Abstract
Chronic myeloid leukemia (CML) is a common malignant disease from hematopoietic system. Aberrant expression of microRNAs (miRNAs) has been found in CML, however, the roles of many miRNAs including miR-15a-5p in CML are still unknown. In this study, the expression and roles of miR-15a-5p in CML were investigated. We found that restoration miR-15a-5p expression in CML cells decreased cell growth, metastasis and enhanced cell apoptosis. Chemokine ligand 10 (CXCL10) was predicted as a target gene of miR-15a-5p, which was verified by luciferase assay. CXCL10 mRNA and protein was down-regulated in the CML cells with miR-15a-5p overexpression by real time RT-PCR and western blotting. We also found that there were low levels of miR-15a-5p companied with high levels of CXCL10 in blood samples from CML patients. In a conclusion, miR-15a-5p suppresses cell survival and metastasis of CML by targeting CXCL10, which is a therapeutic option for CML patients.
Keywords: MiR-15a-5p, chronic myeloid leukemia, CXCL10, metastasis
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder from hematopoietic stem cell or a multipotent progenitor and a remarkable feature of CML is a reciprocal translocation of 9q34 with 22q11 [1-3]. Although there are great achievements on exploring the molecular mechanism and therapeutic methods for CML previously, the chemotherapy resistance and low survival rates are the main questions in the therapy of CML [1-3]. A better understanding for CML in molecular mechanisms may be benefit for finding new therapeutic targets.
MicroRNAs (miRNAs) are regulatory molecules involved in many diseases. MiRNAs are small non-coding RNAs and negatively regulate gene expressionby targeting mRNAs at the 3’-untranslated region (UTR) [3,4]. MiRNAs play key roles in numerous biological processes, including cell growth, cell cycle progression, apoptosis, migration and invasion [5]. Aberrant expression of miRNAs has been found and confirmed in CML like miR-362-5p [6], miR-320a [7], miR-21 [8], miR-486 [9], miR-29 [10], miR-424 [11], miR-370 [12], miR-181c [13] and miR-30a [14]. MiRNAs function as oncogenes or tumor suppressors in CML. For example, miR-362-5p, acting as an oncogene, promotes CML cell growth via down-regulation of GADD45α [6], whereas miRNA-320a acts as a tumor suppressor by targeting BCR/ABL oncogene in CML cells [7]. Lack of miR-15a-5p expression was reported in leukemic cells [15,16], however, its roles and regulation were far from clear.
In this study, we evaluated the roles of miR-15a-5p in CML cell growth, apoptosis, cell cycle distribution and metastasis. Data from the study demonstrated that miR-15a-5p played a suppressing role in CML cell growth and metastasis by targeting CXCL10. In addition, the negative correlation between the expressions of miR-15a-5p and CXCL10 in CML samples was verified.
Materials and methods
Samples from the patients
Blood of patients with CML or the healthy people was collected between 2011 and 2013 at the First Affiliated Hospital of Zhejiang Chinese Medical University (Hangzhou, China). Samples were obtained from patients with diagnosed as CML. Negative control samples were from healthy volunteers.
Cell culture and transfection
The human CML cell line including KU812, K562, BV173, Jurkat, Ball-1 and normal blood cells was maintained in RPMI 1640 containing 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) at 37°C, 95% air and 5% CO2. CML cells were transfected with miR-15a-5p mimics using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Leukocyte isolation
Peripheral blood samples were from patients newly diagnosed as CML before treatment and healthy volunteers. The pooled healthy blood was as the control. Only blood samples containing 67-70% granulocytes (according to routine blood counts) were used. Blood samples, meeting these criteria, were mixed with erythrocyte lysis buffer (Qiagen, Hilden, Germany) and centrifuged at 400 g for 10 min at 4°C. The leukocyte pellets were washed, centrifuged again and frozen as dry pellets or viably frozen in 10% DMSO. For miR-15a-5p expression assays, miRNAs were isolated from frozen cell pellets using the miRNeasy Mini Kit.
Luciferase activity assay
The 3’ untranslated region (3’-UTR) of CXCL10 was amplified from human genomic DNA and cloned into the pmiRGLO vector (Promega, USA) to generate the 3’-UTR wild-type reporter plasmid. A mutation of the CXCL10 3’-UTR sequence was performed using a Quick-change Site-Directed Mutagenesis kit (Stratagene, LaJolla, CA). CML cells were transfected with the pmiRGLO plasmids and miR-15a-5p mimics. The cells were harvested 48 h after transfection and analyzed for luciferase signals using Dual Luciferase Assays Kits (Promega).
RNA extraction and quantitative RT-PCR
The total RNA of sample or cells was extracted using Trizol (Invitrogen, Carlsbad, CA, USA). The expression of miR-15a-5p was detected by quantitative RT-PCR using the TaqManmiRNA assay kit (Applied Biosystems, Foster City, CA, USA) and U6 snRNA was used as a control. Quantitative RT-PCR was performed using ABI7000 sequence detector (Applied Biosystems, Foster City, CA, USA). MiRNA or gene expression was analyzed using the method of 2-ΔΔCt.
Cell survival assay
Cell proliferation was evaluated at the indicated time points using the Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies Inc, Kumamoto, Japan) according to the manufacturer’s protocol. Briefly, cells were incubated in 10% CCK-8 for 2 h at 37°C and the optical density value was then measured at 570 nm with a microplate reader (Sunrise, Tecan, Austria). The data are presented as the means ± SD of three independent experiments.
Western blot analysis
The total protein from CML cells transfected with miR-15a-5p mimics was extracted using RIPA lysis buffer containing proteinase inhibitors (Sigma). Proteins were separated by SDS-PAGE and transferred to PVDF membranes. They were probed with primary antibodies against CXCL10 (1:500) and β-actin overnight at 4°C and then incubated with horseradish peroxidase-labeled antibodies for 2 h at room temperature. The signals were detected by an enhanced chemiluminescence system.
ELISA
The concentrations of CXCL10 in conditioned medium from cells were determined using Human CXCL10/IP-10 Quantikine ELISA Kit (R&D, Minneapolis, USA) according to the manufacturer’s instructions.
Cell migration and invasion assay
The CML cells (2×104) in serum-free DMEM were plated on the top surface of polycarbonate membranes (coated with Matrigel for invasion assay or without Matrigel for migration assay) in Transwell chambers (Costar, Corning Inc., NY, USA). After incubation for 24 h at 37°C, invaded or migrated cells on the lower membrane surface were fixed, stained, photographed and counted under a microscope (5 random fields per well, 100× magnification).
Flow cytometry
CML cells were seeded in 6-well plates and transfected with miR-15a-5p for 48 h. Then 1×106 cells were harvested for each group and washed twice with PBS. The cells were double-stained with FITC-conjugated Annexin V and propidium iodide (PI). Apoptosis and necrosis were analyzed by quadrant statistics. Data were shown as the percentage of apoptotic cells.
Statistical analysis
All statistical analyses were carried out by SPSS13.0 software. Data were presented as mean ± SEM. The miR-15a-5p expression was compared in CML blood and cells by the unpaired Student’s t test. The relationship between miR-15a-5p and CXCL10 expression was analyzed by Spearman’s correlation. A p value lower than 0.05 was considered significant.
Results
MiR-15a-5p expression in CML patient samples and cell lines
In order to evaluate the role of miR-15a-5p in CML, we first investigated the relative miR-15a-5p expression in human CML blood samples from the patients. The results showed that miR-15a-5p expression was significantly down-regulated in blood samples of CML when comparing to miR-15a-5p levels in the healthy blood (Figure 1A and 1B). The results from CML cell lines also showed that the miR-15a-5p expression was significantly decreased in most of CML cell lines comparing it in normal cells (Figure 1C). It was speculated that miR-15a-5p may play as a suppressing role in CML according to the data of low miR-15a-5p expression in CML samples and cells.
Figure 1.
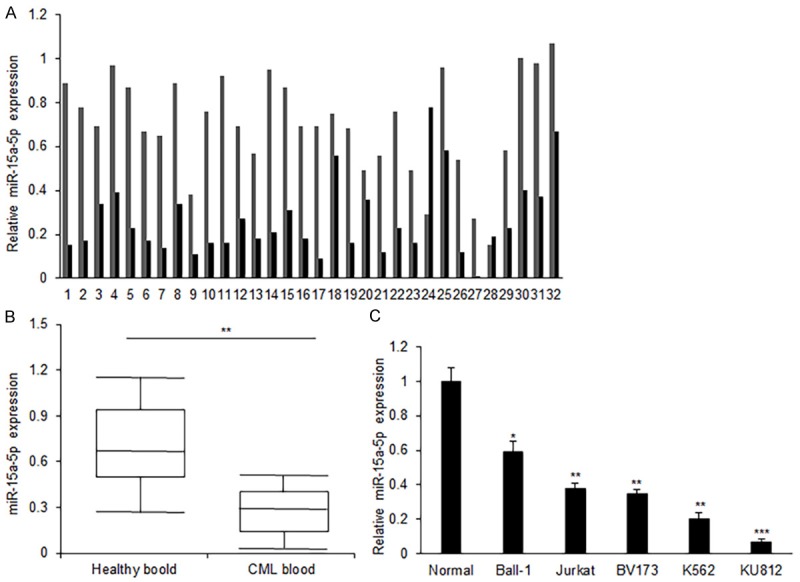
MiR-15a-5p expression in CML patients’ samples and cell lines. (A) MiR-15a-5p expression in the blood of patients with CML. Total RNA was extracted from the blood and miR-15a-5p was measured using real time RT-PCR. (B) Data analysis of miR-15a-5p expression from (A). (C) MiR-15a-5p expression in the CML cell lines including KU812, K562, BV173, Jurkat, Ball-1 and normal blood cells. Total RNA was extracted from the blood and miR-15a-5p was measured using real time RT-PCR. MiR-15a-5p expression was normalized to snoRNA U6. Error bars are SD. Experiments were conducted in triplicates. *P<0.05; **P<0.01.
MiR-15a-5p suppresses CML cell proliferation in vitro
To explore the role of miR-15a-5p in CML cells, K562 and KU812 cells were transfected with miR-15a-5p mimics and scramble mimics to observe cell survival ability using the method of MTT assay. When miR-15a-5p mimics was transfected into K562 and KU812 cells, the expression of miR-15a-5p increased effectively (Figure 2A). MiR-15a-5p mimics transfection inhibited K652 cell survival ability significantly (Figure 2B). MiR-15a-5p could also inhibit KU812 cell proliferation (Figure 2C). The data indicated that miR-15a-5p played an inhibiting role in CML cell growth.
Figure 2.
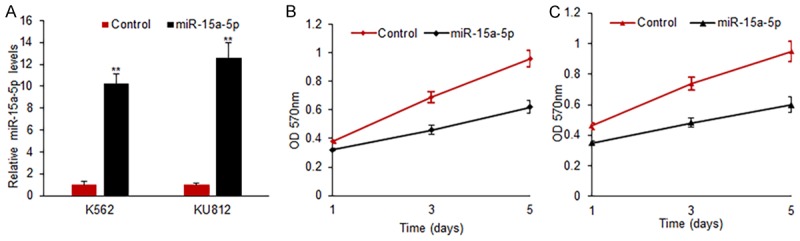
MiR-15a-5p suppresses CML cell proliferation. A. MiR-15a-5p expression in CML cells with miR-15a-5p mimics transfection. K562 and KU812 cells were transfected with miR-15a-5p mimics or scramble mimics and miR-15a-5p levels were examined after 48 h using real time RT-PCR. B. Cell proliferation was examined in K562 cells with miR-15a-5p mimics transfection using MTT method. K562 cells were transfected with miR-15-5p mimics or scramble mimics and cell survival rates were examined at day 1, 3 and 5. C. Cell proliferation was examined in KU182 cells with miR-15a-5p mimics transfection using MTT method. KU812 cells were transfected with miR-15a-5p mimics or scramble mimics and cell survival rates were examined at day 1, 3 and 5. Error bars are SD. Experiments were conducted in triplicates. **P<0.01.
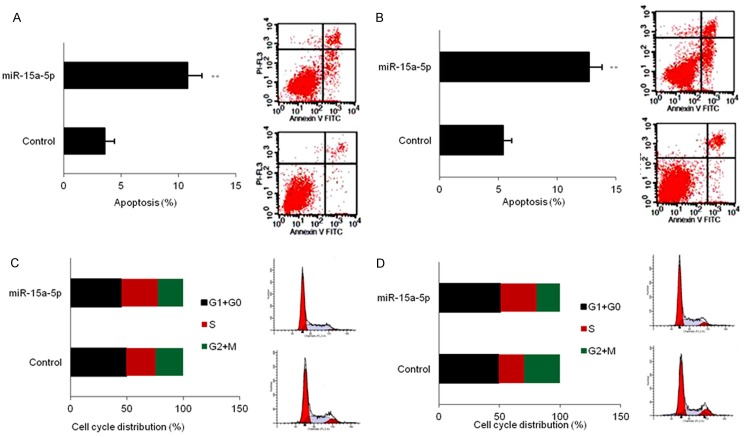
MiR-15a-5p induces apoptosis and cell cycle arrest in CML cells
To elucidate the mechanisms of the role of miR-15a-5p on CML cell apoptosis and cell cycle, K562 and KU812 cells were transfected miR-15a-5p for apoptosis and cell cycle analysis by flow cytometry. MiR-15a-5p significantly increased the apoptosis rates of K562 and KU812 cells when compared to the miRNA controls (Figure 3A and 3B). Moreover, miR-15a-5p greatly reduced the proportion of G0/G1 and G2/M phases in K562 and KU812 cells and increased the proportion of S phase in cells when compared to the controls (Figure 3C and 3D). The data indicated that miR-15a-5p could promote CML cell apoptosis and alter the cell cycle distribution.
Figure 3.
MiR-15a-5p induces apoptosis and cell cycle arrest in CML cells. A, B. Apoptosis of K652 or KU812 cells with miR-15a-5p mimics transfection. K562 or KU812 cells were transfected with miR-15a-5p mimics or scramble mimics for 48 h and cell apoptosis was tested by flow cytometry. The left panel was the analyzed apoptosis rate and the right panel was the photos of flow cytometry. C, D. Cell cycle of K652 or KU812 cells with miR-15a-5p mimics transfection. K562 or KU812 cells were transfected with miR-15a-5p mimics or scramble mimics for 48 h and cell cycle was tested by flow cytometry. The left panel was the analyzed the phases of cell cycle and the right panel was the photos of flow cytometry. Error bars are SD. Experiments were conducted in triplicates. **P<0.01.
MiR-15a-5p inhibits CML cell metastasis
To learn the effect of miR-15a-5p on the CML cell metastasis, K562 and KU812 cells were transfected with miR-15a-5p and migration was analyzed using wound healing method. The founding showed that miR-15a-5p significantly decreased the migration in K562 and KU812 cells with miR-15a-5p transfection comparing with their controls (Figure 4A and 4B). Moreover, miR-15a-5p greatly reduced the invading ability in K562 and KU812 cells (Figure 4C and 4D). The data elucidated that miR-15a-5p suppressed CML metastasis.
Figure 4.
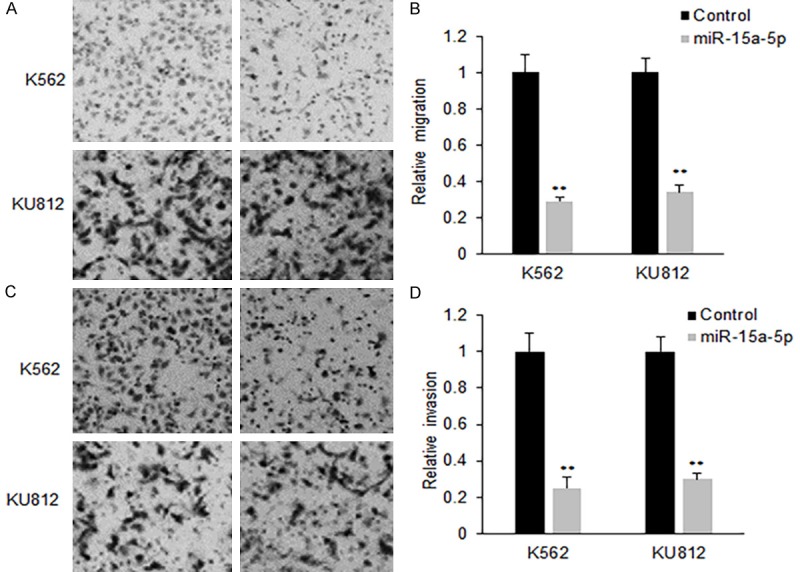
MiR-15a-5p inhibits CML cell metastasis. (A) Cell migration of K562 cells with miR-15a-5p mimics transfection. K562 or KU812 cells were transfected with miR-15a-5p mimics or scramble mimics for 48 h and cell migration was assayed using transwell system with the membrane without Matrigel. The migrated cells were taken photos using a camera linked to the microscope. (B) The analyzed migrated migration rates from (A). (C) Cell invasion of K562 and KU812 cells with miR-15a-5p mimics transfection. K562 or KU812 cells were transfected with miR-15a-5p mimics or scramble mimics for 48 h and cell migration was assayed using transwell system with the membrane with Matrigel. The invaded cells were taken photos using a camera linked to the microscope. (D) The analyzed migrated invasion rates from (C). Error bars are SD. Experiments were conducted in triplicates. **P<0.01.
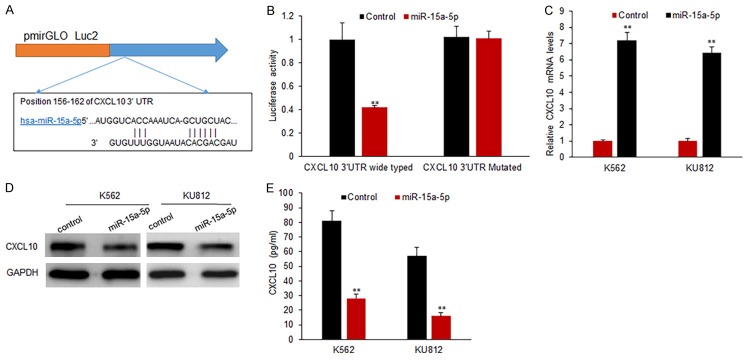
CXCL10 expression is down-regulated by miR-15a-5p
To determine the possible target genes of miR-15a-5p in CML cells, we used bioinformatics to analyze the putative targets of miR-15a-5p. Our analysis identified that CXCL10 was a potential target gene of miR-15a-5p (Figure 5A). Furthermore, the target sequences of CXCL10 3’UTR (wild type, WT) or the mutant sequence (mutant type, MUT) were cloned into the luciferase reporter vector respectively. Our results showed that the miR-15a-5p significantly decreased the firefly luciferase activity of the reporter with wild type 3’UTR, but the activity of mutant 3’UTR vector remained unaffected (Figure 5B). K562 and KU812 cells were transfected with miR-15a-5p for measuring CXCL10 mRNA expression by qRT-PCR. It was found that miR-15a-5p transfection led to an obvious decrease of CXCL10 mRNA (Figure 5C), so did protein levels in K562 and KU812 cells (Figure 5D and 5E).
Figure 5.
CXCL10 expression is down-regulated by miR-15a-5p in CML cells. A. CXCL10 is predicted as a target gene of miR-15a-5p using TargetScan 7.0. B. Luciferase activity of CXCL10 3’UTR and mutated CXCL10 3’UTR in CML cells with miR-15a-5p mimics transfection. K562 cells were transfected with wide type or mutant type of CXCL10 3’UTR combined with miR-15a-5p mimics and luciferase activity was assayed by dual luciferase assay system. C. qRT-PCR was used to analyze CXCL10 mRNA expression in CML cells. K562 or KU812 cells were transfected with miR-15a-5p mimics and CXCL10 mRNA was assayed by qRT-PCR. D. CXCL10 protein levels in CML cells. K562 or KU812 cells were transfected with miR-15a-5p mimics and CXCL10 protein levels was assayed by Western blotting. E. CXCL10 protein levels in CML cells. K562 or KU812 cells were transfected with miR-15a-5p mimics and CXCL10 protein levels was assayed by ELISA. Error bars are SD. Experiments were conducted in triplicates. **P<0.01.
Relationship between CXCL10 and miR-15a-5p expression in blood samples of CML patients
Above data confirmed that CXCL10 is the target gene of miR-15a-5p. To know the correlation of CXCL10 and miR-15a-5p in blood samples of CML patients, CXCL10 protein levels in the blood from 32 patients was measured by ELISA. It was observed that the average CXCL10 protein levels was higher in the blood of CML (Figure 6A). Further analysis showed that CXCL10 increased in most blood samples of CML patients with low levels of miR-15a-5p and CXCL10 expression was negatively correlated to miR-15a-5p expression in the blood of CML patients (Figure 6B).
Figure 6.
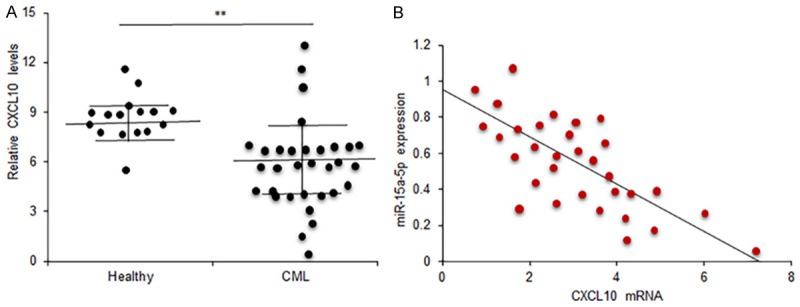
Relationship between CXCL10 and miR-15a-5p expression in CML blood samples. A. CXCL10 expression in CML blood samples was carried out by ELISA and CXCL10 expression was analyzed. Healthy: the blood samples from healthy persons; CML: the blood samples from CML patients. B. Data analysis of the relationship between miR-15a-5p and CXCL10 in CML blood. r=-0.437. Experiments were conducted in triplicates. **P<0.01.
Discussion
In recent years, many miRNAs are studied in various disease using cellular and molecular methods. The illustration of a new discovered miRNA including their target genes may expand the knowledge for molecular mechanism of cancer. MiR-15a-5p is reported in cancer, however, the role of miR-15a-5p in CML is unclear. In this study, we examined miR-15a-5p expression in CML samples and cell lines. Our data showed that miR-15a-5p expression was lower in the blood samples of CML patients and cell lines comparing to the controls. The data from cellular functions indicated that miR-15a-5p significantly suppressed cell proliferation, promoted apoptosis, and decreased migration and invasion of CML cells in vitro. Our study illustrated that miR-15a-5p acts as a novel tumor suppressor which exerts an important effect on CML progression.
It is reported that miR-15a-5p could inhibit human hepatocellular carcinoma cell proliferation and division by down-regulation of BDNF expression [15]. Another report points out that miR-15a-5p is up-regulated in Kaposi’s sarcoma through miRNA array [16]. The previous reports suggest that miR-15a-5p functions as an oncogene or a tumor suppressor depending on the tumor cell background. In our study, we found that miR-15a-5p was significantly down-regulated in CML samples and cell lines. The functional analysis showed that miR-15a-5p inhibited cell survival and metastasis, which suggested that it functions as a tumor suppressor in CML.
CXCL10, also named as interferon-γ inducible protein-10 (IP-10), is a member of chemokine family and has been identified as an oncogene [17]. CXCL10 plays an important role on the attraction and activation of T cells, B cells, mononuclear macrophage, dendritic cells, NK cells and other immune inflammatory cells [18-22]. A recent report shows that CXCL10 plays important roles in the interaction of neuroendocrine differentiation and TAMs, and high expression of CXCL10 indicates the poor prognosis of colorectal cancer. There is a report showing that CXCL10 status can predict tamoxifen treatment response in breast cancer patients [23]. Our study showed that CXCL10 protein levels in the blood of CML patients were significantly higher than it in normal paired ones and CXCL10 is regulated by miR-15a-5p in CML cells. A report showed that CXCL10 is the target gene of miR-21 in the pseudorabies virus amplication [24]. Our data for the first time demonstrate that CXCL10 is regulated by miR-15a-5p in CML cells.
In this study, we found that CXCL10 was a direct target gene of miR-15a-5p in CML cells. The expression of miR-15a-5p was negatively related to CXCL10 expression in CML blood samples. The biological functions showed that miR-15a-5p could inhibit cell proliferation, metastasis and promoted apoptosis of CML cells. These data suggest that the miR-15a-5p/CXCL10 axis could be a key growth regulator in CML cells and miR-15a-5p is an inhibitor of CML. MiR-15a-5p may be a method for CML therapy in the further.
Acknowledgements
This study was funded Natural Science Foundation by Zhejiang Province of China (Y14H2900122014).
Disclosure of conflict of interest
None.
References
- 1.Machova Polakova K, Koblihova J, Stopka T. Role of epigenetics in chronic myeloid leukemia. Curr Hematol Malig Rep. 2013;8:28–36. doi: 10.1007/s11899-012-0152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellicano F, Sinclair A, Holyoake TL. In search of CML stem cells’ deadly weakness. Curr Hematol Malig Rep. 2011;6:82–87. doi: 10.1007/s11899-011-0085-y. [DOI] [PubMed] [Google Scholar]
- 3.Perrotti D, Harb JG. BCR-ABL1 kinase-dependent alteration of mRNA metabolism: potential alternatives for therapeutic intervention. Leuk Lymphom. 2011;52(Suppl 1):30–44. doi: 10.3109/10428194.2010.546914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty C, Sharma AR, Patra BC, Bhattacharya M, Sharma G, Lee SS. MicroRNAs mediated regulation of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget. 2016;7:42683–42697. doi: 10.18632/oncotarget.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertacchini J, Heidari N, Mediani L, Capitani S, Shahjahani M, Ahmadzadeh A, Saki N. Targeting PI3K/AKT/mTOR network for treatment of leukemia. Cell Mol Life Sci. 2015;72:2337–2347. doi: 10.1007/s00018-015-1867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang P, Ni F, Deng RQ, Qiang G, Zhao H, Yang MZ, Wang XY, Xu YZ, Chen L, Chen DL, Chen ZJ, Kan LX, Wang SY. MiR-362-5p promotes the malignancy of chronic myelocytic leukaemia via down-regulation of GADD45α. Mol Cancer. 2015;14:190. doi: 10.1186/s12943-015-0465-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Xishan Z, Ziying L, Jing D, Gang L. MicroRNA-320a acts as a tumor suppressor by targeting BCR/ABL oncogene in chronic myeloid leukemia. Sci Rep. 2015;5:12460. doi: 10.1038/srep12460. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Zhou M, Zeng J, Wang X, Wang X, Huang T, Fu Y, Sun T, Jia J, Chen C. Histone demethylase RBP2 decreases miR-21 in blast crisis of chronic myeloid leukemia. Oncotarget. 2015;6:1249–1261. doi: 10.18632/oncotarget.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang LS, Li L, Li L, Chu S, Shiang KD, Li M, Sun HY, Xu J, Xiao FJ, Sun G, Rossi JJ, Ho Y, Bhatia R. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood. 2015;125:1302–1313. doi: 10.1182/blood-2014-06-581926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kollinerova S, Vassanelli S, Modriansky M. The role of miR-29 family members in malignant hematopoiesis. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2014;158:489–501. doi: 10.5507/bp.2014.029. [DOI] [PubMed] [Google Scholar]
- 11.Hershkovitz-Rokah O, Modai S, Pasmanik-Chor M, Toren A, Shomron N, Raanani P, Shpilberg O, Granot G. Restoration of miR-424 suppresses BCR-ABL activity and sensitizes CML cells to imatinib treatment. Cancer Lett. 2015;360:245–256. doi: 10.1016/j.canlet.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 12.Zhou M, Zeng J, Wang X, Guo Q, Huang T, Shen H, Fu Y, Wang L, Jia J, Chen C. MiR-370 sensitizes chronic myeloid leukemia K562 cells to homoharringtonine by targeting forkhead box M1. J Transl Med. 2013;11:265. doi: 10.1186/1479-5876-11-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosakhani N, Mustjoki S, Knuutila S. Downregulation of miR-181c in imatinib-resistant chronic myeloid leukemia. Mol Cytogenet. 2013;6:27. doi: 10.1186/1755-8166-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Song Y, Ma W, Zheng W, Yin H. Decreased microRNA-30a levels are associated with enhanced ABL1 and BCR-ABL1 expression in chronic myeloid leukemia. Leuk Res. 2013;37:349–356. doi: 10.1016/j.leukres.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Long J, Jiang C, Liu B, Fang S, Kuang M. MicroRNA-15a-5p suppresses cancer proliferation and division in human hepatocellular carcinoma by targeting BDNF. Tumour Biol. 2016;37:5821–5828. doi: 10.1007/s13277-015-4427-6. [DOI] [PubMed] [Google Scholar]
- 16.Wu XJ, Pu XM, Zhao ZF, Zhao YN, Kang XJ, Wu WD, Zou YM, Wu CY, Qu YY, Zhang DZ, Feng YY, Liu JY. The expression profiles of microRNAs in Kaposi’s sarcoma. Tumour Biol. 2015;36:437–446. doi: 10.1007/s13277-014-2626-1. [DOI] [PubMed] [Google Scholar]
- 17.Bai M, Chen X, Ba YI. CXCL10/CXCR3 overexpression as a biomarker of poor prognosis in patients with stage II colorectal cancer. Mol Clin Oncol. 2016;4:23–30. doi: 10.3892/mco.2015.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng YJ, Lai W, Wu H, Liu L, Xu HY, Wang J, Chu ZH. Neuroendocrine-like cells-derived CXCL10 and CXCL11 induce the infiltration of tumorassociated macrophage leading to the poor prognosis of colorectal cancer. Oncotarget. 2016;7:27394–27407. doi: 10.18632/oncotarget.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunardi S, Lim SY, Muschel RJ, Brunner TB. IP-10/CXCL10 attracts regulatory T cells: implication for pancreatic cancer. Oncoimmunology. 2015;4:e1027473. doi: 10.1080/2162402X.2015.1027473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Li F, Ping Y, Wang L, Chen X, Wang D, Cao L, Zhao S, Li B, Kalinski P, Thorne SH, Zhang B, Zhang Y. Local production of the chemokines CCL5 and CXCL10 attracts CD8+ T lymphocytes into esophageal squamous cell carcinoma. Oncotarget. 2015;6:24978–24989. doi: 10.18632/oncotarget.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zumwalt TJ, Arnold M, Goel A, Boland CR. Active secretion of CXCL10 and CCL5 from colorectal cancer microenvironments associates with granzyme B+ CD8+ T-cell infiltration. Oncotarget. 2015;6:2981–2991. doi: 10.18632/oncotarget.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunardi S, Jamieson NB, Lim SY, Griffiths KL, Carvalho-Gaspar M, Al-Assar O, Yameen S, Carter RC, McKay CJ, Spoletini G, D’Ugo S, Silva MA, Sansom OJ, Janssen KP, Muschel RJ, Brunner TB. IP-10/CXCL10 induction in human pancreatic cancer stroma influences lymphocytes recruitment and correlates with poor survival. Oncotarget. 2014;5:11064–11080. doi: 10.18632/oncotarget.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilborn E, Sivik T, Fornander T, Stål O, Nordenskjöld B, Jansson A. C-X-C ligand 10 and C-X-C receptor 3 status can predict tamoxifen treatment response in breast cancer patients. Breast Cancer Res Treat. 2014;145:73–82. doi: 10.1007/s10549-014-2933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Ma G, Fu L, Jia H, Zhu M, Li X, Zhao S. Pseudorabies viral replication is inhibited by a novel target of miR-21. Virology. 2014;456-457:319–328. doi: 10.1016/j.virol.2014.03.032. [DOI] [PubMed] [Google Scholar]