Abstract
The transcription factor, Grainyhead-like 2 (GRHL2), is involved in wound healing, epidermal integrity, and epithelial-to-mesenchymal transition (EMT) in various biological processes; however, the biological function of GRHL2 in non-small cell lung cancer (NSCLC) is unknown. In the current study, we investigated the effect of GRHL2 on cell growth and migration in NSCLC cell lines and clinical tissues. Immunohistochemical analysis of clinical NSCLC specimens revealed that patients with high GRHL2 expression were associated with poor prognosis compared to patients with low GRHL2 expression. GRHL2 overexpression promoted cell growth and colony formation, and simultaneously suppressed cell migration in NSCLC cells. Furthermore, GRHL2 decreased the transcriptional activity of RhoG by directly binding to the RhoG promoter region. These findings confirm that GRHL2 plays an important role in regulating cell proliferation and migration in NSCLC.
Keywords: Non-small lung cancers, GRHL2, RhoG, cell migration, carcinogenesis
Introduction
In 2015, a total of 4,292,000 new cancer cases and 2,814,000 cancer deaths occurred in China. Lung cancer was the most common incident cancer and the leading cause of cancer deaths [1]. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancers [2,3] and is relatively insensitive to chemotherapy compared to small cell carcinoma. Several factors, such as the environment and genetic events, are responsible for the development of NSCLC. Recently, a whole-genome RNAi screen study has shown that GRHL2 is significantly amplified in lung cancer [4].
GRHL2, a mammalian homolog of Drosophila GRHL, was first reported in 2002 by Wilanowski et al. [5] as a novel transcription factor. GRHL2 is not only involved in a battery of processes of life formation [6], such as mammalian embryonic development, epithelial wound healing, epidermal barrier mutation, and central nervous system development, but GRHL2 is also a direct transactivator of apical junctional complex components by associating with Claudin4 (Cldn4) and E-cadherin [7]. In addition, GRHL2 is closely related to diverse neoplasms, such as breast cancer, gastric cancer, hepatocellular carcinoma, human oral squamous carcinoma, and renal cell carcinoma [8-11]. The relationships between GRHL2 and lung cancer have rarely been reported.
RhoG, a member of the Rho family of small GTPases, regulates cell shape, attachment, and motility. RhoG is identified as a growth factor response gene [12] and is involved in cell migration, neurite outgrowth, and cell survival [13-15]. In addition, it has been shown that the RhoG/Cdc42 signaling pathway is necessary for cell spreading and migration, and RhoG may also regulate tubule length [16].
In the present study, we investigated the effect of GRHL2 on cell proliferation and migration in NSCLC. We showed that GRHL2 is critical for NSCLC development and cell migration in vitro. We also demonstrated that GRHL2 regulates the expression of RhoG by binding to the RhoG promoter region in NSCLC.
Materials and methods
Reagents
Mouse monoclonal β-actin antibody was purchased from Proteintech (66009-1-lg). Rabbit polyclonal GRHL2 antibody was purchased from Thermo Fisher (PA5-41639). Mouse monoclonal RhoG was purchased from Abcam (ab76508). Rabbit polyclonal FAK, Cdc42, E-cadherin, and beta-catenin antibodies were purchased from Proteintech (12636-1-AP, 10155-1-AP, 20874-1-AP, and 51067-2-AP, respectively). Crystal violet and other analytical grade chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture
NSCLC cell lines (A549, NCI-H1299, NCI-H292, and NCI-H23) were purchased from the American Type Culture Collection (ATCC) and the SPC-A1 cell line was obtained from the China Centre for Type Culture Collection (China). Cells were grown in RPMI-1640 medium (Gibco), supplemented with 10% fetal bovine serum (Gibco). Human bronchial epithelial (HBE) cells were purchased from Sciencell Company and grown in bronchial epithelial cell medium (3211; Sciencell). All cells were cultured under an atmosphere of 5% CO2 at 37°C.
Patient information and tissue specimens
A total of 66 paraffin-embedded, archived clinical NSCLC specimens, histopathologically-diagnosed at the First Affiliated Hospital of Nanchang University, were used in the current study. Prior patient consent and approval were obtained from the Institutional Research Ethics Committee for use of these clinical materials for research purposes. Clinical information of the samples is summarized in Table 1. Ten fresh human NSCLC and paired normal tissue samples were obtained intra-operatively during surgical procedures performed in the Department of General Surgery of the First Affiliated Hospital of Nanchang University. All samples were collected with informed consent from the patient.
Table 1.
Patient clinical characteristics of expression of GRHL2
| Characteristics | All cases (%) | P value |
|---|---|---|
| Age (year) | ||
| <60 | 40 (81.63%) | 0.303 |
| ≥60 | 26 (72.22%) | |
| Gender | ||
| Male | 51 (78.46%) | 0.745 |
| Female | 15 (75.00%) | |
| Histologic type | ||
| Squamous carcinoma | 30 (81.01%) | 0.505 |
| Adenocarcinoma | 36 (75.00%) | |
| TNM stages | ||
| I+II | 35 (68.62%) | 0.015* |
| III | 31 (91.18%) | |
| Differentiation | ||
| Moderately or highly differentiation | 39 (69.64%) | 0.039* |
| Poorly differentiation | 26 (89.66%) |
p<0.05;
**p<0.01.
DNA constructs
The promoter region of RhoG was isolated by PCR using the gDNA library from NCI-H1299 cells. The construct was cloned into pGL3-Enhancer vector in Hind III and Kpn I restriction sites and verified by sequencing.
Plasmids and siRNA
Expression vectors for GRHL2 were purchased from ViGene. The knocking down of GRHL2 was carried out using two distinct Stealth Select RNAi duplexes (Life Technologies Company). SiRNA transfection or siRNA/DNA co-transfection in cells using SuperFect in siRNA Transfection Reagent (2013-100; Pufei, Shanghai, China) and the relative knock down efficiency was determined by Western blot with GRHL2 antibody. The knocking down of GRHL2 was carried out using two distinct Stealth Select RNAi duplexes (Life Technologies Company). The target oligonucleotide sequences were as follows: siRNA-1, CAGAAGAAGAGUGACAUCACCUACU; and siRNA-2, AGUAGGUGAUGUCACUCUUCUUCUG. Stealth RNAi negative control duplex (Life Technologies Company) was used as a control.
RNA extraction and real-time quantitative PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) and 1 μg of total RNA was used to perform reverse transcription using Prime Script RT reagent kit with gDNA eraser (TaKaRa), according to the manufacturer’s instructions. Quantitative RT-PCR was performed with SYBR Green dye (Applied Biosystems). The relative amount of cDNA was calculated using the comparative Ct method with GAPDH as a control. PCR reactions were performed in triplicate.
Immunohistochemistry (IHC)
Immunohistochemistry and quantification of GRHL2 expression were performed by two observers based on the proportion of positively-stained tumor cells and the intensity of staining. Both sets of results were combined to give a mean score for further comparative evaluations. The IHC score was determined by combining the score for the percentage of positively-stained tumor cells with the grade of the staining intensity.
Western blotting analysis
Protein extracts were prepared using NP-40 lysis buffer containing phosphatase and protease inhibitors. The protein concentration of the cell lysates was determined using a BCA protein assay kit. The cell lysates were then subjected to SDS-PAGE, followed by immunoblot using the indicated antibodies.
Luciferase assay
Luciferase assays were measured with Dual Luciferase Assay kits (Promega) per the manufacturer’s protocol. Reporter activity was normalized to Renilla luciferase values. Each graph represents three biological replicates.
Colony formation
Five hundred cells were seeded in 6-well dishes in RPMI-1640 with 10% FBS. After 10 days of culture, cells were fixed with 4% formaldehyde and stained with 2% crystal violet. The images were obtained using a digital camera (Fujifilm).
Cell migration assay
For the scratch wound healing assay, cells were plated in 6-well plates to create a confluent monolayer of 90%-100% confluence, then the monolayer was scraped in a straight line to create a “scratch” using a 200-μl pipette tip. After removing debris and adding fresh medium containing 1% FBS, cells were photographed using phase contrast microscopy (IX71; Olympus) at the indicated times. The migration distance was assessed using ImageJ software (National Institutes of Health, http://rsb.info.nih.gov/ij/download.html). A relative migration rate was calculated based on the cell relative migration area for each treatment.
The Transwell migration assay was performed using 8-μm Transwell chambers (Corning Costar). A total of 105 cells in 0.2 ml of media supplemented with 1% FBS were plated in the upper chambers. The lower chamber of the Transwell device was filled with 500 μl of RPMI-1640 supplemented with 10% FBS. After incubation at 37°C for 10 h, cells remaining on the upper surface of the membrane were removed. The cells on the lower surface of the membrane were fixed, stained with crystal violet, then counted under an Olympus light phase contrast microscope (IX71). These procedures have been published previously [27].
Statistical analysis
All statistical analyses were carried out using the SPSS13.0 statistical software package. Data are presented as the means ± SD. Comparisons between groups for statistical significance were carried out with a two-tailed paired Student’s t-test. P-values ≤ 0.05 were considered statistically significant.
Results
GRHL2 is up-regulated and closely related to the poor prognosis in NSCLC
To investigate the role of GRHL2 in the progression of NSCLC, we examined the pattern of GRHL2 expression in NSCLC cell lines. Real-time PCR and Western blotting analyses showed that the levels of GRHL2 mRNA and protein expression were remarkable up-regulated in all six cell lines (Figure 1A, 1B). Furthermore, GRHL2 expression was significantly up-regulated in 66NSCLC tissues; the representative paraffin-embedded sections were stained by IHC (Figure 1C). Table 1 shows that the expression of GRHL2 correlated significantly with clinical stage (P=0.015) and differentiation (P=0.039) in NSCLC. Notably, Kaplan-Meier plotter analysis revealed that patients with high GRHL2 expression had poorer overall survival than patients with low GRHL2 expression (P<0.001, Figure 1D), indicating that GRHL2 may have potential as an independent prognostic marker in NSCLC.
Figure 1.
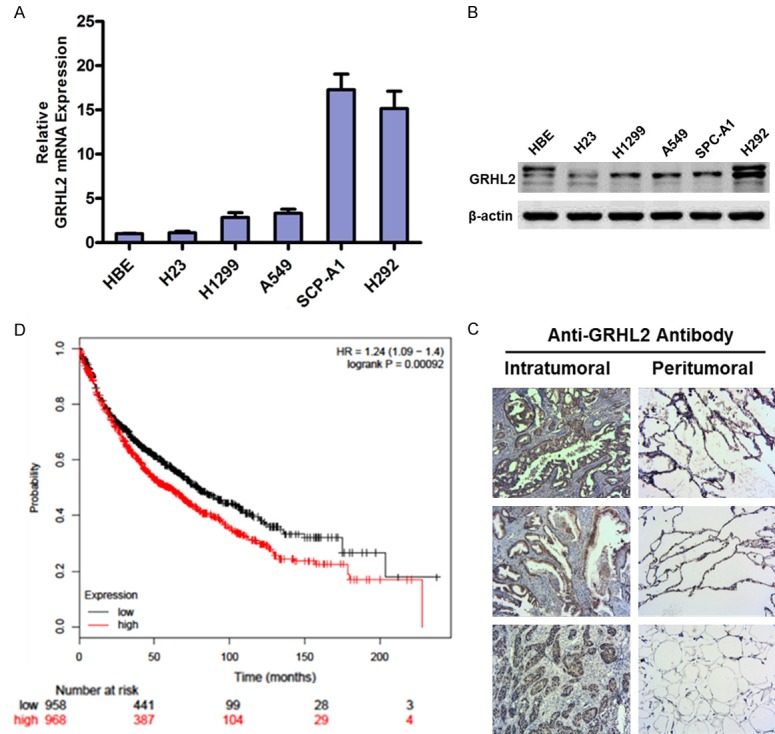
GRHL2 is up-regulated and closely related to the poor prognosis in NSCLC. (A and B) Western blotting (B) and real-time PCR (A) analysis of GRHL2 expression in six NSCLC cell lines; β-actin was used as a loading control. (C) Expression of GRHL2 protein in each of the human primary NSCLC tissues (left panel) and adjacent non-cancerous tissues (right panel) paired from different patients, as determined by IHC. (D) Kaplan-Meier overall survival curves for patients with NSCLC showed that NSCLC patients with GRHL2high tumors had significantly shorter survival than patients with GRHL2low tumors (n=1926, P<0.001 [log-rank]).
GRHL2 promotes NSCLC cell growth and colony formation
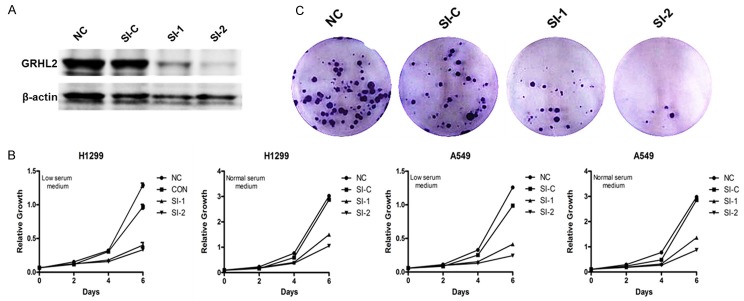
To investigate physiologic function during pathogenesis of NSCLC, we used stealth select RNAi duplexes. These siRNAs were transfected into the indicated cell lines. After 48 h, the level of GRHL2 protein was detected by Western blot. As shown in Figure 2A, both siRNA-1 and siRNA-2 efficiently knocked down GRHL2 in select cell lines compared with control siRNA. The role of GRHL2 in NSCLC cell proliferation was detected by low serum and saturation density assays. As shown in Figure 2B, knocking down GRHL2 significantly blocked the growth of A549 and H1299 cells. Similarly, knocking down GRHL2 greatly decreased colony formation in A549 and H1299 (Figure 2C). Taken together, these results indicate that GRHL2 is essential for NSCLC cell growth and colony formation.
Figure 2.
GRHL2 promotes NSCLC cell growth and colony formation. H1299 cells were transfected with indicated siRNAs or not transfected. A. Western blot of GRHL2 to check the knockdown efficiency of siRNAs in the indicated cell lines; β-actin was used as a loading control. B. Growth in low serum. H1299 cells werecultured in 1640 supplemented with 1% serum (low serum) or 10% serum (normal serum). After the indicated time, cells were trypsinized and counted. Data represent the average of three independent experiments. C. Colony formation assay. After the indicated times, H1299 were fixed and stained with crystal violet. Representative Wells were photographed and shown.
GRHL2 suppress cell migration and invasion in NSCLC cells
To further investigate the biological function of GRHL2, we determined NSCLC cell migration and invasiveness when GRHL2 was knocked down. Similarly, the H1299 cells were transfected with GRHL2 siRNA for scratch wound healing and Transwell migration assays. Indeed, knocking down GRHL2 strongly enhanced cell migration and invasion of H1299 cells (Figure 3A, 3B). Taken together, these results suggest that GRHL2 plays a key and dual role in NSCLC cells.
Figure 3.
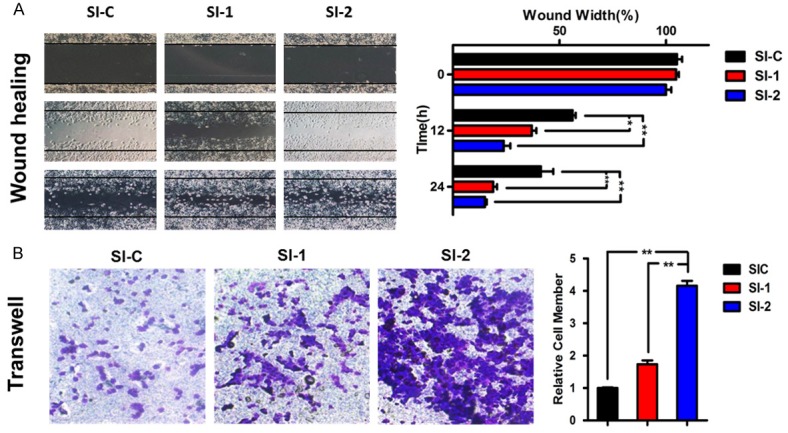
GRHL2 suppress cell migration and invasion in NSCLC cells. H1299 cells were transfected with indicated siRNA or not transfected. A. Scratch wound healing assay. Cell migration rate was quantified by measuring the difference in area between the leading edge at the initiation of the experiment. The wound area was assessed by the ImageJ software. Data represent the average of three independent experiments (mean ± SD). *p<0.05, **p<0.01 vs. control siRNA. B. Transwell migration assay. Cells were trypsinized, counted, and seeded in Transwell chambers 48 h after transfection. After incubation for 10 h, cells were fixed, stained, photographed, and counted in three random views. Data represent the average of three independent experiments (mean ± SD). *p<0.05, **p<0.01 vs. control siRNA.
Inverse correlation between GRHL2 and RhoG expression in NSCLC patients
We checked the cBioPortal database (TCGA, Nature 2012) and found that an inverse correlation existed between the expression of GRHL2 and RhoG in 230 NSCLC patient samples, which was confirmed using Pearson’s (-0.68) and Spearman’s correlations (-0.75; Figure 4A). To further prove the important signal of GRHL2 in NSCLC invasion, we detected a series of relative key regulators involved in cell migration and invasion, such as RhoG, E-cadherin, β-catenin, FAK, and Cdc42. Overexpression and knockdown of GRHL2 significantly decreased and increased the level of E-cadherin protein expression, respectively, but had no effects on the expression of β-catenin and FAK (Figure 4B). We also confirmed the inverse correlation between GRHL2 and RhoG expression in NSCLC.
Figure 4.

Inverse correlation between GRHL2 and RhoG expression in NSCLC patients. A. The association between GRHL2 and RhoGamong 230 NSCLC samples from the cBioPortal database (http://www.cbioportal.org). B. The migration-related protein level was checked by Western blot in H1299, which were or were not transfected with the indicated plasmids or siRNAs; β-actin was used as a loading control.
GRHL2 down-regulates the expression of RhoG via binding to its promoter
GRHL2, as a novel transcription factor, usually binds to the promoter of targeted genes [17-19]. Analysis of the promoter region of RhoG identified three putative GRHL2 binding sequences (Figure 5A). The activity of RhoG promoter was decreased when RGHL2 was overexpressed, and dramatically increased when RGHL2 was knocked down (Figure 4B), as shown by luciferase activity assay in H1299 cells. These data highlight a new mechanism by which GRHL2 can regulate RhoG activity by binding to the promotor of RhoG. Moreover, the level of RhoG protein was up-regulated in GRHL2 overexpression NSCLC cells and down-regulated in GRHL2- silenced NSCLC cells. These results suggest that the biological effects during the progression of NSCLC are may exerted via binding to RhoGpromoter.
Figure 5.

GRHL2 down-regulates the expression of RhoG via binding its promoter. A. Schematic illustration of cloned fragment of the human RhoG promoter. The promoter region was predicted as three fragments (promoters 1-3). B. Transcriptional activity of GRHL2 on three RhoG promoter fragments, as indicated in H1299 cells. The luciferase activities of the promoter constructs were measured after normalization to Renilla luciferase activity. Data represent the average of three independent experiments (mean ± SD). *p<0.05, **p<0.01 vs. control siRNA.
Discussion
Lung cancer is the primary cause of cancer mortality worldwide in both males and females [1]. There is thus an urgent need to elucidate the molecular mechanism underlying lung cancer progression. The majority of evidence suggests that GRHL2 plays a crucial role in development of various carcinomas [8-11,20]. Despite the aforementioned strong clinical relevance, the detailed mechanisms by which GRHL2 regulates oncogenesis of NSCLC are not clear. In the present study, we demonstrated that overexpression of GRHL2 is closely associated with poor prognosis in patients. Silencing of GRHL2 inhibited the growth and colony formation of NSCLC cells in vitro; however, our further study found that GRHL2 suppressed cell migration and invasion in H1299 cells, which indicated a novel GRHL2 function in lung cancer that differs from other types of cancer.
A series of studies have reported that GRHL2 has the potential to serve as a biomarker for predicting the survival rate in patients with cancer [21-23]. The patients with GRHL2low tumors had significantly superior overall survival than patients with GRHL2high tumors using KM-plotters based on TCGA data. Furthermore, as shown by the IHC assay results in the present study, NSCLC patients with high expression of GRHL2 had advanced clinical stages compared with NSCLC patients with low expression of GRHL2. These data further indicate that GRHL2 plays a vital role in NSCLC progression, and GRHL2 may represent a novel prognostic biomarker for survival.
GRHL2 has been reported to be amplified in various types of tumors, such as colorectal cancer, oral cancer cells, and hepatocellular carcinoma [9,24,25]; however, in some studies the level of GRHL2 protein was significantly down-regulated in gastric cancer, cervical cancer, claudin-low subclass breast tumors, and basal-B subclass breast cancer cell lines [17,26,27]. Using a method of the GISTIC algorithm to analyze gene copy number changes in plentiful tumor samples, the copy number for GRHL2 is consistent with significant genomic amplification [4]. Combined with the results of our study in which GRHL2 was shown to be up-regulated in NSCLC tissues and cells, we infer that GRHL2 plays an important and complex role in NSCLC and may be a novel prognostic marker for NSCLC. GRHL2 directly regulates the expression of E-cadherin, which is a key protein for cancer cell invasion and metastasis. N-cadherin, alpha-catenin, beta-catenin, fibronectin1, CITED2, and ERBB3 are thought to be the target genes for GRHL2 in breast cancer [28]. In addition, GRHL2 builds a reciprocal feedback loop with ZEB1 to control epithelial versus mesenchymal phenotypes and epithelial-mesenchymal transition (EMT)-driven tumor progression [29]. To explore further mechanisms underlying GRHL2 function in neoplasms, the inverse correlation between GRHL2 and RhoG was detected from the cBioPortal database in 230 NSCLC samples. Thus, further investigation is warranted to verify the correlation and reveal the mechanism underlying GRHL2 activity in NSCLC.
RhoG is a member of the Rho family of small GTPases, which cycle between inactive GDP-bound and active GTP-bound states, and act as molecular switches in signal transduction cascades. Rho proteins promote reorganization of the actin cytoskeleton and regulate cell shape, attachment, and motility. A study demonstrated that VEGF activates a RhoG/Cdc42 signaling module, which controls lateral filopodia formation that plays a crucial role in the initial stage of the migration of tumor cells [16]. In the current study, ectopic overexpression of GRHL2 dramatically increased, whereas silencing GRHL2 decreased the level of RhoG protein expression in NSCLC cells. In addition, the levels of Cdc42 and E-cadherin protein expression, downstream target genes of RhoG, were also regulated with overexpression and knockdown of GRHL2, respectively.
In summary, this study determined the level of GRHL2 expression in NSCLC cell lines and tissues, and indicates that GRHL2 may have potential as an independent prognostic marker in NSCLC. Additionally, these data suggest that GRHL2 plays a key role in cell metastasis through regulation of the RhoG/Cdc42 signaling pathway in NSCLC. Additional research is need to elucidate the detailed mechanism underlying GRHL2 in the pathogenesis of NSCLC and the dual role of GRHL2 in NSCLC that promotes cell proliferation while inhibiting migration. Perhaps GRHL2 merely acts similar to senescence-associated growth arrest genes that are able to inhibit proliferation, while promoting migration in the tissue injury repair program, such as p16INK4a and PAI-1 [30-33].
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81372823, 31360282).
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Riccardo F, Arigoni M, Buson G, Zago E, Iezzi M, Longo D, Carrara M, Fiore A, Nuzzo S, Bicciato S, Nanni P, Landuzzi L, Cavallo F, Calogero R, Quaglino E. Characterization of a genetic mouse model of lung cancer: a promise to identify Non-Small cell lung cancer therapeutic targets and biomarkers. BMC Genomics. 2014;15(Suppl 3):S1. doi: 10.1186/1471-2164-15-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fidias P, Novello S. Strategies for prolonged therapy in patients with advanced nonsmall-cell lung cancer. J. Clin. Oncol. 2010;28:5116–5123. doi: 10.1200/JCO.2010.30.7074. [DOI] [PubMed] [Google Scholar]
- 4.Dompe N, Rivers CS, Li L, Cordes S, Schwickart M, Punnoose EA, Amler L, Seshagiri S, Tang J, Modrusan Z, Davis DP. A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis. Proc Natl Acad Sci U S A. 2011;108:E943–951. doi: 10.1073/pnas.1100132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilanowski T, Tuckfield A, Cerruti L, O’Connell S, Saint R, Parekh V, Tao J, Cunningham JM, Jane SM. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev. 2002;114:37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 6.Pyrgaki C, Liu A, Niswander L. Grainyheadlike 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev Biol. 2011;353:38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werth M, Walentin K, Aue A, Schonheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M, Barasch J, Rosenbauer F, Luft FC, Schmidt-Ott KM. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137:3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- 8.Singh B, Shamsnia A, Raythatha MR, Milligan RD, Cady AM, Madan S, Lucci A. Highly adaptable triple-negative breast cancer cells as a functional model for testing anticancer agents. PLoS One. 2014;9:e109487. doi: 10.1371/journal.pone.0109487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, Ohta M, Asaoka Y, Seto M, Shiina S, Yoshida H, Kawabe T, Yokosuka O, Ogawa S, Omata M. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol. 2008;49:746–757. doi: 10.1016/j.jhep.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28:565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butz H, Szabo PM, Nofech-Mozes R, Rotondo F, Kovacs K, Mirham L, Girgis H, Boles D, Patocs A, Yousef GM. Integrative bioinformatics analysis reveals new prognostic biomarkers of clear cell renal cell carcinoma. Clin Chem. 2014;60:1314–1326. doi: 10.1373/clinchem.2014.225854. [DOI] [PubMed] [Google Scholar]
- 12.Vincent S, Jeanteur P, Fort P. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol Cell Biol. 1992;12:3138–3148. doi: 10.1128/mcb.12.7.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh H, Yasui H, Yamaguchi Y, Aoki J, Fujita H, Mori K, Negishi M. Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 cells. Mol Cell Biol. 2000;20:7378–7387. doi: 10.1128/mcb.20.19.7378-7387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murga C, Zohar M, Teramoto H, Gutkind JS. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-kappaB. Oncogene. 2002;21:207–216. doi: 10.1038/sj.onc.1205036. [DOI] [PubMed] [Google Scholar]
- 15.Katoh H, Hiramoto K, Negishi M. Activation of Rac1 by RhoG regulates cell migration. J Cell Sci. 2006;119:56–65. doi: 10.1242/jcs.02720. [DOI] [PubMed] [Google Scholar]
- 16.Abraham S, Scarcia M, Bagshaw RD, McMahon K, Grant G, Harvey T, Yeo M, Esteves FO, Thygesen HH, Jones PF, Speirs V, Hanby AM, Selby PJ, Lorger M, Dear TN, Pawson T, Marshall CJ, Mavria G. A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis. Nat Commun. 2015;6:7286. doi: 10.1038/ncomms8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cieply B, Riley Pt, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res. 2012;72:2440–2453. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Xiao Liu Z, Oh JE, Shin KH, Kim RH, Jiang M, Park NH, Kang MK. Grainyheadlike 2 (GRHL2) inhibits keratinocyte differentiation through epigenetic mechanism. Cell Death Dis. 2012;3:e450. doi: 10.1038/cddis.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walentin K, Hinze C, Werth M, Haase N, Varma S, Morell R, Aue A, Potschke E, Warburton D, Qiu A, Barasch J, Purfurst B, Dieterich C, Popova E, Bader M, Dechend R, Staff AC, Yurtdas ZY, Kilic E, Schmidt-Ott KM. A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development. 2015;142:1125–1136. doi: 10.1242/dev.113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng L, Wang P, Yang S, Yang Y, Zhang Q, Zhang W, Xiao H, Gao H, Zhang Q. Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med Genomics. 2012;5:14. doi: 10.1186/1755-8794-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danila DC, Samoila A, Patel C, Schreiber N, Herkal A, Anand A, Bastos D, Heller G, Fleisher M, Scher HI. Clinical validity of detecting circulating tumor cells by AdnaTest assay compared with direct detection of tumor mRNA in stabilized whole blood, as a biomarker predicting overall survival for metastatic castrationresistant prostate cancer patients. Cancer J. 2016;22:315–320. doi: 10.1097/PPO.0000000000000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan Y, Jin R, Huang A, Zhao H, Feng B, Zang L, Zheng M. Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol Ther. 2014;15:878–887. doi: 10.4161/cbt.28877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danila DC, Anand A, Schultz N, Heller G, Wan M, Sung CC, Dai C, Khanin R, Fleisher M, Lilja H, Scher HI. Analytic and clinical validation of a prostate cancer-enhanced messenger RNA detection assay in whole blood as a prognostic biomarker for survival. Eur Urol. 2014;65:1191–1197. doi: 10.1016/j.eururo.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quan Y, Xu M, Cui P, Ye M, Zhuang B, Min Z. Grainyhead-like 2 promotes tumor growth and is associated with poor prognosis in colorectal cancer. J Cancer. 2015;6:342–350. doi: 10.7150/jca.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W, Yi JK, Shimane T, Mehrazarin S, Lin YL, Shin KH, Kim RH, Park NH, Kang MK. Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis. 2016;37:500–510. doi: 10.1093/carcin/bgw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang J, Fu X, Ran W, Chen X, Hang Z, Mao H, Wang Z. Expression and role of grainyheadlike 2 in gastric cancer. Med Oncol. 2013;30:714. doi: 10.1007/s12032-013-0714-5. [DOI] [PubMed] [Google Scholar]
- 27.Torres-Reyes LA, Alvarado-Ruiz L, Pina-Sanchez P, Martinez-Silva MG, Ramos-Solano M, Olimon-Andalon V, Ortiz-Lazareno PC, Hernandez-Flores G, Bravo-Cuellar A, Aguilar-Lemarroy A, Jave-Suarez LF. Expression of transcription factor grainyhead-like 2 is diminished in cervical cancer. Int J Clin Exp Pathol. 2014;7:7409–7418. [PMC free article] [PubMed] [Google Scholar]
- 28.Mlacki M, Kikulska A, Krzywinska E, Pawlak M, Wilanowski T. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp Biol Med (Maywood) 2015;240:1396–1401. doi: 10.1177/1535370215588924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyheadlike-2. Cancer Res. 2013;73:6299–6309. doi: 10.1158/0008-5472.CAN-12-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simone TM, Higgins CE, Czekay RP, Law BK, Higgins SP, Archambeault J, Kutz SM, Higgins PJ. SERPINE1: a molecular switch in the Proliferation-Migration dichotomy in wound-”Activated” Keratinocytes. Adv Wound Care (New Rochelle) 2014;3:281–290. doi: 10.1089/wound.2013.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan JC, Duszczyszyn DA, Castellino FJ, Ploplis VA. Accelerated skin wound healing in plasminogen activator inhibitor-1-deficient mice. Am J Pathol. 2001;159:1681–1688. doi: 10.1016/S0002-9440(10)63015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ploplis VA, Balsara R, Sandoval-Cooper MJ, Yin ZJ, Batten J, Modi N, Gadoua D, Donahue D, Martin JA, Castellino FJ. Enhanced in vitro proliferation of aortic endothelial cells from plasminogen activator inhibitor-1-deficient mice. J Biol Chem. 2004;279:6143–6151. doi: 10.1074/jbc.M307297200. [DOI] [PubMed] [Google Scholar]
- 33.Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]