Abstract
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a broadly expressed lncRNA involved in many aspects of cellular processes. To further delineate the underlying molecular mechanism, we employed a high-throughput strategy to characterize the interacting proteins of MALAT1 by combining RNA pull-down, quantitative proteomics, bioinformatics, and experimental validation. Our approach identified 127 potential MALAT1-interacting proteins and established a highly connected MALAT1 interactome network consisting of 788 connections. Gene ontology annotation and network analysis showed that MALAT1 was highly involved in five biological processes: RNA processing; gene transcription; ribosomal proteins; protein degradation; and metabolism regulation. The interaction between MALAT1 and depleted in breast cancer 1 (DBC1) was validated using RNA pull-down and RNA immunoprecipitation. Further mechanistic studies reveal that MALAT1 binding competes with the interaction between sirtuin1 (SIRT1) and DBC1, which then releases SIRT1 and enhances its deacetylation activity. Consequently, the deacetylation of p53 reduces the transcription of a spectrum of its downstream target genes, promotes cell proliferation and inhibits cell apoptosis. Our results uncover a novel mechanism by which MALAT1 regulates the activity of p53 through the lncRNA–protein interaction.
INTRODUCTION
Over the last decade, transcriptome analysis revealed that only 1–2% of the genome serves as a template for protein biogenesis, whereas up to 80% of the genome is transcribed (1–3). The vast majority of the human genome is pervasively transcribed into non-coding RNAs. Long non-coding RNAs (lncRNAs) are defined as transcripts of more than 200 nucleotides without evident protein coding functionality (4). Increasing numbers of studies have shown that lncRNAs are actively involved in a large spectrum of biological processes. For example, lncRNAs are believed to regulate protein expression at epigenetic, transcriptional, and post-transcriptional levels (5); they also participate in chromatin modification, X-chromosome inactivation, and genomic imprinting (6, 7). Furthermore, alterations in the expression levels of lncRNAs as well as their interactions with other biological molecules clearly regulate a diverse set of physiological and pathological processes, including proliferation (8), apoptosis (9), metastasis (10, 11), metabolism (12), drug-resistance (13), and cancer-related inflammation (14).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a broadly expressed lncRNA with a length of ∼8000 nt. MALAT1 was initially discovered as a prognostic marker for cancer metastasis in non-small cell lung cancer (15). In addition to being involved in regulating cellular differentiation in endothelial cells (16), MALAT1 is up-regulated in various cancer cell types, including hepatocellular carcinoma (HCC) (17–19). MALAT1 overexpression promotes cell proliferation, invasion and migration in vitro and in vivo (17, 20). Moreover, the injection of an antisense oligonucleotide targeting MALAT1 inhibits cancer metastasis in a xenograft mouse model of human lung cancer (21). MALAT1 is believed to act by regulating RNA processing and gene transcription. MALAT1 has been implicated in regulating RNA splicing through interacting with several splicing factors, such as serine/arginine-rich splicing factor 1 (SRSF1) and serine/arginine-rich splicing factor 3 (SRSF3) (22, 23). Furthermore, MALAT1 can interact with unmethylated polycomb 2 (Pc2) protein to control the expression of cell cycle genes in HeLa cells (24). In dedifferentiating breast cancer cells, MALAT1 binds with ELAV-like protein 1 (ELAVL1) and forms a chromatin regulatory complex to repress CD133 expression (25). Therefore, to establish a large-scale lncRNA–protein interaction network associated with MALAT1 would help illuminate its functions and uncover the underlying molecular mechanisms.
In this study, we employed a quantitative proteomics strategy to characterize the lncRNA MALAT1 interactome. The lncRNA–protein complexes were first isolated through RNA pull-down using an in vitro-transcribed MALAT1 RNA chain. Quantitative proteomics, based on stable isotope labeling by amino acids in cell culture (SILAC), was applied to distinguish non-specific binding proteins in the RNA pull-down samples. Next, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) separation and LC–MS/MS analyses were used to identify and quantify the isolated proteins. Bioinformatics analysis was performed to further analyze the mass spectrometry data. Using this approach, we successfully identified 127 potential interacting proteins and established a highly connected interactome network associated with lncRNA MALAT1. We identified and verified a novel MALAT1-interacting protein, depleted in breast cancer 1 (DBC1), and investigated its molecular mechanism.
MATERIALS AND METHODS
Antibodies and reagents
Dithiothreitol (DTT), iodoacetamide (IAA), urea, recombinant His-tagged sirtuin 1 (rSIRT1), and mouse monoclonal against Flag were from Sigma (St. Louis, MO, USA); mouse monoclonal against GAPDH were from Santa Cruz (Santa Cruz, CA, USA); Rabbit polyclonal antibodies against DBC1, heterogeneous nuclear ribonucleoprotein H1 (hnRNP H1), insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), splicing factor 3B subunit 3 (SF3B3), transcriptional activator protein Pur-alpha (PURA), and SIRT1 were from Proteintech (Chicago, IL, USA). Rabbit polyclonal antibody against acetyl-p53 (K382) and p53 were from Cell Signaling Technology (Danvers, MA, USA). His-tagged recombinant DBC1 (rDBC1), DBC1-N3 (aa55–120), and DBC1-N4 (aa120–280) were purified from Escherichia coli by GenScript (Nanjin, China). Recombinant lactate dehydrogenase B (rLDHB) was bought from Abcam (Cambridge, MA, USA). Dynabeads® His-tag isolation kit, Lipofectamine 2000, BCA reagents, Protein A and G magnetic beads were purchased from Invitrogen (Grand Island, NY, USA). Enhanced chemiluminescence (ECL) reagents were purchased from Pierce Biotechnology (Rockford, IL, USA). Protease Inhibitor Cocktail tablets were purchased from Roche Diagnostics (Indianapolis, IN, USA). Sequencing grade modified trypsin was purchased from Promega (Madison, WI, USA). LC–MS grade acetonitrile was from Merck (White-house Station, NJ, USA). Water used in this study was deionized using a Milli-Q purification system (Millipore, Billerica, MA,USA).
Cell culture and SILAC labelling
HepG2 cells were maintained in our laboratory using the SILAC Dulbecco's modified Eagle's medium (DMEM) cell culture kit (ThermoFisher Scientific, Waltham, MA,USA). The cells were cultured in SILAC DMEM supplemented with 10% dialyzed fetal bovine serum and 1% penicillin–streptomycin (100 μg/ml, Invitrogen, NY, USA) at 37°C in a humidified atmosphere with 5% CO2. The light labeled cells were cultured in normal lysine and arginine containing medium, whereas the heavy labeled cells were cultured in medium containing both [13C6]-l-lysine and [13C6]-l-arginine. The cells were cultured for more than seven generations, and the same amount of proteins from light labeled and heavy labeled cells were mixed and analyzed by mass spectrometry to evaluate the labeling efficiency.
Plasmids and cloning
Total RNA was isolated from the HepG2 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and complementary DNA (cDNA) was synthesized using a FastQuant RT kit (TianGen, Beijing, China) according to the manufacturer's instructions. The cDNA of the fragment 6918–8441 nt of MALAT1 was amplified via PCR using the Premix Taq DNA Polymerase (TaKaRa, Otsu, Japan) and cloned into the pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA, USA). The MALAT1 and empty vector plasmids were then transfected into HepG2 cells using Lipofectamine 2000. MALAT1 expression was detected by the quantitative real-time polymerase chain reaction (qRT-PCR). For MALAT1 knockdown, plasmid containing two interfering sequences against human MALAT1 was constructed by GeneSil (Wuhan, China) (sequences as shown in Supplementary Table S1) and transfected into HepG2 cells using Lipofectamine 2000. For experiments using siRNAs, two human MALAT1–siRNA duplexes and non-targeting siRNA were designed and synthesized by RiboBio (Guangzhou, China), and the sequences were shown in Supplementary Table S1. MALAT1 knockdown was confirmed by qRT-PCR.
DNA fragments encoding the complete coding sequence of human DBC1 were amplified by PCR using the Premix Taq DNA Polymerase. The flanking EcoRI and NotI restriction sites were created; then, the DNA fragment was cloned in the pCDH lentivector expression vector (System Biosciences, Palo Alto, CA, USA). The primer sequences used for cloning the full-length DBC1 and its fragments are listed in Supplementary Table S2. Lentiviruses were produced and cells were transfected according to the manufacturer's protocol.
RNA isolation and qRT-PCR
Quantitative mRNA expression analysis targeting gene and the reference gene was performed on a 7500 Fast Real-Time PCR System (ABI, Foster City, CA, USA) using the SuperReal SYBR Green PreMix (TianGen, Beijing, China) following the manufacturer's protocols. The Mean Ct for each sample was normalized using 18s-rRNA as the reference gene (for primer sequences, see Supplementary Table S3).
In vitro transcription
The templates for in vitro transcription were obtained by PCR, and the MALAT1 primers containing the T7 promoter sequence (TAATACGACTCACTATAGGG) were purchased from Invitrogen (Carlsbad, CA, USA). The sequences of the primers for the amplification of MALAT1 sense and antisense chains are shown in Supplementary Table S4. The PCR products were examined with 1% agarose gel electrophoresis. The target bands were removed and purified with an Agarose Gel DNA Extraction Kit Ver.4.0 (TaKaRa, Otsu, Japan). Biotin-labeled sense and antisense chains of MALAT1 were transcribed in vitro using biotin-16-UTP (Epicentre, Madison, WI, USA) with a MEGAscript Kit (Invitrogen, Carlsbad, CA, USA). The synthesized RNAs were purified using a MEGAclear Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturers’ instructions. The integrity and size of the synthesized RNAs were evaluated using agarose gel electrophoresis, and the incorporation of biotin was detected by biotin-HRP dot blot with a chemiluminescent biotin-labeled nucleic acid detection kit (Beyotime, Shanghai, China) according to the manufacturer's instructions.
RNA pull-down
For the proteomics analysis, cellular proteins were extracted from SILAC-labeled HepG2 cells with lysis buffer (40 mM Tris, 120 mM NaCl, 1% Triton X-100, 1 mM NaF, and 1 mM Na3VO4) supplemented with 1 × protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) and 1 U/mL RNase inhibitor (Promega, Madison, WI, USA). The total protein concentration of the extract was measured with a BCA assay. Next, 1 mg of SILAC light-labeled HepG2 cellular proteins were mixed with 50 pmol of biotinylated MALAT1 RNA. Sixty microliters of M-280 Streptavidin Dynabeads (Invitrogen, Carlsbad, CA, USA) were washed with lysis buffer and added to each binding reaction; the beads were incubated at room temperature for 1 h. Then, the beads were washed five times, mixed with SDS-PAGE sample loading buffer, and boiled for 10 min to elute the binding proteins. The same amount of antisense MALAT1 RNA was incubated with the heavy labeled cell extract and processed in parallel as the negative control. Then, the purified interacting proteins were mixed, separated on a 10% SDS-PAGE and visualized using silver staining. Each gel lane was diced into 10 slices and in-gel tryptic digestion was conducted, as previously reported (26). Three biological replicates and two technical replicates were analyzed using mass spectrometry.
LC–MS/MS and data analysis
Samples were desalted using C18 ziptip and loaded on a nanoUPLC system (Waters, Milford, MA, USA) equipped with a self-packed C18 column (C18, 150 × 0.075 mm, 1.7 μm). The peptides were eluted using a 5–40% B gradient (0.1% formic acid in acetonitrile) over 90 min into a nano-electrospray ionization (nESI) Q Exactive mass spectrometer (ThermoFisher Scientific, Waltham, MA, USA). The mass spectrometer was operated in data-dependent mode in which an initial FT scan recorded the mass range of m/z 350–1500. The spray voltage was set between 1.8 and 2.0 kV, and the mass resolution used for MS scan is 70,000. The dynamic exclusion was set to 45 s. The top 20 most intense masses were selected for higher-energy collision dissociation (HCD) with an isolation window of 1.5 m/z and a normalized collision energy of 30 eV. MS/MS spectra were acquired starting at m/z 200 with a resolution of 17,500. The AGC target value and maximum injection time were set as 1E6 and 100 ms respectively for MS scans, as well as 5E4 and 110 ms for MS/MS scans.
Raw data was searched against the Uniprot human protein database containing 129499 sequence entries using the SEQUEST algorithm embedded in the Protein Discoverer 1.4 Software (ThermoFisher Scientific, Waltham, MA, USA). The following parameters were applied during the database search: 10 ppm precursor and fragment mass error tolerance, Arg/Lys (+6.0201 Da, SILAC heavy amino acid) as variable modifications, static modifications of carbamidomethylation for all cysteine residues, flexible modification of oxidation modifications for methionine residues, and one missed cleavage site of trypsin was allowed. False discovery rate (FDR) <0.01 was used as filtering criteria for all identified peptides. In addition, only proteins identified with two or more peptides were considered, and proteins identified with the same set of peptides were grouped and treated as one. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (27) partner repository with the dataset identifier PXD005154.
Quantitation analysis was conducted by the Protein Discoverer 1.4 software (ThermoFisher Scientific, Waltham, MA, USA). Three biological replicates and two technical replicates were analyzed. Ratios between control and MALAT1 pull-down samples were calculated based on the extract ion chromatography areas. Identified proteins with ratiocontrol/MALAT1 ≤ 0.5 (2-fold enrichment) and standard deviation (SD) ≤0.2 were considered as potential MALAT1 interacting proteins. Gene ontology annotation information was acquired through Panther database analysis (http://www.pantherdb.org). Protein domain and signaling pathway analyses were performed using DAVID (https://david.ncifcrf.gov/). Known protein–protein interactions (PPIs) were retrieved from the String 9.0 (http://string90.embl.de/) and GeneMANIA (http://genemania.org/) databases, and integrated in the Cytoscape 3.1.1 for visualization.
RNA immunoprecipitation (RIP)
Cellular proteins from 108 cells were extracted with lysis buffer (40 mM Tris, 120 mM NaCl, 1% Triton X-100, 1 mM NaF, and 1 mM Na3VO4) supplemented with 1 × protease inhibitor cocktail and 1 U/ml RNasin inhibitor. Then, the total protein concentration of the extract was measured with the BCA assay. The proteins of interest, along with the binding RNAs, were isolated with the corresponding primary antibodies using Protein A/G Magnetic Beads. Cellular extract with the same amount of total proteins was simultaneously assayed for the IgG control. Then, the co-precipitated RNAs were extracted with the TRIzol reagent, and the copy number of MALAT1 in the RNA elute was analyzed by absolute quantification via qRT-PCR using the standard-curve quantitation method (28). 18S rRNA and lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) were investigated as irrelevant RNA controls.
Co-immunoprecipitation (Co-IP) and western blot
Cell lysates were prepared with 1% Tris-Triton cell lysis buffer (Cell Signaling Technology) pre-mixed with protease inhibitor cocktail on ice for 30 min and centrifuged at 12 000 g for 30 min. The supernatants were incubated overnight with 30 μl of protein A/G magnetic beads pre-coated with the corresponding primary antibodies. The immunocomplexes were subjected to western blot analysis. The normal corresponding IgG control was simultaneously assayed. For western blot analysis, the proteins were resolved by SDS-PAGE, and transferred to the Immobilon-P membrane (0.45μm pore size, Millipore, Billerica, MA, USA). Primary antibodies were incubated with the membranes at 4°C overnight in 5% skim milk (BD Biosciences, San Jose, CA, USA). Then, the blots were incubated with the horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology, CA, USA) and developed by enhanced chemiluminescence (Millipore, Billerica, MA, USA). GAPDH was used as internal standard. The intensities of the protein bands were measured by the ImageJ software (http://rsb.info.nih.gov/ij/).
SIRT1 activity assay
The decetylase activity of SIRT1 was determined by using a SIRT Activity Assay Kit (Abcam, Cambridge, MA, USA) as recommended by the manufacturer. Briefly, cells were washed with cold PBS and lysed in 1% Tris-Triton cell lysis buffer supplemented with protease inhibitor cocktail. Cell lysates were incubated with protein A magnetic beads pre-coated with anti-SIRT1 antibody for 2 h at 4°C. Immunoprecipitates were incubated with Assay Buffer, SIRT substrate, co-factor and HDAC inhibitor at 37°C for 90 min. The solution was removed, and the detection antibody was added to each well and incubated at 37°C for 60 min. Next, developing solution was added followed by the stop solution. Absorbance at 450 nm was measured using a microplate reader to calculate the activity of SIRT1.
Cell proliferation assays
For the growth curve assay, cells were plated in a 6-well plate with a density of 2 × 105 cells/well, counted manually and replated every other day. The growth curve was plotted based on the fold increase of cell number. For the assessment of colony formation, cells were seeded evenly in a six-well plate (500 cells/well) and cultured for 14 days. The medium was replaced every 3 days. Next, the cells were fixed in methanol for 10 min, and the colonies were visualized by 0.01% crystal violet staining for an additional 10 min. Colonies larger than 0.5 mm in diameter were counted manually.
Cell proliferation capacity was also measured with the cell counting kit-8 (CCK8) assay (Dojindo, Kumamoto, Japan) according to the manufacturer's protocol. Briefly, cells were seeded at a density of 1 × 103 cells/well in a 96-well plate and cultured for 48 h. Ten microliter of CCK8 solution was added to each well, and the samples were incubated at 37°C for 3 h before the absorbance was measured at 450 nm.
The 5-bromo-2′-deoxyuridine (5-BrdU) incorporation assay was performed according to the manufacturer's protocol (BD Biosciences, San Jose, CA, USA). Briefly, cells were seeded on glass coverslips in the 12-well plate with a density of 2 × 104/well. Then, 20 μM of BrdU was added to each well and the samples were incubated at 37°C for 1 h. The cells were washed by PBS three times and fixed with methanol for 10 min at room temperature. Next, the cells were incubated with 0.3% hydrogen peroxide in methanol for 30 min. The cells were then washed twice with PBS and blocked with 5% BSA in PBS for 1 h; then, the cells were incubated with formamide at 100°C for 5min. After cooled, cells were incubated with the anti-BrdU antibody (1:100) at 4°C overnight. After washed with PBS and incubated with the Alexa 546-conjugated secondary anti-mouse IgG antibody (1:400) for 1 h and counter strained with Hoechst 33258 dye, the cells were examined using confocal laser scanning microscopy (Leica, Buffalo Grove, IL, USA). The percentage of 5-BrdU positive cells was calculated as (BrdU add-in cells/Hoechst stained cells) × 100%.
Apoptosis analysis
To assess cell apoptosis, cells were planted at a density of 1 × 105 cells/well in a 12-well plate. After cultured for 24 h, a PE-Annexin V apoptosis detection kit (Sanjian, Tianjin, China) was applied to analyze the cell apoptosis according to the manufacturer's protocol. The data were collected on a BD FACSVerse flow cytometer (BD Biosciences, San Jose, CA, USA), processed with the BD FACSuite software and analyzed by the BD Flow Jo software.
Statistical evaluation
SPSS version 17.0 software were performed for statistical analyses and Prism version 5.0 (GraphPad) were used to plotted to show mean and standard deviation of mean (SD). For the comparisons, Student's t test was performed between two groups and analysis of variance (ANOVA) were performed among multiple groups, and growth curve results were compared by repeated measures ANOVA. All statistical tests were two-sided and P values were considered statistically significant for P ≤ 0.05.
RESULTS
The quantitative proteomics workflow for identifying lncRNA-interacting proteins
To characterize the interacting proteins of MALAT1, a quantitative proteomics strategy was employed by combining SILAC labeling, RNA pull-down, SDS-PAGE separation and LC–MS/MS analyses. As shown in Figure 1, the HCC cell line HepG2 cells used for the control experiment were labeled using SILAC medium containing both 13C6 lysine and 13C6 arginine to ensure quantification coverage for the detected tryptic peptides, while the cells used for MALAT1 pull-down were cultured in cell medium containing normal lysine and arginine. To purify the MALAT1-interacting proteins, we first synthesized MALAT1 with in vitro transcription (Supplementary Figure S1). MALAT1 is highly conserved among different species, and the homologous sequence of MALAT1 is located at the 3′ end region (29). The 6918–8441 nt region from the 3′ end of MALAT1 gene is reported to be an important functional motif of MALAT1 (30). Therefore, we chose to investigate the 6918–8441 nt fragment of MALAT1 in this study. The MALAT1 template was amplified from the HepG2 cDNA via PCR. Biotin-16-UTP was randomly incorporated into the MALAT1 RNA during in vitro transcription. Agarose gel electrophoresis and dot blot analysis confirmed that both sense and antisense MALAT1 RNA chains were successfully synthesized and incorporated with biotin-UTP (Supplementary Figure S1C and D).
Figure 1.

Overview of the experimental workflow. Cells were cultured in the SILAC medium. Sense and antisense MALAT1 chains were randomly incorporated with biotin-16-UTP using in vitro transcription. The synthesized MALAT1 was incubated with cellular proteins extracted from SILAC-labeled HepG2 cells. The antisense MALAT1 was used as a negative control and processed in parallel. The MALAT1-interacting proteins were pulled down using streptavidin-conjugated magnetic beads. Then, the beads were mixed and separated using SDS-PAGE. Each gel lane was divided into ten bands and digested with trypsin for LC–MS/MS analyses.
The synthesized MALAT1 was incubated with cellular proteins extracted from SILAC light-labeled HepG2 cells. To distinguish non-specific binding proteins, the antisense MALAT1 was used as a negative control in this step and incubated with the same amount of cellular proteins extracted from SILAC heavy-labeled HepG2 cells. After incubation, the MALAT1-interacting proteins were pulled down using streptavidin-conjugated magnetic beads. Then, the beads were mixed, and the eluted proteins were separated using SDS-PAGE. Each gel lane was divided into ten bands and digested with trypsin for downstream LC–MS/MS analyses.
Identification of the MALAT1-interacting proteins using quantitative proteomics
The MALAT1-interacting proteins were analyzed using the quantitative proteomics strategy as described above. As a result, 1584 proteins were detected, and 939 proteins were identified with two or more unique peptides. Within these 939 proteins, 929 proteins were quantified and 127 proteins were detected with ratiocontrol/MALAT1 ≤ 0.5 (2-fold enrichment) with SD ≤ 0.2 in the SILAC quantification experiment (Supplementary Table S5). Thus far, only approximately twenty MALAT1-interacting proteins have been reported in the literature (20). The majority of these proteins relate to RNA processing and gene transcription. For example, pre-mRNA-processing factor 6 (PRPF6) is known to bind to MALAT1 and influence its nuclear localization (31). PRPF6 was also identified in this study as a potential MALAT1-interacting protein, along with several other members from the same family, including pre-mRNA-processing factor 6 (PRPF8) and PRP40 pre-mRNA processing factor 40 homolog A (PRPF40A). MALAT1 is also known to interact with serine/arginine-rich splicing factors (22,23). In this study, we detected many splicing factors, including SF3A1, SF3B3, and SRSF7. Another known MALAT1-interacting protein, ELAVL1 (25,32), was also observed in this study. Intriguingly, many novel MALAT1-interacting proteins were discovered, including several isoforms of the heterogeneous nuclear ribonucleoproteins (hnRNPs), transcriptional activator protein Pur-alpha (PURA), etc.
The bioinformatics analysis of MALAT1-interacting proteins
To further understand the potential biological functions of MALAT1, gene ontology analysis was performed on the identified proteins using the Panther database. As shown in Supplementary Figure S2, molecular function analysis revealed that most of the identified proteins related with catalytic activity (40.0%), binding (37.2%), structural molecule activity (6.2%) and enzyme regulator activity (5.5%). Biological process analysis showed that MALAT1-interacting proteins were involved in metabolic processes (38.8%), cellular processes (14.5%), and localization (7.9%). Protein domain analysis was performed using DAVID, and 18 proteins of the detected MALAT1-interacting proteins contain an RNA recognition motif (P = 1.52E–13) (Supplementary Figure S2C). Other enriched domains include the DZF domain, double-stranded RNA binding domain and the 26S proteasome subunit P45. The identified interacting proteins were searched against the KEGG database using DAVID to reveal the related signaling pathways (Supplementary Figure S2D). Four pathways were significantly enriched with P values < 0.05. MALAT1 was significantly involved with the spliceosome (P = 5.28E–13) and proteasome (P = 9.07E–4) pathways. Two amino acid metabolism pathways were also enriched, including the valine, leucine and isoleucine degradation pathway (P = 0.01) as well as the alanine, aspartate and glutamate metabolism pathway (P = 0.04).
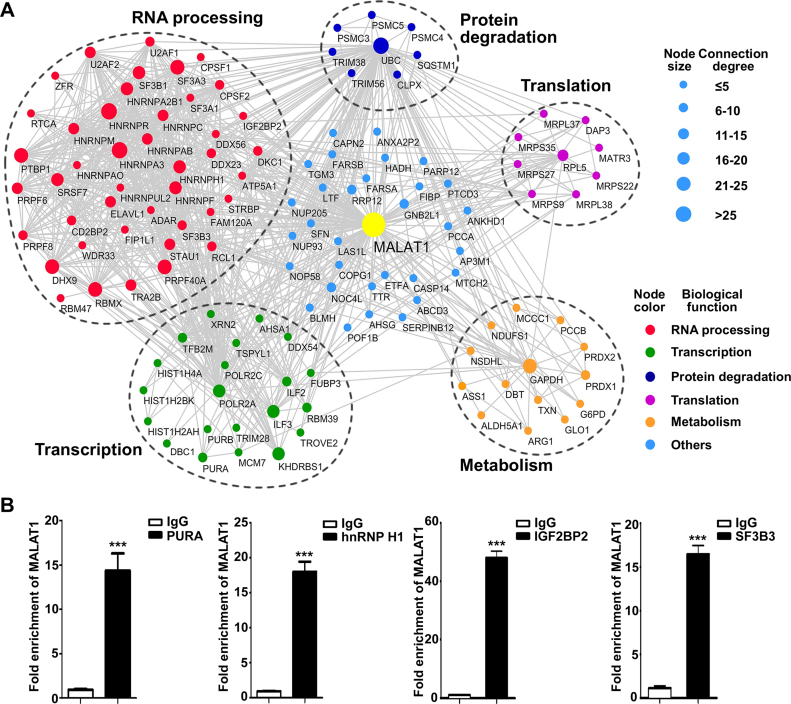
For network construction, the identification data were integrated with interaction information obtained through the PPI database search and imported to Cytoscape for visualization. A highly connected network consisting of 127 proteins and 788 connections was mapped (Figure 2A). These proteins can be divided into five major sub-clusters based on their biological functions: RNA processing; transcription regulation; ribosomal proteins; protein degradation; and metabolism regulation. Some of these proteins are highly connected in the constructed network (the degree of connection is indicated by the node size), such as polyubiquitin-C (UBC) and certain hnRNP proteins. Taken together, the protein interactome analysis provided a more complete picture of the biological functions of MALAT1, suggesting its potential roles in diverse biological processes.
Figure 2.
Network representation of the MALAT1-interacting proteins and validation of selected identified MALAT1-interacting proteins using RIP. (A) The PPI information was obtained through a database search using String 9.0 and GeneMANIA, incorporated with lncRNA–protein interactions identified in this study, and imported into Cytoscape 3.1.1 for network construction. Proteins and their interactions are shown as nodes and edges. The node size reflects the interaction degree. The proteins are grouped based on their biological functions, as represented by the node color. (B) RIP enrichment was determined as the relative levels of MALAT1 in the immunoprecipitates of PURA, hnRNP H1, IGF2BP2, and SF3B3 compared with the IgG control, and 18S rRNA was used as internal standard. Data represent the means ± SD of triplicate independent analyses (***P < 0.001, by Student's t-test).
To evaluate the results from the quantitative proteomics analysis, the interactions between MALAT1 and four newly identified proteins were validated using RIP assay and examined by qRT-PCR. We observed significant enrichment of MALAT1 in the immunoprecipitates of PURA, hnRNP H1, IGF2BP2, and SF3B3 compared with the normal IgG control (Figure 2B and Supplementary Figure S2E). 18S rRNA, an abundant RNA in the cell, was used as sample loading control. An irrelevant nuclear-localized lncRNA, nuclear paraspeckle assembly transcript 1 (NEAT1) was also examined to evaluate the specificity of the identified interactions (33). No enrichment of NEAT1 was observed in the immunoprecipitates of the selected proteins (Supplementary Figure S2E).
MALAT1 physically interacts with the aa120-280 region of DBC1
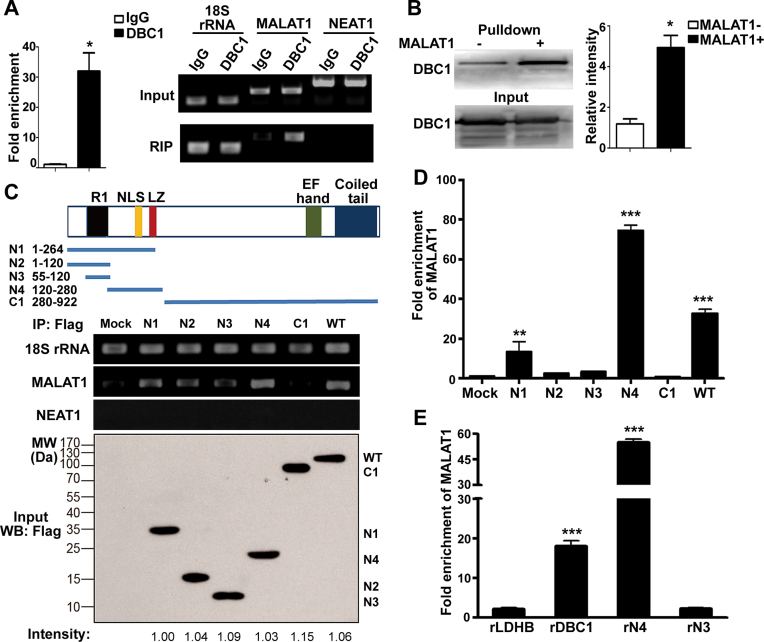
Among all the interacting proteins identified by the SILAC quantitative proteomic analysis in our study, we chose DBC1 for further investigation. Human DBC1 (also named CCAR2 or KIAA1967) is a nuclear protein that plays an important role in gene expression regulation (34). The RIP assay was performed to confirm the interaction between MALAT1 and DBC1. The results show that MALAT1 was enriched with the DBC1 antibody with greater than 30-fold enrichment compared with the IgG control (Figure 3A). Reciprocally, RNA pull-down and western blot showed that DBC1 could bind with MALAT1 (Figure 3B). These results suggest that MALAT1 is physically associated with DBC1.
Figure 3.
MALAT1 directly interacts with the aa120–280 region of DBC1. (A) qRT-PCR analysis of MALAT1 in the immunoprecipitates of DBC1 as compared to the IgG control, and 18S rRNA was used as internal standard (left panel). Data represent the means ± SD of triplicate independent analyses (*P < 0.05, by Student's t-test). Agarose gel electrophoresis analysis of the qRT-PCR products of MALAT1, 18S rRNA and NEAT1 in the cell lysates and immunoprecipitates of DBC1 (right panel). (B) RNA pull-down experiment showed the interaction between DBC1 and MALAT1. Biotin-labeled MALAT1 fragment (6918–8441 nt) was incubated with HepG2 cell lysates, and the enriched DBC1 was detected by western blot. The antisense MALAT1 was used as control. The right panel shows the relative intensities of DBC1 in the immunoblots of MALAT1 pull-down as compared to the antisense control (*P < 0.05, by Student's t-test). (C) Schematic diagram of the plasmids encoding Flag-tagged full-length or the fragments of DBC1 (upper panel). Lower panel: plasmids encoding Flag-tagged full-length or fragments of DBC1 were transfected in the HepG2 cells, and their expression levels were detected by western blot. RIP of each fragment was performed using the anti-Flag antibody, and qRT-PCR was used to determine the enriched levels of MALAT1, 18S rRNA, and NEAT1 in the immunoprecipitates. Normal HepG2 cells were used as control. (D) Histogram represents the fold enrichment of MALAT1 in the Flag immunoprecipitates as measured by qRT-PCR. Data represent the means ± SD of triplicate independent analyses (**P < 0.01; ***P < 0.001, by one-way ANOVA). (E) In vitro His-tag pull-down assay using rDBC1, DBC1-N3, and DBC1-N4. 2 μg of recombinant protein purified from E. coli was incubated with 1 μg of in vitro transcribed sense MALAT1 or antisense MALAT1 for 1 h at 4 °C. The recombinant proteins were isolated using Dynabeads® His-tag isolation magnetic beads. The enriched MALAT1 was examined using qRT-PCR. An irrelevant protein rLDHB was used as negative control. Data represent the means ± SD of triplicate independent experiments (***P < 0.001, by Student's t-test).
To identify the regions of DBC1 that are responsible for binding with MALAT1, we generated five fragments of DBC1 (Figure 3C and Supplementary Figure S3). DBC1 has five major functional domains: an S1-like domain (R1) on the N-terminus; a nuclear location signal domain (NLS); a centrally located leucine zipper (LZ) domain; an EF-Hand domain; and a coiled tail region on the C-terminus (35–37). The N-terminal region of DBC1 (aa1–264) is required for its binding to epigenetic modifiers, such as deacetylases sirtuin 1 (SIRT1), histone deacetylase 3 (HADC3), and methyltransferase SUV39H1 (34,38). The R1 domain near the N terminus has been reported to bind with RNAs (37,39). The results from the RIP assay showed that DBC1 fragments containing residues aa1–264 (N1), residues aa120–280 (N4) and the full-length DBC1 could interact with MALAT1 (Figure 3C and D). Interestingly, aa55–120 (N3) containing the R1 region showed no enrichment of MALAT1, while aa120–280 (N4) showed the highest enrichment of MALAT1. To investigate whether MALAT1 could directly bind to DBC1, in vitro His-tag pull-down assay was performed by using recombinant proteins purified from E. coli. Both of the full-length DBC1 and DBC1-N4 fragment can bind to MALAT1 in vitro (Figure 3E and Figure S3C). Overall, these results suggest that the aa120–280 region of DBC1, containing the NLS and LZ motifs, is required for its direct interaction with MALAT1.
The interaction between MALAT1 and DBC1 regulates the activities of SIRT1 and p53
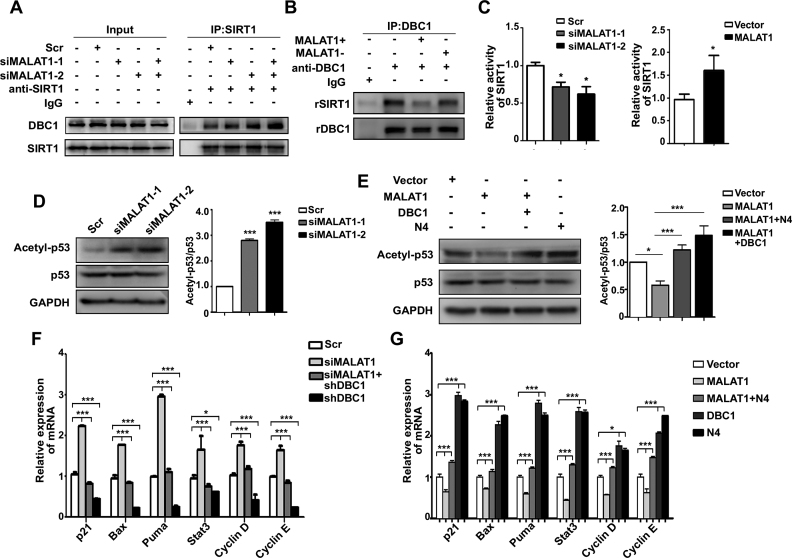
Next, we sought to determine whether the association between MALAT1 and DBC1 had functional significance. Because the LZ motif of DBC1 is responsible for its binding to the catalytic domain of epigenetic modifier SIRT1, and the binding of DBC1 suppress the deacetylation activity of SIRT1 (38,40), we investigated whether the interaction between MALAT1 and DBC1 could regulate the functions of SIRT1. MALAT1 downregulation with siRNAs increased the interaction between DBC1 and SIRT1 (Figure 4A and Supplementary Figure S4A), suggesting that MALAT1 may compete with SIRT1 for DBC1 binding. In addition, incubation of MALAT1 instead of the antisense MALAT1 with rDBC1 and rSIRT1 reduced the interactions of these two proteins in vitro (Figure 4B). Moreover, we observed that SIRT1 activity was enhanced with MALAT1 overexpression and decreased with MALAT1 downregulation (Figure 4C and Supplementary Figure S4A, B). In addition, the level of MALAT1 doesn’t affect the expressions of both DBC1 and SIRT1 (Supplementary Figure S4C). These results indicate that MALAT1 regulates the SIRT1 activity through binding DBC1.
Figure 4.
MALAT1 regulates the activities of p53 via DBC1. (A) Co-IP of SIRT1 in control and MALAT1-knockdown HepG2 cells. SIRT1 and DBC1 were detected by western blot analysis. (B) Co-IP of SIRT1 and DBC1 in vitro. 2 μg of rDBC1 and rSIRT1 were incubated with 1 μg of in vitro transcribed sense MALAT1 or antisense MALAT1 for 1 h at 4°C. Next, DBC1 was isolated with anti-DBC1 antibody, and SIRT1 and DBC1 in the immunoprecipitates were analyzed by western blot. (C) The deacetylation activity of SIRT1 in MALAT1-knockdown (left panel) and MALAT1-overexpressing (right panel) HepG2 cells. The data are means ± SD of triplicate independent experiments (*P < 0.05, by Student's t-test). (D) Western blot of acetyl-p53 and p53 in MALAT1-downregulated cells (left panel). The histogram shows the relative intensities of p53 acetylation verses total p53 levels (right panel) measured by the ImageJ software (http://rsb.info.nih.gov/ij/index.html). Data represent the means ± SD (***P < 0.001, by one-way ANOVA). (E) Western blot analysis of acetyl-p53 and p53 in the HepG2 cells transfected with MALAT1 expression plasmid, co-transfected with expression plasmids encoding MALAT1 and full-length DBC1 or DBC1-N4 fragment (left panel). Histogram shows the relative intensities of p53 acetylation verses total p53 levels measured by ImageJ (right panel). Data represent the means ± SD (*P < 0.05; ***P < 0.001, by one-way ANOVA). qRT-PCR analysis of p53 target genes in MALAT1-knockdown cells (F) and MALAT1-overexpressing cells (G). Data represent means ± SD of four independent experiments (*P < 0.05 and ***P < 0.001, by one-way ANOVA).
p53 has been reported as one of the deacetylation substrates of SIRT1 (38), we further examined whether p53 acetylation is regulated by MALAT1. Indeed, the p53 acetylation level was increased in MALAT1-knockdown cells and decreased with MALAT1 overexpression compared with the corresponding control cells (Figure 4D and E). Moreover, the co-transfection of full-length DBC1 or DBC1-N4 fragment reversed the decrease in p53 acetylation in response to MALAT1 overexpression, suggesting that MALAT1 regulates p53 acetylation through DBC1 (Figure 4E and Supplementary Figure S4D). Furthermore, we investigated the role of MALAT1 in p53-regulated transcription. Six canonical target genes of p53, p21, Bax, Puma, Stat3, Cyclin D and Cyclin E, were examined using qRT-PCR. The results showed that the mRNA levels of those selected genes were increased in MALAT1-knockdown cells, and knockdown of DBC1 reduced the transcription level of the studied genes in the siMALAT1 cells (Figure 4F and Supplementary Figure S4E). On the other hand, p53 regulated transcription was reduced with the overexpression of the 6918–8441 nt fragment of MALAT1, and DBC1-N4 overexpression increased the mRNA levels of the studied genes in the MALAT1-overexpression cells to similar levels as compared to the control cells (Figure 4G and Supplementary Figure S4F). Taken together, the results suggest that MALAT1 decreases the acetylation of p53 and regulates the transcription activities of p53 by interacting with DBC1.
MALAT1 regulates cell proliferation and apoptosis through DBC1
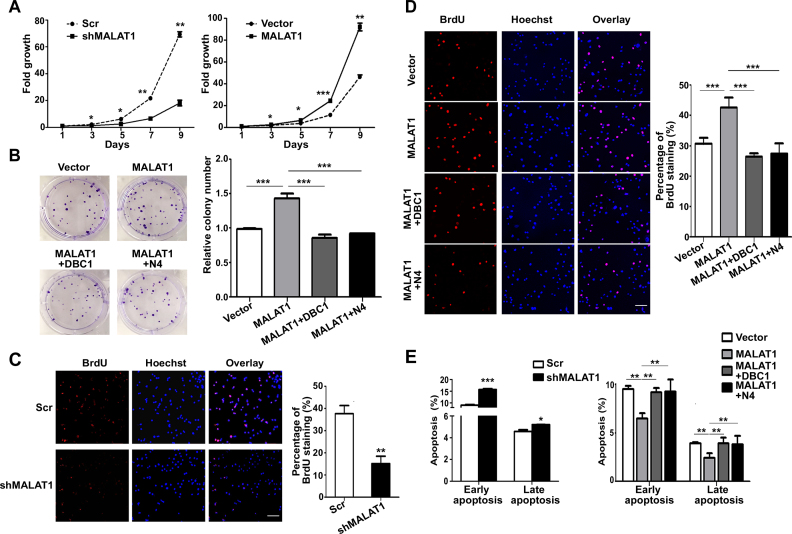
To further understand the cellular functions of MALAT1, we performed a set of in vitro experiments to investigate its roles in HepG2 cell proliferation and apoptosis. The roles of MALAT1 in cell growth and viability were first evaluated using a growth curve assay. MALAT1 downregulation greatly decreased cell viability over 9 days in culture. Conversely, the proliferation ability was increased by the overexpression of the 6918–8441 nt fragment of MALAT1 (Figure 5A). The colony-formation assay also indicated that MALAT1 overexpression promoted cell proliferation (Figure 5B). To explore the mechanism by which MALAT1 regulates cell proliferation, we performed a BrdU-incorporation assay. The results showed that the ratio of BrdU-positive cells was significantly reduced after MALAT1 knockdown and increased upon the transfection of the 6918–8441 nt fragment of MALAT1 (Figure 5C and D), indicating that MALAT1 may play an important role in the entry of HepG2 cells into the S phase of the cell cycle, promoting cell proliferation. To examine the role of MALAT1 in regulating cell apoptosis, an Annexin V assay was performed and examined via flow cytometry analysis. The results showed that the percentage of apoptotic cells was significantly increased in shMALAT1 cells for both early- and late-stage apoptosis, and MALAT1 overexpression greatly decreased cell apoptosis in HepG2 cells (Figure 5E), suggesting that MALAT1 plays an important role in protecting the cells against apoptosis.
Figure 5.
MALAT1 regulates cell proliferation and apoptosis in HepG2 cells through binding with DBC1. (A) The growth curve of MALAT1-downregulated cells (left panel) and MALAT1-overexpressed cells (right panel) over 9 days. Cells (2 × 105) were seeded in each well, and the cell numbers were counted every 48 h. Data are from three independent assays. Bar, mean; error bar, SD (*P < 0.05, **P < 0.01, ***P < 0.001, by repeated measures ANOVA). (B) Colony-formation assay of HepG2-MALAT1 and HepG2 cells co-transfected with both MALAT1 and DBC1 or DBC1-N4. Representative images show the results of the colony-formation assay (left panel). Histogram shows the relative colony numbers (right panel). Data are from three independent assays. Bar, mean; error bar, SD (***P < 0.001, by one-way ANOVA). BrdU incorporation analysis of MALAT1-knockdown HepG2 cells (C) and MALAT1-overexpressing cells with or without the overexpression of full-length or N4 fragment of DBC1 respectively (D). Representative images showing alterations in S phase distribution (left panel). Histogram shows the percentage of BrdU staining (right panel). Data are from three independent assays. Bar, mean; error bar, SD (**P < 0.01, ***P < 0.001, by Student's t-test or one-way ANOVA). (E) Annexin V assay for analyzing cell apoptosis in HepG2 cells transfected with shMALAT1 plasmid (left panel) or MALAT1-overexpressing plasmid with or without co-transfection of full-length or N4 fragment of DBC1 respectively (right panel). Data are from three independent assays. Bar, mean; error bar, SD (*P < 0.05, **P < 0.01, ***P < 0.001, by Student's t-test or one-way ANOVA).
Furthermore, we investigated whether MALAT1 plays its role in regulating cell proliferation through binding with DBC1. Firstly, we examined the functional significance of full-length and N4 fragment of DBC1 expression in HepG2 cells. The results showed that the overexpression of the full-length DBC1 and DBC1-N4 can both impaired cell proliferation and increased cell apoptosis (Supplementary Figure S5). Rescue experiments were performed by using both full-length DBC1 and DBC1-N4. The colony-formation assay showed that the co-transfection of DBC1 or DBC1-N4 significantly reduced the cell proliferation that was promoted by MALAT1 overexpression (Figure 5B). In addition, cells co-transfected with MALAT1 and DBC1 or DBC1-N4 had a similar BrdU-positive rate as control cells (Figure 5D). Moreover, co-transfection of DBC1 or DBC1-N4 increased the apoptosis rates in MALAT1-overexpression cells (Figure 5E). Taken together, these results suggest that MALAT1 promotes cell proliferation and inhibits apoptosis via interacting with DBC1.
DISCUSSION
MALAT1 and protein interactions in previous reports are mostly discovered by using protein-centric methods, such as RIP and crosslinking-immunoprecipitation (CLIP) combined with RNA-sequencing (41), therefore, will not provide a comprehensive profile of the MALAT1 interactome. To better understand the biological functions of MALAT1 and the molecular mechanism, we applied a high-throughput and lncRNA-centric method that provides comprehensive information regarding the lncRNA protein interactome. Both known and novel potential interacting proteins of MALAT1 were identified in this study. Overall, the results greatly expand the current knowledge of MALAT1 interacting proteins, laying the foundation for further clarifying the biological functions and related molecular mechanism of MALAT1. However, several known MALAT1 interacting proteins were not observed here, such as TAR DNA-binding protein 43 (TDP-43), and DGCR8 microprocessor complex subunit (DGCR8) (20). This is probably due to three reasons: first, as mentioned above, different investigation methods were applied in the earlier studies, such as CLIP, leading to differences in the detected interactions; second, the 6918 -8441 nt region of MALAT1 was investigated in this study, and some proteins may interact with other regions of MALAT1. For example, DGCR8 was reported to bind to the 1–500 nt region of MALAT1 (42); finally, the molecular functions of ncRNAs are believed to be highly cell- and tissue-type specific. All the known interacting proteins were identified in other types of cells, but not in HCC cells. Of note, the MALAT1 interacting protein data obtained via the high throughput quantitative proteomics analysis in this study may require further validation via other techniques, such as RIP, electrophoretic mobility shift assay (ESMA), surface plasmon resonance (SPR), etc.
Gene ontology analysis and network construction show that MALAT1-interacting proteins are enriched for five major biological functions. These sub-clusters are highly connected with each other, suggesting that MALAT1 may regulate the crosstalk between different biological processes. It is not surprising that the majority of the identified proteins are involved in RNA processing and gene transcription regulation because numerous studies have implicated MALAT1 in these two cellular processes (20,23,24). This study further confirms this theory and provides a substantially more detailed picture regarding the mechanism by which MALAT1 regulates protein biosynthesis from transcription to translation. Interestingly, our results show that MALAT1 may also regulate protein degradation. For example, UBC, conjugated to target proteins undergoing degradation via proteasome, was identified in this study as a novel MALAT1-interacting protein (unique peptides = 9, Ratiocontrol/MALAT = 0.12). UBC was connected with 117 proteins in the network of the MALAT1 interactome, suggesting it is very likely that MALAT1 can regulate the degradation of its interacting proteins through UBC. Furthermore, some MALAT1-interacting proteins are involved in regulating metabolism. For example, eight proteins were enriched in the amino acid metabolism pathways, including hydroxyacyl-coenzyme A dehydrogenase (HADH), methylcrotonoyl-CoA carboxylase subunit alpha (MCCC1), and succinate-semialdehyde dehydrogenase (ALDH5A1). Several studies have implicated lncRNAs in the regulation of several aspects of cellular metabolism. For example, lincRNA-p21 was reported to interact with HIF-1α and modulate the Warburg effect (12). Li et al reported that a liver-enriched long non-coding RNA (lncLSTR) could regulate systemic lipid metabolism in mice (43). Our results show that MALAT1 may also be involved in regulating cellular metabolism. Taken together, bioinformatics analysis of the MALAT1 interactome suggests important roles of MALAT1 in regulating metabolism and protein degradation. Further investigation is needed to confirm such observation and delineate the underlying molecular mechanisms.
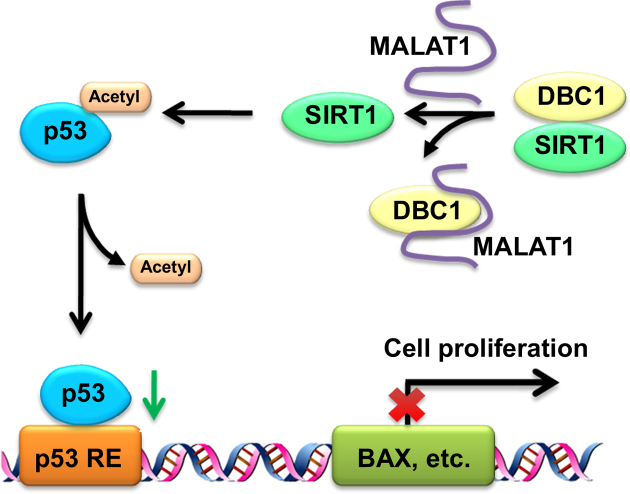
Among all interacting proteins identified by the SILAC quantitative proteomic analysis in our study, DBC1 was selected for further investigation. DBC1 is a nuclear protein involved in many biological processes, such as the DNA damage response, apoptosis, metabolism, epigenetics, tumorigenesis and transcription regulation (34). DBC1 has been reported to play a crucial physiological role as a modulator of epigenetics through binding multiple epigenetic modifiers, such as SIRT1, HDAC3 and the methyltransferase SUV39H1 (38,40,44,45). The interaction between DBC1 and SIRT1 has been extensively studied. Kim et al. observed a direct interaction between the LZ domain of DBC1 and the catalytic domain of SIRT1, and this interaction inhibits the ability of SIRT1 to deacetylate its substrate p53 (38). Intriguingly, we find that MALAT1 binds to the aa120–280 region of DBC1 containing the LZ domain and blocks the interaction between SIRT1 and DBC1. Moreover, MALAT1 up-regulates the deacetylation activity of SIRT1, decreases the acetylation of p53, and reduces the transcription levels of p53 target genes. It is well known that p53 transcriptionally activates numerous genes and induces cell cycle arrest, cellular senescence and apoptosis (46). Acetylation increases the stability of p53 and its binding to low-affinity promoters, and it is required for its checkpoint responses to DNA damage and activated anti-oncogenes (47). Moreover, acetylation blocks the interaction of p53 with its cognate repressors (Mdm2 and Mdmx) on DNA, which directly result in p53 activation regardless of its phosphorylation status (47). Taken together, our results suggest that MALAT can regulate p53 acetylation and modulate its transcription activity through inhibiting the interaction between DBC1 and SIRT1 (Figure 6).
Figure 6.
The proposed mechanism of MALAT1 regulating p53 through interacting with DBC1. MALAT1 binds with DBC1, inhibits the interaction between DBC1 and SIRT1, and enhances the deacetylation activity of SIRT1. The upregulation of MALAT1 reduces p53 acetylation as well as impairs its transcription activity, and thus inhibits the functions of p53.
In this study, we mainly focused on the interaction between DBC1 and SIRT1 as well as their downstream substrate p53. However, the biological roles of DBC1 and SIRT1 are extremely complex. For example, DBC1 also regulates nuclear receptors, such as estrogen receptor α (ERα), estrogen receptor β (ERβ), androgen receptor (AR), and retinoic acid receptor (PAR), and DBC1 can bind with these receptors and regulate gene transcription (48–50). Depending on the binding partners, DBC1 may play many different roles in diverse biological processes. It is only reasonable to believe that the interaction between MALAT1 and DBC1 has substantially broader biological impacts than described in this study, which will be explored in future experiments.
Several previous studies have implicated lncRNAs in the p53 pathway. For example, lincRNA-p21 is reported to repress the expression of many p53-targeted gene (9). As another lncRNA, PANDA, negatively regulates the expression of pro-apoptotic genes upon DNA damage as well as controls apoptosis in a p53-dependent manner (51). With regard to MALAT1, a recent study demonstrated that MALAT1 modulates the expression of cell cycle genes and is required for G1/S and mitotic progression in normal human diploid fibroblasts (52). It is proposed that MALAT1 and p53 share a synthetically lethal relationship, where MALAT1 silencing is lethal in the context of mutation of the p53 gene (53). More interestingly, a recent report showed that p53 could regulate MALAT1 expression in erythroid myeloid lymphoid cells (31). There are two p53 binding motifs on the promoter region of MALAT1, and p53 could directly bind to the promoter of MALAT1 as a transcription repressor (29,31). Therefore, it is very likely that there is an inhibitory feedback loop between p53 and MALAT1. MALAT1 promotes cell proliferation and inhibits cell apoptosis by inhibiting p53 activation through decreasing its acetylation level. Conversely, the enforced expression of the p53 gene suppresses the transcription level of MALAT1, inhibits cell proliferation and induces cell apoptosis.
Taken together, this study greatly expands our knowledge of the MALAT1 protein interactome. Many novel MALAT1-interacting proteins were identified, suggesting novel biological functions of MALAT1, laying the foundation for further clarifying the biological roles of MALAT1. Furthermore, our study establishes MALAT1 as an epigenetic regulator through interacting with DBC1 as well as reveals a new regulatory mechanism of p53.
Supplementary Material
ACKNOWLEDGEMENTS
L.L. acknowledges a Vilas Distinguished Achievement Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy, Tianjin 1000 Talent Plan from Tianjin China and Changjiang Professorship from the Chinese Ministry of Education.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
863 program [2015AA020403 to N.Z.]; National Key Research and Development Program [2016YFC0900100 to R.C.]; National Natural Science Foundation of China [21575103 to R.C., 81472683 to N.Z., 31671421 to N.Z.]; National Key Scientific Instrument and Equipment Development Project [2013YQ16055106 to N.Z.]; NSFC-FRQS program [81661128009 to N.Z.]; Basic and Advanced Technology Research Foundation from Science and Technology Department of Henan Province [152300410162 to H.Z.]. The Q-Exactive Orbitrap instrument was purchased through the support of an NIH shared instrument grant [S10RR029531 to L.L.]. Funding for open access charge: 863 program.
Conflict of interest statement. None declared.
REFERENCES
- 1. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponting C.P., Belgard T.G.. Transcribed dark matter: meaning or myth? Hum. Mol. Genet. 2010; 19:R162–R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007; 447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batista P.J., Chang H.Y.. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013; 152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guttman M., Rinn J.L.. Modular regulatory principles of large non-coding RNAs. Nature. 2012; 482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. An S., Song J.J.. The coded functions of noncoding RNAs for gene regulation. Mol. Cells. 2011; 31:491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilusz J.E., Sunwoo H., Spector D.L.. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009; 23:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D., Zhang J. et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013; 500:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010; 142:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang F., Huo X.S., Yuan S.X., Zhang L., Zhou W.P., Wang F., Sun S.H.. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell. 2013; 49:1083–1096. [DOI] [PubMed] [Google Scholar]
- 12. Yang F., Zhang H., Mei Y., Wu M.. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell. 2014; 53:88–100. [DOI] [PubMed] [Google Scholar]
- 13. Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y.. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014; 281:1750–1758. [DOI] [PubMed] [Google Scholar]
- 14. Liu B., Sun L., Liu Q., Gong C., Yao Y., Lv X., Lin L., Yao H., Su F., Li D. et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015; 27:370–381. [DOI] [PubMed] [Google Scholar]
- 15. Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041. [DOI] [PubMed] [Google Scholar]
- 16. Michalik K.M., You X., Manavski Y., Doddaballapur A., Zornig M., Braun T., John D., Ponomareva Y., Chen W., Uchida S. et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014; 114:1389–1397. [DOI] [PubMed] [Google Scholar]
- 17. Yoshimoto R., Mayeda A., Yoshida M., Nakagawa S.. MALAT1 long non-coding RNA in cancer. Biochim. Biophys. Acta. 2016; 1859:192–199. [DOI] [PubMed] [Google Scholar]
- 18. Liu W.T., Lu X., Tang G.H., Ren J.J., Liao W.J., Ge P.L., Huang J.F.. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J. Gastroenterol. 2014; 20:6314–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin R., Maeda S., Liu C., Karin M., Edgington T.S.. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007; 26:851–858. [DOI] [PubMed] [Google Scholar]
- 20. Gutschner T., Hammerle M., Diederichs S.. MALAT1 – a paradigm for long noncoding RNA function in cancer. J. Mol. Med. (Berl.). 2013; 91:791–801. [DOI] [PubMed] [Google Scholar]
- 21. Gutschner T., Hammerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R.. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007; 14:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010; 39:925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L., Lin C., Liu W., Zhang J., Ohgi K.A., Grinstein J.D., Dorrestein P.C., Rosenfeld M.G.. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011; 147:773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Latorre E., Carelli S., Raimondi I., D’Agostino V., Castiglioni I., Zucal C., Moro G., Luciani A., Ghilardi G., Monti E. et al. The ribonucleic complex HuR-MALAT1 represses CD133 expression and suppresses epithelial-mesenchymal transition in breast cancer. Cancer Res. 2016; 76:2626–2636. [DOI] [PubMed] [Google Scholar]
- 26. Wang Y., Yue D., Xiao M., Qi C., Chen Y., Sun D., Zhang N., Chen R.. C1QBP negatively regulates the activation of oncoprotein YBX1 in the renal cell carcinoma as revealed by interactomics analysis. J. Proteome Res. 2015; 14:804–813. [DOI] [PubMed] [Google Scholar]
- 27. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rutledge R.G., Cote C.. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 2003; 31:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma X.Y., Wang J.H., Wang J.L., Ma C.X., Wang X.C., Liu F.S.. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics. 2015; 16:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu C., Yang M., Tian J., Wang X., Li Z.. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 2011; 39:169–175. [DOI] [PubMed] [Google Scholar]
- 31. Miyagawa R., Tano K., Mizuno R., Nakamura Y., Ijiri K., Rakwal R., Shibato J., Masuo Y., Mayeda A., Hirose T. et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012; 18:738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., Rajewsky N.. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011; 43:340–352. [DOI] [PubMed] [Google Scholar]
- 33. Sasaki Y.T., Ideue T., Sano M., Mituyama T., Hirose T.. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chini E.N., Chini C.C., Nin V., Escande C.. Deleted in breast cancer-1 (DBC-1) in the interface between metabolism, aging and cancer. Biosci. Rep. 2013; 33:e00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu E.J., Kim S.H., Heo K., Ou C.Y., Stallcup M.R., Kim J.H.. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor alpha activity and co-activator synergy. Nucleic Acids Res. 2011; 39:6932–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011; 39:D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brunquell J., Yuan J., Erwin A., Westerheide S.D., Xue B.. DBC1/CCAR2 and CCAR1 are largely disordered proteins that have evolved from one common ancestor. Biomed. Res. Int. 2014; 2014:418458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J.E., Chen J., Lou Z.. DBC1 is a negative regulator of SIRT1. Nature. 2008; 451:583–586. [DOI] [PubMed] [Google Scholar]
- 39. Mannen T., Yamashita S., Tomita K., Goshima N., Hirose T.. The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J. Cell Biol. 2016; 214:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao W., Kruse J.P., Tang Y., Jung S.Y., Qin J., Gu W.. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008; 451:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McHugh C.A., Russell P., Guttman M.. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014; 15:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macias S., Plass M., Stajuda A., Michlewski G., Eyras E., Caceres J.F.. DGCR8 HITS-CLIP reveals novel functions for the microprocessor. Nat. Struct. Mol. Biol. 2012; 19:760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li P., Ruan X., Yang L., Kiesewetter K., Zhao Y., Luo H., Chen Y., Gucek M., Zhu J., Cao H.. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015; 21:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chini C.C., Escande C., Nin V., Chini E.N.. HDAC3 is negatively regulated by the nuclear protein DBC1. J. Biol. Chem. 2010; 285:40830–40837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li Z., Chen L., Kabra N., Wang C., Fang J., Chen J.. Inhibition of SUV39H1 methyltransferase activity by DBC1. J. Biol. Chem. 2009; 284:10361–10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vousden K.H., Prives C.. Blinded by the light: the growing complexity of p53. Cell. 2009; 137:413–431. [DOI] [PubMed] [Google Scholar]
- 47. Tang Y., Zhao W., Chen Y., Zhao Y., Gu W.. Acetylation is indispensable for p53 activation. Cell. 2008; 133:612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garapaty S., Xu C.F., Trojer P., Mahajan M.A., Neubert T.A., Samuels H.H.. Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation. J. Biol. Chem. 2009; 284:7542–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu J., Jiang J., Li J., Wang S., Shi G., Feng Q., White E., Qin J., Wong J.. Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J. Biol. Chem. 2009; 284:6832–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu E.J., Kim S.H., Heo K., Ou C.Y., Stallcup M.R., Kim J.H.. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor alpha activity and co-activator synergy. Nucleic Acids Res. 2011; 39:6932–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D., Horlings H.M., Shah N., Umbricht C., Wang P. et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011; 43:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tripathi V., Shen Z., Chakraborty A., Giri S., Freier S.M., Wu X., Zhang Y., Gorospe M., Prasanth S.G., Lal A. et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013; 9:e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.