Abstract
Termination of transcription is important for establishing gene punctuation marks. It is also critical for suppressing many of the pervasive transcription events occurring throughout eukaryotic genomes and coupling their RNA products to efficient decay. In human cells, the ARS2 protein has been implicated in such function as its depletion causes transcriptional read-through of selected gene terminators and because it physically interacts with the ribonucleolytic nuclear RNA exosome. Here, we study the role of ARS2 on transcription and RNA metabolism genome wide. We show that ARS2 depletion negatively impacts levels of promoter-proximal RNA polymerase II at protein-coding (pc) genes. Moreover, our results reveal a general role of ARS2 in transcription termination-coupled RNA turnover at short transcription units like snRNA-, replication-dependent histone-, promoter upstream transcript- and enhancer RNA-loci. Depletion of the ARS2 interaction partner ZC3H18 mimics the ARS2 depletion, although to a milder extent, whereas depletion of the exosome core subunit RRP40 only impacts RNA abundance post-transcriptionally. Interestingly, ARS2 is also involved in transcription termination events within first introns of pc genes. Our work therefore establishes ARS2 as a general suppressor of pervasive transcription with the potential to regulate pc gene expression.
INTRODUCTION
Termination of transcription is essential for avoiding RNA polymerases (RNAPs) from invading neighboring transcription units (TUs) (for recent reviews see (1,2)). It is also a prerequisite for liberating RNAPs from DNA for additional rounds of transcription. In mammalian cells, transcription termination of RNAPII, which synthesizes all cellular m7G-capped RNA, is often coupled to the co-transcriptional 3′ end processing of the nascent transcript. This is illustrated by the dependence of proper transcription termination on: (i) transcript cleavage by the 3′ end cleavage and polyadenylation (CPA) complex at polyadenylation (pA) sites of protein-coding (pc) RNAs, (ii) the cleavage of nascent snRNA transcripts by the CPSF73L endonuclease of the integrator complex or (iii) cleavage by the CPSF73 endonuclease of the CPA complex at conserved stem-loops and histone downstream elements of non-polyadenylated replication-dependent histone (RDH) RNAs (3,4). Transcription often terminates hundreds to thousands of nucleotides beyond these sites, probably due to the time needed for RNA cleavage to occur (1). For both short and long TUs, this has complicated an exact determination of the molecular mechanism(s) by which RNAPII transcription is finally terminated, because these long termination regions tend to contain a diverse repertoire of signals, possibly providing a combination of events contributing to transcription termination (1,2,5). Still, it is suggested that RNAPII passage across RNA processing sites induces its transcriptional slow down and that factor exchanges and structural rearrangements within the transcription complex result in the gradual halt of RNAPII, whose release from the DNA template may be aided by the 5′-3′ degradation of the uncapped transcript emanating from the enzyme (1,2). Other features of the genomic template, such as its chromatin state and its sequence composition are also likely effectors of the transcription termination process (6). For example, recent data provide evidence that RNAPII release sites coincide with DNA motifs predicted to favor RNAPII backtracking and arrest (5).
Providing a considerable complication to the overall transcriptional landscape, eukaryotic genomes are pervasively transcribed, both outside of conventional genic regions but also markedly overlapping with traditional gene units in both sense and antisense transcriptional directions (1,2,7). This puts an added pressure on transcription to terminate, to avoid a transcriptional chaos with possible RNAP interference and collision as well as unwanted production of double-stranded RNA. It also necessitates that many of these pervasive transcription events are not linked to the formation of stable RNA, but rather that transcripts are rapidly targeted for decay. Examples here are promoter upstream transcripts (PROMPTs) and enhancer RNAs (eRNAs), whose RNA 3′ end formation is coupled to the 3′-5′ exonucleolytic degradation by the RNA exosome complex (8–11). Although, the CPA and integrator complexes have both been implicated in the termination of PROMPT and eRNA transcription (12–14), the exact mechanism remains to be described, as does why such transcription termination events do not link to stable RNA production.
Coupling of pervasive transcription termination to RNA degradation is better understood in Saccharomyces cerevisiae, where the RNAPII-associating Nrd1p/Nab3p/Sen1p (NNS) complex can be recruited to short sequence motifs in the nascent RNA to elicit RNAPII termination (2). A direct interaction between Nrd1p and the RNA exosome, via its Trf4p/5p-Air1p/2p-Mtr4p polyadenylation (TRAMP) co-factor, then establishes an efficient ‘hand-over’ of the terminated transcript to the nuclear decay machinery (15–19). The NNS complex works most efficiently on short TUs, like those producing cryptic unstable transcripts (CUTs) and small nuclear- and nucleolar-RNAs (sn(o)RNAs) (20). This specificity can be attributed to a generally higher density of NNS complex binding sites within these RNAs as compared to e.g. mRNA 5′ end regions (21), as well as an interaction of Nrd1p with the Ser5-phosphorylated (Ser5p) C-terminal domain (CTD) of RNAPII, which is prevalent early during the transcription process (20).
Mammalian cells appear to lack functionally conserved homologs of Nrd1p and Nab3p. However, the RNA binding ARS2 protein was recently discovered to impact transcription termination of a few selected short mammalian TUs, like U2 snRNA and PROMPT genes (22,23). RNA analyses of ARS2 depleted cells were compatible with a role of the protein in transcription termination (22–24) and extensive affinity capture analysis identified strong physical links of ARS2 with the m7G-cap binding complex (CBC), forming the CBC-ARS2 (CBCA) complex, as well as with the Zn-finger protein ZC3H18 and the nuclear exosome targeting (NEXT) complex, forming the CBC-NEXT (CBCN) complex (22,23,25). Thus, like the NNS complex, ARS2 is physically connected to the RNA exosome and its tight association with the CBC might explain its predominant activity on shorter TUs (23). Unlike the NNS complex, however, ARS2 does not display any apparent selectivity for specific RNA motifs in vivo (26) and its role in transcription termination and 3′ end processing/degradation is presumably coupled to transcript cleavage (22,23), whereas the transcription termination activity of the NNS complex is triggered by the Sen1p helicase without prior breakage of the nascent RNA chain (27). Given these conceptual links to the S. cerevisiae system, a description of any global role of ARS2 in transcription termination-coupled RNA decay has been warranted.
To examine directly the generality and specificity of ARS2 in transcription termination, we employed chromatin immunoprecipitation sequencing (ChIP-seq) experiments of RNAPII in HeLa cells depleted for ARS2 or its interaction partners ZC3H18 and the core exosome subunit RRP40. In addition to a decline in promoter-proximal associated RNAPII at pc genes in ARS2-depleted cells, our experiments uncovered a general genome-wide role of ARS2 in transcription termination downstream of short snRNA, RDH, PROMPT and eRNA TUs. This result was corroborated by RNA sequencing (RNA-seq) data from the same knock-down samples. Our data also revealed that ARS2, together with its interaction partner ZC3H18, is involved in premature transcription termination events within early regions of pc genes, often giving rise to RNA 3′ ends within the first introns of such TUs. These RNAs are exosome substrates and we conclude that ARS2 generally aids in coupling transcription termination of pervasive transcripts to their rapid turnover.
MATERIALS AND METHODS
RNAi
All experiments were carried out in HeLa Kyoto cells treated with siRNAs for 36 h, repeating the initial transfection after 24 h.
ChIP and ChIP-seq
ChIP was performed as described in (28), mapped to human genome release 19 (hg19) and coverage computed using the R package Pasha as described in (28,29). Further details are available as Supplementary Data.
RNA and RNA-seq
RNA was prepared from siRNA-treated HeLa Kyoto cells, RNA-seq libraries prepared and mapped to hg19 using HISAT as described (30). Further details are available as Supplementary Data.
Bioinformatics
Further details of the bioinformatics analysis are available as Supplementary Data.
RESULTS
ARS2 depletion affects the levels of promoter-proximal RNAPII
To interrogate the genome-wide effect of ARS2 on transcription, we conducted RNAPII ChIP-seq analysis of chromatin derived from duplicate HeLa cell samples depleted by RNA interference (RNAi) of ARS2 (siARS2), ZC3H18 (siZC3H18) or RRP40 (siRRP40) (Supplementary Figure S1A and Table S1). As control, ChIP data were collected from cells treated with siRNAs targeting Firefly Luciferase RNA (siFFL). All replicate experiments were generally well correlated (Supplementary Figure S1B). In parallel to the ChIP-seq analysis, HeLa cells subjected to ARS2 or ZC3H18 siRNA-mediated depletion (Supplementary Figure S1C) were subjected to triplicate rRNA-depleted total RNA-seq. Here, RNA samples from cells administered with siRNAs against EGFP (siEGFP) were used as controls. Similar triplicate RRP40-depletion RNA-seq data, and their corresponding siEGFP control samples, collected previously (30), were included for comparison in downstream analysis. All RNA-seq samples displayed good reproducibility and distinct phenotypes for the different factor depletions (Supplementary Figure S1D).
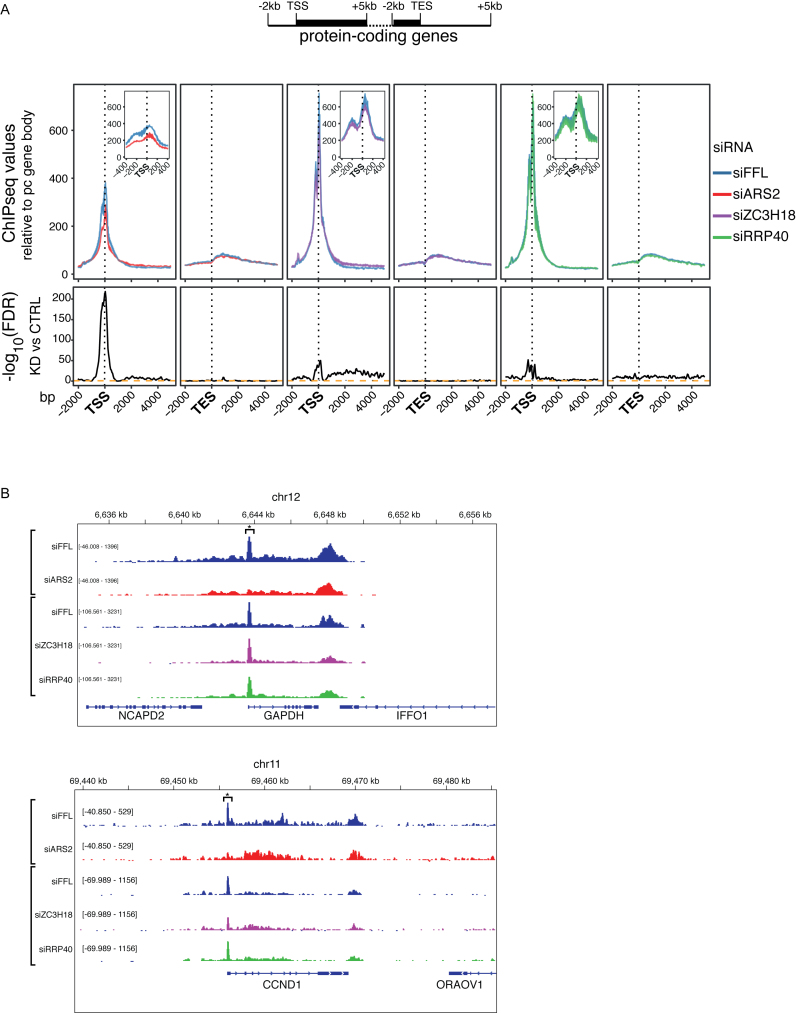
To overview the RNAPII ChIP-seq data, we first plotted average signals from the respective libraries and their controls over regions from 2 kb upstream to 5 kb downstream of the annotated transcription start sites (TSSs) and transcript end sites (TESs), respectively (Figure 1A, top schematics), of the most highly expressed quintile of Refseq annotated pc genes. Data were background subtracted and scaled to the mean signal within gene bodies (Supplementary Figure S1E and ‘Materials and Methods’ section). This revealed the typical pattern of a robust TSS-proximal ChIP peak (31,32) and a weaker accumulation of RNAPII downstream of the TES (Figure 1A and Supplementary Figure S1E). A zoom-in of the TSS region exposed a bimodal RNAPII enrichment, reflecting promoter-proximal stalling of forward (peak at ∼+50 bp) and reverse (peak at ∼−250 bp) transcription complexes (Figure 1A, insets). Reduced RNAPII occupancy at the TSS-proximal positions compared to gene bodies was highly significant in the siARS2 sample, whereas the siZC3H18- and siRRP40 samples showed only weak phenotypes (Figure 1A, bottom panels). This result was observed for many of the expressed genes (Figure 1B, Supplementary Figure S1F and G).
Figure 1.
(A) RNAPII chromatin immunoprecipitation sequencing (ChIP-seq) values from the libraries indicated to the right and plotted in regions 2-kb upstream and 5-kb downstream of TSSs or TESs (see schematics on top with black bars denoting the gene body). The 20% of protein-coding (pc) genes with the highest ChIP-seq promoter signal in the siRNAs targeting Firefly Luciferase (siFFL) library and longer than 7 kb were employed in the analysis (n = 1766). Data from 50 bp bins were background subtracted and scaled to the mean signal within gene bodies (Supplementary Figure S1E). Values shown are mean signals per position computed for each replicate independently with the area in between the two replicates colored. Insets show zoom-ins of data for regions −400 to +400 bp around TSSs. The broader TSS-proximal peaks in the siARS2 experiments are due to somewhat longer DNA fragments sizes (Supplementary Table S1). Lower panel shows the –log10 false discovery rate (FDR), using Benjamini–Hochberg correction of P-values from a two-sided paired Wilcoxon rank sum test performed for each position, comparing the factor depletion library to its respective control. (B) Genome browser screenshots of the pc genes GAPDH and CCND1 and their surrounding annotations. RNAPII ChIP-seq data from single replicates of the siARS2, siZC3H18 and siRRP40 libraries are shown together with a replicate of the siFFL control from the same experiment. Data are color coded as in (A). All tracks are scaled relative to genome coverage and tracks within groups are scaled to the same range. Promoter-proximal stalling peaks are marked with an asterisk.
Since we did not employ external spike-ins, the gene body normalization approach only allowed for relative comparisons. Hence, the siARS2 effect could be interpreted as a decrease in RNAPII levels at gene promoters, as an increase inside gene bodies or both. To distinguish between these possibilities, we took advantage of our RNA-seq data and used changes in intronic reads as a proxy for changes in RNAPII transcription activity (33,34). Hence, we compared log2-fold changes (log2-FCs) between RNAPII ChIP signals for genes showing significantly up or downregulated levels of intronic reads in the corresponding RNA-seq samples. This provided validation of the employed ChIP-seq scaling procedure by demonstrating a strong correlation between RNAPII ChIP-seq and RNA-seq intronic read analyses in the siARS2 and siZC3H18 samples, in that genes found to be down- or upregulated by RNA-seq were also on average down- or upregulated by ChIP-seq analysis (Supplementary Figure S1H). Moreover, consistent with a specific post-transcriptional role of the exosome, the siRRP40 sample showed no significant correlation between ChIP-seq and RNA-seq data. We therefore conclude that ARS2 depletion impacts levels of TSS-associated RNAPII at pc genes, presumably by negatively affecting its promoter loading or its residence time at promoter-proximal stall sites.
snRNA TUs
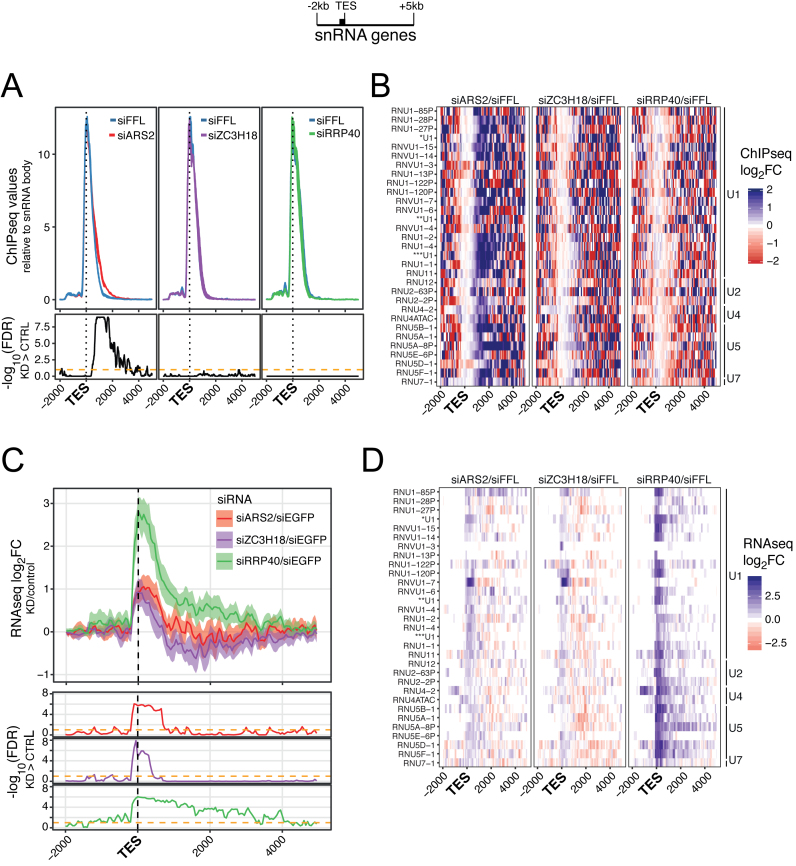
ARS2 was previously implicated in transcription termination downstream of the TESs of the U2 and U4 snRNA genes (22,23). We therefore plotted RNAPII ChIP-seq signal, scaled using snRNA gene bodies for normalization, around the annotated TESs of 31 expressed and non-overlapping snRNA genes (Supplementary Table S2). This revealed a significantly increased RNAPII density, upon ARS2 depletion, in a region app. 1–2 kb downstream of snRNA TESs (Figure 2A, left image). No significant effects were visible upon ZC3H18 or RRP40 depletion (Figure 2A). Anchoring the ChIP-seq data to annotated snRNA TSSs yielded an indistinguishable result and showed that ARS2 depletion had only minor impact on RNAPII peak signals at these genes (Supplementary Figure S2A). The results were confirmed for individual genes (Supplementary Figure S2B) and by heat map representations of the same data (Figure 2B). A few snRNA genes appeared to also display a transcription termination phenotype in siZC3H18 samples, whereas RRP40 depletion did not affect RNAPII profiles.
Figure 2.
(A) RNAPII ChIP-seq values plotted as in Figure 1A but for regions 2 kb upstream and 5 kb downstream the TESs of selected active snRNA genes (n = 31, see schematics on top with black bar denoting the gene body). Values were scaled to gene body signals within the selected snRNA genes (see ‘Materials and Methods’ section). Experiment display as in Figure 1A. Calculation of −log10 FDRs was done as described in Figure 1A, except that a one-sided, paired Wilcoxon rank sum test was used, testing whether ChIP values from depletion samples were higher than the control. (B) Heat map representations of log2FC between ChIP-seq signals of the indicated factor depletion libraries versus their controls. Data were scaled as in (A). The identities of analyzed snRNA genes are shown to the left. The three U1 annotations refer to Gencode annotations ENSG00000277918.1(*), ENSG00000273727.1(**) and ENSG00000273768.1(***). (C) Profile plots of log2FC between mean signals of the indicated triplicate RNA-seq libraries and their controls for the sense strand. Genes and regions as in (A). Values were calculated in 50 bp bins and displayed with their 95% confidence intervals (shaded areas). Lower panels show the −log10 (FDR) as described in (A). (D) Heat map representations and display as in (B) but for RNA-seq libraries, that were tpm normalized as in (C).
We next examined the RNA-seq data and plotted the sensitivity of depleting a given factor as the ratio between relevant read counts from the respective siRNA-treated libraries and their siEGFP control. Using snRNA TESs as data anchoring points, this revealed a strongly stabilized RNA signal from snRNA gene termination regions of siRRP40-treated cells, consistent with previous reports that 3′ extended snRNAs are substrates of the RNA exosome (22,25,26,30) (Figure 2C). Notably, this occurs in the absence of a discernable transcription termination defect in siRRP40 cells (Figure 2A, right image). Heat map representation of the data demonstrated that such exosome-sensitive RNAs are most abundant within 1 kb from the TES, but at times extend downstream of the TES for thousands of nucleotides (Figure 2D). Consistent with previous results, ARS2 and ZC3H18 depletion also led to elevated levels of 3′ extended snRNAs, although more modestly than for the RRP40 depletion (Figure 2C and D). While this confirms a functional relevance for the CBCN linkage in targeting these RNA species, it also suggests that there are alternative ways for delivering these substrates to the exosome. Finally, the siARS2 sensitivity extended further downstream in the snRNA gene termination regions than the siZC3H18 sensitivity (Figure 2C, lower tracks, ∼900 versus ∼450 bp), probably reflecting the dual role of ARS2 in transcription termination and RNA decay. We conclude that ARS2 generally acts in transcription termination downstream of snRNA gene TESs and that this likely leads to RNA exosome targeting.
RDH TUs
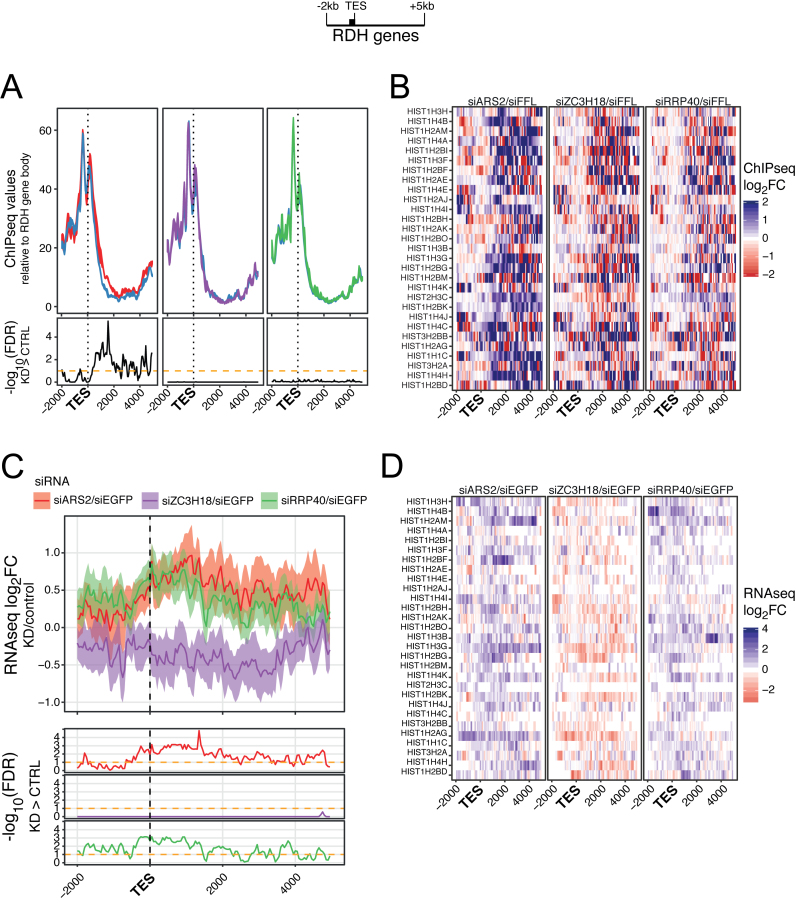
We then turned to RDH genes, another class of short TUs, where the absence of an intact CBCA complex has been shown to increase the abundance of RNAs extending beyond their annotated TESs, often leading to their targeting by the RNA exosome (22–25). To visualize a potential transcription termination defect, we selected 29 expressed and non-overlapping RDH TUs (Supplementary Table S3) and scaled the ChIP data to their gene body regions. Anchoring data to these gene TESs revealed a significant increase of RNAPII in an ∼2-kb downstream region upon ARS2 depletion (Figure 3A). Like for snRNA genes, siZC3H18 and siRRP40 administration displayed only minor or no effect and anchoring ChIP-seq data to annotated TSSs revealed no impact on RNAPII peak signals (Supplementary Figure S3A). Finally, heat map representation of individual RDH genes and single gene inspection demonstrated that the siARS2 effect on transcription termination was general (Figure 3B and Supplementary Figure S3B).
Figure 3.
(A) RNAPII ChIP-seq values plotted as in Figure 1A but for regions 2 kb upstream and 5 kb downstream the TESs of selected active RDH genes (n = 29, see schematics on top with black bar denoting the gene body). Values were scaled to the gene body signal within the selected RDH gene. Experiment display and FDR calculation as in Figure 2A. (B) Heat map representations of log2FC between ChIP-seq signals of the indicated factor depletion libraries versus their controls. Data were scaled as in (A). The identities of analyzed RDH genes are shown to the left. (C) Profile plots of log2FC between mean signals of the indicated triplicate RNA-seq libraries and their controls. Genes and regions as in (A). Calculation and display as in Figure 2C. (D) Heat map representations and display as in (B) but for RNA-seq libraries, that were tpm normalized as in (C).
RNA-seq data displaying the sensitivity of factor depletion mirrored the siARS2 effect seen by RNAPII ChIP (Figure 3C and D). Moreover, they showed that ZC3H18 is probably irrelevant for the turnover of these extended RDH RNA species and that the exosome plays a more modest role than in the case of 3′ extended snRNAs. We conclude that transcription termination within RDH gene terminator regions generally responds to the presence of ARS2 although less strongly than at snRNA genes.
PROMPT and eRNA TUs
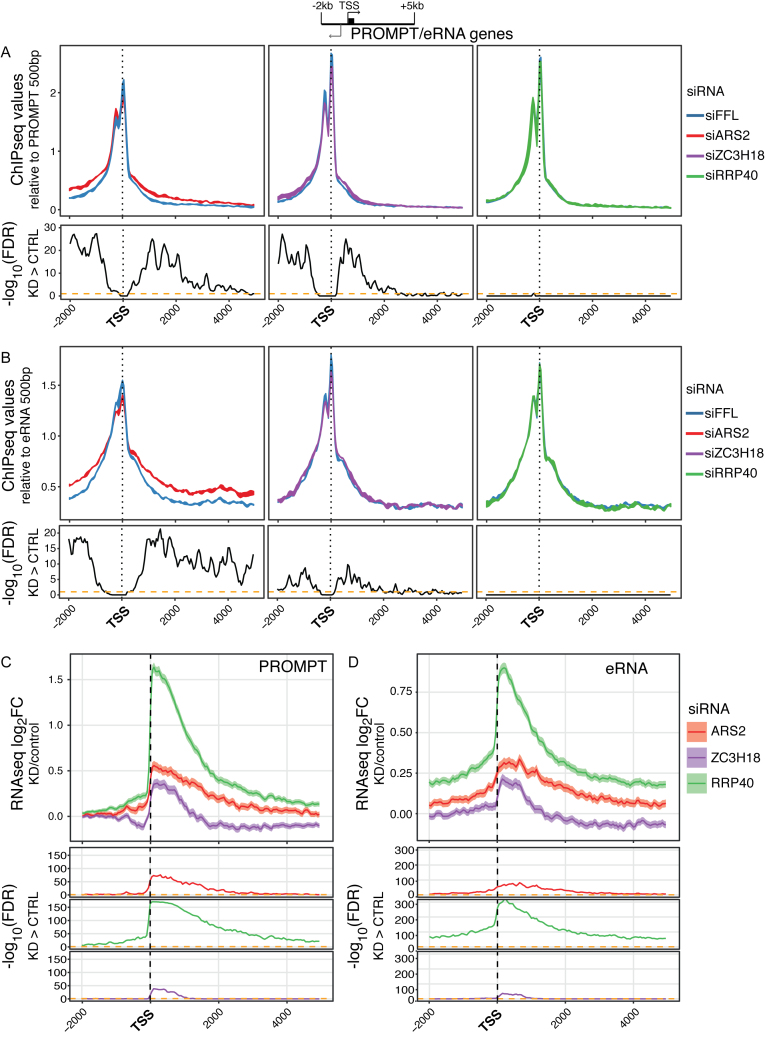
As a final collection of short TUs, we examined regions expressing PROMPTs and eRNAs. Although the biogenesis of these transcripts is normally efficiently coupled to degradation by the RNA exosome (8–11), their transcription termination does not appear to follow a uniform mechanism as both the CPA and integrator complexes have been implicated (12–14). This probably reflects non-uniform terminator/3′ end processing mechanisms for these very labile RNAs. In addition, PROMPT and eRNA 3′ ends are heterogeneous in nature and hence not well annotated (12–14). We therefore anchored the data to the TSSs of 1097 PROMPT and 2552 eRNA TUs as defined by cap analysis of gene expression (9) and scaled the data relative to RNAPII occupancy within the 500 bp TSS-proximal regions. Like for snRNA and RDH genes, depletion of ARS2 resulted in the increased density of RNAPII in a region of a few kb downstream of PROMPT TSSs (Figure 4A, left image). While siRRP40 treatment had no impact on RNAPII levels in such regions, depletion of ZC3H18 yielded a lower but significant effect (Figure 4A, right and middle images). The ARS2-depletion phenotype, which was previously observed for one native and one artificially constructed PROMPT TU (22), was largely visible for all analyzed cases (Supplementary Figure S4A). Moreover, a strikingly similar result was obtained when visualizing RNAPII ChIP sequence reads anchored to eRNA TSSs, which were not previously interrogated in ARS2-depletion conditions (Figure 4B and Supplementary Figure S4B). As for PROMPTs, termination of these divergently transcribed TUs was affected to a lower extent by ZC3H18 depletion while RRP40 depletion had no effect.
Figure 4.
(A and B) RNAPII ChIP-seq values plotted as in Figure 2A but for regions 2 kb upstream and 5 kb downstream the TSSs of selected PROMPTs (A, n = 1097) and eRNAs (B, n = 2552) (see schematics on top). Values were scaled to the ChIP signal within the first 500 bp from the TSS. Experiment displays and FDR calculations as in Figure 2A. (C and D) Profile plots of log2FC between mean signals of the indicated triplicate RNA-seq libraries and their controls. Regions as in (A) and (B). Calculation and display as in Figure 2C.
In contrast, RNA-seq sensitivity data revealed a marked RRP40-sensitivity of both transcript types as previously reported (8–11) (Figure 4C and D; Supplementary Figure S4C and D). A more modest, yet significant, effect was observed for both ARS2 and ZC3H18 depletions. In line with ARS2 exercising a more pronounced effect on RNAPII termination, its depletion led to more elongated transcript stabilization profiles, extending 600–800 bp further than what was observed after ZC3H18 and RRP40 depletion (Supplementary Figure S4E). This is the reminiscent of RNA-seq sensitivity profiles downstream of snRNA TESs (Figure 2C), although much clearer for PROMPT and eRNAs TUs, likely because of their higher sample sizes. Taken together, this confirms a dual role for ARS2 in transcription termination and RNA decay. Interestingly, ZC3H18 also seems to play a similar, though less prominent, role at these TUs.
Protein-coding TUs
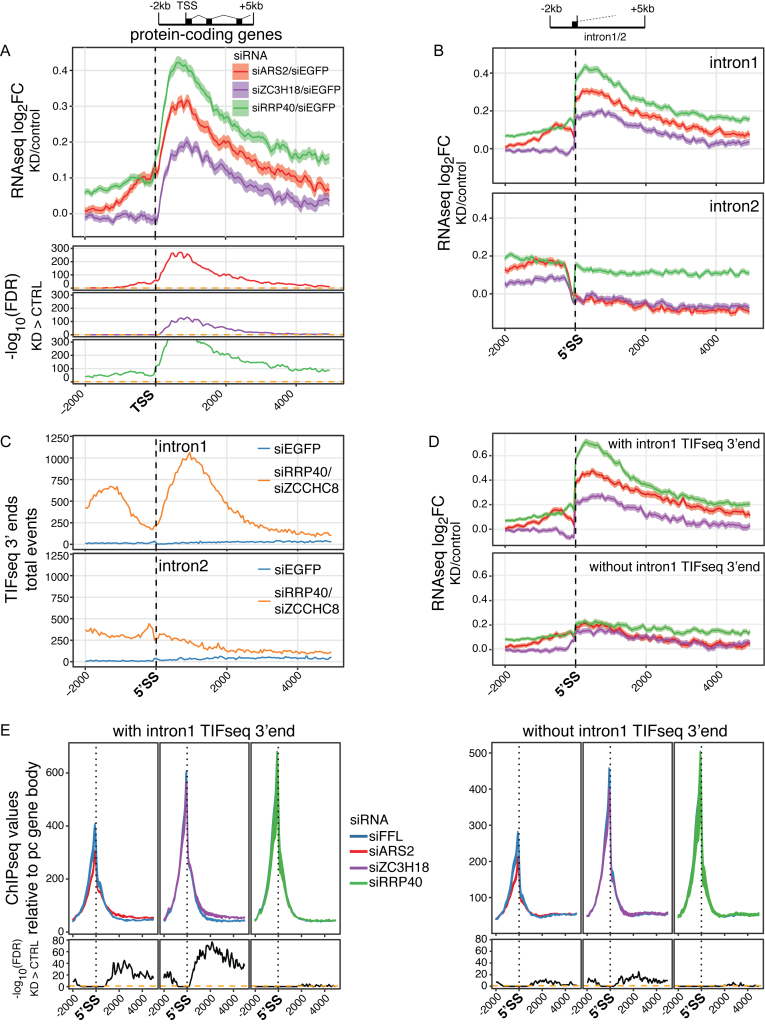
While reverse-oriented PROMPT transcription from mammalian gene promoters is usually terminated rapidly, forward (e.g. mRNA) transcription is generally more elongation competent, which has been attributed to a decreased density of pA-sites downstream of mRNA compared to PROMPT TSSs (12,14,35,36). Moreover, 5′ splice site (5′SS) sequences, capable of binding U1 snRNA and suppressing pA site usage (37), are over-represented in promoter proximal regions downstream of mRNA TSSs largely due to the relatively short length of first exons. However, despite these precautions to delimit transcription termination within pc TUs, cryptic pA sites are to some extent still being utilized (12,37), giving rise to the detection of exosome-sensitive RNA 3′ ends (14). Hence, we decided to examine this phenomenon in more detail using our new datasets. Indeed, RNA-seq sensitivity plots supported a ‘PROMPT-like’ phenotype in these TSS-proximal regions, with prominent siRRP40-, and less pronounced siARS2- and siZC3H18-RNA sensitivity phenotypes largely restricted to a few kb downstream of the mRNA TSSs (Figure 5A and Supplementary Figure S5A). The ‘beginning’ of these sensitivity profiles occurred at distances downstream of the TSS, in contrast to the rather sharp TSS-anchored sensitivities observed for PROMPTs and eRNAs (Figure 4C and D). This is consistent with most spliced mRNAs not being targeted by the RNA exosome, and instead suggested that the sensitive RNA species harbor intronic sequence. To elaborate on this notion, the data were anchored to the 5′ SSs of first and second introns, respectively, which revealed a sharply increased sensitivity at the 5′SS of intron 1 for all three factor depletions, declining with increasing distance from the 5′SS (Figure 5B, top panel). In contrast, neither ARS2 nor ZC3H18 depletion impacted RNA sensitivity at, or downstream of, the 5′SS of intron 2 (Figure 5B, bottom panel). The modest and uniform stabilization of intron 2 sequence upon RRP40 depletion most likely reflects the activity of the RNA exosome in degradation of some pre-mRNA and/or excised introns (38,39). Heat maps sorted by intron lengths (Supplementary Figure S5B and C), profiles of introns scaled by length (Supplementary Figure S5D) and screenshots of individual examples (Supplementary Figure S5E) further confirmed that the RNA sensitivity effect was specific for intron 1 and biased toward RNA 5′ends. The sensitivity was considerably more skewed toward 5′ ends in siRRP40- compared to siARS2 and siZC3H18 samples (Supplementary Figure S5D, left panel), possibly reflecting an additional transcription termination phenotype and/or a deficiency in first intron splicing upon depletion of the latter two factors.
Figure 5.
(A) Profile plots of log2FC between mean signals of the indicated triplicate RNA-seq libraries and their controls for regions 2 kb upstream and 5 kb downstream the TSSs of selected multi-exonic pc genes (n = 7233, see schematics on top with black bars denoting exons). Calculation and display as in Figure 2C. (B) Profile plots as in (A) but anchored at the 5′SSs of first (top) or second (bottom) introns of the set of genes shown in (A). (C) Profile plots of transcript isoform sequencing data (TIF-seq)-defined RNA 3′ends in control (siEGFP) or siRRP40/siZCCHC8 double-deplete libraries over the same regions as in (B). Data were collapsed with single genomic positions only counted once. (D) Profile plots as in top panel of (B) and stratified for pc genes that contain (top) or lack (bottom) RNA 3′ends in the siRRP40/siZCCHC8 TIF-seq data. (E) RNAPII ChIP-seq values plotted as in Figure 1A but anchored to first 5′SS and stratified for pc genes that contain (top) or lack (bottom) first intron RNA 3′ends in the siRRP40/siZCCHC8 TIF-seq data. ChIP values were scaled and treated as in Figure 1A.
To address whether RNA-seq sensitivity in the TSS-proximal parts of first introns derives from available RNA 3′ends, we analyzed transcript isoform sequencing data (TIF-seq) (40) from HeLa cells treated with control siRNA (siEGFP) or cells co-depleted of RRP40 and the NEXT complex component ZCCHC8 (9,25). Consistent with exosome targeting of RNAs arising from premature transcription termination events, the peak of 3′end tags from the siRRP40/siZCCHC8 TIF-seq library, downstream of first intron 5′SSs, fell in the region of declining RNA-seq sensitivity (Figure 5C, top panel; compare to Figure 5B, top panel). In contrast, the region downstream of the second intron 5′SS yielded much fewer TIF-seq 3′tags (Figure 5C, bottom panel). Moreover, TIF-seq reads from the control library did not show any enrichment of 3′ends within introns 1 or 2. This implies that the RRP40-sensitivity profile visualized by RNA-seq data reflects exosome activity toward transcripts ending mostly within the beginning of first introns. Overall, these results are consistent with previous suggestions that cryptic pA sites reside primarily in intronic sequence within 5 kb of pc gene TSSs (37).
Since TIF-seq yields both 5′- and 3′-end identities of individual transcripts due to its dual cap and poly(A) tail enrichment steps, we were able to address whether the exosome-sensitive RNA 3′ends within first introns resulted from transcription events initiating at annotated pc gene TSSs or whether they might derive from overlapping short TUs; e.g. local enhancer-like activities. We therefore compared TIF-seq derived 5′ends of fragments with mate 3′ends mapping at the annotated TES ±100 bp, reflecting full length transcripts (Supplementary Figure S5F), to those with 3′end mates inside first introns, reflecting premature transcription termination (Supplementary Figure S5G). This analysis showed that 5′ends for both classes stem commonly from annotated pc gene TSSs, although downstream starts sites also contribute to prematurely terminated and exosome-sensitive transcripts. Moreover, only transcripts with intronic 3′ends were exosome-sensitive (Supplementary Figure S5F and G). Thus, a subset of correctly initiated transcription events terminate prematurely and yield exosome-sensitive transcripts.
To analyze whether ARS2 is involved in such premature transcription termination, we divided the analyzed pc genes into those exhibiting TIF-seq 3′ends within first introns and those that did not. RNA-seq sensitivity profiles confirmed the production of sensitive transcripts from the former and to an only minor extent from the latter of these gene groups (Figure 5D). Interestingly, similarly anchored RNAPII ChIP-seq data demonstrated increased levels of RNAPII downstream the first 5′SS upon ARS2 and ZC3H18 depletions specifically for the gene set with TIF-seq 3′ends in intron 1 (Figure 5E). We therefore conclude that ARS2 and ZC3H18 partake in early premature transcription termination events of pc genes, giving rise to RNAs, whose intronic 3′ends are targeted by the RNA exosome.
DISCUSSION
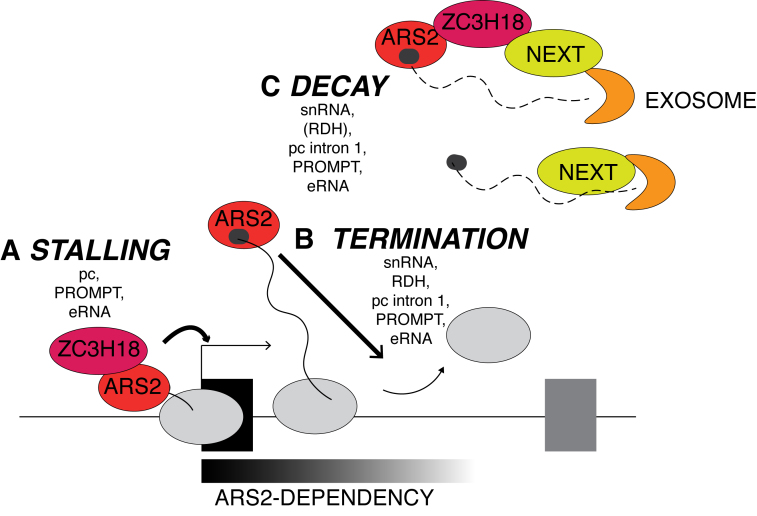
Limiting the pervasive transcription of eukaryotic genomes necessitates efficient transcription termination and its swift coupling to RNA turnover in cases where longer transcript half-lives are not warranted. The ability of ARS2 to interact both with the CBC of nascent transcripts and with the RNA exosome targeting NEXT complex, positions the protein favorably to play an active role in such coupling. Indeed, ARS2 depletion was previously shown to lead to elevated levels of otherwise labile PROMPTs and to reduce the efficiency of transcription termination of a few individual short TUs (22–24). Here, we demonstrate that such ARS2 depletion phenotype extends to a broad variety of short TUs and we therefore suggest that the protein is a general suppressor of pervasive transcription as well as of pervasive transcripts (Figure 6).
Figure 6.
Roles of ARS2 in suppressing pervasive RNA production as described in this paper. (A) ARS2 and ZC3H18 impact promoter-proximal stalling of RNAPII at pc genes as well as PROMPT and eRNA TUs. (B) ARS2 impacts transcription termination downstream of snRNA, RDH, PROMPT and eRNA TUs as well as inside intron 1 of pc genes. (C) ARS2, ZC3H18 and the RNA exosome cooperate in the decay of 3′extended snRNAs, PROMPTs, eRNAs and RNAs with 3′ends inside first introns of pc genes. As indicated, decay of some species may also occur via direct exosome targeting by the NEXT complex. Note also that ZC3H18 appears dispensable for targeting of RDH gene-derived species. The fact that ARS2 appears to preferentially act on short TUs is indicated at the bottom of the figure. See text for details.
Our ChIP data revealed that ARS2 depletion also results in diminished levels of RNAPII at promoter proximal positions of pc genes (Figure 6A). However, the data do not allow discriminating whether this is due to reduced promoter-proximal pausing, increasing elongation or a decrease in RNAPII loading at the promoter. Regarding the latter possibility, we note that a transcription-activating role of ARS2 was previously reported for the Sox2 gene in neural stem cells (41). Due to its impact on transcription termination of even very short TUs, ARS2 presumably associates with the nascent RNA immediately after its 5′end capping and CBC binding. By inference, ARS2 is likely to be present in RNA protein particles (RNPs) forming at capped transcript 5′ends during early stalling of RNAPII (42). Absence of ARS2 here might, directly or indirectly, affect the activity of factors involved in RNAPII stalling or stall site-release. It is for example, interesting to note that ARS2 associates with 7SK RNA (24), which sequesters the early transcription elongation factor P-TEFB in its inactive form (43). P-TEFB has also been reported to associate with the CBC (44). Hence, various models, involving either direct contacts of ARS2 with stalling/elongation factors or the mutual competition of relevant factors for CBC binding (26) can be formulated and are testable to delineate the exact mechanisms underlying the observed effect of ARS2 depletion. It will also be important to elucidate whether this impact of ARS2 on early RNAPII activity contributes to the ensuing reduction of premature termination of pc gene transcription induced by ARS2 depletion. The probability of factors to bind the nascent 5′RNP is expected to depend on RNAPII promoter-proximal residence time and thus likely to influence downstream events (45).
ARS2 and ZC3H18 associate in the CBCN complex, which further recruits the RNA exosome (22). To address any phenotypic relationships of these factors, we therefore included depletions of ZC3H18 and the exosome subunit RRP40 in our experimentation. For all gene classes tested, RRP40 depletion yielded no, or only marginal, changes of RNAPII ChIP signals. Instead, levels of 3′extended snRNAs and RDH RNAs, PROMPTs, eRNAs and short promoter-proximal transcripts within pc TUs all increased upon RRP40 depletion. This is consistent with previous analysis (8,10,11,22,25,46) and establishes the exosome as a strict post-transcriptional player in the suppression of these transcripts, which presumably can be targeted in CBC-dependent and -independent ways (Figure 6C). For PROMPTs, eRNAs and within pc gene units, ZC3H18 depletion mimicked both transcriptional and post-transcriptional effects of ARS2 depletion (Figure 6A), although to a lesser extent, and for snRNA genes, ZC3H18 depletion only yielded a detectable effect at the RNA level. Since ARS2 protein levels are not affected by ZC3H18 knock-down (22), we suggest that the ZC3H18 depletion phenotype is related to its direct physical association with ARS2 (26), and that its more moderate impact might be due to the presence of functionally redundant factors or the possible shielding of its depletion phenotype by compensatory mechanisms.
As demonstrated in this paper, and consistent with earlier findings, ARS2 depletion-induced transcription termination phenotypes appear to be restricted to rather short TUs (22,23) (Figure 6B). That is, no noticeable transcription defects could be detected downstream of annotated mRNA TESs generally positioned quite distal from their partner TSSs (data not shown). Perhaps ARS2 function is more effective in TSS-proximal regions owing to its intimate link to the CBC. In addition, 3′end processing and transcription termination at the ends of conventional pc genes are efficiently aided by an optimally modified CTD of the largest RNAPII subunit, perhaps making ARS2 activity inconsequential in these cases. Such ability of more efficient 3′end processing reactions to cancel out ARS2 activity may also be reflected by the relatively modest effect of ARS2 depletion on both RNA and RNAPII levels downstream the TESs of our analyzed RDH genes. These often occur in clusters requiring efficient 3′end formation/transcription termination to avoid cross-interference and RDH RNA 3′end processing factors locally concentrate to facilitate effective 3′end formation of these transcripts (47), which may compensate for the short stature of these genes.
The above considerations beg the question: which DNA/RNA signals do ARS2 actually react to? Although a precise answer is still elusive, it is interesting to note that: (i) ARS2 does not appear to be a bona fide member of any known 3′end processing machinery, (ii) ARS2 depletion affects TUs that are presumably employing diverse terminators and (iii) ARS2 depletion also impacts transcript decay by coupling to the NEXT complex. Thus, it is tempting to speculate that the protein exercises its function at 3′end processing sites/transcription terminators that are positioned in non-optimal settings. In the case of PROMPTs, harboring TSS-proximal pA sites, ARS2 may respond to these because of an inefficient CPA complex, functioning non-optimally due to unfavorable RNAPII post-translational modifications. For eRNAs, harboring TSS-proximal integrator sites, these may not operate ideally due to inefficient promoter-terminator pairing, which is known to be important for proper integrator processing at e.g. snRNA 3′ends (48). In such a model, ARS2 would facilitate transcription termination and transcript turnover early in the transcription process, where RNAPII has not yet gained sufficient transcription elongation capacity and where terminator-like signals are not presented in a context yielding stable RNA production (Figure 6). An accessory role of ARS2 is also consistent with the fact that the transcription termination phenotype triggered by ARS2-depletion is mild compared to the phenotypes instigated by CPSF73- or CPSF73L-depletions at their respective targets (13,49). Such ‘fragile’ RNAPII complexes have been suggested to exist in S. cerevisiae (50). In this context, RNA splicing may be viewed as an event improving RNAPII elongation capacity, consistent with our observation that ARS2 responsiveness within pc genes decreases upon splicing of the first intron; i.e. RRP40-sensitive RNA 3′ends are more plentiful in first than second intronic regions.
This last point again highlights, that transcription, even in the context of pc gene sequences, quite frequently terminates prematurely (12,14,37). We demonstrate here that the derived exosome-sensitive RNA 3′ends most often arise from transcription events initiating at, or close to, annotated TSSs, ruling out any cryptic intergenic transcription. This phenomenon of transcription attenuation is well known from S. cerevisiae where e.g. the NNS complex autoregulates Nrd1p levels by terminating NRD1 gene transcription prematurely when Nrd1p levels are high (51). Such gene regulation also appears to play a more general role upon changing growth conditions in S. cerevisiae (52). Determining the extent to which this occurs in higher eukaryotes and whether ARS2 is a central factor in such regulation will be a matter of future analysis.
AVAILABILITY
Code for all bioinformatics analysis is available at GitHub (https://github.com/manschmi/arsRtools).
ACCESSION NUMBERS
All NGS data are available at GEO. Published RNA-seq and TIF-seq data were obtained from GSE84172 and GSE75183. ChIP-seq data and RNA-seq data from this study were deposited to GSE99344 and GSE99059, respectively.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
ERC (339953 to T.H.J.); Danish National Research Council (to THJ); Lundbeck Foundation (to THJ); Novo Nordisk Foundation (to THJ); FRM (to J-C.A.); Ligue nationale contre le cancer (to J-C.A.); INCA (to J-C.A.); Ligue Nationale Contre le Cancer (to E.B.). Funding for open access charge: FP7 Ideas: European Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Proudfoot N.J. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science. 2016; 352:6291–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Porrua O., Boudvillain M., Libri D.. Transcription termination: variations on common themes. Trends Genet. 2016; 32:508–522. [DOI] [PubMed] [Google Scholar]
- 3. Baillat D., Wagner E.J.. Integrator: surprisingly diverse functions in gene expression. Trends Biochem. Sci. 2015; 40:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duronio R.J., Marzluff W.F.. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 2017; 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwalb B., Michel M., Zacher B., Fruhauf K., Demel C., Tresch A., Gagneur J., Cramer P.. TT-seq maps the human transient transcriptome. Science. 2016; 352:1225–1228. [DOI] [PubMed] [Google Scholar]
- 6. O’Reilly D., Kuznetsova O.V., Laitem C., Zaborowska J., Dienstbier M., Murphy S.. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res. 2014; 42:264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jensen T.H., Jacquier A., Libri D.. Dealing with pervasive transcription. Mol. Cell. 2013; 52:473–484. [DOI] [PubMed] [Google Scholar]
- 8. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y., Pai A.A., Herudek J., Lubas M., Meola N., Jarvelin A.I., Andersson R., Pelechano V., Steinmetz L.M., Jensen T.H. et al. Principles for RNA metabolism and alternative transcription initiation within closely spaced promoters. Nat. Genet. 2016; 48:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flynn R.A., Almada A.E., Zamudio J.R., Sharp P.A.. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M.S., Mapendano C.K., Schierup M.H., Jensen T.H.. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008; 322:1851–1854. [DOI] [PubMed] [Google Scholar]
- 12. Almada A.E., Wu X., Kriz A.J., Burge C.B., Sharp P.A.. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013; 499:360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lai F., Gardini A., Zhang A., Shiekhattar R.. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015; 525:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ntini E., Jarvelin A.I., Bornholdt J., Chen Y., Boyd M., Jorgensen M., Andersson R., Hoof I., Schein A., Andersen P.R. et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013; 20:923–928. [DOI] [PubMed] [Google Scholar]
- 15. Arigo J.T., Eyler D.E., Carroll K.L., Corden J.L.. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006; 23:841–851. [DOI] [PubMed] [Google Scholar]
- 16. Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D.. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006; 23:853–864. [DOI] [PubMed] [Google Scholar]
- 17. Tudek A., Porrua O., Kabzinski T., Lidschreiber M., Kubicek K., Fortova A., Lacroute F., Vanacova S., Cramer P., Stefl R. et al. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol. Cell. 2014; 55:467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vasiljeva L., Buratowski S.. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell. 2006; 21:239–248. [DOI] [PubMed] [Google Scholar]
- 19. Wlotzka W., Kudla G., Granneman S., Tollervey D.. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011; 30:1790–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Porrua O., Libri D.. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol. 2015; 16:190–202. [DOI] [PubMed] [Google Scholar]
- 21. Cakiroglu S.A., Zaugg J.B., Luscombe N.M.. Backmasking in the yeast genome: encoding overlapping information for protein-coding and RNA degradation. Nucleic Acids Res. 2016; 44:8065–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andersen P.R., Domanski M., Kristiansen M.S., Storvall H., Ntini E., Verheggen C., Schein A., Bunkenborg J., Poser I., Hallais M. et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 2013; 20:1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hallais M., Pontvianne F., Andersen P.R., Clerici M., Lener D., Benbahouche Nel H., Gostan T., Vandermoere F., Robert M.C., Cusack S. et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013; 20:1358–1366. [DOI] [PubMed] [Google Scholar]
- 24. Gruber J.J., Olejniczak S.H., Yong J., La Rocca G., Dreyfuss G., Thompson C.B.. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell. 2012; 45:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H.. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 2011; 43:624–637. [DOI] [PubMed] [Google Scholar]
- 26. Giacometti S., Benbahouche N.E., Domanski M., Robert M.C., Meola N., Lubas M., Bukenborg J., Andersen J.S., Schulze W.M., Verheggen C. et al. Mutually exclusive CBC-containing complexes contribute to RNA fate. Cell Rep. 2017; 18:2635–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Porrua O., Libri D.. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat. Struct. Mol. Biol. 2013; 20:884–891. [DOI] [PubMed] [Google Scholar]
- 28. Fenouil R., Cauchy P., Koch F., Descostes N., Cabeza J.Z., Innocenti C., Ferrier P., Spicuglia S., Gut M., Gut I. et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012; 22:2399–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fenouil R., Descostes N., Spinelli L., Koch F., Maqbool M.A., Benoukraf T., Cauchy P., Innocenti C., Ferrier P., Andrau J.C.. Pasha: a versatile R package for piling chromatin HTS data. Bioinformatics. 2016; 32:2528–2530. [DOI] [PubMed] [Google Scholar]
- 30. Meola N., Domanski M., Karadoulama E., Chen Y., Gentil C., Pultz D., Vitting-Seerup K., Lykke-Andersen S., Andersen J.S., Sandelin A. et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell. 2016; 64:520–533. [DOI] [PubMed] [Google Scholar]
- 31. Koch F., Fenouil R., Gut M., Cauchy P., Albert T.K., Zacarias-Cabeza J., Spicuglia S., de la Chapelle A.L., Heidemann M., Hintermair C. et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 2011; 18:956–963. [DOI] [PubMed] [Google Scholar]
- 32. Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A.. c-Myc regulates transcriptional pause release. Cell. 2010; 141:432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gaidatzis D., Burger L., Florescu M., Stadler M.B.. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 2015; 33:722–729. [DOI] [PubMed] [Google Scholar]
- 34. Jonkers I., Kwak H., Lis J.T.. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014; 3:e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Core L.J., Martins A.L., Danko C.G., Waters C.T., Siepel A., Lis J.T.. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014; 46:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duttke S.H., Lacadie S.A., Ibrahim M.M., Glass C.K., Corcoran D.L., Benner C., Heinz S., Kadonaga J.T., Ohler U.. Human promoters are intrinsically directional. Mol. Cell. 2015; 57:674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G.. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010; 468:664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lubas M., Andersen P.R., Schein A., Dziembowski A., Kudla G., Jensen T.H.. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 2015; 10:178–192. [DOI] [PubMed] [Google Scholar]
- 39. Valen E., Preker P., Andersen P.R., Zhao X., Chen Y., Ender C., Dueck A., Meister G., Sandelin A., Jensen T.H.. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat. Struct. Mol. Biol. 2011; 18:1075–1082. [DOI] [PubMed] [Google Scholar]
- 40. Pelechano V., Wei W., Steinmetz L.M.. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013; 497:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andreu-Agullo C., Maurin T., Thompson C.B., Lai E.C.. Ars2 maintains neural stem-cell identity through direct transcriptional activation of Sox2. Nature. 2011; 481:195–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mandal S.S., Chu C., Wada T., Handa H., Shatkin A.J., Reinberg D.. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:7572–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y., Liu M., Chen L.F., Chen R.. P-TEFb: finding its ways to release promoter-proximally paused RNA Polymerase II. Transcription. 2017; doi:10.1080/21541264.2017.1281864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lenasi T., Peterlin B.M., Barboric M.. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb). J. Biol. Chem. 2011; 286:22758–22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H.. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007; 26:349–365. [DOI] [PubMed] [Google Scholar]
- 46. Preker P., Almvig K., Christensen M.S., Valen E., Mapendano C.K., Sandelin A., Jensen T.H.. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011; 39:7179–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tatomer D.C., Terzo E., Curry K.P., Salzler H., Sabath I., Zapotoczny G., McKay D.J., Dominski Z., Marzluff W.F., Duronio R.J.. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J. Cell Biol. 2016; 213:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Egloff S., O’Reilly D., Murphy S.. Expression of human snRNA genes from beginning to end. Biochem. Soc. Trans. 2008; 36:590–594. [DOI] [PubMed] [Google Scholar]
- 49. Nojima T., Gomes T., Grosso A.R., Kimura H., Dye M.J., Dhir S., Carmo-Fonseca M., Proudfoot N.J.. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell. 2015; 161:526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Milligan L., Huynh-Thu V.A., Delan-Forino C., Tuck A., Petfalski E., Lombrana R., Sanguinetti G., Kudla G., Tollervey D.. Strand-specific, high-resolution mapping of modified RNA polymerase II. Mol. Syst. Biol. 2016; 12:874–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arigo J.T., Carroll K.L., Ames J.M., Corden J.L.. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell. 2006; 21:641–651. [DOI] [PubMed] [Google Scholar]
- 52. Bresson S., Tuck A., Staneva D., Tollervey D.. Nuclear RNA decay pathways aid rapid remodeling of gene expression in yeast. Mol. Cell. 2017; 65:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.