Abstract
Hi-C experiments generate data in form of large genome contact maps (Hi-C maps). These show that chromosomes are arranged in a hierarchy of three-dimensional compartments. But to understand how these compartments form and by how much they affect genetic processes such as gene regulation, biologists and bioinformaticians need efficient tools to visualize and analyze Hi-C data. However, this is technically challenging because these maps are big. In this paper, we remedied this problem, partly by implementing an efficient file format and developed the genome contact map explorer platform. Apart from tools to process Hi-C data, such as normalization methods and a programmable interface, we made a graphical interface that let users browse, scroll and zoom Hi-C maps to visually search for patterns in the Hi-C data. In the software, it is also possible to browse several maps simultaneously and plot related genomic data. The software is openly accessible to the scientific community.
INTRODUCTION
It is well known that the chromosomes in eukaryotic nuclei occupy separate territories, but their exact three-dimensional (3D) organization remains unclear. Previous studies have shown that genes and their regulatory elements can interact even if they are located far apart on the linear chromosome and that these interactions are necessary for effective gene expression (1–3). Chromosome conformation capture techniques (denoted by 3C) have been developed to understand the relationship between chromosome folding and gene regulation; the Hi-C variant can provide spatial contact frequencies between chromosomes at a genome-wide scale (3–6). These Hi-C contact maps have revealed that chromosomes have organized configurations rather than random spatial arrangements. Recently, Hi-C maps have been produced from several species and cell types (6–10). Analyses of these maps could be used to understand the 3D organization of a genome, the mechanism of its formation and its relationship with gene regulation. For example, certain human proteins, such as CCCTC-binding factor (CTCF) and the cohesin complex, seem to play a role in establishing the 3D structure of a genome (11), but the underlying mechanisms are largely unknown.
The full utilization of the large datasets produced by 3C-based techniques, such as Hi-C, requires specific software that can visualize contact frequencies between chromosomal regions alongside genomic datasets, e.g. chromatin immunoprecipitation sequencing (ChIP-seq) mapping data of chromatin factors. These visualizations can also serve to identify artefacts in the contact maps that were introduced during pre- or post-processing calculations. Several visualization software solutions have been developed, such as my5C (12), HiTC (13), HiBrowse (14), WashU epigenome browser (15) and Juicebox (16). Among these, WashU browser and Juicebox can be used for interactive visualization of Hi-C maps in real time along with genomic datasets, such as those produced by ChIP-seq or RNA-seq.
To better understand the mechanisms underlying eukaryotic genome folding, chromosome conformations of different cell types, as well as cells with wild-type and mutant genomes need to be compared. This requires a software that can simultaneously visualize and browse more than one Hi-C map. Currently, HiCPlotter (17) and the WashU epigenome browser (15) can plot multiple maps alongside additional genomic tracks, such as ChIP-seq data. However, HiCPlotter only generates static plots. In the WashU browser, maps are displayed along its diagonal as a half triangle. Since this triangle is severely truncated, maps cannot be browsed to distant coordinates in the off-diagonal direction and additionally, the coordinate on the map does not vertically corresponds to the respective genomic track coordinate. Although in Juicebox (16), multiple maps can be synchronously browsed between multiple independent windows, comparing maps is impractical because these windows must be manually organized on the screen and the syncing is often slow. Therefore, an interactive browser where the user can freely navigate through multiple maps in real time would clearly be preferable for the exploration and comparison of different ccmaps.
Apart from contact map visualization, the analysis of these maps alongside 2D genomic datasets is challenging due to the immense size of the maps. Additionally, a platform is required to develop and implement new methods to analyze these huge Hi-C maps through programming. Therefore, a community supported open-source integrated platform is necessary for both programmers and non-programmers to interactively visualize, develop new methods and analyze Hi-C maps along with genomic track datasets. The Hi-C contact maps, despite their size, should be easily read, processed and analyzed using the platform. The processing will also ensure that the maps used for comparative analysis have been obtained through the same procedure.
An important hurdle for developing such software is the rapid, real-time reading of the contact map, because the maps can reach sizes of tens to hundreds of gigabytes and reading an entire map at once can easily exceed the available computer memory. The current software packages implement various file formats, most of which are flat text files (8,9,18). The browsing of a map only requires a small segment of the entire map, but reading a segment from a large flat file in real time is rather slow. Moreover, these files cannot be used to perform calculations because map reading is time-consuming and loading an entire map into computer memory is impractical. Therefore, there is a clear need for an indexed, simple, easy-to-read/write and portable file format that could be accessible through multiple programming languages.
For these reasons, we have developed genome contact map explorer (gcMapExplorer), a platform that enables a user to browse and analyze genome contact maps with reference to genomic datasets. The package includes an interactive visualizer that facilitates the browsing of several contact maps alongside various datasets, such as those obtained through DNase-seq, ChIP-seq or RNA-seq. We have included an automatic resolution interchanger that allows the user to browse contact maps from the finest available resolution to the whole chromosome level. Additionally, the package includes several normalization methods so that all maps can be normalized with an identical method and can be browsed for comparative study. The implementation of the normalization methods has been designed in such a way that even a large map can be normalized on a desktop computer. To address the file format problem, we used the popular HDF5 format (https://support.hdfgroup.org/HDF5/) for both contact maps and genomic datasets. It is important to note that the HDF5 format is used in several computational genomics packages (19–21). Additionally, we used a memory mapped matrix file to perform calculations on huge contact maps to bypass the need to load these maps into computer memory (22). The chosen file format solved the problems of mathematical operations, browsing, storage and portability, which are commonly associated with large contact maps. Most importantly, when gcMapExplorer is used as a Python module, the user can access contact maps easily and perform custom analyses by writing Python scripts. Additionally, the package contains several interfaces that make it accessible for experimental biologists with no prior programming experience.
MATERIALS AND METHODS
Requirements and installation
gcMapExplorer is written purely in Python3, and uses several standard and external Python modules. The external modules are Numpy, Scipy, matplotlib, appdirs, h5py, Cython and PyQt5. All modules, except for PyQt5, are available from the Python Package Index (PyPI) repository. Python3 and Cython must be installed before the gcMapExplorer package is installed and PyQt5 is only required for graphical user interface (GUI) applications.
On a Linux platform, Python3 and python3-qt5 can be easily installed through software package managers. On a Mac platform, both Python3 and python3-qt5 should be installed using the Homebrew package manager. On a windows platform, we recommend users to use the WinPython3-Qt5 package because it already contains all the dependencies of gcMapExplorer. After Python3 has been installed, gcMapExplorer can be directly installed by using the command ‘pip3 install gcMapExplorer’. All of the depending packages, except for PyQt5, will be automatically installed. The GUI applications (browser, importer, normalizer) cannot be executed if PyQt5 is not installed; however, all commands and the gcMapExplorer library will still be functional for their respective tasks.
Implementation
Our main objective was to make gcMapExplorer a platform for both visualization and analysis. Therefore, the gcMapExplorer package includes three types of user interfaces, the GUI, command line (CLI) and application programming (API) interfaces.
PyQt5 was used to build the GUIs because Qt is one of the most popular GUI libraries. We note that PyQt5 is essential only for GUI applications, while CLI applications and the gcMapExplorer Python module (APIs) can be used without PyQt5. The interactive visualizer’s features were inherited from the Python plotting library Matplotlib. The chromosomal contact map (ccmap) file format was generated using the numpy memmap module (https://docs.scipy.org/doc/numpy/reference/generated/numpy.memmap.html). Both genome contact map (gcmap) and genomic dataset h5 files are in the HDF5 format, and were generated and read through the h5py module. Matrix operations and normalizations were implemented using Numpy and Scipy modules. The Knight and Ruiz (KR) normalization algorithm was ported to Python from the original MATLAB code supplied with the publication (23).
Availability
The gcMapExplorer source code is openly available under the GPL v3 license on Github (https://github.com/rjdkmr/gcMapExplorer) and can be directly downloaded from this site. However, the PyPI repository can also be used to directly install the platform without downloading the package as written above. The manual for gcMapExplorer is freely available on http://gcmapexplorer.readthedocs.io/. This website describes all of the available tools and their purposes. Additionally, detailed documentation regarding the gcMapExplorer Python module exists on this website. We have also included several examples on the website.
RESULTS AND DISCUSSION
We developed the gcMapExplorer, as a platform for the interactive visualization and analysis of genomic contact maps. This platform includes three types of user interfaces, the GUI, CLI and API interfaces that can be used to get the most out of the package’s functionality. The GUIs and CLIs are listed in Table 1.
Table 1. Tools, along with their respective function, presently available in the gcMapExplorer package.
| Tools | Function |
|---|---|
| Graphical User Interfaces (GUI) | |
| browser | To browse contact maps with genomic datasets |
| cmapImporter | To import or convert data files compatible with gcMapExplorer |
| cmapNormalizer | To normalize contact maps with various methods |
| h5Converter | To convert bigwig/wig/bed file to browser compatible hdf5 format |
| Commands for normalization of Hi-C maps | |
| normKR | Knight-Ruiz matrix balancing |
| normIC | Iterative-correction matrix balancing |
| normMCFS | Median contact frequency scaling |
| Commands to import contact map files | |
| coo2cmap | COO sparse matrix format to ccmap or gcmap formats |
| homer2cmap | HOMER Hi-C matrix format to ccmap or gcmap files |
| bc2cmap | Bin-Contact files pair to ccmap or gcmap format |
| pairCoo2cmap | Paired COO sparse matrix format to ccmap or gcmap formats |
| Commands to convert genomic track files | |
| bigwig2h5 | bigwig format to browser compatible hdf5 format |
| wig2h5 | wig format to browser compatible hdf5 format |
| bed2h5 | bed format to browser compatible hdf5 format |
| encode2h5 | Download and convert files from ENCODE portal |
Users of gcMapExplorer will most likely need to convert their own data files into the file format that gcMapExplorer uses internally. Therefore, we developed a GUI and several command interfaces for these operations (Table 1). We also developed similar interfaces for normalizing Hi-C maps because this is an essential step in the contact map analysis (see below for details).
The GUIs and CLIs were developed to provide easy access to the functions offered by the gcMapExplorer package. These tools broaden the scope of our package because no prior programming experience is required to run these commands. In this way, the package provides an easy-to-use interface that allows experimental biologists to process browse and compare genomic contact maps with genomic datasets. On the other hand, the API can be used to write custom analysis programs. The details of these interfaces and commands are discussed below.
Browser for contact maps with genomic datasets
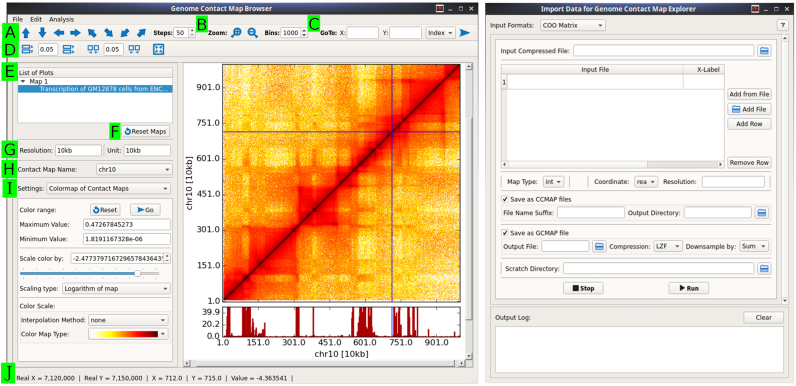
The GUI for interactive visualization is termed browser and was developed to browse both contact maps and genomic datasets, such as those provided by DNase-seq, ChIP-seq or RNA-seq. The browser has a rich, intuitive interface with several options for browsing maps and datasets (Figure 1). To load data inside the browser, gcmap/ccmap/h5 files (see below for details) can be opened through the ‘File’ option on the menu bar. The toolbars at the top contain multiple options for browsing maps and controlling the white space between plots (Figure 1A–D). The left control panel displays a list of maps and datasets, along with other visualization settings, such as color scaling, colormaps, Y-axis scaling for 2D datasets and marker drawing. (Figure 1E–I). In the following section, we describe how to browse one map, as well as two or more maps, along with genomic datasets.
Figure 1.
Browser (left) and importer (right) GUI applications for browsing Hi-C maps and importing data, respectively. Several options in the browser are indicated as follows: (A) Browse along, up, down, left, right, diagonal and off-diagonal of maps; (B) Zoom in and out; (C) Go to a specific coordinate on the map; (D) Change spacing between plots; (E) List all plots; (F) Reset all maps; (G) Resolution and unit of currently active map; (H) Name of contact map; (I) Selectable settings for several options. In the presented state, a colormap has been selected and thus, its options are displayed on the interface. Other available options which are not shown here can be used for genomic dataset plots and markers. (J) Real time status of mouse pointer with contact frequency. At the right side, a graphical user interface (GUI) interface for importing data are shown, with options for converting files with a COO format to either genome contact map (gcmap) or chromosomal contact map (ccmap) files. A menu for selecting the input format is displayed at the top of the window.
After opening a map in the browser, only the first 1000 × 1000 bins of the map are rendered by default. Subsequently, the map can be browsed in any direction by either dragging the map with the mouse button or using the toolbar’s arrow buttons (Figure 1A). The stride with which the arrow buttons scroll can be precisely set through the ‘Steps’ option. Moreover, the user can set precise coordinates, either location on the chromosome or axis units, in the ‘Go To’ option to visualize a specific region of the map (Figure 1C). The user can zoom in and out of the maps in real time by rotating the mouse wheel or using the toolbar’s zoom in and out buttons (Figure 1B). Additionally, a precise zoom level can be attained for a constant resolution by setting values in the ‘Bins’ option. In the case of a gcmap file that contains maps of various resolutions (see below for details), map resolution automatically changes to coarser and finer in real time when bin number is >1000 or <500, respectively. Therefore, the contact map of a whole chromosome can be visualized at the finest to coarsest available resolution in the browser. However, this feature is unavailable when a ccmap file is loaded in the browser as this file contains only a single map (see below for details). The current resolution and bin-size are displayed in the left control panel (Figure 1G). When a gcmap file, which contains several maps for different chromosomes, is opened, the maps can be interchanged by selecting a map based on its given name (Figure 1H). When the mouse pointer hovers over the contact map, the exact X- and Y- coordinates, as well as the contact value, of the pointer’s current position will be displayed in the status bar at the bottom of the browser (Figure 1J).
An essential part of map visualization is using different colors to represent different contact frequencies; in this way, certain features can be highlighted and noise can be suppressed by modifications in the mapping limits. In the browser, the color of contact frequencies changes linearly from a minimum to a maximum value according to a selected colormap. To control this color mapping, we have included several options that can be used to modify the colormap limits in real time (Figure 1I). Additionally, the contact map can be colored in logarithmic space. The user can set the minimum and maximum colormap values and then use a slider to modify the latter value in real time. Moreover, the browser includes more than 30 colormap choices for clear and coherent visualization. Additional colormaps can be created and modified by clicking on ‘Add/modify colormap’ option in the ‘Edit’ menu option.
The browser can also visualize multiple contact maps side-by-side. The browsing is synchronized and therefore, when one map is dragged or zoomed, all other maps will follow the same movements in real time. All of the options discussed above for a single map are also available for multiple maps. Additionally, we have included a marker with horizontal and/or vertical lines that spans all contact maps and simultaneously indicates the precise location of the mouse pointer on all maps (shown as the horizontal and vertical blue lines in Figure 1). This marker eases comparison because it allows the user to pinpoint an interesting region on all of the maps simultaneously by selecting a region on only one of the maps. Although map browsing is synchronized, the color scaling options are different for each map and the user is therefore free to change the colormap limits for each map separately.
The user can compare a contact map with other genomic experimental data by plotting these data above or below the contact map. This feature allows the user to browse both the contact map and genomic data in real-time. The user can also change the Y-axis scale, color and width of the vertical bar through the left panel (selectable setting in Figure 1I). The marker is also extended to these genomic data plots, allowing the user to pinpoint the exact location of peaks.
The browser can be used to produce high-quality publication images. Additionally, we included a rich interface that allows the user to customize axis properties, such as labels, tick labels and fonts. The axis properties dialog box can be accessed by right clicking on a plot. Moreover, page size and layout can be changed through the menu bar options. These interfaces help to provide publication quality plots that can be further saved in several formats, such as png, eps, svg and pdf. Thus, the browser offers both comparative interactive visualization of contact maps and the ability to produce publication quality images.
Detecting functional genomics loci with Hi-C map browsing
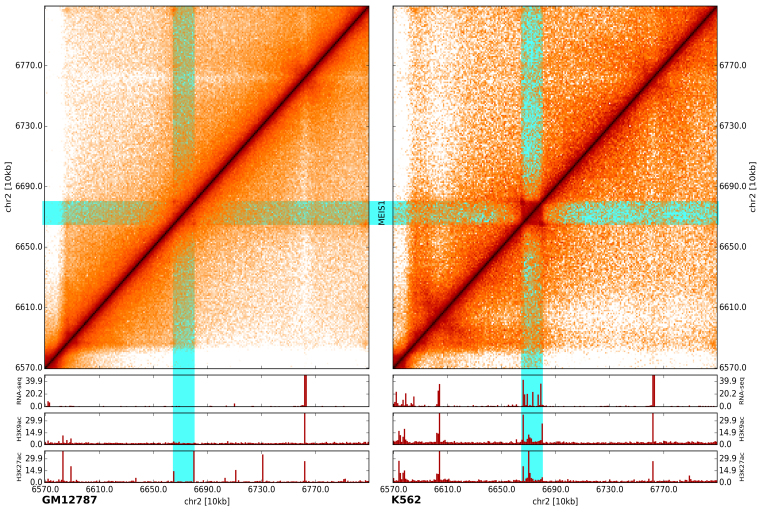
To demonstrate the utility of the developed browser, we compared two 10-kb resolution Hi-C maps from GM12878 and K562 human cell lines together with gene expression data (RNA-seq) and two histone modification (ChIP-seq) datasets. During browsing we were able to easily observe several locations by visual inspection where the cell lines have different chromatin contact patterns and, more importantly, where these differences are associated with gene expression changes and altered histone modification.
For example, Figure 2 shows how chromatin contact patterns in the region surrounding the MEIS1 gene [MEIS1 is linked with acute lymphoblastic leukemia (24) and is expressed in K562, which is an immortalized lymphoblastoid human erythroleukemia cell line.] differ between K562 and GM12878 cell lines. A box-like pattern is present in the K562 cell line (right upper panel), but this pattern is nearly lost in the GM12878 cell line (left upper panel). When this observation is compared with RNA-seq and ChIP-seq data, it shows that MEIS1 is expressed in K562 cells and associated with the active marks H3K9ac and H3K27ac, but inactive in GM128781 cells.
Figure 2.
A comparison of two maps by visualization. Differences in the contact map patterns at the MEIS1 gene locus in GM12787 (left) and K562 (right) cell lines are shown. Gene expression (RNA-seq) and active histone modifications (H3K9ac and H3K27ac, ChIP-seq) (shown below) also differ between the two cell lines at the same locus.
This result illustrates how our browser can help users find potentially interesting genomic loci by browsing entire chromosomes or genomes and visually compare different Hi-C maps, along with any type of genomic track datasets. As compared with Juicebox, that also has synchronized browsing, gcMapExplorer’s browser synchronize the plots faster, in real time. Also, all maps are automatically arranged side-by-side in a single application window and movements of the marker is synced between the maps. Furthermore, flexible customization of color mapping makes it easier to visualize the maps in accordance with user’s comfortability and utility. Therefore, gcMapExplorer provides several advantages over existing program packages for browsing multiple maps and to identify differences between the maps by visual inspection.
Normalization of contact map
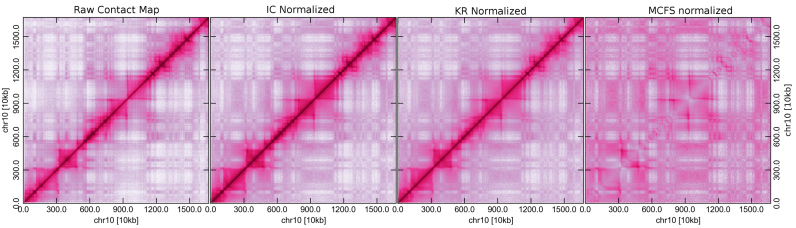
A raw contact map contains systematic biases, which mostly include mappability, GC content and restriction fragment lengths (25). The map should be normalized prior to any analyses to eliminate these biases. Several normalization methods have been developed and are discussed in multiple reviews (26,27). We selected two popular matrix balancing methods because they do not require any additional information other than the raw contact map. The two methods are Iterative Corrections (IC) (28) and an algorithm by KR (23), which can be performed using the normIC and normKR tools (Table 1), respectively. In the matrix balancing method, the sums of rows and columns are equalized through iterative operations. Figure 3 demonstrates the differences between a raw map and maps that are obtained from the IC and KR methods. Normalizations are currently only implemented for cis- or intra-ccmaps.
Figure 3.
A comparison of normalization methods. An example is shown to demonstrate differences between normalized maps and raw maps at a resolution of 20 kb.
KR normalization produces a doubly stochastic matrix in which the sums of rows and columns are equal to one while IC normalization requires an additional step to get a matrix that is similar to those resulting from KR method. Moreover, KR normalization is able to achieve a high degree of accuracy in the sum of rows and columns because sums are equal to one at a precision of more than six digits. We have also included two cut-off schemes to discard very sparse rows and columns during normalizations. In the first scheme, the fraction of data occupancy in a given row or column is calculated, and if the amount of missing data are above the threshold fraction, then the respective row and column is ignored during the calculation. In the second scheme, a percentile of missing-data count is calculated for rows and columns and a row or column will be discarded if the missing-data percentile exceeds the input threshold percentile. From our experience, the first scheme is suitable for coarser resolutions, >100 kb, while the second scheme is suitable for finer resolutions, such as 10, 5 or 1 kb.
In addition to the two normalization methods described above, we have introduced another method which considers the median contact frequency (MCFS) as a function of genomic distance to scale the contact frequency of each locus pair. In this method, the diagonal and near-diagonal loci approach a value of one, a characteristic which can be used to highlight off-diagonal contact frequencies (Figure 3). This method could be especially useful for between contact map comparisons of regions which are distant from the diagonal. This normalization can be performed using the command normMCFS (Table 1).
All of the normalization methods included in our platform are also applicable to large maps. Specifically, KR normalization can be performed using either RAM or disk memory; however, the latter option noticeably slows down the calculation. A GUI normalizer, which allows the user to select and perform any of the three normalization methods, was also developed (Table 1).
Contact map file format
As discussed earlier, contact maps can be immense in size and performing operations on these maps is unfeasible because it may require tens to hundreds of gigabytes of memory/RAM. This type of problem is often addressed by storing the data in a file and reading it in real time during operations. Reading a file during operations is always time-consuming and the file should be in optimal format to reduce the delay in performing operations. To address this problem, we decided to use two different formats for different functions. The ccmap format stores one map in a file while the second gcmap format stores several maps in an individual file. In this way, these two formats address two different problems; ccmap is used when rapid calculations are performed whereas gcmap is used when the main objective is browsing and storage.
The ccmap format is characterized by a 2D memory-mapped matrix file and this format is often used in the numpy python module (22). This file can be used as a 2D matrix and can be used directly in numpy, scipy and related Python modules for any mathematical operations. This file can undergo slicing operations and fancy indexing for the rapid retrieval of a particular matrix segment. As this file can be used as a numpy array, the calculation speed can be maximized by employing functions that use this array as an input. However, this file can be huge without compression and is also only readable through the Python language.
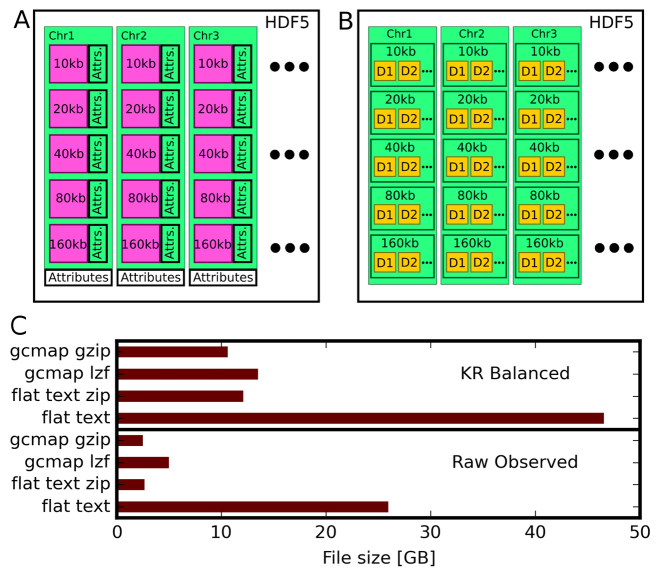
The gcmap file is based on the HDF5 format and contains contact maps in a hierarchical order (Figure 4A). For each chromosome, maps are stored as compressed 2D arrays at several resolutions. Additionally, certain attributes of each map, such as minimum and maximum contact values, resolution and map shape, are stored. We used Lempel-Ziv-Free (LZF) compression to reduce the storage size without a significant impact on reading or writing speed. Although the standard GNU-zip (GZIP) compression was also implemented, its use is not recommended because under this compression both reading and writing are comparatively slow. This format enables us to rapidly access any segment of the contact map for a chromosome at the available resolution. The HDF5 library is available for C, C++, Java, Perl, Python and R languages; therefore, gcmap files can be read through any programming language .
Figure 4.
File format of genome contact maps and genomic datasets. (A) Contact maps (2D-array as magenta) for different chromosomes at various resolutions, along with their attributes. Each map includes attributes such as minimum and maximum contact values, resolution, map shape, while the chromosome’s attributes are names along the X- and Y-axes. (B) Genomic dataset file containing several datasets (1D-array as yellow) at different resolutions. (C) A comparison of contact map file sizes that are in different formats. Sizes for Knight and Ruiz (KR) normalized maps and raw observed maps are shown separately.
As described earlier, the size of conventional files containing contact maps limits their portability. The compression of the 2D arrays within gcmap files was able to overcome this limitation. We compared the size of this gcmap file with the size of a flat text file that uses a popular and memory-efficient coordinate list (COO) sparse matrix format. As shown in Figure 4C, the gcmap file is almost five-times smaller than the flat text file with COO format. Thus, the gcmap file is advantageous for both browsing and storage because the contact map can be read efficiently in real time despite the data compression and reduced size.
We have designed several CLIs, such as coo2cmap, homer2cmap and bc2cmap, that enable the conversion of contact maps into either gcmap or ccmap format (Table 1). Additionally, we developed cmapImporter, a GUI application for easy conversion of contact maps into ccmap and gcmap (Figure 2).
Genomic track dataset file format
For the simultaneous browsing of genomic datasets and contact maps, we needed to choose a file format that could read the data for a given input chromosome segment. We used the HDF5 format again because the popular bigWig format is written in C language and a Python interface is not yet available. As shown in Figure 4B, a single HDF5 file may contain several datasets at various resolutions for all of the chromosomes. It contains precomputed data at several resolutions and any segment of a dataset can be accessed directly from this file, which makes it suitable for the browser.
We have implemented several command-line utilities bigwig2h5, wig2h5 and bed2h5 to convert bigWig, wig and bed format files to gcMapExplorer compatible HDF5 format, respectively. Additionally, a GUI application h5Converter is also implemented for easy conversion of these files to HDF5 format. During conversion, the data are down-sampled to several resolutions through six methods, which are arithmetic mean, geometric mean, harmonic mean, maximum, minimum and median. When plotting, any resolution and down sampling method can be selected for visualization.
Analysis using APIs
The gcMapExplorer is a Python library or module, which allows the scientific community to make valuable contributions to its further development. Several APIs that can be used to perform a wide array of tasks through Python programming are available in this library through submodules. A detailed documentation of the Python modules is provided at http://gcmapexplorer.readthedocs.io. The website also provides several examples for new users. These APIs allow users to perform extensive analyses by writing their own Phython scripts. Several external Python modules, such as numpy, scipy and scikit-learn, can be used to perform mathematical and statistical calculations.
To demonstrate the applicability of the gcMapExplorer Python module, we considered the problem of how to computationally compare two contact maps and identify differences. As an example, we implemented a correlation-based method. We used a sliding box approach where a certain sized block moves along the contact map’s diagonal. For each slide of the block, the correlation between two maps can be calculated through either the Spearman’s or Pearson’s correlation coefficient. The correlation between maps along the diagonal is then expressed as a function of the block’s center. It can be used to identify uncorrelated regions in the maps, which could represent interesting biological differences. The implementation of this method can be found in the online documentation (https://gcmapexplorer.readthedocs.io). We used the gcMapExplorer module to read the contact maps from gcmap and ccmap files, and performed the correlation calculations using numpy and scipy modules.
The gcMapExplorer provides a platform that the scientific community can use to develop and integrate new processing and analytical methods. The APIs are a useful starting point for users with programming experience.
CONCLUSION
Hi-C contact maps have revealed that chromosomes are folded in particular 3D configurations. These maps are now available for several species and cell types. However, software that enable the comparative visualization of these maps alongside their respective genomic datasets are necessary to study genome organization, the mechanisms of its formation and its impact on gene regulation. Although software for processing and visualization exist, none of them offer the comparison of multiple maps through real-time interactive, efficient and fast browsing in a single application instance. Additionally, an integrated platform is necessary for developing new methods, and to analyze Hi-C datasets along with genomic datasets through programming and user interfaces. For this reason, we developed gcMapExplorer, a platform for the normalization, analysis and visualization of contact maps. It contains several interfaces that enable both experimental biologists and programmers to harness its functionality.
The browser is a rich interface that allows the user to browse multiple contact maps side-by-side with their respective genomic datasets. It contains several options and settings that make the browsing experience easy and comfortable. Other interfaces can be used to perform tasks such as the normalization of contact maps or the importing of data to gcMapExplorer compatible formats. The popular HDF5 file format was chosen for fast browsing and storage because this format is portable and data can be rapidly retrieved in real-time. Moreover, this format is readable through several other programming languages. A different file format, ccmap, was chosen for processing and analysis so that optimum speed could be achieved during calculations. This file format allows a computer with normal memory to execute the included normalization methods despite the large size of contact maps.
The gcMapExplorer package is useful for the interactive and comparative visualizations of contact maps. Additionally, it provides a platform that programmers can use to develop new methods to analyze contact maps and genomic datasets. These methods can then be integrated into the package. For example, the processing and generation of Hi-C maps, as well as a method for identifying topological associated domains could be developed for the package in future. The gcMapExplorer presents a platform that researchers can use to study how the 3D organization of a genome influences gene regulation.
AVAILABILITY
The source code of gcMapExplorer is available at https://rjdkmr.github.io/gcMapExplorer under GNU GPL v3 license and documentation is available at http://gcmapexplorer.readthedocs.io.
FUNDING
Kempe Foundation [JCK-1347 to L.L., P.S.]; Knut and Alice Wallenberg Foundation [2014–0018 to EpiCoN, co-PI, P.S.]. Funding for open access charge: Knut and Alice Wallenberg foundation [2014-0018 to EpiCoN].
Conflict of interest statement. None declared.
REFERENCES
- 1. Belmont A.S. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr. Opin. Cell Biol. 2014; 26:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dixon J.R., Gorkin D.U., Ren B.. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016; 62:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pombo A., Dillon N.. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015; 16:245–257. [DOI] [PubMed] [Google Scholar]
- 4. de Wit E., de Laat W.. A decade of 3C technologies: insights into nuclear organization. Gene Dev. 2012; 26:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dekker J., Rippe K., Dekker M., Kleckner N.. Capturing chromosome conformation. Science. 2002; 295:1306–1311. [DOI] [PubMed] [Google Scholar]
- 6. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crane E., Bian Q., McCord R.P., Lajoie B.R., Wheeler B.S., Ralston E.J., Uzawa S., Dekker J., Meyer B.J.. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015; 523:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- 10. Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., Hadjur S.. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015; 10:1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghirlando R., Felsenfeld G.. CTCF: making the right connections. Genes Dev. 2016; 30:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lajoie B.R., van Berkum N.L., Sanyal A., Dekker J.. My5C: web tools for chromosome conformation capture studies. Nat. Methods. 2009; 6:690–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Servant N., Lajoie B.R., Nora E.P., Giorgetti L., Chen C.J., Heard E., Dekker J., Barillot E.. HiTC: exploration of high-throughput ‘C’ experiments. Bioinformatics. 2012; 28:2843–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paulsen J., Sandve G.K., Gundersen S., Lien T.G., Trengereid K., Hovig E.. HiBrowse: multi-purpose statistical analysis of genome-wide chromatin 3D organization. Bioinformatics. 2014; 30:1620–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou X., Lowdon R.F., Li D., Lawson H.A., Madden P.A., Costello J.F., Wang T.. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat. Methods. 2013; 10:375–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Durand N.C., Robinson J.T., Shamim M.S., Machol I., Mesirov J.P., Lander E.S., Aiden E.L.. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016; 3:99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Akdemir K.C., Chin L.. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 2015; 16:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E.. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015; 16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoffman M.M., Buske O.J., Noble W.S.. The Genomedata format for storing large-scale functional genomics data. Bioinformatics. 2010; 26:1458–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T. et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat. Methods. 2015; 12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pyl P.T., Gehring J., Fischer B., Huber W.. h5vc: scalable nucleotide tallies with HDF5. Bioinformatics. 2014; 30:1464–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Der Walt S., Colbert S.C., Varoquaux G.. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011; 13:22–30. [Google Scholar]
- 23. Knight P.A., Ruiz D.. A fast algorithm for matrix balancing. IMA J. Numer. Anal. 2012; 33:1029–1047. [Google Scholar]
- 24. Wong P., Iwasaki M., Somervaille T.C., So C.W., Cleary M.L.. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007; 21:2762–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yaffe E., Tanay A.. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011; 43:1059–1065. [DOI] [PubMed] [Google Scholar]
- 26. Ay F., Noble W.S.. Analysis methods for studying the 3D architecture of the genome. Genome Biol. 2015; 16:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lajoie B.R., Dekker J., Kaplan N.. The Hitchhiker's guide to Hi-C analysis: practical guidelines. Methods. 2015; 72:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A.. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012; 9:999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]