Abstract
MicroRNAs (miRNAs) have been described to simultaneously inhibit hundreds of targets, albeit to a modest extent. It was recently proposed that there could exist more specific, exceptionally strong binding to a subgroup of targets. However, it is unknown, whether this is the case and how such targets can be identified. Using Argonaute2-ribonucleoprotein immunoprecipitation and in vivo competitive binding assays, we demonstrate for miRNAs-21, -199–3p and let-7 exceptional regulation of a subset of targets, which are characterized by preferential miRNA binding. We confirm this finding by analysis of independent quantitative proteome and transcriptome datasets obtained after miRNA silencing. Our data suggest that mammalian miRNA activity is guided by preferential binding of a small set of 3′-untranslated regions, thereby shaping a steep gradient of regulation between potential targets. Our approach can be applied for transcriptome-wide identification of such targets independently of the presence of seed complementary sequences or other predictors.
INTRODUCTION
MicroRNAs (miRNAs) are RNA molecules with a length of 19–25 nts, which act in complex with Argonaute (Ago) proteins to guide the activity of the RNA-induced silencing complex (1). MiRNAs typically recognize sites localized in 3′-untranslated regions (3′-UTRs) of mRNA molecules and their binding leads to translational repression and increased decay of target RNAs (2). Canonical target sites contain sequences complementary to miRNA nucleotides 2–7 (miRNA 6-mer seed), with additional complementarity to nucleotides 1 and/or 8 or an A opposite nucleotide 1 increasing target regulation (7- or 8-mer target sites). Due to the high abundance of such phylogenetically conserved 7–8 mer sequences in the transcriptome, miRNAs are assumed to typically regulate large numbers of targets, however to a moderate extent. Thereby one miRNA targets dozens to hundreds of targets and most mRNAs are targeted by one or more miRNAs (1–3). While widespread target regulation has indeed been demonstrated for some of the most highly expressed miRNAs and in overexpression experiments, it requires sufficiently high abundance of an miRNA compared with the total abundance of its targets (4–6). However, this condition was suggested to be met only by a small number of miRNAs in a given cell type (6). It appears therefore that in many cases the free and active miRNA concentration is the limiting factor for target regulation and that the multitude of endogenous targets is in a state of constant competition for miRNA binding (7). This view is at odds with recent findings that some individual endogenous RNAs can functionally inhibit miRNAs through regular miRNA–target interactions (competing endogenous RNA or ceRNA hypothesis) (7–9). Furthermore, it was recently observed that for many mRNAs the amount of miRNA-conferred regulation is relatively low compared with the baseline interindividual variation, suggesting that most predicted targets do not contribute to miRNA function (10).
In a competitive environment binding affinities can determine the distribution of macromolecules between the available targets. The parameters currently known to influence the affinity of an miRNA to a specific target are seed complementarity (8-mer > 7-mer > 6-mer), supplementary 3′ pairing and site accessibility (11,12). Predictions based on in vitro measurements of miRNA-target affinity have challenged the ceRNA hypothesis early on (11). So far however, it remains unclear whether the binding constants measured in this way do effectively predict the endogenous fraction of targets bound at a given miRNA level. In fact several processes have been proposed to influence target association in vivo, such as recruitment of other RNA binding proteins, the formation of P-bodies, changes in subcellular localization (7,9) or more recently regulation of Argonaute protein phosphorylation (13). These factors cannot currently be accounted for, which hinders precise modeling of in vivo association kinetics between a given miRNA and target. Furthermore, it remains unclear how to identify targets subject to such modified binding.
We hypothesized that such processes, if take place, will be reflected in notably increased association of an miRNA with specific targets compared with others. Supplementary Figure S1A depicts a possible setting in which a protein ‘X’ binds to the miRNA–target complex and strongly inhibits miRNA dissociation from the target, thus decreasing the apparent KD. Although this is only one possible scenario, we expect that a similar apparent increase in target association should be noticeable in many settings (such as sequestration in P-bodies, enrichment in subcellular compartments etc.). Several observations indirectly support the presence of such a mechanism. For example, it was observed that AU-rich sequences in binding site proximity are a strong predictor for target regulation independent of site accessibility (14). Such sequences may therefore enhance regulation by promoting the interaction with AU-rich element binding proteins (15).
Also, it has been consistently observed that the curves of cumulative distributions obtained for target derepression upon miRNA inhibition typically diverge most from non-targets in the region of strong derepression ((14), see also Figure 3B). Thus, the distribution of derepressions often appears skewed for targets versus non-targets. We suggest that this results from a subpopulation of targets with overproportionally strong regulation, compared with the average value (as modeled in Supplementary Figure S1B).
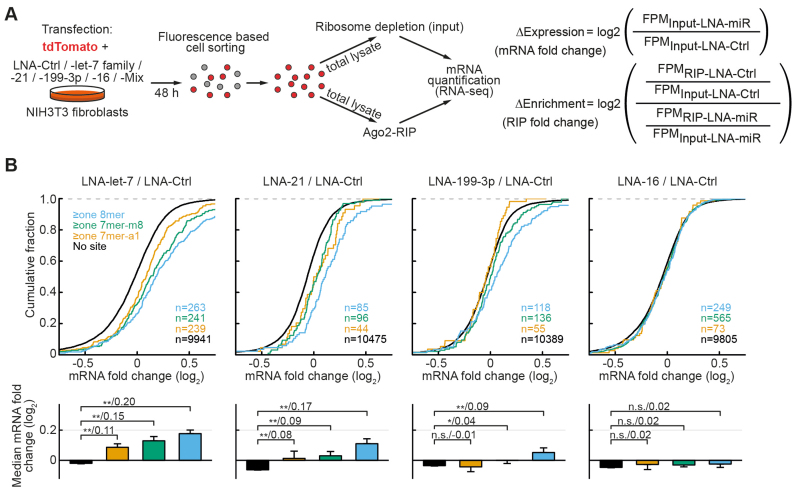
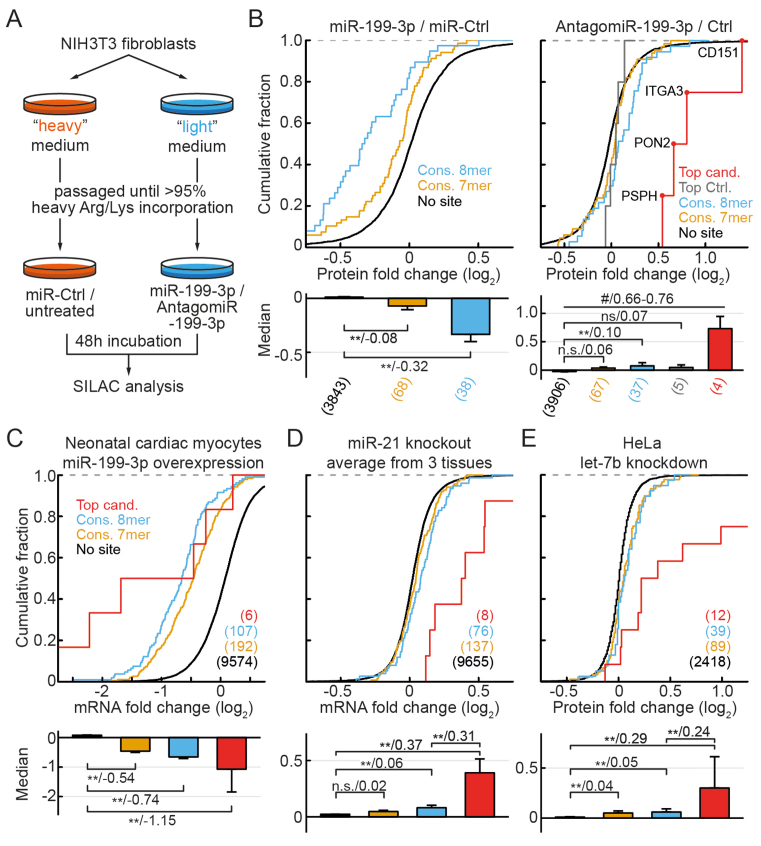
Figure 3.
Experimental design and distributions of mRNA fold changes upon miRNA inhibition. (A) Experimental design for simultaneous measurement of antimiR-mediated depletion from Ago2-complexes (RIP-enrichment changes) and mRNA expression changes for the four miRNAs (or miRNA families) in NIH3T3 fibroblasts. (B) Non-targets and conserved targets were grouped as indicated on the left. Panels below depict median mRNA fold changes ± bootstrap standard errors. For the contrasts P-values (*P < 0.05, **P < 0.01 in a Mann–Whitney U-Test) and the difference of medians of log2 transformed data are indicated separated by a slash. N-numbers are indicated in parentheses.
In this study, we assess the competitive binding potencies of exemplary miRNA targets in living cells and transcriptome Ago2 associations mediated by specific miRNAs. We thereby find strong direct evidence that supports the existence of a process of preferential binding and regulation of a small subpopulation of targets. Furthermore, our approach can be employed to identify such targets among the hundreds of potential candidates for studies addressing miRNA functions and target binding.
MATERIALS AND METHODS
Detailed methods are provided in the ‘Supplementary Materials and Methods’ section.
Reporter assays
NIH3T3 cells were transfected in 96-well plates using a total amount of 200 ng DNA and (for antimiR experiments) 50 nM of locked nucleic acid modified antimiRs (LNAs, Exiqon) using Lipofectamine 2000 (ThermoFisher) and fixed 48 h later. Cells were stained with diamidino-phenylindole (DAPI), fluorescence signals were automatically acquired using microscopy imaging and single-cell analysis performed as further described in the ‘Supplementary Materials and Methods’ section.
Ago2-ribonucleoprotein immunoprecipitation (RIP)
Argonaute2-ribonucleoprotein immunoprecipitation (Ago2-RIP) was performed as described previously (16) using a monoclonal antibody against endogenous mouse Ago2 (clone 2D4, WAKO).
RNA-seq library preparation and analysis
RNA-seq libraries were prepared from input and Ago2-RIP RNA from transfected cells which were isolated using fluorescence activated cell sorting (FACS). First, the RNA was isolated using RNApure (VWR) and the contaminating DNA digested using DNase I (NEB). Ribosomal RNA depletion was performed for input samples using RiboMinus Eukaryote Kit v2 (ThermoFisher) and the RNA was fragmented at 94°C using a magnesium-containing buffer. Then the first strand synthesis (with Superscript III, ThermoFisher) and second strand synthesis (DNA polymerase I, ThermoFisher) were performed. For further steps, the NEBNext Ultra II DNA library prep kit for Illumina (NEB) was applied. Sequencing was performed on an Illumina HiSeq 4000 machine with paired reads of 100-bp length. Two independent biological replicates were thus analyzed for each condition.
Reads were mapped to the mouse genome (mm10) using TopHat2. For each mRNA, we selected a unique representative transcript annotation based on Gencode VM3 and 3P-seq tags (14). The uniquely overlapping reads for each transcript were calculated using htseq-count. Fragments per million mapped reads were then calculated for each transcript (after adding +1 to eliminate 0 values) by normalizing to the total number of reads mapped to the representative transcripts in this sample (divided by 1 million).
SILAC experiments
For proteome analysis, NIH3T3 cells were cultured in medium containing either regular (‘light’) or labelled (‘heavy’) lysine and arginine. Light cells were treated with either miR-199–3p mimic or antagomiR, while heavy cells served as controls.
After transfection cells were grown for 48 h, then lysed and combined at equal protein ratios of treated with respective control lysates. Liquid chromatography–mass spectrometry was then performed in a setup coupling an Eksigent nanoLC-Ultra 1D+ (Eksigent) and an Orbitrap Velos instrument (Thermo Scientific). The data were processed using MaxQuant software. Further details are described in the ‘Supplementary Materials and Methods’ section.
RESULTS
A fluorescence microscopy-based assay for low variability measurement of in vivo miRNA competitive binding and activity
Classically dual-luciferase assays have been used to measure changes in miRNA activity. More recently dual-fluorescent assays have been introduced, which allowed single-cell measurements of fluorescent reporters in a similar way using FACS (6,17). These measurements have shown that at high reporter concentrations the activity of many miRNAs is increasingly inhibited. Therefore, variations in transfection efficiency may contribute to the relatively high variability in luciferase assays, especially since strongly transfected cells express more luciferase and therefore contribute more to the readout.
We adopted a dual fluorescent approach for automated microscopic assessment of miRNA function, which allowed us to combine the advantage of single-cell measurements with the throughput of luciferase assays. We generated a dual-fluorescent reporter with a green (EGFP) and red fluorescence (tdTomato) driven by identical promoters (CMV, Figure 1A and B). The cDNA of EGFP is followed by the 3′-UTR of interest and allows readouts based on miRNA activity on the target (Figure 1C). Assays are thereby performed on all relevant control and treatment groups in technical replicates simultaneously on a 96-well plate, which reduces inter-assay variability. Plates are automatically acquired using fluorescence microscopy, the images are background corrected and morphology filters applied to extract red and green fluorescence values for individual cells, similar to a FACS measurement (Figure 1D). This allows the selection of a defined range of cells based on the red fluorescence (RFP) value (red fluorescent protein, tdTomato, used as control for transfection efficiency), allowing us to select a region where the extent of target regulation is highest due to low miRNA inhibition by the reporter (Figure 1D, right panel).
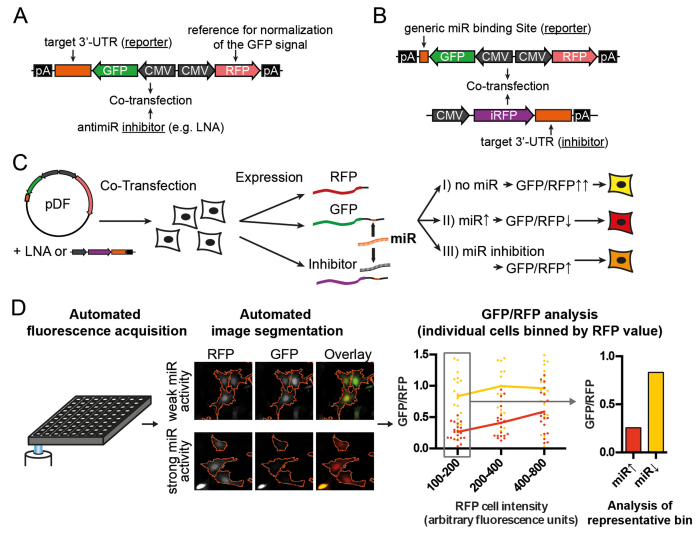
Figure 1.
Schematic representation of dual-fluorescent reporter assays. (A and B) Reporter constructs for the determination of miRNA activity in cells are cotransfected with two types of inhibitors: the effect of an miRNA on a target is assessed by cloning the UTR of interest downstream of GFP and treatment with small molecule inhibitors (LNA-antimiRs, A). Conversely, the inhibitory effect of a UTR on an miRNA can be assessed by cloning generic binding sites downstream of GFP and co-transfecting plasmids expressing the UTR of interest downstream of the non-interfering fluorophore iRFP (infrared fluorescent protein (36), B). (C) MiRNA activity is assessed by measuring the GFP normalized to the RFP (tdTomato) signal after reporter transfection into cells. (D) Readouts for GFP and RFP values for individual cells are obtained by automated microscopic image acquisition and high content image analysis. Selection of cells from a narrow range with little miRNA inhibition by the reporter reduces variability due to different transfection strength between cells.
We successfully confirmed the validity and very low inter-assay variability of this approach using reporters with generic binding sites for let-7 and miR-21 in NIH3T3 cells (Supplementary Figure S2). For further experiments, we applied this assay to either measure the effect of an miRNA on a target (Figure 1A, target as reporter, LNA-antimiR as inhibitor) or the effect of the target on the miRNA (competitive potency, Figure 1B, target as inhibitor, generic miR-binding site(s) as reporter).
Strong target regulation is associated with increased competitive potency for the 3′-UTRs of CNTFR and TMEM2
We hypothesized that preferential miRNA association for specific targets will result in exceptionally strong target regulation.
We used NIH3T3 cells, in which we established the dual-fluorescent assay above, as a model system and performed miRNA quantification using small RNA-seq. Cells were transfected with an RFP reporter and sorted prior to sequencing to mimic the conditions of a transfection experiment as used further below for RNA-seq assays (Supplementary Figure S3A).
We thereby found miR-21, to be strongly expressed (Supplementary Figure S3B) and selected previously described targets of this miRNA for comparison of the regulation of their 3′-UTRs: PDCD4 (18), SPRY1 (19), SPRY2 (20), BTG2 (21) and PTEN (22). We furthermore performed a custom bioinformatic search for conserved targets with 3′ supplementary pairing, which was reported to increase affinity (11). We thereby obtained the candidate target CNTFR as the top hit and included it in the analysis, despite it not being well studied so far (Figure 2A).
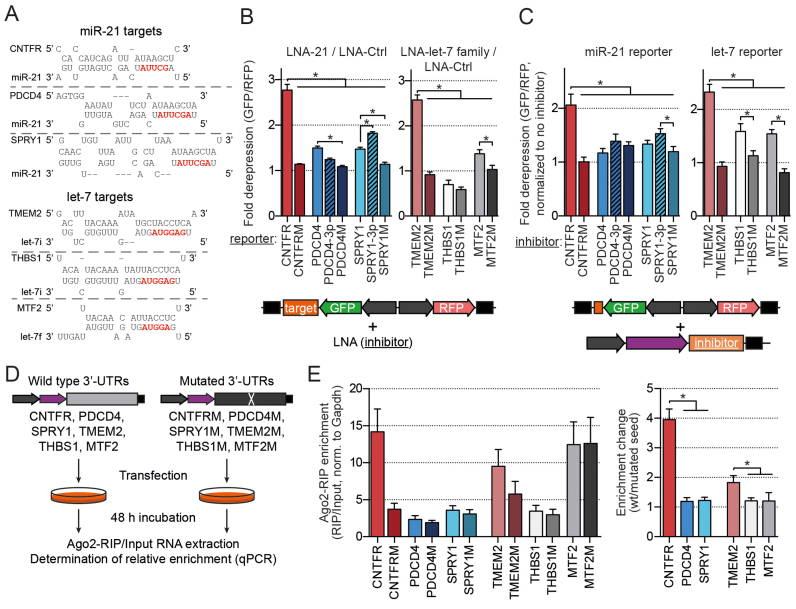
Figure 2.
Superior regulation of CNTFR and TMEM2 3′-UTRs is mediated by strong miRNA binding to single target sites. (A) Predicted hybridization of the binding sites of selected targets. (Mutant and modified sites are depicted in Supplementary Figure S4B). (B) Results of the dual-fluorescent reporter assays. Measurements with the antimiRs LNA-21 (n = 8) or LNA-let-7 family (n = 5) were normalized to LNA-Ctrl. Depicted are values from cells in the bin with 100–200 AFU RFP values (as in Figure 1D). (C) Results of the competitive binding assay. Dual fluorescent reporters specific for miR-21 or let-7 activity (binding sites are depicted in Supplementary Figure S2) were co-transfected with the analyzed UTRs inserted 3′ of iRFP to compare their potencies as inhibitors of the respective miRNAs. Transfection ratios of reporter to inhibitor were 1:1 for miR-21 (n = 4) and 1:10 for let-7 (n = 7) targets. Depicted is the bin with 400–800 AFU for miR-21 and 100–200 AFU for let-7. (D and E) Design and results of the Ago2-RIP assay to compare changes in RIP-fraction upon seed mutation (n = 5). *P < 0.05 in ANOVA for repeated measures with a post-hoc Tukey’s test.
We cloned full-length 3′-UTRs of these targets downstream of EGFP into dual-fluorescent reporters as either wild-type (wt) or with a mutated seed (Figure 2A, Supplementary Figure S4B) and performed assays as in Figure 1A using LNA-21 and LNA-Ctrl to determine their regulation. Among the known targets PDCD4 and SPRY1 showed the strongest upregulation (≈1.5-fold, Figure 2B and Supplementary Figure S4A). Interestingly, we observed the most prominent de-repression for the 3′-UTR of CNTFR (>2.5-fold), which was completely abolished after seed mutation (Figure 2B).
To test whether this was solely due to the presence of 3′ supplementary pairing, we inserted the respective sequence 5′ of the seed match into the UTRs of PDCD4 and SPRY1 target sites (‘PDCD4–3p’ and ‘SPRY1–3p’, Supplementary Figure S4B). This only led to a minor increase in regulation for SPRY1 but not for PDCD4 and did not match that of CNTFR (Figure 2B). Yet, the mutation of this region in CNTFR strongly reduced its regulation (Supplementary Figure S4C), suggesting that the effect of supplementary pairing is important for CNTFR but context dependent.
To test whether increased regulation was indeed associated with stronger miRNA binding, we performed fluorescence-based competitive binding assays as described in Figure 1B at a transfection ratio reporter:inhibitor of 1:1. We thereby expect that increased target association will be reflected in comparably stronger inhibition of miR-21 in vivo by the respective 3′-UTR, as further explained in the ‘Supplementary Materials and Methods’ section. Interestingly the results mirrored those obtained for target regulation, with CNTFR showing a significantly stronger inhibition of miR-21 (Figure 2C). These findings suggest that strong endogenous target regulation correlates with exceptional miRNA association and led us to test the hypothesis that this may represent a general phenomenon.
We therefore performed a search for conserved targets of another miRNA, the let-7 family in a similar way as for CNTFR and selected TMEM2, THBS1 and MTF2 (Figure 2A and Supplementary Figure S4B, selection parameters further described in ‘Supplementary Materials and Methods’ section). Of these only the 3′-UTR of TMEM2 showed strong de-repression upon let-7 family silencing (Figure 2B). In the competitive assay, no measureable inhibition of let-7 activity could be achieved at a 1:1 transfection ratio with either of the targets (not shown), we therefore increased the ratio of reporter:inhibitor to 1:10. At this ratio, we did observe significant reporter upregulation, with TMEM2 again showing the strongest effect (Figure 2C).
Determination of site-specific association with Ago2 confirms preferential binding of CNTFR and TMEM2
To more directly assess target-site occupancy, we performed Ago2-RIP assays. We thereby co-transfected a mix of either wt or seed-mutated plasmids, each mix containing all of the six analyzed full-length 3′-UTRs (Figure 2D). The relative enrichment of the different targets in the RIP compared with the input fractions was then assessed using quantitative polymerase chain reaction (Figure 2E, left panel). RIP enrichment values for wt were then normalized to the respective mutated transcripts to obtain relative change in RIP enrichment due to the respective single miRNA-binding sites (Figure 2E, right panel). Similar to the competitive binding assay, we expect the RIP assay to be influenced by changes in apparent KD, as further described in ‘Supplementary Materials and Methods’ section.
In this assay, we indeed observed markedly higher changes in the enrichment fraction (wt/mutated) for CNTFR versus PDCD4 and SPRY1 (miR-21 targets) and for TMEM2 versus THBS1 and MTF2 (let-7 targets, Figure 2E). This confirms the notion that stronger regulation is tightly correlated with a higher extent of target association of an miRNA for these targets.
Since neither the inclusion of supplementary pairing in PDCD4 and SPRY1, nor the selection of let-7 targets based on conserved supplementary pairing reliably resulted in strong regulation, we concluded that supplementary pairing alone does not sufficiently explain preferential binding. This is in agreement with previous reports that found it on average to be associated with only modest added target regulation (23). Interestingly, apart from supplementary pairing and shorter UTR length (CNTFR: 0.64 kbp versus PDCD4: 1.92 kbp and SPRY1: 1.21 kbp) we could not observe any major predictor that would clearly distinguish CNTFR. Rather, the context++ scores (14) were superior for PDCD4 (score of −0.48) and SPRY1 (−0.41) due to 8-mer seed complementarity rather than 7-mer for CNTFR (−0.27).
RNA-seq using Ago2-RIP and miRNA inhibition reveals common association of preferential binding with strong target regulation
The clear distinction of CNTFR and TMEM2 from other targets in RIP assays suggests that this approach can be applied for transcriptome-wide identification of preferential targeting by combining Ago2-RIP with deep sequencing of the RNA (RIP-seq). However, since Ago-association can occur through different miRNAs, or even in an miRNA-independent way through co-localization in RNP-complexes (24), this assay requires specific inhibition of miRNA binding. This was achieved above through target-site mutation and can potentially be achieved for all targets of an miRNA by specific silencing with antimiRs.
Although Ago-RIP assays have been first introduced some time ago (25,26), they have been classically used with miRNA overexpression. We were unable to identify any reports systematically correlating changes in gene expression with Ago-RIP enrichment upon miRNA silencing using RNA-seq. Yet, one group previously observed a correlation between target depletion in Ago-RIP and target upregulation for a prespecified subset of predicted miR-122 targets (27).
To systematically assess this question, we therefore devised an experimental procedure as described in Figure 3A. Hereby, antimiRs with LNA chemistry are co-transfected with a fluorescent reporter plasmid (tdTomato) and 2 days later the cells are sorted using FACS to purify transfected cells prior to lysis. As shown in Supplementary Figure S2, such co-transfection in NIH3T3 cells resulted in highly efficient miRNA silencing. RNA-seq of the input and endogenous Ago2-RIP then allows simultaneous, transcriptome-wide analysis of the expression and RIP-fraction changes for each mRNA (Figure 3A).
For our analysis, we selected the top three expressed miRNA-families: the let-7 family, miR-21 and miR-199–3p (Supplementary Figure S3B) and performed parallel Ago2-RIP experiments with and without miRNA silencing. As an additional negative control, we used an miRNA which was weakly expressed under these conditions (miR-16, Supplementary Figure S3B) and a mix of all antimiRs was used as an additional positive control for filtering of top candidates (see below). In a first step, we performed a conventional analysis of mRNA regulation based on the presence of conserved seeds and their complementarity. As expected, we found modest upregulation of targets, confirming successful inhibition for the three miRNAs of interest and no regulation for miR-16 (Figure 3B).
To test whether increased target regulation is in fact associated with preferential binding, we then binned messages based on their (log2) fold changes upon miRNA silencing separately for predicted targets (conserved 7- or 8-mer site) and non-targets (Figure 4A). We thereby selected the cutoffs of <0.15 for no or weak regulation, 0.15–0.5 for average regulation, >0.5 for strong regulation (for let-7 further separated into 0.5–1 and >1 due to sufficiently high number of regulated targets). This resulted in homogenous distributions of observed fold changes for targets and non-targets in each bin (Figure 4A, bottom panels). Interestingly, the Ago2-RIP fold changes showed marked (median change >0.8) and highly significant differences for targets versus non-targets preferentially in the bin(s) that group the most strongly regulated targets. This observation was consistent for all three highly expressed miRNAs, but not for the poorly expressed miR-16 (Figure 4A, top panels). Furthermore, a clear progression in RIP fold changes was observable from the ‘0.15–0.5’ to ‘>0.5’ bins specifically for targets (median change >0.7). This confirms that strong target de-repression is associated with markedly higher association with the respective miRNA. Targets with conserved sites were consistently enriched in the highly regulated bins (compared with mRNAs without sites) for all miRNAs (pχ2 < 0.01), again with the exception of miR-16.
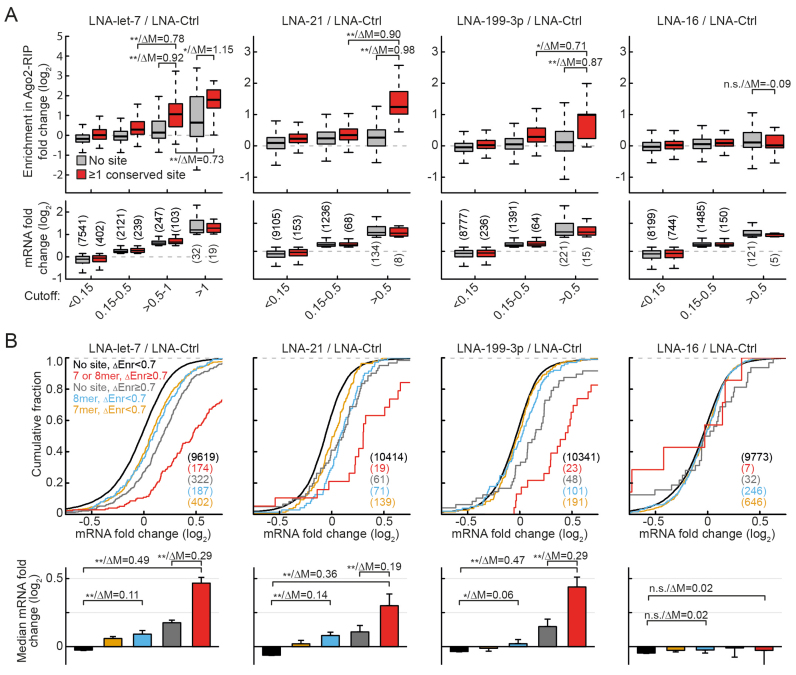
Figure 4.
Transcriptome-wide analysis shows strong correlation between preferential regulation and preferential binding. (A) Boxplots of fold changes in RIP-enrichment (ΔEnrichment) upon miRNA inhibition depicted separately for conserved targets and non-targets (no 7- or 8-mer seed match in the 3′-UTR). MRNAs were binned based on their expression fold changes (depicted as boxplots in the lower row). (B) Distributions of mRNA fold changes for the indicated conserved target and non-target groups as described in the main text. Panels below depict median mRNA fold changes with bootstrap standard errors. For group comparisons P-value (*P < 0.05, **P < 0.01 in a Mann–Whitney U-Test) and the difference of medians of log2 transformed data are indicated separated by a slash. N-numbers are indicated in parentheses.
This parallel experimental design and the way of data representation used in Figure 4A allowed us to make several important observations. First, although the inhibition of miR-16 does not induce a measurable shift in target level distribution (Figure 3B and 4B), there are some predicted targets and non-targets meeting the cutoff for strong regulation (Figure 4A, LNA-16/LNA-Ctrl panels). We expect that such messages are false positives resulting from selecting extreme values from a distribution of a total of >10 000 interrogated candidates. This is confirmed by the fact that as mentioned above targets are not enriched in the highest bin compared with non-targets for miR-16. Importantly, at the same time only minimal deviation from 0 (≤0.11 median deviation) is observed for fold changes in RIP, showing that the two parameters are technically highly independent (the influence of extreme measurements in mRNA fold change on the outcome of RIP fold change is minimal). Furthermore, this observation suggests that false positives can also be expected in higher bins for the remaining miRNAs, leading to an underestimation of the actual change in Ago2-RIP for strongly regulated targets.
The strong and consistent difference between predicted targets and non-targets with a cutoff >0.5 for the top three miRNAs also suggests that preferential binding and regulation occurs primarily at canonical seed-complementary sites.
Changes in Ago2-RIP predict regulation for canonical and non-canonical targets
To test whether changes in Ago2-RIP can be used to predict strong mRNA regulation, we grouped targets and non-targets based on the RIP value. We selected 0.7 as a threshold, as it corresponds to the top 2.5 percentile and furthermore is in the range of what is observed as median change between average and strongly regulated targets (see Figure 4A). To further reduce the number of false positives we applied additional filters based on RIP values in the LNA-Mix and LNA-16 groups as described in ‘Supplementary Materials and Methods’ section.
This resulted in clear separation of a group of strongly regulated conserved canonical targets for each of the three top miRNAs (Figure 4B, red curves). Also for messages without canonical sites, we observed a substantial right-shift in distribution (Figure 4B, gray curves). Again for miR-16, although messages meeting the threshold were present, they showed no significant shift, suggesting that they were in fact false positives. This implies that also the curves for the other miRNAs are a result of an overlay of truly bound targets and of false positives, which leads to an underestimation of the actual regulation of both preferentially bound canonical and even more so for non-canonical targets. Interestingly, despite higher expression (Supplementary Figure S3B) and measured activity of let-7 compared with miR-21 and -199–3p their average regulation of strongly bound targets was highly similar (Figure 4B bottom panels). This suggests that less active miRNAs are in fact capable of strong target regulation comparable with that of let-7, however for a lower number of targets.
Overall these data confirm preferential regulation of canonical and to some extent non-canonical targets with higher miRNA-bound fraction and provide evidence that under native conditions in living cells there exist widely differing endogenous apparent affinities for targets of the same miRNA. Notably, a prediction of strong regulation based on the presence of conserved sites combined with Ago2-RIP enrichment changes (red curves in Figure 4B) is on par with top predictions obtained so far for target upregulation upon miRNA silencing (i.e. combination of TargetScan 7 and dCLIP in wt versus miR-155 knockout lymphocytes (14)).
Quantitative proteomics and transcriptomics from miR-manipulated cells and tissues confirm exceptional regulation of preferentially bound targets
As observed in Figure 4, both a selection based on mRNA as well as RIP values results in a considerable number of false positives (up to 20% for canonical and >50% for non-canonical targets). As both parameters are likely independent predictors of preferential binding, we selected a subgroup (termed ‘top candidates’), based on a combination of the two. For this, we used a threshold of 0.7 for both and additional quality filters as described in ‘Supplementary Materials and Methods’ section. Notably, the presence of seed complementarity was disregarded. None of the mRNAs passed these filters as a potential target of miR-16, confirming high specificity as miR-16 was de facto non-functional under these conditions.
We next assessed, whether the pattern of preferential regulation of these candidates is reflected on the protein level. We used quantitative proteomics (“stable isotope labeling with amino acids in cell culture" or SILAC) analysis after inhibition and overexpression of miR-199–3p in NIH3T3 cells (Figure 5A), thereby detecting ≈4000 proteins for each condition. While in the overexpression experiments, a marked downregulation of a large number of predicted targets was observed (Figure 5B, left panel), their upregulation upon inhibition of miR-199–3p was much weaker (Figure 5B, blue and yellow curves in the right panel), matching the modest de-repression observed on the mRNA level (Figure 3B). This confirms that the overall relative saturation of miR-199–3p targets is low under baseline conditions, as it can be strongly increased by mimic transfection. However, for the top candidates we observed an exceptionally high de-repression (red curves in Figure 5B). This applied to each of the four top candidates (out of nine total) detected in this assay, resulting in a median upregulation 10- to 12-fold higher than for other targets. As an additional control, we selected mRNAs, which showed strong regulation (mRNA fold-change >0.7), yet a RIP-enrichment change below threshold (<0.7). Of these, five were detected and showed very weak protein regulation (Figure 5B, gray curve in the right panel), making them likely false positives. This confirms, that the simultaneous assessment of RIP-enrichment changes and mRNA de-repression effectively predicts exceptionally strongly regulated targets.
Figure 5.
Proteome and tissue analysis confirms preferential regulation of predicted preferential targets at endogenous miRNA levels. (A) Experimental design for SILAC proteomics analyses of miR-199–3p targets in NIH3T3 fibroblasts. (B) Protein fold changes for the indicated target and control groups after miR-199–3p overexpression (miR-mimic, left) or knockdown (antagomiR, right). Predicted top candidates, which were identified in the knockdown assay, are indicated in red. Top Ctrl. indicates targets with an mRNA fold change above and RIP-enrichment fold change below the threshold. (C) As in (B) for mRNA fold changes after introduction of miR-199–3p in neonatal mouse cardiac myocytes (28). (D) As in (B) for average mRNA fold changes between wild-type (wt) and miR-21 knockout mice, measured by RNA-seq in mammary gland (29) or microarray in blood neutrophils and macrophages (30). Averages were calculated after quantile normalization. Graphs for individual experiments depicted in Supplementary Figure S5. (E) As in (B) for protein fold changes following let-7b knockdown in HeLa cells, measured by pSILAC (31). N-numbers, P-values and median differences indicated as in Figure 4. #P < 0.05 for Top candidates versus Top Ctrl. and P < 0.01 versus all other groups.
Since only two of the top candidates were detected in the SILAC overexpression experiments (compare Supplementary Table S1C), we further analyzed mRNA changes after miR-199–3p overexpression in neonatal mouse cardiac myocytes (28). We thereby observed that the median fold changes of top target mRNAs showed little distinction from other predicted targets in this setting of high miRNA abundance due to overexpression (Figure 5C).
To assess, whether our selection of top candidates can be used to predict strongly regulated targets in complex tissues and different contexts, we analyzed gene regulation in response to miR-21-ablation in mammary gland tissue (mouse, RNA-Seq, (29)), neutrophils and macrophages (mouse, microarray, (30)). We thereby observed consistently stronger upregulation of top candidates compared with other targets (Supplementary Figure S5). Although, as previously reported (30), no significant regulation of regular 8-mer targets was present in neutrophils, interestingly top candidates were still significantly regulated, some of them quite strongly. To reduce variation, we calculated an average of quantile normalized results from the three tissues, which further confirmed preferential regulation of the top candidates (Figure 5D and Supplementary Table S1B).
Finally, we analyzed target regulation following let-7b-inhibition in a human cell line (HeLa, pSILAC, from (31)). Indeed, top candidates of this miRNA again showed stronger de-repression than other target groups (Figure 5E and Supplementary Table S1A). This was especially prominent for higher values of mRNA fold changes, where strong diversion of the curves could be observed. These results suggest some extent of context-specific target regulation, yet confirm the notion that preferential regulation of a low number of strongly bound targets is a widespread phenomenon.
Having two independent transcriptome or proteome readouts for each miRNA allowed us to compare the contribution of the two main predictive parameters (i.e. expression and Ago2-RIP-enrichment fold changes) as predictors of strong target regulation. To this end, we fit logistic regression models to our datasets (Supplementary Table S2), thereby observing consistently superior fits for RIP-enrichment changes, while in all cases the best model incorporated both parameters (see additional legend in Supplementary Table S2).
Preferential regulation cannot be explained by multiple binding sites for the same miRNA
Exceptionally strong regulation by one miRNA is currently often explained by the presence of multiple functional binding sites for the same miRNA on one target. We therefore analyzed whether preferential regulation of the top candidates could be explained in this way. To this end, we quantified the number of seed complementary sites (conserved and non-conserved) in the 3′-UTRs of all groups (Supplementary Figure S6). However, we did non see an enrichment for multiple binding sites within the top candidates, with the exception of a small signal for let-7. We did however observe that the vast majority of the top candidates indeed contained one or two canonical sites, although site content was not taken into account during the selection. This confirms that preferential binding is largely mediated through canonical interactions at specific sites.
To further confirm direct miRNA-mediated binding, we analyzed sequencing coverage in a cross-linking immunoprecipitation assay (iCLIP, (32)). Interestingly we thereby observed unique peaks in iCLIP coverage overlapping the predicted binding site for many of the top candidates (Supplementary Figures S7 and 8). In the absence of other clear coverage peaks on these same transcripts, this further supports that the strong Ago2 association is mediated through preferential binding rather than through additive action of multiple regular sites.
To additionally exclude that preferential binding of top candidates was due to exceptionally strong expression of these mRNAs at baseline, we analyzed their expression in NIH3T3 cells. We thereby observed similar or tendentially decreased expression (Supplementary Figure S9A).
A previous report observed that changes in Ago-RIP were lower for targets containing more binding sites for different miRNAs (27). To exclude that our findings could be explained by a compensatory effect from non-inhibited miRNAs, we plotted the distributions of baseline Ago2-RIP enrichment of targets and non-targets. We thereby observed that the distributions were not truncated for higher values of RIP enrichment (non-targets in Supplementary Figure S9B). Therefore, under the conditions used in our assays binding of additional Ago2 should be detectable even for targets already bound at a different position. Furthermore, top candidates showed significantly higher baseline RIP-enrichment than the remaining 8-mer or 7-mer targets (Supplementary Figure S9B), making it unlikely that the latter two groups are bound by more Ago complexes than the top candidates. Rather, these results suggest that preferential binding is responsible for increased Ago2 association.
DISCUSSION
Taken together, our data provide strong evidence that in vivo, miRNA target sites widely differ in the extent of interaction strength with some sites being bound exceptionally tightly. We propose that this has important consequences on how miRNA and target site abundance interact to determine miRNA function. For exceptionally active miRNAs, which are present in a free and active state in necessary quantities with regard to the average KD, many targets will be sufficiently bound and regulated. We observed this upon overexpression of miR-199–3p or inhibition of let-7, as has been reported for many other miRNAs upon their overexpression. In contrast, the low proficiency typically encountered for most endogenous miRNAs results in a steep gradient of regulation between targets of high and low apparent affinity. We suggest that this gradient ensures preferential regulation of a small portion of the targetome, enabling high specificity of miRNA function.
We see no explanation for such exceptional regulation through current models based on free and unassisted diffusion and dissociation kinetics. Rather our findings strongly argue that one (or possibly several) processes assist in maintaining the miRNA binding to such preferential targets (or reducing the binding to other targets).
The hypothesis that such interactions may be influencing miRNA targeting has been proposed previously (2) and our work successfully addresses the question of how such targets can be systematically identified. An important concept underlying our study is the separation of values for target regulation into at least two different groups, with one group being responsible for the enrichment of strongly regulated targets (Supplementary Figure S1B). In contrast, many previous studies implicitly view the values obtained for target regulation as being drawn from one single distribution. Identifying these strongly regulated targets is however quite difficult if their number (prevalence) is low. In this case, choosing messages solely based on strong regulation (e.g. in RNA-seq) will result in a considerable number of false positives (as observed for miR-16 in Figure 4A, see also Supplementary Table S2, model 1).
As demonstrated in Supplementary Table S2 (model 2), changes in Ago2-RIP were in fact more closely associated with true predictions of strong regulation than mRNA regulation measurement itself, when disregarding canonical seed presence. This is likely due to lower relative variability of Ago2-RIP, and strongly supports the notion that preferential binding determines regulation. Adding mRNA fold change as additional predictive parameter further increased specificity. Importantly, this enabled the identification of targets irrespective of specific sequence features, such as the presence of canonical seed sites. Consequently, some messages without canonical sites passed our thresholds for preferential targeting. A notable example is Dicer1, which has been previously described to be targeted by let-7 (33,34), yet in mouse has no canonical sites. Thus, we conclude that our approach of combining Ago2-RIP and mRNA-fold changes after miRNA inhibition allows for the efficient identification of both canonical and non-canonical strongly regulated targets. The identification of such exceptional targets should enable further studies to investigate the underlying processes of preferential binding.
The analysis of preferential miRNA-target interactions is additionally relevant for the controversy surrounding the validity of the ceRNA hypothesis (5,6,8,35). An important question in this respect is the number of added (or removed) target sites required to induce a measurable change in the expression of other targets of the same miRNA. The reported estimates for 8-mer target sites have ranged between 3000 (6) and >10 000 (5,35) copies per cell, making physiological ceRNA effects highly unlikely and reserved for drastic changes within the top expressed genes in a cell. However, these estimates were calculated under the assumption of homogenous binding between the competitor (ceRNA) and the remaining targets with a similar seed type. Our finding of preferential binding for specific targets could therefore lead to identification of messages which are capable of acting as ceRNAs at endogenous concentrations.
AVAILABILITY AND ACCESSIONS
RNA-seq and proteomics datasets are available through ArrayExpress (E-MTAB-5386) and ProteomeXchange (PXD005569) respectively, as further described in ‘Supplementary Materials and Methods’ section.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Thomas Meitinger and Tim-Matthias Strom from Helmholtz Zentrum München for sequencing RNA-Seq libraries, Angela Weigert Muñoz for help with RIP assays and Alessandro Grodziecki for help with reporter assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Bavarian Ministry of Sciences, Research and the Arts in the framework of the Bavarian Molecular Biosystems Research Network; DZHK (German Center for Cardiovascular Research). Funding for open access charge: DZHK Project Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Krol J., Loedige I., Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P.. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011; 18:1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M.. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014; 54:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosson A.D., Zamudio J.R., Sharp P.A.. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014; 56:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jens M., Rajewsky N.. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2015; 16:113–126. [DOI] [PubMed] [Google Scholar]
- 8. Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P.. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011; 146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomson D.W., Dinger M.E.. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016; 17:272–283. [DOI] [PubMed] [Google Scholar]
- 10. Pinzón N., Li B., Martinez L., Sergeeva A., Presumey J., Apparailly F., Seitz H.. microRNA target prediction programs predict many false positives. Genome Res. 2017; 27:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides Its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V.. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015; 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Golden R.J., Chen B., Li T., Braun J., Manjunath H., Chen X., Wu J., Schmid V., Chang T.-C., Kopp F. et al. . An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017; 542:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Agarwal V., Bell G.W., Nam J.-W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B.. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007; 13:1894–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Werfel S., Nothjunge S., Schwarzmayr T., Strom T.M., Meitinger T., Engelhardt S.. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016; 98:103–107. [DOI] [PubMed] [Google Scholar]
- 17. Mukherji S., Ebert M.S., Zheng G.X.Y., Tsang J.S., Sharp P.A, van Oudenaarden A.. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011; 43:854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu Z., Liu M., Stribinskis V., Klinge C.M., Ramos K.S., Colburn N.H., Li Y.. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008; 27:4373–4379. [DOI] [PubMed] [Google Scholar]
- 19. Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S. et al. . MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008; 456:980–984. [DOI] [PubMed] [Google Scholar]
- 20. Sayed D., Rane S., Lypowy J., He M., Chen I., Vashistha H., Yan L., Malhotra A., Vatner D., Abdellatif M.. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell. 2008; 19:3272–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu M., Wu H., Liu T., Li Y., Wang F., Wan H., Li X., Tang H.. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009; 19:828–837. [DOI] [PubMed] [Google Scholar]
- 22. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T.. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007; 133:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pinder B.D., Smibert C.A.. microRNA-independent recruitment of Argonaute 1 to nanos mRNA through the Smaug RNA-binding protein. EMBO Rep. 2013; 14:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Easow G., Teleman A.A., Cohen S.M.. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007; 13:1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karginov F. V, Conaco C., Xuan Z., Schmidt B.H., Parker J.S., Mandel G., Hannon G.J.. A biochemical approach to identifying microRNA targets. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19291–19296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Androsavich J.R., Chau B.N.. Non-inhibited miRNAs shape the cellular response to anti-miR. Nucleic Acids Res. 2014; 42:6945–6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eulalio A., Mano M., Ferro M.D., Zentilin L., Sinagra G., Zacchigna S., Giacca M.. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012; 492:376–381. [DOI] [PubMed] [Google Scholar]
- 29. Feuermann Y., Kang K., Shamay A., Robinson G.W., Hennighausen L.. MiR-21 is under control of STAT5 but is dispensable for mammary development and lactation. PLoS One. 2014; 9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eichhorn S.W., Guo H., McGeary S.E., Rodriguez-Mias R.A., Shin C., Baek D., Hsu S.hao, Ghoshal K., Villén J., Bartel D.P.. MRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell. 2014; 56:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N.. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008; 455:58–63. [DOI] [PubMed] [Google Scholar]
- 32. Gurtan A.M., Ravi A., Rahl P.B., Bosson A.D., Jnbaptiste C.K., Bhutkar A., Whittaker C.A., Young R.A., Sharp P.A.. Let-7 represses Nr6a1 and a mid-gestation developmental program in adult fibroblasts. Genes Dev. 2013; 27:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tokumaru S., Suzuki M., Yamada H., Nagino M., Takahashi T.. let-7 regulates dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008; 29:2073–2077. [DOI] [PubMed] [Google Scholar]
- 34. Jakymiw A., Patel R.S., Deming N., Bhattacharyya I., Shah P., Lamont R.J., Stewart C.M., Cohen D.M., Chan E.K.L.. Overexpression of dicer as a result of reduced let-7 microRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010; 49:549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M.. Impact of MicroRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell. 2016; 64:565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Filonov G.S., Piatkevich K.D., Ting L.-M., Zhang J., Kim K., Verkhusha V. V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011; 29:759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.