Abstract
Mitochondrial aprataxin (APTX) protects the mitochondrial genome from the consequence of ligase failure by removing the abortive ligation product, i.e. the 5′-adenylate (5′-AMP) group, during DNA replication and repair. In the absence of APTX activity, blocked base excision repair (BER) intermediates containing the 5′-AMP or 5′-adenylated-deoxyribose phosphate (5′-AMP-dRP) lesions may accumulate. In the current study, we examined DNA polymerase (pol) γ and pol β as possible complementing enzymes in the case of APTX deficiency. The activities of pol β lyase and FEN1 nucleotide excision were able to remove the 5′-AMP-dRP group in mitochondrial extracts from APTX−/− cells. However, the lyase activity of purified pol γ was weak against the 5′-AMP-dRP block in a model BER substrate, and this activity was not able to complement APTX deficiency in mitochondrial extracts from APTX−/−Pol β−/− cells. FEN1 also failed to provide excision of the 5′-adenylated BER intermediate in mitochondrial extracts. These results illustrate the potential role of pol β in complementing APTX deficiency in mitochondria.
INTRODUCTION
Aprataxin (APTX) removes the 5′-adenylate (5′-AMP) lesion from abortive ligation products and base excision repair (BER) intermediates (1,2). APTX has been associated with the BER and single-strand break repair (SSBR) pathways (3,4), and protein–protein interactions between APTX and BER/SSBR factors, such as X-ray repair cross-complementing protein 1 (XRCC1) and poly[ADP-ribose] polymerase 1 (PARP-1), were reported (5–7). The APTX protein was also recovered in a DNA polymerase (pol) β immunoaffinity pull down fraction along with other BER/SSBR factors (8). Mutations in the APTX gene (aptx) are linked to the autosomal recessive neurodegenerative disorder Ataxia with oculomotor apraxia type 1 (AOA1) (9). However, APTX null cells fail to show a hypersensitivity phenotype when treated with DNA damage-inducing agents (10,11). Similarly, normal repair of base lesions and strand breaks have been reported in an APTX deficient mouse model (12). One explanation for these observations could be that the APTX deficiency is complemented by other enzymes or repair pathways. In this respect, we previously identified complementation roles of the pol β lyase activity in single-nucleotide (SN) BER and flap endonculease 1 (FEN1) strand excision in the long patch (LP) sub-pathway of BER (13,14). In this previous study, whole cell extracts derived from both AOA1 patients and APTX−/− DT40 cells were used, and repair of a stalled BER intermediate was demonstrated with the 5′-adenylated-deoxyribose phosphate (5′-AMP-dRP) group via pol β and FEN1 (14).
APTX is localized in mitochondria and mitochondrial dysfunction in the AOA1 mouse model and in APTX−/− HeLa cells was reported (15–17). In mitochondria, pol γ is the main DNA polymerase capable of functioning in mitochondrial DNA replication and BER (18,19). Pol γ has 5′-dRP lyase activity that appears to contribute to mitochondrial SN BER (20). Moreover, LP BER mediated by pol γ, along with required nuclease activities, has been identified in mitochondrial extracts, and LP BER was significantly diminished after FEN1 immunodepletion (21,22). The questions of APTX complementing activity in mitochondria and association with mitochondrial DNA repair in AOA1 pathobiology are under investigation. In an important recent development, however, pol β has been found to have a role in mitochondrial DNA repair (23).
In the present study, we questioned whether the pol β lyase is capable of serving a back up role during mitochondrial BER in APTX deficient cells. We also tested the activity of purified pol γ against a 5′-adenylated BER intermediate. Purified pol γ had only very weak lyase activity for removal of the 5′-AMP-dRP group, and the accessory subunit of pol γ (pol γB) had no effect on this activity. In mitochondrial extracts from APTX−/− DT40 cells, pol β lyase activity and FEN1-mediated strand excision were able to remove the 5′-AMP-dRP blocking group. However, in mitochondrial extracts from APTX−/− Pol β−/− cells, no activity was observed against the 5′-adenylated BER intermediates. These results highlight the potential of APTX deficiency for harmful effects on mitochondrial genome integrity in the absence of pol β.
MATERIALS AND METHODS
Materials
Recombinant wild-type human DNA ligase III, pol β, FEN1 and uracil–DNA glycosylase (UDG), with 84 amino acids deleted from the N-terminus, were purified as described (13,14). The exonuclease deficient form of the recombinant wild-type human pol γ catalytic subunit (pol γA) and the accessory subunit (pol γB) were purified as described (18,19,24). Recombinant human APTX and the 5′-DNA adenylation kit were from Fitzgerald and New England BioLabs, respectively. Oligodeoxyribonucleotides with a 6-carboxyfluorescein (FAM) label at the 3′-end were from Integrated DNA Technology. The gapped and nicked DNA substrates used in this study are illustrated in Supplementary Table S1. The DNA substrates were 5′-adenylated using DNA adenylation kit as described (13).
Cell lines and growth conditions
The wild-type and pol β single knockout (Pol β−/−) DT40 cell lines used in this study were reported previously (25). APTX single (APTX−/−) and APTX/Pol β double (APTX−/−Pol β−/−) knockout DT40 cells were constructed as reported (25). Briefly, APTX-ecogptR was generated from genomic PCR products combined with the ECOGPTR selection marker gene. Genomic DNA sequences were amplified using the primers 1–4 (Supplementary Table S2) that were inserted into NheI and AflII sites, respectively, of DT-A-pA/loxP/PGK-ECOGPT-pA/loxP to create APTX-ECOGPTR (Supplementary Figure S1A), using the GENEART seamless cloning and assembly kit (Life Technologies). APTX−/− and APTX−/− Pol β−/− DT40 cells were generated using the clustered, regularly interspaced short palindromic repeat (CRISPR) method as reported (26). Briefly, the guide sequence (5′-GAGCAGGGGAATCAAAATCT-3′) was inserted into the pX330 vector (APTX-CRISPR), and DT40 cells were transfected with 2 μg targeting vector (APTX-ECOGPTR) and 6 μg of the guide sequence-containing pX330 vector (APTX-CRISPR), using the NEON Transfection System (Life Technologies) and according to the manufacture's instructions. After 16 h, the cells were plated in 96-well plates and then subjected to ecogpt (30 μg/ml) selection. The drug-resistant cell colonies were picked on days 6–7 after transfection. The loss of the APTX transcript in these cells was confirmed by RT-PCR using the primers 5 and 6 (Supplementary Table S2). The β-actin transcript was analyzed as a positive control for the RT-PCR analysis using the primers 7 and 8 (Supplementary Figure S1B). DT40 cells used in this study were cultured and maintained as reported (14).
Preparation of mitochondrial extracts
Mitochondria were isolated from the wild-type, APTX−/−, Pol β−/− and APTX−/− Pol β−/− DT40 cells (1.05 g wet pellet) using a two-step discontinuous sucrose gradient method as described (27,28). Briefly, the mitochondria from the sucrose gradient fraction were pelleted and lysed in 1% Triton X-100, 10% glycerol, 20 mM Tris–HCl, pH 8.0, 0.2 mM EDTA, 14 mM 2-mercaptoethanol and protease inhibitors. The broken mitochondria were centrifuged for 2 h at 50 000 g, and the supernatant fraction was aliquoted, frozen and stored at −80°C.
Immunoblot analysis of mitochondrial extracts
Proteins (25 μg) in purified mitochondrial extract samples were separated by SDS-PAGE using Nu-PAGE 4–12% Bis-Tris minigel electrophoresis and transferred onto nitrocellulose membranes. The membranes were incubated in 5% nonfat dry milk in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBS-T) and then probed with either affinity purified polyclonal antibody to pol β, FEN1 or rabbit monoclonal antibody to pol γ (Abcam). Goat anti-rabbit or goat anti-mouse IgG conjugated to horseradish peroxidase (1:10 000 dilution) was used as secondary antibody (Abcam), and the immobilized horseradish peroxidase activity was detected by SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). Membranes were stripped by incubation in Restore Western Blot Stripping Buffer (Thermo Scientific) for 30 min at room temperature, followed by three washes with TBS-T. Then, the membranes were used for probing with another antibody as above.
Enzymatic assays using purified proteins
For DNA ligation assays, the single-nucleotide gapped DNA substrate including tetrahydrofuran (THF), an abasic site analog, at the 5′-end of the DNA in the gap was used (Supplementary Table S1). The ligation reaction by DNA ligase III was measured as described (13). Briefly, the reaction mixture (10 μl) contained DNA substrate (140 nM) and DNA ligase III (100 nM), 50 mM NaMOPS, pH 7.5, 1 mM DTT, 0.05 mg/ml BSA, 100 mM NaCl, 0.1 mM ATP and 1 mM MgCl2. The reaction mixture was incubated at 37°C for the indicated times, and then the reaction was terminated by mixing with gel loading buffer (95% formamide, 20 mM EDTA, 0.02% bromphenol blue, and 0.02% xylene cyanol). The reaction products were separated by electrophoresis in a 15% polyacrylamide gel, and detected by a PhosphorImager as reported (13).
For dRP lyase activity measurements, the single-nucleotide gapped DNA substrates with a 5′-uracil base, either with adenylation (5′-AMP-dRP) or without adenylation (5′-dRP), were used (Supplementary Table S1). The lyase assays with purified exonuclease deficient catalytic subunit of pol γ (pol γA) or pol β were performed as reported (13). Briefly, the reaction mixture (10 μl) contained 50 mM HEPES, pH 7.5, 20 mM KCl, 0.5 mM EDTA, 2 mM DTT, and the indicated concentrations of pol γA or pol β. The reaction was initiated by addition of UDG-pretreated DNA substrates to a final concentration of 100 nM, and the incubation was at 37°C for the indicated times. The reaction products were stabilized by addition of 100 mM NaBH4, incubated on ice for 30 min, and then mixed with gel loading buffer. For the single-turnover kinetic measurements, the dRP lyase assays were performed with enzyme in 5-fold excess over the DNA substrate. For the dRP lyase assays in the presence of the purified accessory subunit of pol γ (pol γB), the holoenzyme, including pol γA:pol γB mixture at a 1:4 ratio, was reconstituted as reported (24). The lyase activity was measured as described above. The reaction products were separated, and the data were analyzed as described above.
The enzymatic assays for APTX deadenylation and FEN1 nucleotide excision activities were as reported (13). For these assays, the nicked DNA substrate containing 5′-AMP was used (Supplementary Table S1). Briefly, APTX activity was determined in a reaction mixture (10 μl) including 50 mM HEPES, pH 7.5, 20 mM KCl, 0.5 mM EDTA, 2 mM DTT and 100 nM DNA. FEN1 activity was determined in a reaction mixture (10 μl) including 50 mM HEPES, pH 7.5, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA and 100 nM DNA. In both cases, the reaction was initiated by addition of 100 nM APTX or FEN1, and the reaction mixture was incubated at 37°C for 15 min. The reactions were stopped by adding gel loading buffer.
For the nicked 3 nucleotide (nt) flap-including DNA substrate (Supplementary Table S1), FEN1 excision activity was determined as reported (29). Briefly, the reaction mixture (10 μl) included 50 mM Tris–HCl, pH 7.5, 50 mM KCl, 0.2 mM EDTA, 5 mM MgCl2 and 0.1 mg/ml BSA. The reaction was initiated by addition of 100 nM FEN1, and the reaction mixture was incubated at 37°C for 15 min. The reaction products were separated, and the data were analyzed as described above.
Enzymatic assays in mitochondrial extracts
The dRP lyase, FEN1 excision and APTX deadenylation assays for reference reactions were performed as described above. The enzymatic activity measurements for the reaction mixtures with mitochondrial extracts were performed as reported (14). Briefly, the reaction was initiated by addition of mitochondrial extract (15 μg), and the reaction mixture was incubated at 37°C for the indicated times. For the dRP lyase assays with the gapped 5′-dRP or 5′-AMP-dRP DNA substrates, the reactions were terminated by the addition of 100 mM NaBH4. For the enzymatic activity measurements with the DNA substrates 5′-AMP or 3 nt-flap, the reactions were stopped by mixing with gel loading buffer. The reaction products were separated, and the data were analyzed as described above.
RESULTS
DNA ligation by mitochondrial DNA ligase III
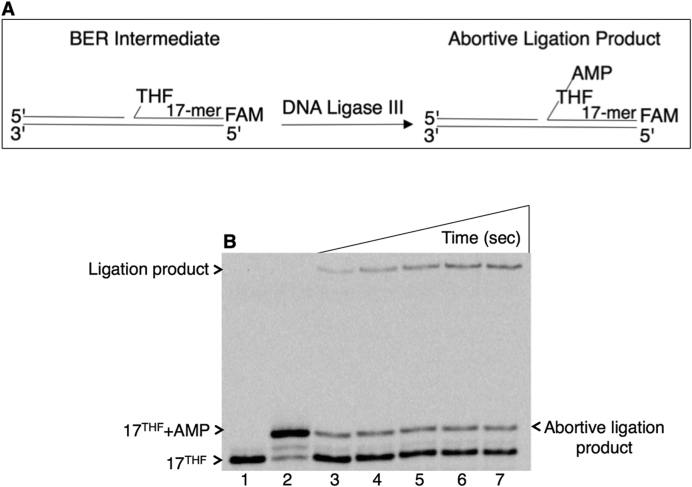
To evaluate abortive ligation in mitochondria, the DNA ligation reaction by mitochondrial DNA ligase III was first examined on the single-nucleotide gapped DNA substrate with a 5′-tetrahydrofuran (THF) group in place of the deoxyribose (dRP) abasic site (Supplementary Table S1). This model ligase substrate mimics a BER intermediate that can be formed during mitochondrial base lesion repair. Ligase failure on such an intermediate could result in formation of the abortive ligation product with an adenylate (AMP) group attached to the 5′-end of the intermediate (Figure 1A). The results revealed that abortive ligation occurs with this substrate leading to formation of the 5′-adenylated-THF-containing BER intermediate (Figure 1B, lanes 3–7). The enzyme also produced the successful ligation product (Figure 1B, lanes 3–7) that was weaker than the abortive ligation product (Figure 1B, lanes 3–5).
Figure 1.
Ligation failure by DNA ligase III on the 5′-dRP-including BER intermediate. (A) Illustrations of the single-nucleotide gapped DNA substrate with 5′-THF (17THF) and abortive ligation product (17THF+AMP). (B) Lane 1 is the minus enzyme control, and lane 2 is the adenylated oligonucleotide used as a size marker. Lanes 3–7 illustrate abortive ligation products and correspond to time points 10, 20, 30, 40 and 60 s. A representative gel of three independent experiments is presented.
Pol γ activity against the 5′-adenylated-dRP-containing BER intermediate
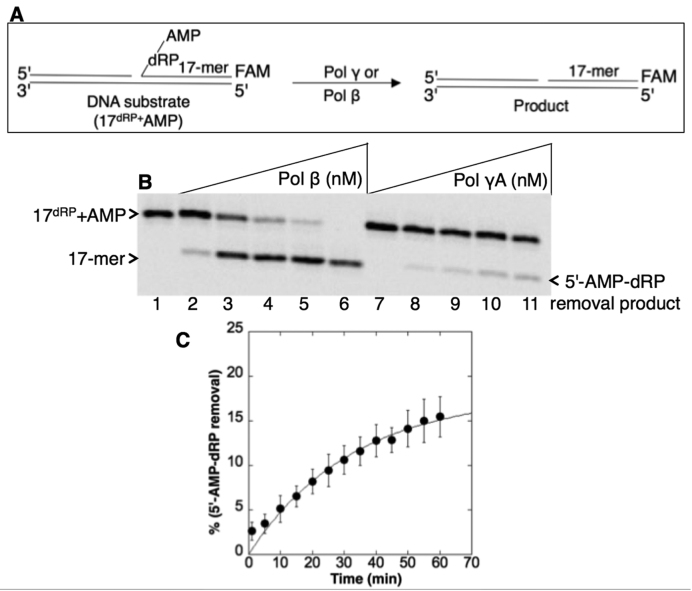
Next the pol γ enzymatic activity against the BER intermediates mimicking abortive ligation products was examined. For this purpose, we constructed a DNA substrate including 5′-AMP-dRP (Supplementary Table S1). The lyase activity of purified pol γ was compared with that of purified pol β (Figure 2A). Pol γ was able to remove the 5′-AMP-dRP group (Figure 2B, lanes 8–11), but the activity of pol β was much stronger (Figure 2B, lanes 2–6), as expected (13). In control experiments, we confirmed pol γ lyase activity against the regular BER dRP lyase substrate, i.e., a single-nucleotide gapped DNA with the 5′-dRP group (Supplementary Table S1), and compared the activity with that of pol β (Supplementary Figure S2A). Pol γ was active against this substrate (Supplementary Figure S2B, lanes 8–11); the activity was weaker than that of pol β (Supplementary Figure S2B, lanes 2–6), consistent with previous results (20). Importantly, however, the removal of the 5′-AMP-dRP group by the pol γ lyase activity was much weaker than for 5′-dRP removal, cf. compare lanes 7–11 in Figure 2B with lanes 7–11 in Supplementary Figure S2B. To extend these observations, we measured the rate of pol γ lyase activity for 5′-AMP-dRP removal under single-turnover kinetic conditions with enzyme in excess over substrate. The data for product formation fit to an exponential curve with a rate constant of 0.03 min−1 (Figure 2C).
Figure 2.
Purified pol γ lyase activity on the 5′-AMP-dRP-including BER intermediate. (A) Illustrations of the single-nucleotide gapped DNA substrate with 5′-AMP-dRP (17dRP+AMP) and the dRP lyase reaction product (17-mer). (B) Lane 1 is the minus enzyme control. Lanes 2–6 are the positive control reaction products of pol β (20–500 nM), and lanes 7–11 are the reaction products of pol γ (20–500 nM) for removal of the 5′-AMP-dRP group. (C) Rate of 5′-AMP-dRP removal by pol γ. The dRP lyase assay was performed under single-turnover conditions with purified pol γ (500 nM) in excess over the DNA substrate (100 nM). A plot of product formation was obtained by fitting the data to an exponential time course, yielding the rate of pol γ as kobs of 0.03 min−1. The data represent the average of three independent experiments ± SD.
As a negative control, purified pol γ enzymatic activity was tested against another BER intermediate mimicking an abortive ligation product. For this purpose, a DNA substrate including the 5′-AMP group was constructed (Supplementary Table S1), and a comparison of pol γ and the activities of APTX and FEN1 were made (Supplementary Figure S3A). Pol γ failed to function on this substrate (Supplementary Figure S3B, lanes 5–10), and this also was true for pol β (Supplementary Figure S3B, lane 4), while APTX and FEN1 were active on the (Supplementary Figure S3B, lanes 2 and 3, respectively), as reported (13).
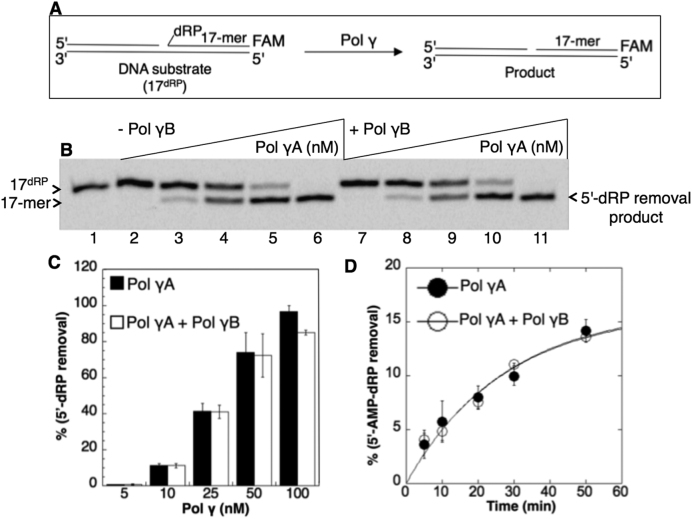
Effect of the pol γ accessory subunit on the pol γ lyase activity
The experiments shown in Figure 2 and Supplementary Figure S2 and Supplementary Figure S3 were conducted with the catalytic subunit of pol γ (pol γA). In order to test the holoenzyme containing the pol γ accessory subunit (pol γB), the holoenzyme was reconstituted by preincubating the purified subunits, and then the lyase activities of the holoenzyme and catalytic subunit alone were compared (Figure 3). For the 5′-dRP-containing substrate (Figure 3A), the lyase activity was tested in a concentration range (5–100 nM) of the catalytic subunit, pol γA, in the absence (Figure 3B, lanes 2–6) and presence (Figure 3B, lanes 7–11) of pol γB. The catalytic subunit and holoenzyme exhibited similar lyase activities for 5′-dRP removal (Figure 3C). For the 5′-AMP-dRP-containing substrate, we compared 5′-AMP-dRP removal under single-turnover conditions (Figure 3D), and the results revealed similar rate constants for the two enzyme forms: 0.036 min−1 (pol γA) and 0.04 min−1 (pol γA+pol γB).
Figure 3.
Lack of influence of the pol γB subunit on the pol γA dRP lyase activity. (A) Illustrations of the single-nucleotide gapped DNA substrate (17dRP) and the dRP lyase reaction product (17-mer). (B) Lane 1 is the minus enzyme control. Lanes 2–6 are the reaction products of 5′-dRP removal by pol γA alone (5–100 nM). Lanes 7–11 are the reaction products of 5′-dRP removal by the holoenzyme including pol γA (5–100 nM) plus pol γB (20–400 nM). (C) The graph shows pol γ concentration-dependent changes in the products of 5′-dRP removal in the absence and presence of the pol γB subunit. The data represent the average of three independent experiments ± SD. (D) The rates of the 5′-AMP-dRP removal are 0.036 min−1 (pol γA) and 0.04 min−1 (pol γA+pol γB). The lyase assays were performed in the absence and presence of pol γ accessory subunit (pol γB) under same single-turnover conditions. The data represent the average of three independent experiments ± SD.
Characterization of mitochondrial extracts from DT40 cells
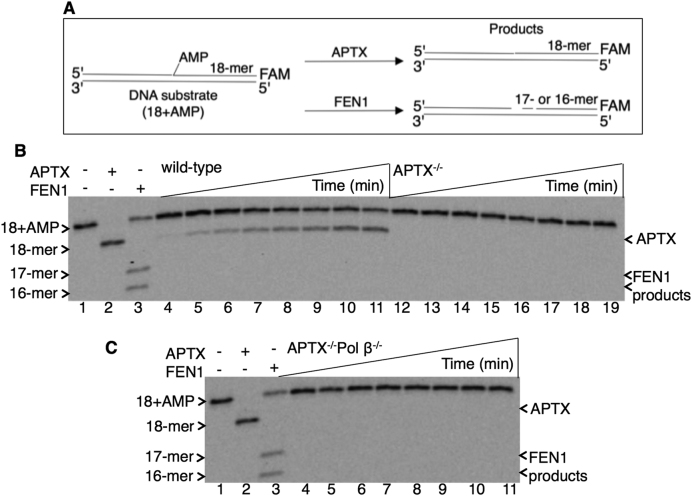
In control experiments, the presence or absence of pol γ, pol β, and FEN1 in the mitochondrial extracts was verified by immunoblotting (Supplementary Figure S4), and the deficiency in APTX activity for 5′-AMP removal was confirmed using whole cell extracts from APTX−/− and APTX−/−Pol β−/− DT40 cells (Supplementary Figure S5). Purified pol γ is able to remove the 5′-AMP-dRP group (Figure 2), but we questioned whether this weak lyase function of pol γ could be strong enough to provide complementation in the case of APTX deficiency during mitochondrial BER. For this purpose, extracts from mitochondria isolated from wild-type, APTX−/−, Pol β−/− and APTX−/−Pol β -/− DT40 cells were prepared. Pol β null cells were used to enable the interpretation of results on the pol γ lyase activity (30). APTX activity was first analyzed in the mitochondrial extracts (Figure 4A), and a regular APTX substrate, i.e. nicked DNA with a 5′-AMP group, was used (Supplementary Table S1). In reference reactions with purified APTX and FEN1, APTX removed the 5′-AMP group (lane 2, Figure 4B and C), and FEN1 removed the 5′-AMP group along with the adjacent first and/or second nucleotide (lane 3, Figure 4B and C). APTX activity, i.e., 5′-AMP group removal, in the mitochondrial extract from wild-type cells was observed (Figure 4B, lanes 4–11). In contrast, mitochondrial extracts from both APTX−/− (Figure 4B, lanes 12–19) and APTX−/−Pol β−/− (Figure 4C, lanes 4–11) cells were devoid of this activity. In both of these APTX null mitochondrial extracts, FEN1 activity was not detected. This was not in agreement with the strong FEN1 activity observed in whole cell extracts from wild-type and APTX null cells (Supplementary Figure S5).
Figure 4.
Processing of the 5′-AMP-containing BER intermediate in mitochondrial extracts. (A) Illustrations of the nicked DNA substrate with 5′-AMP (18+AMP) and the reaction products of APTX (18-mer) and FEN1 (17- and 16-mer). In panels B and C, lane 1 is the minus enzyme control; lanes 2 and 3 are the reference reaction products of APTX and FEN1, respectively. (B) Lanes 4–11 and 12–19 are the reaction products observed in mitochondrial extracts from wild-type and APTX−/− cells, respectively, and correspond to time points 1, 3, 5, 10, 15, 20, 25 and 30 min. (C) Lanes 4–11 are the reaction products observed in mitochondrial extracts from APTX−/−Pol β−/− cells and correspond to time points 1, 3, 5, 10, 15, 20, 25 and 30 min. A representative gel of three independent experiments is presented.
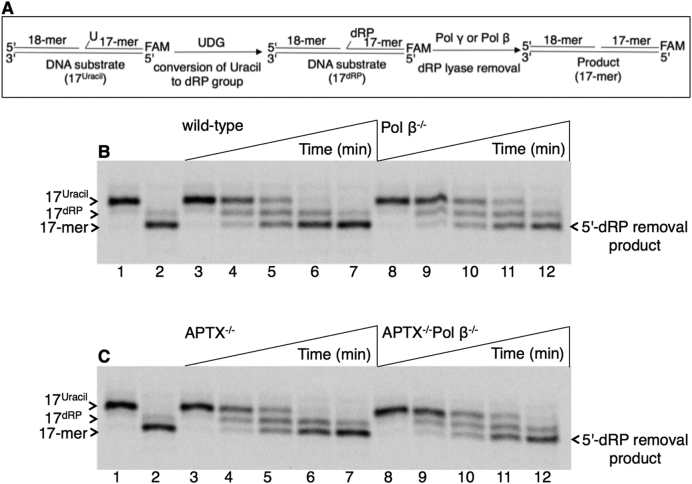
The dRP lyase activity was next examined in the mitochondrial extracts (Figure 5A). For these assays, the regular BER dRP lyase substrate was used, i.e. a single-nucleotide gapped DNA with the 5′-dRP group (Supplementary Table S1). In reference reactions with purified pol β, 5′-dRP group removal with this substrate was verified (lane 2, Figure 5B and C). In both wild-type (Figure 5B, lanes 3–7) and APTX−/− (Figure 5C, lanes 3–7) mitochondrial extracts, a similar amount of product formation for the 5′-dRP group removal was observed. This could be due to 5′-dRP group removal via the pol β lyase activity. However, in mitochondrial extracts from Pol β−/− (Figure 5B, lanes 8–12) and APTX−/−Pol β−/− (Figure 5C, lanes 8–12) cells, comparable amounts of 5′-dRP removal were observed. These results confirmed the presence of pol γ dRP lyase activity in the DT40 mitochondrial extracts.
Figure 5.
Processing of the 5′-dRP-containing BER intermediate in mitochondrial extracts. (A) Illustrations of the single-nucleotide gapped DNA substrate before (17Uracil) and after (17dRP) UDG treatment and the reaction product after 5′-dRP group removal (17-mer). In panels B and C, lane 1 is the minus enzyme control, and lane 2 is the reference reaction product for 5′-dRP removal by purified pol β. (B) Lanes 3–7 and 8–12 are the reaction products observed in mitochondrial extracts from wild-type and Pol β−/− cells, respectively, and correspond to time points 5, 15, 30, 45 and 60 min. (C) Lanes 3–7 and 8–12 are the reaction products observed in mitochondrial extracts from APTX−/− and APTX−/−Pol β−/− cells, respectively, and correspond to time points 5, 15, 30, 45 and 60 min. A representative gel of three independent experiments is presented.
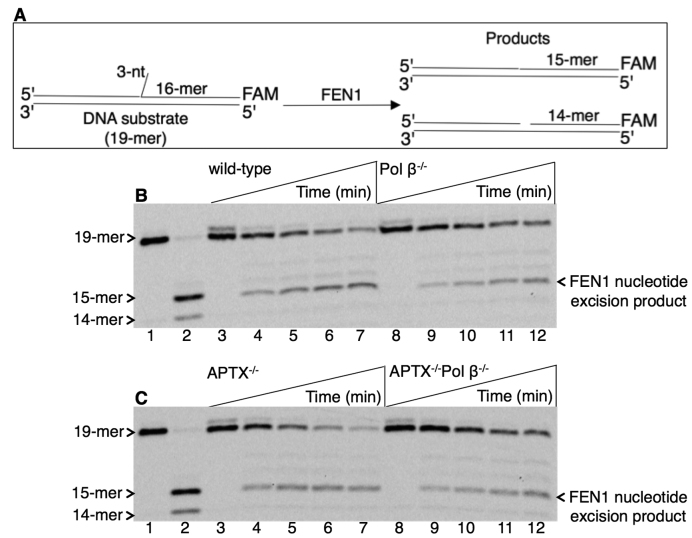
Mitochondrial extracts were next examined for the presence of FEN1 activity via LP BER (Figure 6A). For this purpose, a regular FEN1 substrate, i.e. a 3 nt flap-containing nicked DNA, was used (Supplementary Table S1). In reference reactions with purified FEN1, the expected nucleotide excision products were observed (lane 2, Figure 6B and C). With mitochondrial extract from wild-type (Figure 6B, lanes 3–7) and APTX−/− (Figure 6C, lanes 3–7) cells, flap removal along with the adjacent first nucleotide excision was observed. In mitochondrial extracts from Pol β−/− (Figure 6B, lanes 8–12) and APTX−/−Pol β−/− (Figure 6C, lanes 8–12) cells, slightly weaker FEN1 excision products also were observed. This could be due to a stimulatory effect of pol β on FEN1 activity, as reported (29). In order to examine this notion, FEN1 activity was tested in the absence and presence of purified APTX or pol β using this flap-containing DNA substrate (Supplementary Figure S6). FEN1-mediated nucleotide excision products were observed with increasing concentrations of pol β (Supplementary Figure S6A, compare lane 2 with lanes 3–7), while an effect of APTX on FEN1 activity was not detected (Supplementary Figure S6B, compare lanes 2–7 with lanes 8–13).
Figure 6.
Processing of the flap-including BER intermediate in mitochondrial extracts. (A) Illustrations of the DNA substrate (19-mer) and reaction products of FEN1 excision (15- and 14-mer). In panels B and C, lane 1 is the minus enzyme control, and lane 2 is the reference reaction product by purified FEN1. (B) Lanes 3–7 and 8–12 are the reaction products observed in mitochondrial extracts from wild-type and Pol β−/− cells, respectively, and correspond to time points 5, 15, 30, 45 and 60 min. (C) Lanes 3–7 and 8–12 are the reaction products observed in mitochondrial extracts from APTX−/− and APTX−/−Pol β−/− cells, respectively, and correspond to time points 5, 15, 30, 45 and 60 min. A representative gel of three independent experiments is presented.
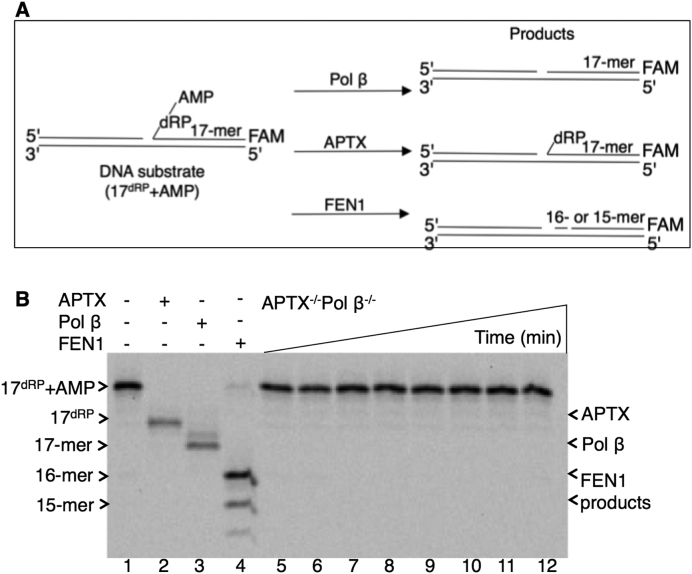
Processing of the BER intermediate with the 5′-adenylated-dRP block in mitochondrial extracts
Potential APTX deficiency complementation roles of the pol γ lyase and FEN1 nucleotide excision activities were examined using mitochondrial extracts from APTX−/− Pol β−/− DT40 cells (Figure 7). For these assays, the substrate was single-nucleotide gapped DNA with the 5′-AMP-dRP group (Supplementary Table S1). In reference reactions with purified enzymes, the expected products of APTX, pol β and FEN1 (Figure 7B; lanes 2, 3 and 4, respectively) were observed, confirming the status of the substrate (Figure 7A). Surprisingly, products of 5′-AMP-dRP group removal and FEN1 excision in this mitochondrial extract were not found (Figure 7B, lanes 5–12), even with increased amounts of the mitochondrial extract (Supplementary Figure S7A). Note that the mitochondrial extract from APTX−/− Pol β−/− cells was positive for pol γ lyase and FEN1 activities when measured against their regular substrates (Figures 5 and 6). Thus, with the substrate containing the 5′-AMP-dRP group, the activities of pol γ and FEN1 were not strong enough to provide APTX deficiency complementation. In the mitochondrial extract from Pol β−/− cells, we confirmed 5′-AMP group removal only from this substrate (Supplementary Figure S7B).
Figure 7.
Processing of the 5′-AMP-dRP-containing BER intermediate in mitochondrial extracts from APTX−/−Pol β−/− cells. (A) Illustrations of the single-nucleotide gapped 5′-AMP-dRP-containing DNA substrate (17dRP+AMP) and reaction products after pol β 5′-AMP-dRP removal (17-mer), APTX 5′-AMP removal (17dRP), and FEN1 removal of 5′-AMP plus one or two nucleotides (16- or 15-mer). (B) Lane 1 is the minus enzyme control, lanes 2, 3, and 4 are the reference reaction products of purified enzymes APTX, pol β and FEN1, respectively. Lanes 5–12 are the reaction products observed in mitochondrial extracts from APTX−/− Pol β−/− cells and correspond to time points 1, 5, 10, 15, 20, 30, 45 and 60 min. A representative gel of three independent experiments is presented.
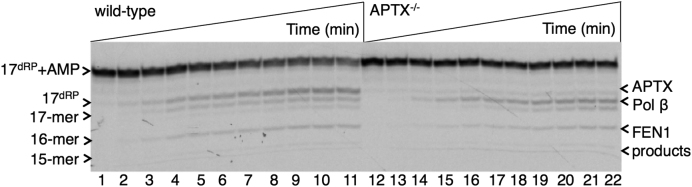
Complementation of APTX deficiency by mitochondrial pol β
Finally, using DT40 cell mitochondrial extracts, we examined a potential APTX deficiency complementation role for mitochondrial pol β (23). The substrate containing the 5′-AMP-dRP group was used, and mitochondrial extracts from wild-type and APTX−/− DT40 cells were compared (Figure 8). For the mitochondrial extract from wild-type cells, formation of the APTX, pol β and FEN1 products was observed (Figure 8, lanes 1–11). For the reaction in the mitochondrial extract from APTX−/− cells, APTX activity was deficient and the product corresponding to pol β removal of the 5′-AMP-dRP group was stronger, yet the product of FEN1 excision was also observed (Figure 8, lanes 12–22). These results indicate that the pol β lyase and FEN1 excision activities are strong enough in the mitochondrial extract to complement the deficiency in APTX activity. The explanation for the FEN1 activity found in the wild-type and APTX−/− mitochondrial extracts (Figure 8), but not in APTX−/− Pol β−/− mitochondrial extracts (Figure 7), could be due to a stimulatory effect of pol β on FEN1 activity, as illustrated (Supplementary Figure S6).
Figure 8.
Complementation of APTX deficiency by pol β and FEN1 for processing of the 5′-AMP-dRP-containing BER intermediate in mitochondrial extracts. Lanes 1–11 and 12–22 are the reaction products observed in mitochondrial extracts from wild-type and APTX−/− cells, respectively and correspond to time points 1, 3, 5, 10, 15, 20, 25, 30, 40, 50, and 60 min. Illustrations of the single-nucleotide gapped 5′-AMP-dRP-containing DNA substrate and reaction products of pol β, APTX, and FEN1 are as presented in Figure 7A.
DISCUSSION
APTX deficient cells fail to show hypersensitivity to genotoxic agents that generate base lesions and DNA strand breaks, and APTX null mice lack an overt phenotype (10–12,16,31,32). These results point to the presence of repair pathways that can compensate for APTX deficiency. For example, regarding stability of the nuclear genome, it was previously demonstrated that pol β and FEN1 could function as back up mechanisms for APTX deficiency in repair of blocks with the 5′-AMP-dRP and 5′-AMP groups (14).
A lack of coordination during BER can enable premature action by a DNA ligase prior to 5′-dRP removal and gap filling. In this case, 5′-adenylation of the dRP group containing BER intermediate can occur. In a previous study, it was found that nuclear DNA ligases, DNA ligase III/XRCC1 complex and DNA ligase I, can fail when processing this BER intermediate, leading to formation of the 5′-adenylated-dRP block, i.e. 5′-AMP-dRP (13). Here, the mitochondrial DNA ligase III also failed during repair of abasic sites (33,34). Similarly, it has been shown that a premature DNA ligation reaction by mitochondrial DNA ligase III can occur when the enzyme attempts to seal a nick with an oxidatively modified 3′-DNA end (35). Nevertheless, 5′-adenylation of the dRP group stabilizes the BER intermediate against the polymerase lyase reaction in repair of the 5′-AMP-dRP group. Therefore, this DNA intermediate may pose a block to BER, especially in the case of the involvement of a polymerase with weak lyase activity. In this regard, it was known that the activity of mitochondrial pol γ for 5′-dRP group removal is much weaker (∼40-fold) than that of pol β (20), but the pol γ lyase in repair of the 5′-AMP-dRP group has not been determined.
At the outset of this work, two main complementing activities for mitochondrial APTX deficiency were envisioned: dRP lyase by pol γ or pol β and FEN1-mediated LP BER. We found that the lyase activity of pol γ is very weak against the 5′-AMP stabilized dRP group and this pol γ activity was not strong enough to remove the AMP block during BER. Similarly, we also found that the FEN1-mediated strand excision reaction of the LP BER sub-pathway in mitochondrial extracts was not active against the 5′-AMP block. However, in the mitochondrial extracts from APTX−/− cells, the strong pol β lyase and FEN1 activities appear able to repair the blocked BER intermediates with 5′-AMP-dRP groups. Overall, these results suggest that neither pol γ lyase nor FEN1 is able to complement APTX deficiency in the absence of pol β.
A stimulatory role of the pol γ accessory subunit (pol γB) in processive DNA synthesis, as well as an enhancement in DNA binding and affinity for DNA primer-templates have been reported (24,36). However, we observed similar lyase activity for the catalytic subunit of pol γ (pol γA), i.e. for either 5′-dRP or 5′-AMP-dRP removal, in the absence or presence of pol γB subunit. The difference between these results and an earlier study that showed pol γB stimulation of the pol γ lyase (36) could be due to measurement of protein-dRP intermediate cross-linking in the earlier study instead of 5′-dRP group release from the substrate, as shown here.
Previously, a slow rate of DNA repair was reported in the mitochondrial extracts from human AOA1 cells for the 5′-AMP-containing substrate (37). Our current results demonstrate that this could have been due to a lack of back up enzymes or repair mechanisms, especially those of the pol γ lyase or LP BER. These findings regarding the inability of pol γ to complement APTX deficiency in mitochondria could be useful for understanding the potential for mitochondrial repair mechanisms to modulate AOA1 pathobiology. A potential role of mitochondrial BER deficiency in the pathobiology of neurological diseases such as Alzheimer's disease, Huntington's disease, Ataxia Telangiectasia, and AOA1 has been proposed (37–42), and DNA damage-induced mitochondrial dysfunction has been suggested to be associated with neurological phenotypes of AOA1 patients (37). Therefore, roles of mitochondrial BER in modifying neurodegenerative diseases are important to consider.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Robert M. Petrovich (Protein Expression Core Facility, NIEHS) for assistance with cell growth. We thank Julie K. Horton for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the NIH; National Institute of Environmental Health Sciences [Z01 ES050158, ES050159 to S.H.W., Z01 ES065078 to W.C.C.]; JSPS KAKENHI [JP15K00537 to K.T.]; JSPS Core-to-Core Program; A. Advanced Research Networks and JSPS KAKENHI Grant-in-Aid for Scientific Research [JP16H06306 to S.T.]. Funding for open access charge: Intramural Research Program of the NIH, National Institute of Environmental Health Sciences [Z01 ES050158, ES050159 to S.H.W., Z01 ES065078 to W.C.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ahel I., Rass U., El-Khamisy S.F., Katyal S., Clements P.M., McKinnon P.J., Caldecott K.W., West S.C.. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006; 443:713–716. [DOI] [PubMed] [Google Scholar]
- 2. Rass U., Ahel I., West S.C.. Defective DNA repair and neurodegenerative disease. Cell. 2007; 130:991–1004. [DOI] [PubMed] [Google Scholar]
- 3. Mosesso P., Piane M., Paliitti F., Pepe G., Penna S., Chessa L.. The novel human gene aprataxin is directly involved in DNA single-strand-break repair. Cell Mol. Life Sci. 2005; 62:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sano Y., Date H., Igarashi S., Onodera O., Oyake M., Takahashi T., Hayashi S., Morimatsu M., Takahashi H., Makifuchi T. et al. the causative protein for EAOH is a nuclear protein with a potential role as a DNA repair protein. Ann. Neurol. 2004; 55:241–249. [DOI] [PubMed] [Google Scholar]
- 5. Clements P.M., Breslin C., Deeks E.D., Byrd P.J., Ju L., Bleganowski P., Brenner C., Moreira M.C., Taylor A.M., Caldecott K.W.. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair. 2004; 3:1493–1502. [DOI] [PubMed] [Google Scholar]
- 6. Hirona M., Yamamoto A., Mori T., Lan L., Iwamoto T., Aoki M., Shimmada K., Furiya Y., Kariya S., Asai H. et al. DNA single-strand break repair is impaired in Aprataxin-related ataxia. Ann. Neurol. 2007; 61:162–174. [DOI] [PubMed] [Google Scholar]
- 7. Harris J.L., Jakob B., Taucher-Scholz G., Dianov G.L., Becherel O.J., Lavin M.F. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum. Mol. Genet. 2009; 18:4102–4117. [DOI] [PubMed] [Google Scholar]
- 8. Prasad R., Williams J.G., Hou E.W., Wilson S.H.. Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 2002; 40:11571–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Date H., Onodera O., Iwabuchi K., Uekawa K., Igarashi S., Kolke R., Hiro T., Yuasa T., Awaya Y., Sakai T. et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat. Genet. 2001; 29:184–188. [DOI] [PubMed] [Google Scholar]
- 10. Reynolds J.J., El-Khamisy S.F., Katyal S., Clements P., McKinnon P.J., Caldecott K.W.. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia-1. Mol. Cell Biol. 2009; 29:1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Becherel O.J., Jakob B., Cherry A.L., Gueven N., Fusser M., Kijas A.W., Peng C., Katyal S., McKinnon P.J., Chen J. et al. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2009; 38:1489–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El-Khamisy S.F., Katyal S., Patel P., Ju L., McKinnon P.J., Caldecott K.W.. Synergic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair. 2009; 8:760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Çağlayan M., Batra V.K., Sassa A., Prasad R., Wilson S.H.. Role of polymerase β in complementing aprataxin deficiency during abasic-site base excision repair. Nat. Struct. Mol. Biol. 2014; 21:497–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Çağlayan M., Horton J.K., Prasad R., Wilson S.H.. Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Res. 2015; 43:2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sykora P., Croteau D.L., Bohr V.A., Wilson D.M.. Aprataxin localized to mitochondria and preserves mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7437–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gueven N., Becherel O.J., Kijas A.W., Chen P., Howe O., Rudolph J.H., Gatti R., Date H., Onodera O., Taucher-Scholz G. et al. Aprataxin, a novel protein that protects against genotoxic stress. Hum. Mol. Genet. 2004; 13:1081–1093. [DOI] [PubMed] [Google Scholar]
- 17. Garcia-Diaz B., Barca E., Balreira A., Lopez L.C., Tadesse S., Krishna S., Naini A., Mariotti C., Castellotti B., Quinzii C.M.. Lack of aprataxin impairs mitochondrial functions via downregulation of the APE1/NRF1/NRF2 pathway. Hum. Mol. Gen. 2015; 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Longley M.J., Ropp P.A., Copeland W.C.. Characterization of the native and recombinant catalytic subunit of human DNA polymerase γ: Identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998; 37:10529–10539. [DOI] [PubMed] [Google Scholar]
- 19. Kasiviswanathan R., Longley M.J., Young M.J., Copeland W.C.. Purification and functional characterization of human mitochondrial DNA polymerase gamma harboring disease mutations. Methods. 2010; 51:379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Longley M.J., Prasad R., Srivastava D.K., Wilson S.H., Copeland W.C.. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase γ and its role in mitochondrial base excision repair in vitro. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12244–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szczesny B., Tann A.W., Longley M.L., Copeland W.C., Mitra S.. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008; 283:26349–26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu P., Qian L., Sung J., Souza-Pinto N.C., Zheng L., Bogenhagen D.F., Bohr V.A., Wilson D.M., Shen B., Demple B.. Removal of oxidative DNA damage via FEN1-dependent long-path base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008; 28:4975–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sykora P., Kanno S., Akbari M., Kulikowicz T., Baptiste B.A., Leandro G.S., Hu H., Tian J., May A., Becker K.A. et al. DNA polymerase beta participates in mitochondrial DNA repair. Mol. Cell. Biol. 2017; doi:10.1128/MCB.00237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim S.E., Longley M.J., Copeland W.C.. The mitochondrial p55 accessory subunit of human DNA polymerase γ enhances DNA binding, promotes processive DNA synthesis, and confers N-Ethylmaleimide resistance. J. Biol. Chem. 1999; 274:38197–38203. [DOI] [PubMed] [Google Scholar]
- 25. Tano K., Nakamura J., Asagoshi K., Arakawa H., Sonoda E., Braithwaite E.K., Prasad R., Buerstedde J., Takeda S., Watanaba M. et al. Interplay between DNA polymerase β and λ in repair of oxidation DNA damage in chicken DT40 cells. DNA Repair. 2007; 6:869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bogenhagen D., Clayton D.A.. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 1974; 249:7991–7995. [PubMed] [Google Scholar]
- 28. Ropp P.A., Copeland W.C.. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996; 36:449–458. [DOI] [PubMed] [Google Scholar]
- 29. Liu Y., Beard W. A., Shock D.D., Prasad R., Hou E.W., Wilson S.H.. DNA polymerase β and Flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J. Biol. Chem. 2005; 280:3665–3674. [DOI] [PubMed] [Google Scholar]
- 30. Hansen A.B., Griner N.B., Anderson J.P., Kujoth G.C., Prolla T.A., Loeb L.A., Glick E.. Mitochondrial DNA integrity is not dependent on DNA polymerase β activity. DNA Repair. 2006; 5:71–79. [DOI] [PubMed] [Google Scholar]
- 31. Gueven N., Chen P., Nakamura J., Becherel O.J., Kijas A.W., Grattan-Smith P., Lavin M.F.. A subgroup of spinocerebellar ataxias defective in DNA damage responses. Neuroscience. 2007; 145:1418–1425. [DOI] [PubMed] [Google Scholar]
- 32. Luo H., Chan D.W., Yang T., Rodriguez M., Chen P., Leng M., Mu J., Chen D., Songyang Z., Qin J.. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol. Cell Biol. 2004; 24:8356–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levin C.J., Zimmerman S.B.. A DNA ligase from mitochondria of rat liver. Biochem. Biophys. Res. Commun. 1976; 69:514–520. [DOI] [PubMed] [Google Scholar]
- 34. Pinz K.G., Bogenhagen D.F.. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol. Cell Biol. 1998; 18:1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ellenberg T., Tomkinson A.E.. Eukaryotic DNA polymerases: structural and functional insights. Annu. Rev. Biochem. 2008; 77:313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pinz K.G., Bogenhagen D.F.. The influence of the DNA polymerase γ accessory subunit on base excision repair by the catalytic subunit. DNA Repair. 2006; 5:121–128. [DOI] [PubMed] [Google Scholar]
- 37. Akbari M., Sykora P., Bohr V.A.. Slow mitochondrial repair of 5′-AMP renders mtDNA susceptible to damage in APTX deficient cells. Sci. Rep. 2015; 5:12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meagher M., Lightowlers R.N.. The role of TDP1 and APTX in mitochondrial DNA repair. Biochimie. 2014; 100:121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Çağlayan M., Wilson S.H.. Oxidant and environmental toxicant-induced effects compromise DNA ligation during base excision DNA repair. DNA Repair. 2015; 35:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Canugovi C., Shamanna R.A., Croteau D.L., Bohr V.A.. Base excision DNA repair levels in mitochondrial lysates of Alzheimer's disease. Neurobiol. Aging. 2014; 35:1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharma N.K., Lebedeva M., Thomas T., Kovalenko O.A., Stumpf J.D., Shadel G.S., Santos J.H. Intrinsic mitochondrial DNA repair defects in Ataxia Telangiectasia. DNA Repair. 2014; 13:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ross C.A., Truant R.. DNA repair: a unifying mechanism in neurodegeneration. Nature. 2017; 541:34–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.