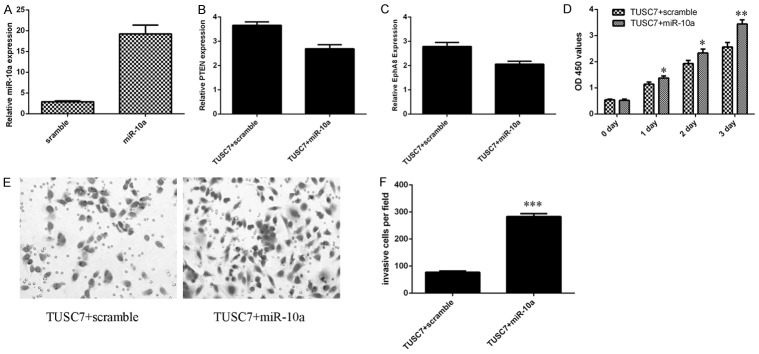
The expression of miR-10a was significantly upregulated in SW480 cell after treated with miR-10a mimic (Figure 7A). To study whether miR-10a was involved in the function of TUSC7 in SW480 cells, we restored miR-10a expression in the TUSC7 overexpressing SW480 cell. Overexpression of miR-10a suppressed the expression of PTEN (Figure 7B) and EphA8 (Figure 7C) in the TUSC7 overexpressing SW480 cell. Ecoptic expression of miR-10a promoted SW480 cell proliferation, reversing TUSC7-inhibition of proliferation (Figure 7D). Furthermore, evelated expression of miR-10a promoted the TUSC7 overexpressing SW480 cell invasion (Figure 7E and 7F).
Figure 7.
TUSC7 suppressed the colorectal cancer cell proliferation and invasion through targeting the miR-10a expression. A: The expression of miR-10a in the SW480 cell was determined by qRT-PCR. U6 was performed as the control. B: The expression of PTEN was determined by qRT-PCR. C: The expression of EphA8 was determined by qRT-PCR. D: The cell proliferation was determined by MTT analysis. E: Evelated expression of miR-10a promoted the TUSC7 overexpressing SW480 cell invasion. F: The relative invasive cells were shown. *P<0.05, **P<0.01 and ***P<0.001.
Discussion
In this study, we indicated that TUSC7 expression was downregulated in colorectal cancer cell lines and tissues. Moreover, the expression of TUSC7 was lower in the high-grade (Dukes C and D) colorectal cancer patients compared to in that in the low-grade colorectal cancer patients (Dukes A and B). The colorectal cancer patients with a lower level of TUSC7 expression had worse overall survival rate compared with those who had high TUSC7 expression. Elevated expression of TUSC7 suppressed colorectal cancer cell proliferation and invasion. In addition, we demonstrated that overexpression of TUSC7 inhibited miR-10a expression and enhanced the expression of PTEN and EphA8, which were the direct target genes of miR-10a. Furthermore, we showed that the expression of miR-10a was upregulated in colorectal cancer cell lines and tissues. TUSC7 suppressed colorectal cancer cell proliferation and invasion partly through targeting miR-10a. These results suggested that TUSC7 played as a tumor suppresor gene in colorectal cancer partly through inhibiting miR-10a expression.
TUSC7, previous named as LSAMP-AS3 (LOC285194), was originally indentified in the osteosarcoma [29]. Previous studies indicated that TUSC7 played important roles in the several tumors [26-31]. It was located at the chromosome 3q13.31 and played as a p53-regulated tumor suppressor gene. For example, Wang et al. [28] showed that TUSC7 expression was downregulated in non-small-cell lung cancer (NSCLC) tissues and cells. Lower TUSC7 expression was related with higher TNM and larger tumor size. Patients with higher TUSC7 expression had better overall survival in NSCLC. Overexpression of TUSC7 suppressed lung cancer cell proliferation. Wang et al. [30] indicated that TUSC7 expression was downregulated in hepatocellular carcinoma (HCC) and served as a risk factor for HCC cases. Elevated expression of TUSC7 suppressed HCC cell invasion, epithelial-to-mesenchymal transformation (EMT) and metastasis through regulating miR-10a expression. Shang et al. [31] demonstrated that TUSC7 expression was downregulated in glioma cell lines and tissues and lower expression of TUSC7 was associated with worse histological grade. Overexpression of TUSC7 inhibited glioma cell proliferation and invasion and enhanced glioma cell apoptosis partly through targeting TUSC7. Qi et al. [26] showed that TUSC7 expression was downregulated in gastric cancer tissues and overexpression of TUSC7 decreased gastric cancer cell growth in vivo and in vitro by repressing miR-23b expression. In line with these, we showed that the TUSC7 expression was lower in colorectal cancer cell lines (HT29, SW480, SW620 and DLD-1) compared to in the normal colon epithelium cell line (FHC). Of 40 colorectal cancer samples, TUSC7 expression was downregulated in 29 cases (27/40; 67.5%) compared with adjacent non-cancerous tissues. The expression of TUSC7 was also lower in the colorectal cancer samples than in the matched non-cancerous tissues. Moreover, compared to colorectal cancer samples with low-grade (Dukes A and B), high-grade (Dukes C and D) samples expressed lower TUSC7. The colorectal cancer patients with a lower level of TUSC7 expression had worse overall survival rate compared with those with higher TUSC7 expression. Elevated expression of TUSC7 suppressed cell proliferation and invasion in colorectal cancer cells. These results indicated that TUSC7 acted as a tumor suppressor gene in colorectal cancer.
Increasing studies suggested that lncRNA played as a ceRNA to modulate miRNAs in tumor development [16,32-34]. Previous studies demonstrated that TUSC7 played as a tumor suppressor gene in several tumors by inhibiting the expression of some miRNAs such as miR-10a, miR-211 and miR-23b [26,30,31]. In our study, we focused on the role of miR-10a since it was proved to act crucial roles in the development of tumors [35-38]. Yu et al. [39] showed that miRNA-10a expression was upregulated in non-small cell lung cancer tissues compared with corresponding normal samples and the expression of miR-10a was correlated with lymph node metastasis and tumor node metastasis. Ecoptic expression of miR-10a enhanced cell proliferation, invasion and migration through targeting the PTEN in the non-small cell lung cancer. In addition, Yan et al. [36] demonstrated that miR-10a promoted glioma cell invasion and migration and EMT through inhibiting EphA8 expression. In line with these, we proved that overexpression of TUSC7 suppressed the miR-10a expression and promoted the expression of PTEN and EphA8, which were the direct target genes of miR-10a. Moreover, we demonstrated that miR-10a expression was upregulated in colorectal cancer cell lines and samples. Of 40 colorectal cancer samples, miR-10a expression was upregulated in 28 cases (28/40; 70%) compared with adjacent non-cancerous tissues. The expression of TUSC7 was negatively correlated with the expression of miR-10a in colorectal cancer samples. Furthermore, we showed that TUSC7 suppressed the colorectal cancer cell proliferation and invasion through targeting miR-10a.
In conclusion, we reported that TUSC7 expression was downregulated in colorectal cancer tissues and cell lines. In addition, TUSC7 expression was associated with tumor grade and overall survival rate. Overexpression of TUSC7 suppressed colorectal cancer cell proliferation and invasion through inhibiting miR-10a expression. These results suggested that TUSC7 might represent a new therapeutic target in colorectal cancer.
Disclosure of conflict of interest
None.
References
- 1.Li P, Xue WJ, Feng Y, Mao QS. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1) Am J Transl Res. 2015;7:2053–2059. [PMC free article] [PubMed] [Google Scholar]
- 2.Yu X, Li Z, Yu J, Chan MT, Wu WK. MicroRNAs predict and modulate responses to chemotherapy in colorectal cancer. Cell Prolif. 2015;48:503–510. doi: 10.1111/cpr.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, Li D, Li Z, Ding Y, Chen L. MiR-124 radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014;9:e93917. doi: 10.1371/journal.pone.0093917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perilli L, Vicentini C, Agostini M, Pizzini S, Pizzi M, D’Angelo E, Bortoluzzi S, Mandruzzato S, Mammano E, Rugge M, Nitti D, Scarpa A, Fassan M, Zanovello P. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget. 2014;5:6611–6619. doi: 10.18632/oncotarget.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Liu J, Wang X, Wu R, Lin M, Laddha SV, Yang Q, Chan CS, Feng Z. MicroRNA-339-5p inhibits colorectal tumorigenesis through regulation of the MDM2/p53 signaling. Oncotarget. 2014;5:9106–9117. doi: 10.18632/oncotarget.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z, Protivankova M, Vycital O, Liska V, Schwarzova L, Vodickova L, Vodicka P. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014 doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 8.Kalimutho M, Di Cecilia S, Del Vecchio Blanco G, Roviello F, Sileri P, Cretella M, Formosa A, Corso G, Marrelli D, Pallone F, Federici G, Bernardini S. Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer. Br J Cancer. 2011;104:1770–1778. doi: 10.1038/bjc.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamimae S, Yamamoto E, Yamano HO, Nojima M, Suzuki H, Ashida M, Hatahira T, Sato A, Kimura T, Yoshikawa K, Harada T, Hayashi S, Takamaru H, Maruyama R, Kai M, Nishiwaki M, Sugai T, Sasaki Y, Tokino T, Shinomura Y, Imai K, Toyota M. Epigenetic alteration of DNA in mucosal wash fluid predicts invasiveness of colorectal tumors. Cancer Prev Res (Phila) 2011;4:674–683. doi: 10.1158/1940-6207.CAPR-10-0214. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J, Thomas GJ, Packham G, Mirnezami AH. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73:6435–6447. doi: 10.1158/0008-5472.CAN-12-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013;42:219–228. doi: 10.3892/ijo.2012.1707. [DOI] [PubMed] [Google Scholar]
- 12.Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37:4025–33. doi: 10.1007/s13277-015-4227-z. [DOI] [PubMed] [Google Scholar]
- 13.Lu H, He Y, Lin L, Qi Z, Ma L, Li L, Su Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016;37:1683–91. doi: 10.1007/s13277-015-3946-5. [DOI] [PubMed] [Google Scholar]
- 14.Tang Y, Jin X, Xiang Y, Chen Y, Shen CX, Zhang YC, Li YG. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett. 2015;589:3189–3196. doi: 10.1016/j.febslet.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 15.He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 16.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C, Lu J. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:5427–5434. [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W, Yan L, Wang K, Feng J. Downregulated long noncoding RNA BANCR promotes the proliferation of colorectal cancer cells via downregualtion of p21 expression. PLoS One. 2015;10:e0122679. doi: 10.1371/journal.pone.0122679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Shen ED, Liao MM, Hu YB, Wu K, Yang P, Zhou L, Chen WD. Expression and mechanisms of long non-coding RNA genes MEG3 and ANRIL in gallbladder cancer. Tumour Biol. 2016;37:9875–9886. doi: 10.1007/s13277-016-4863-y. [DOI] [PubMed] [Google Scholar]
- 20.Su S, Gao J, Wang T, Wang J, Li H, Wang Z. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol. 2015;36:7205–7211. doi: 10.1007/s13277-015-3413-3. [DOI] [PubMed] [Google Scholar]
- 21.Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46:2497–2505. doi: 10.3892/ijo.2015.2943. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, Yu SZ, Pu PY, Liu N, You YP, Jiang T, Kang CS. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013;15:1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Huang T, Luo G, Huang C, Xiao XY, Wang L, Jiang GS, Zeng FQ. Long non-coding RNA MEG3 induces renal cell carcinoma cells apoptosis by activating the mitochondrial pathway. J Huazhong Univ Sci Technolog Med Sci. 2015;35:541–545. doi: 10.1007/s11596-015-1467-5. [DOI] [PubMed] [Google Scholar]
- 25.Cao X, Zhao R, Chen Q, Zhao Y, Zhang B, Zhang Y, Yu J, Han G, Cao W, Li J, Chen X. MALAT1 might be a predictive marker of poor prognosis in patients who underwent radical resection of middle thoracic esophageal squamous cell carcinoma. Cancer Biomark. 2015;15:717–723. doi: 10.3233/CBM-150513. [DOI] [PubMed] [Google Scholar]
- 26.Qi P, Xu MD, Shen XH, Ni SJ, Huang D, Tan C, Weng WW, Sheng WQ, Zhou XY, Du X. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer. 2015;137:1269–1278. doi: 10.1002/ijc.29516. [DOI] [PubMed] [Google Scholar]
- 27.Coccaro N, Zagaria A, Tota G, Anelli L, Orsini P, Casieri P, Cellamare A, Minervini A, Impera L, Minervini CF, Brunetti C, Mestice A, Carluccio P, Cumbo C, Specchia G, Albano F. Overexpression of the LSAMP and TUSC7 genes in acute myeloid leukemia following microdeletion/duplication of chromosome 3. Cancer Genet. 2015;208:517–522. doi: 10.1016/j.cancergen.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Wang ZW, Jin YY, Ren HT, Ma XL, Wang BF, Wang YL. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am J Transl Res. 2016;8:680–U1079. [PMC free article] [PubMed] [Google Scholar]
- 29.Cong ML, Li JM, Jing R, Li ZZ. Long noncoding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumor Biol. 2016;37:9441–9450. doi: 10.1007/s13277-015-4414-y. [DOI] [PubMed] [Google Scholar]
- 30.Wang YF, Liu ZK, Yao BW, Dou CW, Xu M, Xue YM, Ding LL, Jia YL, Zhang HY, Li Q, Tu KS, Jiao Y, Liu QG, Guo C. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumor Biol. 2016;37:11429–11441. doi: 10.1007/s13277-016-4892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang C, Guo Y, Hong Y, Xue YX. Long noncoding RNA TUSC7, a target of miR-23b, plays tumor-suppressing roles in human gliomas. Front Cell Neurosci. 2016;10:235. doi: 10.3389/fncel.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, Li H, Pan Y, Li A, Ye Z, Wang J, Xu H, Huang Q. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JQ, Hu SY, Wang ZY, Lin J, Jian S, Dong YC, Wu XF, Dai L, Cao LJ. Long non-coding RNA MEG3 inhibits microRNA-125a-5p expression and induces immune imbalance of Treg/Th17 in immune thrombocytopenic purpura. Biomed Pharmacother. 2016;83:905–911. doi: 10.1016/j.biopha.2016.07.057. [DOI] [PubMed] [Google Scholar]
- 35.Sun W, Ma YP, Chen P, Wang D. MicroRNA-10a silencing reverses cisplatin resistance in the A549/cisplatin human lung cancer cell line via the transforming growth factor-beta/Smad2/STAT3/STAT5 pathway. Mol Med Rep. 2015;11:3854–3859. doi: 10.3892/mmr.2015.3181. [DOI] [PubMed] [Google Scholar]
- 36.Yan Y, Wang Q, Yan XL, Zhang Y, Li W, Tang F, Li X, Yang P. miR-10a controls glioma migration and invasion through regulating epithelialmesenchymal transition via EphA8. FEBS Lett. 2015;589:756–765. doi: 10.1016/j.febslet.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Huang HB, Fan LP, Zhan R, Wu SQ, Niu WY. Expression of microRNA-10a, microRNA-342-3p and their predicted target gene TIAM1 in extranodal NK/T-cell lymphoma, nasal type. Oncol Lett. 2016;11:345–351. doi: 10.3892/ol.2015.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An YB, Zhao HX, Zhang J, Lu T, Jia JL, Zhao B. Up-regulation of miR-10a and down-regulation of miR-148b serve as potential prognostic biomarkers for osteosarcoma. Int J Clin Exp Pathol. 2016;9:186–190. [Google Scholar]
- 39.Yu T, Liu L, Li J, Yan MX, Lin HC, Liu Y, Chu DD, Tu H, Gu AQ, Yao M. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget. 2015;6:30239–30250. doi: 10.18632/oncotarget.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]