Abstract
Chondrogenic differentiation of mesenchymal stem cells is regulated by many different pathways. Recent studies have established that hypoxia and epigenetic alterations potently affect expression of chondrogenesis marker genes. Sox9 is generally regarded as a master regulator of chondrogenesis and microRNA-124 (miRNA-124) regulates gene expression in murine bone marrow-derived mesenchymal stem cells. Therefore, in this study we investigated whether epigenetic regulation of miRNA-124 could affect the expression of Sox9 and thereby regulate chondrogenesis. A cell pellet culture model was used to induce chondrogenesis in C3H10T1/2 cells under hypoxic conditions (2% O2) to determine the effects of hypoxia on miR-124 expression and DNA methylation. The expression of miR-124 was significantly downregulated under hypoxic conditions compared to normoxic conditions (21% O2). The expression of chondrogenesis marker genes was significantly increased under hypoxic conditions. Bisulfite sequencing of the CpG islands in the promoter region of miR-124-3 showed that CpG methylation was significantly increased under hypoxic conditions. Treating the cells with the DNA demethylating agent 5’-AZA significantly increased miR-124 expression and decreased expression of markers of chondrogenesis. Overexpressing miR-124 under hypoxic conditions inhibited NFATc1 reporter activity. NFATc1 was shown to bind to the promoter region of Sox9. Taken together, our data provide evidence that miR-124 acts as an inhibitor of NFATc1. Under hypoxic conditions when miR-124 is downregulated by methylation of CpG islands in the promoter, NFATc1 can bind to the Sox9 promoter and induce the expression of Sox9 leading to chondrogenesis. These results support the role of epigenetic regulation in establishing and maintaining a chondrogenic phenotype.
Keywords: Hypoxia, miR-124, DNA methylation, chondrogeneic differentiation
Introduction
Chondrogenesis from stem cells can be divided into three distinct stages: cell lodging, proliferation and differentiation, and differentiation and hypertrophy [1]. Chondrocyte hypertrophy is marked by a 10-fold increase in cell volume [2], extracellular matrix (ECM) remodeling, and expression of the terminal differentiation marker collagen X. Due to its avascular nature, cartilage tissue is regarded as a hypoxic microenvironment and has limited regenerative capacity following injury. Hypoxia has been shown to enhance chondrogenic differentiation [3,4], maintain cartilage phenotypes [5,6], and induce DNA hypo- or hyper-methylation [7]. In the murine cell line C3H10T1/2, hypoxia has been shown to enhance chondro-specific differentiation based on the expression of collagen II and aggrecan, key ECM components of functional cartilage [7,8]. Hypoxia also promotes the expression of Sox9, a transcription factors that is one of the master regulators involved in initiating chondrogenesis [9]. Thus, the hypoxic microenvironment is implicated in regulating many cellular events leading to cell survival or differentiation during chondrogenesis.
Under hypoxic conditions, increased calcineurin activity enhances the functionality of nuclear factor of activated T-cells (NFAT). Like hypoxia, calcium [Ca2+] signaling cascades have been linked to vertebrate development and the growth and differentiation of multiple cell types [10,11]. [Ca2+] acts as a switch for NFAT activity by regulating its phosphorylation status. In resting cells, phosphorylated NFAT is retained in the cytoplasm, while dephosphorylated NFAT (mediated by [Ca2+]) translocates to the nucleus and binds to its target promoter regions [12,13]. Previous research indicated that calcineurin/NFAT pathway is primarily linked to cell differentiation, and also influences chondrogenic differentiation [10,12]. NFAT4 and NFATc1 both induce cartilage gene expression [10,14]. Interestingly, recent studies have suggested that Sox9 contains an NFAT binding element in its promoter region [15]. In contrast, NFAT1/NFATc2 is generally regarded as an inhibitor of chondrogenesis based primarily on neoplastic cartilage proliferation in and around articular cartilage of adult Nfat-/- mouse appendicular joints [16,17]. Given the diverse roles of NFAT signaling in chondrogenesis, understanding its regulation is critical to better understanding its role in establishing and maintaining healthy cartilage.
MicroRNAs (miRNA) are approximately 22 nucleotides in length and involved in a wide variety of cellular processes. As previous study, miR-124 suppressed NFATc1 expression by directly targeting its 3’-UTR in many different kinds of cells [18-20]. Other studies have highlighted the importance of miR-124 in regulating gene expression in human bone marrow-derived mesenchymal stem cells and its transcriptional regulation during differentiation [21]. MiR-124 contains CpG islands, which are sequences with a high frequency of cytosine and guanine [22]. The CpG islands are frequently targets for DNA hypermethylation, which can suppress transcription. In pellets of growing cells, hypoxia has been shown to cause alterations in DNA hypo- and hyper-methylation at the CpG islands that regulate gene expression and control fundamental cellular processes such as proliferation and differentiation [5,23]. Similarly, in tumor cells, DNA methylation regulates expression of miRNAs by methylating their promoter CpG islands [24]. COL10A1, the gene encoding collagen Xa1, was also found to be methylated in cartilage chondrocytes [25]. Thus, we hypothesize that miR-124 may be regulated by DNA methylation during hypoxic conditions. Furthermore, methylation changes regulating the expression of miR-124 may affect the activity of NFATc1 and its interaction with Sox9, which drives aggrecan and collagen II expression during hypoxia.
Materials and methods
Cell culture methods
The murine mesenchymal cell line C3H10T1/2 was purchased from ATCC (Manassas, VA, USA) and maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA) at 37°C in a humidified 5% CO2 atmosphere. The medium was replaced every three days. Cells were passaged using 0.05% trypsin (Invitrogen, Carlsbad, CA) when they reached 80% confluence. All of the experiments were conducted using cells that had been passaged less than 15 times. Chondrogenesis was induced as previously described [26], briefly cells were initially grown as a monolayer, then detached by trypsin and collected into 15 mL conical polypropylene tube in a concentration of 5×105 cells per tube. Cells were centrifuged at 1,200 rpm for 5 minutes to form into a pellet of 1 mm diameter approximately. The pellets were then cultured in 2 mL of chondrogenic medium (high glucose DMEM containing 2% FBS, 100 nM dexamethasone [Sigma, Dorset, UK], 50 mM L-ascorbic acid-2 phosphate [Sigma, Dorset, UK], BDTM ITS+Premix 1:100 [6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 ng/mL selenious acid, 1.25 mg/mL bovine serum albumin, and 5.35 mg/mL linoleic acid; BD Biosciences Discovery Labware, Bedford, MA, USA] and 10 ng/mL transforming growth factor-β3 [TGF-β3; PeproTech EC, London, UK].
Hypoxic culture condition
To perform the hypoxic culture, the cell pellets were incubated in a hypoxic chamber (Binder BD 53, BINDER GmbH, Germany). The reduced level of oxygen was ensured by flushing the chamber with 98% N2 and 2% O2 from an outside gas-tank through a cylinder containing water. The oxygen level within the chamber was maintained at 2% O2. The gas exchange was calibrated to a flow rate that would prevent evaporation of the medium, which was replaced every three days. To reduce transient “normoxic” episodes, culture medium was pre-incubated in hypoxic chamber for one hour before replacement. Medium replacement procedure was handled within 10 min.
Bisulfite treatment of DNA samples
Genomic DNA was isolated by standard phenol chloroform extraction and ethanol precipitation using a commercially available DNA extraction Kit (Magen, China). The DNA was modified with sodium bisulfite using the EpiTectTM Bisulfite Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The bisulfite modified DNA was resuspended in 25 uL of distilled water.
Methylation-specific PCR and sequencing
The PCR reaction was carried out in a final volume of 25 μL. The reaction contained 100 ng of bisulfite-treated template DNA, 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 2 mM MgCl2, 0.2 mM of each dNTP, 0.5 μM of each primer, and 0.5 units of Ampli-Taq (TakaRa Japan). To amplify the CpG islands in the promoter region of the miR-124-3 gene, the sequence was amplified from position -131 to +29 relative to the transcriptional starting point using the primers: (F) 5’-GGATTAGGGTAGTATTGTTAGTTTAG-3’ and (R) 5’-TACCTTAATTATATAAACATTAAATCAAAA-3’. The bisulfite-treated PCR products were purified with a commercially available extraction kit (Magen, China) and cloned into the pMD-18T vector (TaKaRa, Japan). Ten clones from each sample were sequenced. The number of total CpG sites in each sample was used as the denominator to determine the percentage change in methylation.
Total RNA extraction and reverse transcription quantitative PCR analysis
Total RNA was extracted from the C3H10T1/2 cell pellets using TRIzol (Invitrogen, CA, USA), according to the manufacturer’s instructions. cDNA was synthesized from 0.5 μg of total RNA with an oligo-dT primer using a commercially available kit according to the manufacturer’s protocol (TaKaLa, Japan). The oligonucleotide primers used to amplify the target mRNA are listed in Table 1. The mRNA expression level of each target gene was normalized to ribosomal 18 s RNA expression from the same sample. The PCR reaction conditions were: an initial denaturation step at 95°C for 10 min; and 40 cycles of 95°C for 10 s and 60°C for 30 s. Each experiment was performed in a real-time PCR system (CFX ConnectTM Real-time system, BIO-RAD, USA). The expression levels of mature miR-124 were detected using a microRNA RT kit (TaKaLa, Japan). For the RT-reaction, 0.5 μg of total RNA was used and no-template reaction was performed for miR-124, according to the manufacturer’s protocol (TaKaLa, Japan). MiR-124 expression was normalized to U6 expression. The relative expression levels of all the genes were calculated using the 2-ΔΔCt method.
Table 1.
Nucleotide sequences of the RT-PCR primers
| Gene | Size of PCR product (bp) | Primer sequence |
|---|---|---|
| 18 s | 112 | 5’-CCTGGATACCGCAGCTAGGA-3’ (F) |
| 5’-GCGGCGCAATACGAATGCCCC-3’ (R) | ||
| NFATc1 | 93 | 5’-ATCCTTGCCTGCCCTTGACT-3’ (F) |
| 5’-TGAGCCCTGTGGTGAGACTT-3’ (R) | ||
| Sox9 | 108 | 5’-CGGCTCCAGCAAGAACAAG-3’ (F) |
| 5’-TTGTGCAGATGCGGGTACTG-3’ (R) | ||
| COL IIA1 | 174 | 5’-ATGTCCATGGGTGCGATGTC-3’ (F) |
| 5’-CATCCAGGGCTCCAATGATGTA-3’ (R) | ||
| AGGRECAN | 105 | 5’-GCTGCAGTGATCTCAGAAGAAG-3’ (F) |
| 5’-GATGGTGAGGGAAGACCCTA-3’ (R) | ||
| COL Xa1 | 190 | 5’-GCAGCATTACGACCCAAGAT-3’ (F) |
| 5’-CATGATTGCACTCCCTGAAG-3’ (R) |
NFATc1 inhibition
NFATc1 expression was suppressed in the pelleted cell culture using a double-stranded small interfering (si) RNA, targeted to the murine NFATc1 mRNA sequence (RIBOBIO, Guangzhou, China). A scrambled 19-mer siRNA with no homology to any known mouse sequence was used as the negative control. The siRNA for NFATc1 and the scrambled siRNA were transfected at a 50 nM concentration into 80% confluent C3H10T1/2 cells using a commercial transfection reagent (riboFECTTM CP Reagent, RIBOBIO, China) according to the manufacturer’s protocol. Transfected cells were incubated for 8 h and then centrifuged into pellets. After pellet forming, siRNA was transfected every 3 days before changing medium.
MiR-124 overexpression and inhibition
pENTR/CMV-EGFP-miR124 vectors (Geneup, Shenzhen, China) were used to overexpress miR-124 in HEK293 cells. A chemically synthesized miRNA mimic and inhibitor (RIBOBIO, Guangzhou, China) were used to mimic or inhibit miR-124. A miRNA mimic for miR-223 was used as the negative control for overexpression of the miR-124 mimic (mimic-con; RIBOBIO). Eighty (80) percent confluent C3H10T1/2 cells were cultured overnight on petri-dishes and then transfected with the miR-223 mimic (50 nM) or miR-124 mimic. The miR-124 inhibitor and inhibitor control (see below) were transfected every 3 days into C3H10T1/2 cells that were cultured as pellets. Transfections were performed using Lipofectamine 2000 (Invitrogen). The media was replaced 6 h after the transfection. Two days later the cells were harvested for Western blot to detect NFATc1 levels. The sequences of the miR-124 mimic were: 5’-UAAGGCACGCGGUGAAUGCC-3’ and 3’-AUUCCGUGCGCCACUUACGG-5’. The control sequences were: 5’-UUUGUACUACACAAAAGUACUG-3’ and 3’-AAACAUGAUGUGUUUUCAUGAC-5’.
Histochemistry
Cell pellets that had been maintained for 3-weeks were fixed in 4% phosphate buffered formalin for 10 min at room temperature. The fixed pellet was dehydrated by treatment with a series of graded alcohols, cleared by treatment with xylene and xylene substitute, and infiltrated with paraffin wax. Then, it sectioned into 5 μm sections and stained with toluidine blue (1% toluidine blue) to detect sulfated proteoglycan matrix deposition. The sections were washed three times with water and then dehydrated with 95-100% graded ethanol. The sections were permanently mounted with Permount TM Mounting Medium. Sulfated proteoglycan was visualized by staining with toluidine blue for 5 min at 60°C. Images were captured using a Leica DMI4000B microscope fitted with an Optixcam summit series 5 MP digital camera. The photographs were assembled in Adobe Photoshop 6.0.
Chromatin immunoprecipitation assay
A chromatin immunoprecipitation (ChIP) assay was performed to evaluate the physical interaction between NFATc1 and the endogenous Sox9 promoter region. Immunoprecipitation of NFATc1 with the Sox9 promoter was performed using the monoclonal mouse anti-NFATc1 antibody (ChIP grade; Abcam) and the Chromatin Immunoprecipitation Kit (EZ-ChIP™ Millipore). Quantitative RT-PCR was utilized to assess the fold enrichment of the immunoprecipitated protein-DNA complex for Sox9. Primers were designed to span the 5’ upstream region of the Sox9 gene including the NFATc1 binding motif. The amplicon produced was 170 bp in length. The primers were: (F) 5’-AAAGCGAAGCTTTGCAAGAA-3’; (R) 5’-AAGGTTGGCTAAGGGAGGAA-3’. Two additional non-specific primers were used as the negative control to assess the specificity of the immunoprecipitated protein-DNA complex for Sox9: (F) 5’-GCTATGAGGAATGGCTGCAT-3’ and (R) 5’-CTGAGCAGGTCACAACAGGC-3’.
Protein extraction and western blot analysis
Lysates were generated from C3H10T1/2 cell pellets using RIPA buffer (50 mM Tris-HCl, Ph 7.5; 150 mM NaCl; 1% NP-40; 0.25% sodium deoxycholate, and 1 mM EDTA) supplemented with a protease inhibitor cocktail (Sigma-Aldrich). The total protein concentration was determined using the Coomassie Bradford protein assay kit (Bio-Rad). After protein quantification, 1 μL of β-mercaptoethanol and bromphenol blue was added to each sample. Twenty [20] micrograms of each lysate was separated by electrophoresis on an 8% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) and then transferred onto a 0.45 μm PVDF membrane (ImmobilonTM, Millipore Corp., Bedford, MA). The membranes were blocked using a buffer containing TBS, 0.25% Tween-20, 0.1% serum from the species that the secondary antibody was raised in, and 5% degreased milk for 1 h at room temperature. The membranes were probed overnight with mouse monoclonal anti-NFATc1 (1:200, Santa Cruz Biotechnology) or rabbit polyclonal β-actin (1:2000, Abcam). They were then incubated with horseradish peroxidase-conjugated secondary antibodies (Jackson Immuno Research). The protein bands were visualized using the Super-Signal chemiluminescent detection module (Pierce) and exposed to X-ray film. Quantitation of protein expression was performed using ImageJ software and normalized by the same protein in control group.
3’-UTR luciferase reporter assay
The TargetScan algorithm (http://www.targetscan.org) was used to predict the miRNA binding sites in the 3’-UTR of NFATc1. The full length 3’-UTR (1789 bp) of NFATc1 was amplified from a mouse NFATc1 plasmid (Open Biosystems) using the primers (F) 5’-GAGAATTCATACGTAACGACCTC-3’ and (R) 5’-GGATCTAGAACTGCTTTATTGGATTCATCTC-3’. The 3’-UTR of NFATc1 was PCR amplified and inserted into the pFLuc-3’-UTR (Geneup, Shenzhen, China). To construct vectors that had mutated miRNA target sequences in the 3’-UTR the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used. HEK293 cells were seeded in 24-well plates and grown until they reached approximately 70-80% confluence. The cells were then transfected with 50 ng of the 3’-UTR reporter vectors or the mutated 3’-UTR reporter vectors, 250 ng of plasmid expressing miR-124 (pENTR/CMV-EGFP-miR-124), 5 ng of phRL-TK, and 2 µL of Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The pENTR/CMV-EGFP vector without the miRNA sequences was used as the negative control. Two days after transfection, the cells were harvested to measure luciferase activity. The wells were measured independently using a Lumat LB9508 luminometer (Berthold, Bad Wildbad, Germany).
Statistical analysis
Each experiment was repeated at least three times. Data are presented as the mean ± SD. Comparisons between groups were performed using the Mann-Whitney test unless otherwise indicated. Statistical significance was determined using a two-tailed distribution assumption. Statistically significant results are denoted using *P<0.05 and **P<0.01.
Results
MiR-124 was downregulated and NFATc1 was upregulated in C3H10T1/2 cell pellets undergoing chondrogenesis in hypoxic conditions
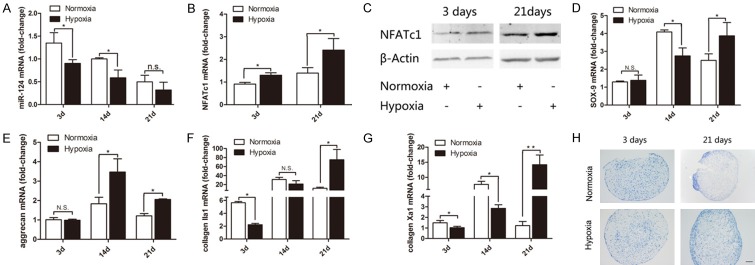
We first assessed the expression of miR-124, Sox9, and chondrogenesis related genes in the C3H10T1/2 cell pellet model under normoxic and hypoxic conditions to establish whether hypoxia affected chondrogenesis in this model. Cell pellets were collected either 3, 14, or 21 days after differentiation was induced and assayed for expression of miR-124, NFATc1, Sox9, collagen IIa1, aggrecan, and collagen Xa1 by RT-PCR. At day 3 and 14, the expression of miR-124 was significantly reduced in pellets under hypoxic conditions compared to normoxic pellets. Notably, while expression of miR-124 was still reduced in the hypoxic pellets compared to normoxic pellets at day 21, the difference was not significant; and miR-124 expression was lower overall on day 21 than on day 3 (Figure 1A). We then assessed the expression of NFATc1 at the gene and protein level. Expression of NFATc1 mRNA (Figure 1B) and protein (Figure 1C) was significantly upregulated on days 3 and 21 in hypoxic pellets compared to normoxic pellets.
Figure 1.
Expression of genes associated with chondrogenic differentiation under normoxic or hypoxic conditions. C3H10T1/2 cells were cultured as cell pellets under normoxic (21% O2) or hypoxic (2% O2) conditions for 3-21 days to induce chondrogenesis. The expression of chondrogenesis related marker genes was assessed by PCR and normalized to the expression of ribosomal 18sRNA. The fold change in gene expression was calculated using the 2-ΔΔCt method comparing with undifferentiated cells in normoxic conditions. The time points when gene expression was assessed are indicated on the X-axis. The mRNA expression levels of (A) microRNA-124 (miR-124); (B) NFATc1; (D) Sox9; (E) aggrecan; (F) collagen IIa1; and (G) collagen Xa1 are shown. The bars represent means ± SD (n=3; *P<0.05; **P<0.01; N.S.: not significant). (C) NFATc1 expression at the protein level was assessed by Western blot. (H) Serial paraffin sections of pellet after 3 and 21 days were stained with toluidine blue; representative images are shown. Scale bar =100 μm.
The onset of chondrogenesis was determined by expression of Sox9 (Figure 1D), aggrecan (Figure 1E), and collagen IIa1 (Figure 1F). The expression of NFATc1 increased from day 3 to day 21 while Sox9 and aggrecan increased since day 14. The expression levels of Sox9, aggrecan and collagen IIa1 tended to be significantly higher in the hypoxic pellets than in the normoxic pellets at day 21. Expecially, the expression of Sox9, collagen IIa1 and collagen Xa1 at day 14 (Figure 1D, 1F and 1G) were significantly lower in the hypoxic pellets compared to the normoxic pellets. To confirm that differentiation reached the stage of hypertrophic chondrocytes, the expression of hypertrophy marker, collagen Xa1, was assessed. The collagen Xa1 level was increased from day 3 to day 21 in hypoxic pellets and reached maximum transiently on day 14 in normoxic pellets. To study onset mediators in epigenetic level, we defined day 3 as the very early onset of chondrogenesis (Figure 1G) and day 21 as hypertrophic chondrocytes in this model. Deposition of ECM in the cell pellets was assessed by toluidine blue staining at 3 and 21 days respectively after differentiation. While the pellets appeared similar at day 3, intense staining was observed in the hypoxic, but not normoxic pellets on day 21. The positive material was fully filled of the periphery of hypoxic pellets (Figure 1H). Together, our results strongly suggested that hypoxia contributed to maintaining a chondrogenic phenotype in C3H10T1/2 cells.
Hypoxia promoted DNA methylation of miR-124-3
Given the significant reduction in miR-124 expression in hypoxic pellets and the role of hypoxia in epigenetic regulation of gene expression through DNA methylation, we next investigated whether DNA methylation regulated the expression of miR-124 during the onset of chondrogenesis under normoxic or hypoxic conditions. The day 3 pellets were harvested for analysis. We evaluated the methylation status of miR-124-3 because its CpG sites are the most concentrated [20]. In the promoter region of miR-124-3 is a 140 bp DNA fragment that contains 23 CpG sites. The sequence was showed in Figure 2A. The DNA was collected from 3 paired cell pellet cultures and cloned for amplification. The methylation status of 10 clones was determined. The black and white circles in Figure 2B represent the methylated and unmethylated CpG islands, respectively. Compar-ed to the levels of DNA methylation in normoxic cell pellets, the promoter region of miR-124-3 in cell pellets from hypoxic conditions was hypermethylated (Figure 2C). To confirm that DNA methylation was being measured, we treated cells with the DNA-demethylating agent 5’-AZA-2’-deoxycytidine (5’-AZA) under normoxic and hypoxic conditions. Cell pellets were treated with 1 μM 5’-AZA and incubated under normoxic or hypoxic conditions. When 5’-AZA was present, the percentage of methylated DNA sites decreased significantly (Figure 2C). These results suggested that hypermethylation of the CpG rich region of miR-124-3 may be associated with hypoxia.
Figure 2.
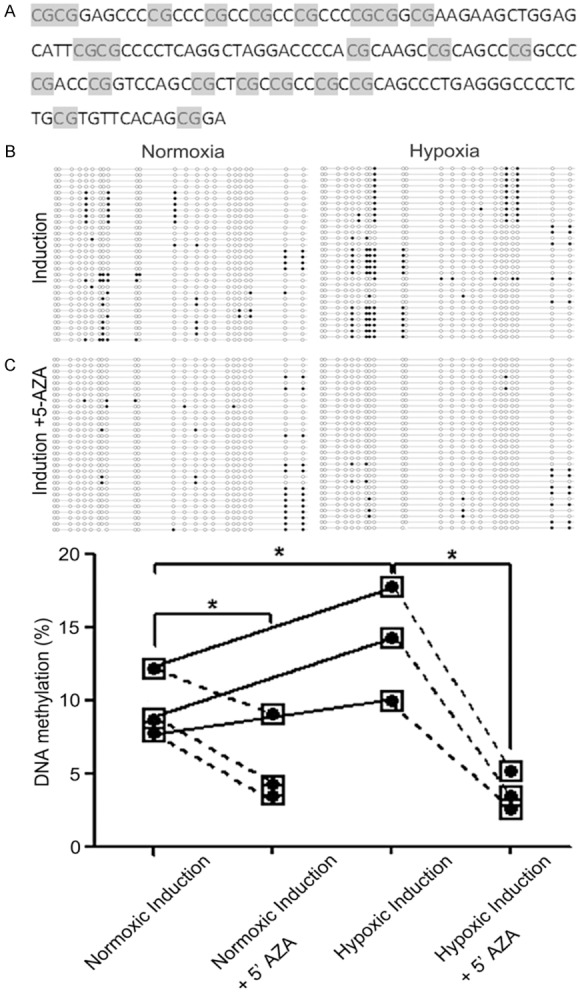
CpG Island methylation in the miR-124-3 promoter region under normoxic and hypoxic conditions. The percentage of CpG islands that were methylated was determined by PCR of bisulfite-treated genomic DNA extracted from C3H10T1/2 cells grown in pellets under normoxic (21% O2) or hypoxic (2% O2) conditions. A: The 140 bp DNA fragment in the promoter region of miR-124-3 that contains 23 CpG sites is shown. B: BGS analysis was conducted using 3 paired normoxic and hypoxic pellets. The promoter region was cloned into a pMD-18T vector for amplification and 10 clones from each pellet were sequenced of the miR-124-3 CpG island in 3 pairs of pellet samples. Each horizontal line indicates a single clone and each vertical column is an individual CpG site. Methylated sites are shown by a black circle and the demethylated sites are shown by a white circle. The number of methylated sites is shown in the absence and presence of the DNA demethylating agent 5’-AZA. C: Graphical representation of the percentage of methylated sites in paired samples under normoxic and hypoxic conditions in the presence and absence of 5’-AZA. Methylated CpG sites are shown by black circles. Each point indicates the mean. The differences in DNA methylation levels in different groups were analysis by Kruskal-Wallis test. *P<0.05.
Inhibiting DNA methylation with 5’-AZA delayed initiation of chondrogenesis
Having established that the promoter region of miR-124-3 was hypermethylated under hypoxic conditions, the effect of DNA methylation on miR-124 expression and chondrogenesis was evaluated. Cell pellets were cultured in the presence and absence of 5’-AZA (1 μM) for 21 days. Total RNA was extracted from the pellets and the relative expression of miR-124, Sox9, collagen IIa1, NFATc1, aggrecan, and collagen Xa1 was assessed. The expression of miR-124 was increased significantly in presence of 5’-AZA during the onset of chondrogenesis (Figure 3A). In contrast, expressions of NFATc1 (Figure 3B), Sox9 (Figure 3C), aggrean (Figure 3D), collagen IIa1 (Figure 3E), and collagen Xa1 (Figure 3F) were all significantly downregulated at day 3 in the presence of 5’-AZA. However, only collagen Xa1 was reduced at 21 days. These results suggested that a reduction in DNA methylation levels significantly increased miR-124 expression; and that inhibiting DNA methylation could delay the initiation of chondrogenesis and hypertrophy (Supplementary Figure 1).
Figure 3.
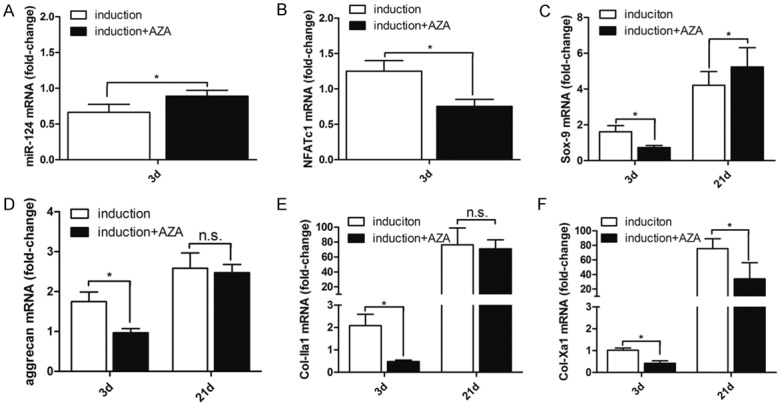
DNA demethylation with 5’-AZA increased miR-124 gene expression at the onset of chondrogenesis under hypoxic conditions. C3H10T1/2 cells were cultured as cell pellets under hypoxic (2% O2) conditions in the presence or absence of 5’-AZA for 3-21 days. The expression of chondrogenesis related marker genes was assessed by PCR and normalized to the expression of ribosomal 18sRNA. The fold change in gene expression was calculated using the 2-ΔΔCt method. The time points when gene expression was assessed are indicated on the X-axis. The mRNA expression levels of (A) miR-124; (B) NFATc1; (C) Sox9; (D) aggrecan; (E) collagen IIa1; and (F) collagen Xa1 are shown. The bars represent means ± SD (n=3). *P<0.05 compared to the condition without 5’-AZA.
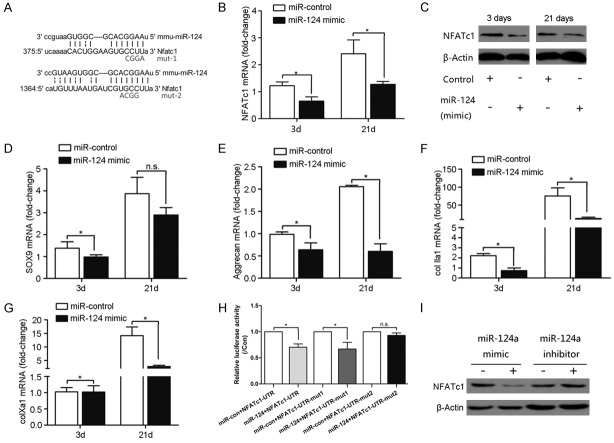
NFATc1 was verified as target of miR-124
To verify our hypothesis that NFATc1 was a target of miR-124, the TargetScan program was used to identify sites in the 3’-UTR of NFATc1 that were complementary to miR-124 (Figure 4A); indicating that NFATc1 mRNA contained sites that were potentially susceptible to miR-124 regulation. To understand the effects of miR-124 downregulation during hypoxia we transfected a miR-124 mimic or negative control miRNA mimic into C3H10T1/2 cells and cultured cells under hypoxic conditions. When the miR-124 mimic was transfected into the cells, the mRNA expression (Figure 4B) and protein level (Figure 4C) of NFATc1 were significantly decreased compared to the negative control at day 3 and 21. We next assessed the expression of chondrogenesis markers in hypoxic cells transfected with the miR-124 mimic or negative control. Sox9 (Figure 4D), aggrecan (Figure 4E), and collagen IIa1 (Figure 4F) were all significantly downregulated compared to the negative control at day 3 and 21. Likewise, the miR-124 mimic also significantly decreased expression of collagen Xa1, a marker of chondrocyte hypertrophy, at both 3 and 21 days (Figure 4G). Taken together, these data indicated that miR-124 is likely an inhibitor of NFATc1 expression, which impairs the induction of chondrogenesis under hypoxic conditions.
Figure 4.
Verification of NFATc1 as a target of miR-124 regulation. (A) The 3’-UTR region of NFATc1 is shown with the miR-124 target sequence highlighted. Two mutations were made to the NFATc1 3’-UTR to disrupt the putative miR-124 target sequence for use as controls in a luciferase assay. C3H10T1/2 cells grown in pellets under hypoxic conditions were transfected with miR-124 mimics or negative control (N.C.) sequences. The expression of NFATc1 was assessed at the mRNA level by RT-PCR (B) and the protein level by Western blot (C). The expression of chondrogenesis marker genes was assessed by RT-PCR assays in cells transfected with either the miR-124 mimic or the N.C. sequence at the time points indicated. (D) Sox9; (E) aggrecan; (F) collagen IIa1; (G) collagen Xa1. (H) Dual luciferase reporter assays were performed in HEK293 cells to test whether there was an interaction between miR-124 and the wildtype NFATc1 3’-UTR or either mutant NFATc1 3’-UTR. (I) The miR-124 inhibitor increased expression of NFATc1 at the protein level. MiR-124 effects were analyzed in parallel as a positive control. (pGL3-Rmut1, pGL3-Rmut2). The bars represent means ± SD (n=3). *P<0.05.
To confirm that the miR-124 mimic could inhibit NFATc1 expression under hypoxic conditions, we constructed a luciferase reporter using the mouse NFATc1 3’-UTR and cotransfected the reporter with either the miR-124 mimic or the negative control. Two mutants of the NFATc1 promoter region were generated for use as controls in the luciferase activity to determine the specificity of miR-124 binding (Figure 4A). As shown in Figure 4H, the miR-124 mimic significantly reduced the luciferase reporter activity with the wildtype (left most bars) or mutant 1 (middle pair of bars) NFATc1 3’-UTR. However, the miR-124 mimic did not affect NFATc1 expression with the mutant 2 3’-UTR (right most bars). Consistent with these findings, Western blot analysis showed that the level of NFATc1 protein was reduced statistically in cells transfected with miR-124 mimic, but increased in cells transfected with the miR-124 inhibitor (Figure 4I). These findings provide evidence that miR-124 negatively regulates expression of NFATc1 through its 3’-UTR.
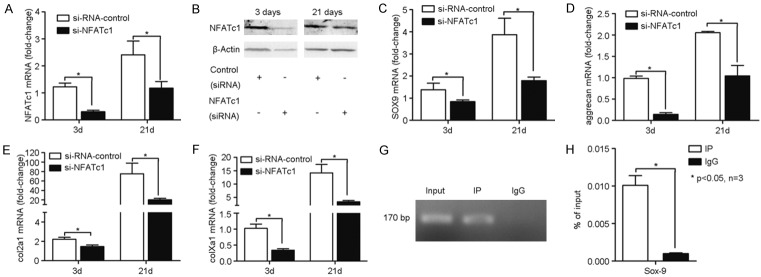
NFATc1 knockdown reduced chondrogenesis in hypoxic C3H10T1/2 cells through its interaction with the Sox9 promoter
Having established that expression of miR-124 negatively regulated the expression of NFATc1, we sought to determine the downstream effects of NFATc1 inhibition on the expression of Sox9, a master regulator of chondrogenesis. To test whether Sox9 transactivation was regulated by NFATc1 during the onset of chondrogenesis, siRNA was used to knockdown NFATc1 expression in C3H10T1/2 cells and then cultured cells under hypoxic conditions. The NFATc1 specific siRNA (si-NFATc1) caused statistically significant reduction of the mRNA level (Figure 5A) and relevant reduction of protein level (Figure 5B) of NFATc1 compared to the scrambled negative control siRNA (si-RNA-control) at both 3 and 21 days. We then determined the expression of Sox9, aggrecan, collagen IIa1, and collagen Xa1 in the presence (si-RNA-control) or absence (si-NFATc1) of NFATc1 expression. As expected, knockdown of NFATc1 expression significantly reduced the mRNA levels of Sox9 at day 3 and 21 compared to the si-RNA-control (Figure 5C). Similarly, aggrecan (Figure 5D), collagen IIa1 (Figure 5E), and collagen Xa1 (Figure 5F) were also significantly reduced in the absence of NFATc1. To ascertain whether the interaction between NFATc1 and Sox9 was direct or indirect, we used a ChIP assay to test whether a 170 bp fragment of the Sox9 promoter containing the NFATc1 biding site was enriched when NFATc1 protein was pulled down. Lysates from cell pellets were used for the ChIP assay. The 170 bp fragment of Sox9 was clearly amplified from the proteins pulled down using an NFATc1 specific antibody, but not when control IgG was used for the ChIP assay (Figure 5G, 5H). Thus, NFATc1 expression is clearly involved in chondrogenic differentiation and hypotrophy under hypoxic conditions (Supplementary Figure 1), and directly regulates the expression of Sox9 by binding to its promoter region.
Figure 5.
Effects of NFATc1 knockdown in C3H10T1/2 cells under hypoxic conditions. NFATc1 expression was suppressed in pelleted C3H10T1/2 cells grown in pellets under hypoxic conditions using a double-stranded small interfering (si) RNA targeted to the murine NFATc1 mRNA sequence (si-NFATc1). A scrambled 19-mer siRNA with no homology to any known mouse sequence was used as the negative control (si-RNA-control). Expression of NFATc1 was determined at the mRNA level by RT-PCR (A) and protein level by western blot (B) at the time points indicated to determine the efficacy of the siRNA knockdown. The mRNA expression levels of (C) Sox9; (D) aggrecan; (E) collagen IIa1; and (F) collagen Xa1 were determined by RT-PCR in cells transfected with the si-NFATc1 or si-RNA-control sequences by RT-PCR at the times indicated. To confirm that NFATc1 bound to the promoter region of Sox9, a ChIP assay was performed pulling down the protein/nucleic acid complex using an anti-NFATc1 antibody or control rabbit IgG. (G) Representative image showing that DNA immune-precipitated with the anti-NFATc1 antibody contained a Sox9 fragment. The input lane was a 10-fold dilution of the cell lysate. (H) The difference in the amount of Sox9 that was pulled down with the NFATc1 or control IgG antibodies were quantified. The bars represent means ± SD (n=3). *0.05, compared with the IgG group.
Discussion
While several studies have suggested that a hypoxic environment can promote and maintain chondrogenesis, the molecular mechanism has remained unclear [27-29]. Here, we utilized a high cell density cell pellet culture system to study the role of hypoxia in chondrogenesis in murine C3H10T1/2 cells. We were particularly interested in understanding the role of hypoxia-induced epigenetic regulation during chondrogenesis. Under hypoxic conditions, the chondrogenesis marker genes collagen IIa1, aggrecan, and collagen Xa1 were all upregulated compared to normoxic cells, indicating that in this system hypoxia helped maintain the chondrogenic phenotypes. We also found that hypoxia downregulated expression of miR-124 which is a putative regulator of Sox9. Interestingly, as the CpG methylation of the miR-124-3 promoter increased, the expression of the chondrogenesis markers increased. The inverse was observed when the DNA demethylating agent 5’-AZA was used to artificially reduce the level of CpG methylation. We provided evidence that miR-124 inhibited NFATc1 expression, thereby inhibiting expression of Sox9. We also documented a direct interaction between NFATc1 and the promoter region of Sox9 that increased expression of Sox9.
A common theme in the analysis of Sox9 in many differentiation pathways is that it functions at an early stage [30,31], possibly to control the proliferation and differentiation of progenitor cells. During chondrogenesis, Sox9 is required for mesenchymal cell condensation and the expression of cartilage-specific ECM genes, including collagen IIa1 and aggrecan. Consistent with previous studies [30,31], this is precisely the role suggested for Sox9 during chondrogenesis in early stage. Epigenetic regulation of Sox9 has broad applications beyond chondrogenesis, especially indirect regulation via miRNAs. The epigenome can be modulated by a variety of environmental factors including oxygen levels, nutrition, and the early environment [31-33]. Epigenetic downregulation of miR-124 as a key modification at the onset of chondrogenesis would be consistent with a corresponding increase in other transcripts, including Sox9, as part of a complex regulatory network.
We also identified a proximal regulator of Sox9 under hypoxic conditions as NFATc1, which directly binds to the promoter of Sox9. The NFAT family of transcription factors is critically involved in vertebrate development and in the growth and differentiation of multiple cell types. Knockdown of NFATc1 significantly downregulated the mRNA levels of Sox9 and its downstream target genes. Consistent with our findings, in vitro studies have shown that constitutively active NFAT induced chondrogenesis and induced the expression of Sox9 in chondroprogenitor cells [15]. However, we cannot exclude that miR-124 was directly regulating the expression of Sox9 in chondrogenesis. During spermiogenesis, miR-124 increased levels of several transcripts including Sox9, which was related to establishing a distinct, heritable chromatin structure in the promoter region of Sox9 [34].
Recent studies have linked NFAT regulation to miRNA expression, for example: miR-184 targets NFATc2 [35]; miR-133a targets NFATc4 [36]; and miR-199b targets the unclear kinase Dyrkla [37], which limits calcineurin/NFAT signal transduction by phosphorylating NFAT. Here, we have shown that miR-124 likely regulates expression of NFATc1 by binding to the promoter region leading to reduced mRNA and protein expression levels; suggesting a mechanism for NFATc1 regulation at the post-transcriptional level.
Given that hypoxia is known to induce epigenetic modifications [5,23] and that expression of miR-124-1, 2, 3 are regulated by methylation changes in their promoter sequences [23], we hypothesized that hypoxia was leading to downregulation of miR-124 expression and thereby increasing chondrogenesis. Consistent with this hypothesis, miR-124-3 was hypermethylated in hypoxic cells, and forced hypomethylation of miR-124-3 coincided with significantly decreased expression of chondrogenesis genes in hypoxic cells. Therefore, our results suggested that environmental changes such as hypoxia do induce epigenetic changes that effect chondrogenesis. While DNA methylation is involved in regulating miR-124 expression, we cannot exclude the possibility that hypoxia inducible factor-1 alpha (HIF-1α) is also involved in reducing miR-124 expression. The balance between the levels of DNA methylation and HIF-1α expression are likely specific to different oxygen conditions or with the amount of time spent in hypoxic conditions. The extent to which DNA methylation contributes to suppressing miR-124 expression under different oxygen conditions warrants further investigation.
Based on our findings, we propose the following working model. Under hypoxic conditions, DNA methylation is induced in the miR-124-3 promoter, repressing its expression. Hypermethylation inhibits the transcription of miR-124. In the absence of an inhibitor, NFATc1 expression increases and the NFATc1 protein is able to bind to the promoter region of Sox9, initiating transcription. The increased expression of Sox9 triggers the downstream events in chondrogenesis leading to the eventual expression of the marker genes collagen IIa1, aggrecan, and collagen Xa1. Given the heritable nature of the epigenetic modification, this may be one mechanism that supports the long-term differentiation and maintenance of chondrogenic cells in the hypoxic microenvironment of the cartilage.
Acknowledgements
This study was supported by Key Program of National Natural Science Foundation of China (No. 31430030); National Basic Research Program of China (973 Program, No. 2012CB619100); The Province Natural Science Foundation of Guangdong (No. 2016A030313031), The Science and Technology Foundation of Shenzhen (JCYJ20150402145016008), and Jiangxi Province Science and Technology Support Project (No. 2010BSA14800). We specially thank Pro. Guangqian Zhou, Pro. Deming Gou and Dr. Yanxia Zhu for expert assistance.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Goldring MB. Chondrogenesis, chondrocyte differentiation, and articular cartilage metabolism in health and osteoarthritis. Ther Adv Musculoskelet Dis. 2012;4:269–285. doi: 10.1177/1759720X12448454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush PG, Pritchard M, Loqman MY, Damron TA, Hall AC. A key role for membrane transporter NKCC1 in mediating chondrocyte volume increase in the mammalian growth plate. J Bone Miner Res. 2010;25:1594–1603. doi: 10.1002/jbmr.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markway BD, Tan GK, Brooke G, Hudson JE, Cooper-White JJ, Doran MR. Enhanced chondrogenic differentiation of human bone marrow-derived mesenchymal stem cells in low oxygen environment micropellet cultures. Cell Transplant. 2010;19:29–42. doi: 10.3727/096368909X478560. [DOI] [PubMed] [Google Scholar]
- 4.Hirao M, Tamai N, Tsumaki N, Yoshikawa H, Myoui A. Oxygen tension regulates chondrocyte differentiation and function during endochondral ossification. J Biol Chem. 2006;281:31079–31092. doi: 10.1074/jbc.M602296200. [DOI] [PubMed] [Google Scholar]
- 5.Watson CJ, Collier P, Tea I, Neary R, Watson JA, Robinson C, Phelan D, Ledwidge MT, McDonald KM, McCann A, Sharaf O, Baugh JA. Hypoxia-induced epigenetic modifications are associated with cardiac tissue fibrosis and the development of a myofibroblast-like phenotype. Hum Mol Genet. 2014;23:2176–2188. doi: 10.1093/hmg/ddt614. [DOI] [PubMed] [Google Scholar]
- 6.Robins JC, Akeno N, Mukherjee A, Dalal RR, Aronow BJ, Koopman P, Clemens TL. Hypoxia induces chondrocyte-specific gene expression in mesenchymal cells in association with transcriptional activation of Sox9. Bone. 2005;37:313–322. doi: 10.1016/j.bone.2005.04.040. [DOI] [PubMed] [Google Scholar]
- 7.An HJ, Lee H, Paik SG. Silencing of BNIP3 results from promoter methylation by DNA methyltransferase 1 induced by the mitogenactivated protein kinase pathway. Mol Cells. 2011;31:579–583. doi: 10.1007/s10059-011-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adesida AB, Mulet-Sierra A, Jomha NM. Hypoxia mediated isolation and expansion enhances the chondrogenic capacity of bone marrow mesenchymal stromal cells. Stem Cell Res Ther. 2012;3:9. doi: 10.1186/scrt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tew SR, Li Y, Pothacharoen P, Tweats LM, Hawkins RE, Hardingham TE. Retroviral transduction with SOX9 enhances re-expression of the chondrocyte phenotype in passaged osteoarthritic human articular chondrocytes. Osteoarthritis Cartilage. 2005;13:80–89. doi: 10.1016/j.joca.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Tomita M, Reinhold MI, Molkentin JD, Naski MC. Calcineurin and NFAT4 induce chondrogenesis. J Biol Chem. 2002;277:42214–42218. doi: 10.1074/jbc.C200504200. [DOI] [PubMed] [Google Scholar]
- 11.Gwack Y, Feske S, Srikanth S, Hogan PG, Rao A. Signalling to transcription: store-operated Ca2+ entry and NFAT activation in lymphocytes. Cell Calcium. 2007;42:145–156. doi: 10.1016/j.ceca.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 13.Viola JP, Carvalho LD, Fonseca BP, Teixeira LK. NFAT transcription factors: from cell cycle to tumor development. Braz J Med Biol Res. 2005;38:335–344. doi: 10.1590/s0100-879x2005000300003. [DOI] [PubMed] [Google Scholar]
- 14.Zanotti S, Canalis E. Notch suppresses nuclear factor of activated T cells (NFAT) transactivation and Nfatc1 expression in chondrocytes. Endocrinology. 2013;154:762–772. doi: 10.1210/en.2012-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SS, Tzeng BH, Lee KR, Smith RJ, Campbell KP, Chen CC. Cav3.2 T-type calcium channel is required for the NFAT-dependent Sox9 expression in tracheal cartilage. Proc Natl Acad Sci U S A. 2014;111:E1990–E1998. doi: 10.1073/pnas.1323112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranger AM, Gerstenfeld LC, Wang J, Kon T, Bae H, Gravallese EM, Glimcher MJ, Glimcher LH. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp Med. 2000;191:9–22. doi: 10.1084/jem.191.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenblatt MB, Ritter SY, Wright J, Tsang K, Hu D, Glimcher LH, Aliprantis AO. NFATc1 and NFATc2 repress spontaneous osteoarthritis. Proc Natl Acad Sci U S A. 2013;110:19914–19919. doi: 10.1073/pnas.1320036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang K, Peng X, Zhang X, Wang Y, Zhang L, Gao L, Weng T, Zhang H, Ramchandran R, Raj JU, Gou D, Liu L. MicroRNA-124 suppresses the transactivation of nuclear factor of activated T cells by targeting multiple genes and inhibits the proliferation of pulmonary artery smooth muscle cells. J Biol Chem. 2013;288:25414–25427. doi: 10.1074/jbc.M113.460287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamachi Y, Ohnuma K, Uto K, Noguchi Y, Saegusa J, Kawano S. MicroRNA-124 inhibits the progression of adjuvant-induced arthritis in rats. Ann Rheum Dis. 2016;75:601–608. doi: 10.1136/annrheumdis-2014-206417. [DOI] [PubMed] [Google Scholar]
- 20.Kim JH, Kim N. Regulation of NFATc1 in osteoclast differentiation. J Bone Metab. 2014;21:233–241. doi: 10.11005/jbm.2014.21.4.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laine SK, Alm JJ, Virtanen SP, Aro HT, Laitala-Leinonen TK. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2012;113:2687–2695. doi: 10.1002/jcb.24144. [DOI] [PubMed] [Google Scholar]
- 22.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 23.Xiong L, Wang F, Huang X, Liu ZH, Zhao T, Wu LY, Wu K, Ding X, Liu S, Wu Y, Zhao Y, Zhu LL, Fan M. DNA demethylation regulates the expression of miR-210 in neural progenitor cells subjected to hypoxia. FEBS J. 2012;279:4318–4326. doi: 10.1111/febs.12021. [DOI] [PubMed] [Google Scholar]
- 24.Gebauer K, Peters I, Dubrowinskaja N, Hennenlotter J, Abbas M, Scherer R, Tezval H, Merseburger AS, Stenzl A, Kuczyk MA, Serth J. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer. 2013;108:131–138. doi: 10.1038/bjc.2012.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann P, Boeuf S, Dickhut A, Boehmer S, Olek S, Richter W. Correlation of COL10A1 induction during chondrogenesis of mesenchymal stem cells with demethylation of two CpG sites in the COL10A1 promoter. Arthritis Rheum. 2008;58:2743–2753. doi: 10.1002/art.23736. [DOI] [PubMed] [Google Scholar]
- 26.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 27.Kanichai M, Ferguson D, Prendergast PJ, Campbell VA. Hypoxia promotes chondrogenesis in rat mesenchymal stem cells: a role for AKT and hypoxia-inducible factor (HIF)-1alpha. J Cell Physiol. 2008;216:708–715. doi: 10.1002/jcp.21446. [DOI] [PubMed] [Google Scholar]
- 28.Maes C, Carmeliet G, Schipani E. Hypoxiadriven pathways in bone development, regeneration and disease. Nat Rev Rheumatol. 2012;8:358–366. doi: 10.1038/nrrheum.2012.36. [DOI] [PubMed] [Google Scholar]
- 29.Saito T, Fukai A, Mabuchi A, Ikeda T, Yano F, Ohba S, Nishida N, Akune T, Yoshimura N, Nakagawa T, Nakamura K, Tokunaga K, Chung UI, Kawaguchi H. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med. 2010;16:678–686. doi: 10.1038/nm.2146. [DOI] [PubMed] [Google Scholar]
- 30.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topol L, Chen W, Song H, Day TF, Yang Y. Sox9 inhibits Wnt signaling by promoting betacatenin phosphorylation in the nucleus. J Biol Chem. 2009;284:3323–3333. doi: 10.1074/jbc.M808048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 33.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 34.Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 35.Weitzel RP, Lesniewski ML, Haviernik P, Kadereit S, Leahy P, Greco NJ, Laughlin MJ. microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood. 2009;113:6648–6657. doi: 10.1182/blood-2008-09-181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Lin X, Yang X, Chang J. NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression. Am J Physiol Heart Circ Physiol. 2010;298:H1340–H1347. doi: 10.1152/ajpheart.00592.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, Hansen A, Coenen-de Roo CJ, Bierhuizen MF, van der Nagel R, van Kuik J, de Weger R, de Bruin A, Condorelli G, Arbones ML, Eschenhagen T, De Windt LJ. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol. 2010;12:1220–1227. doi: 10.1038/ncb2126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.