Acute lymphoblastic leukemia (ALL) is characterized by an outgrowth of malignant lymphoblasts that occupy the bone marrow microenvironment where they reside with mesenchymal stromal cells (MSCs).1 Leukemic cells are able to disrupt the healthy microenvironment and create a leukemic niche that shelters malignant cells from elimination through cytostatic treatment and immune responses.2,3 Disrupting intercellular communication in this malignant microenvironment sensitizes leukemic cells to chemotherapeutic drugs.4 Homing of ALL cells toward the bone marrow microenvironment is thought to be similar to that of hematopoietic stem cells (HSCs).5 Human CD34+ progenitor cells, are attracted by stromal cell-derived factor 1 (SDF-1/CXCL12), a chemoattractant actively produced by MSCs in the bone marrow.5,6 CXCL12 binds to the CXCR4 receptor. Disturbance of the CXCR4/CXCL12 axis by treatment with cytokines and/or CXCR4 antagonists mobilizes HSCs to the peripheral blood7 and shows promising pre-clinical results in acute myeloid leukemia patients.8 Like HSCs, ALL cells have high CXCR4 surface expression and disruption of CXCR4/CXCL12 interaction reduces ALL engraftment in animal models.9,10 However, in mice suffering from ALL, CXCR4/CXCL12 inhibition as mono-treatment did not reduce the leukemic cell number in the bone marrow.9,11 In addition, CXCL12 expression was remarkably down-regulated in murine bone marrow regions of extensive ALL growth.1 Likewise, CXCL12 levels in the bone marrow of patients diagnosed with BCP-ALL were lower compared to those in the same patients at the time of remission and healthy controls.6 This suggests that ALL cells use additional mechanisms to retain a protective microenvironment.
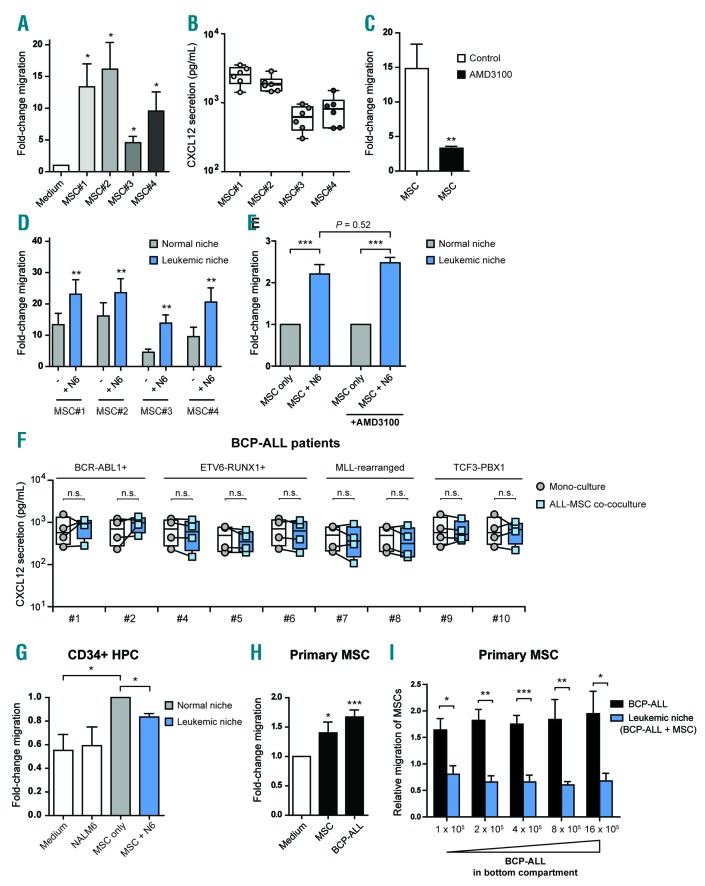
In this study, we used an ex vivo co-culture model in which we cultured B-cell precursor ALL (BCP-ALL) cells with bone marrow-derived primary MSCs. Several studies show that ALL cells home toward the bone marrow microenvironment in a CXCR4/CXCL12-dependent manner.1,9,10 We confirmed these findings in our model by assessing the migration of GFP-positive BCP-ALL cells (NALM6) toward MSCs using a transwell system. MSCs induced migration of BCP-ALL cells by 5-16-fold compared to medium controls (P<0.05) (Figure 1A and Online Supplementary Figure S1A and B). The induction of migration was correlated with the level of CXCL12 produced by the primary MSCs (Figure 1B). Inhibition of the CXCR4/CXCL12 axis with AMD3100 significantly impaired migration of BCP-ALL cells toward MSCs (4.5-fold reduction; P<0.01) (Figure 1C).
Figure 1.
Acute lymphoblastic leukemia (ALL) mesenchymal stromal cell (MSC) co-cultures induce leukemic cell migration without affecting the CXCR4/CXCL12 axis. (A) Fold-change in migration of GFP-positive B-cell precursor (BCP)-ALL cells (NALM6) toward four distinct primary bone marrow-derived MSCs compared to background migration toward culture medium after 48 hours (h) in a 3.0 μm transwell system (n=5 for each MSC; one-tailed t-test, unpaired). We used MSCs derived from healthy controls (MSC#1 and MSC#2) and MSCs derived from leukemia patients at diagnosis (MSC#3 and MSC#4); see also Online Supplementary Table S3. Migration towards culture medium was used to calculate the fold-change migration. (B) Graph showing the secretion of CXCL12 by primary MSCs as determined using a fluorescent bead-based immunoassay (n=6 for each MSC). (C) Graph showing the effect of CXCR4 blockade with AMD3100 (10 μM) on the migration of NALM6 cells toward primary MSCs after 48 h in a 3.0 μm transwell system (n=4; one-tailed t-test, paired). Migration towards culture medium was used to calculate the fold-change migration. For this experiment, we combined the data of four distinct primary MSCs (MSC#1-4). (D) Migration of NALM6 cells toward MSCs in mono-culture (normal niche, gray bars) or toward ALL-MSC co-cultures (leukemic niche, blue bars) after 48 h in a 3.0 μm transwell system (n=5 for each MSC; one-tailed t-test, paired). Migration towards culture medium was used to calculate the fold-change migration. Experiments were performed with MSCs from 4 donors. (E) Graph showing that the leukemic niche affects the migration of BCP-ALL cells independent of CXCR4/CXCL12 signaling. Migration of NALM6 cells toward NALM6-MSC co-cultures (leukemic niche, blue bars) is compared to migration toward MSC mono-cultures (normal niche, gray bars) in absence or presence of AMD3100 (10 μM) after 48 h using a 3.0 μm transwell system (n=4; one-tailed t-test, paired). For this experiment, we combined the data of four distinct primary MSCs (MSC#1-4). Data are shown as fold-induction compared to migration towards MSC mono-culture. (F) Plots show the secreted levels of CXCL12 by primary BCP-ALL cells and MSCs in mono-culture (circles represent the sum of cytokine secretion in mono-culture of BCP-ALL and mono-culture of MSC) compared to ALL-MSC co-culture (squares). Data were determined using a multiplexed fluorescent bead-based immunoassay. Boxes represent p25–p75 intervals. (G) Graph showing the migration of umbilical cord blood-derived healthy CD34+ progenitor cells after 48 h in a 3.0 μm transwell system. Bars represent migration toward culture medium, NALM6 cells, MSCs and NALM6-MSC cultures (n=3; one-tailed t-test; paired). Migration towards MSC mono-culture was used to calculate the fold-change migration. Data are mean±Standard Error of Mean (SEM); *P≤0.05. (H and I) Primary MSCs (experiments performed with MSC#1 and MSC#3) were allowed to migrate to the other side of a 8.0 μm transwell insert for 16 h. Cells were fixed and subsequently stained with crystal violet. MSCs migrated toward a bottom compartment containing culture medium, MSCs or BCP-ALL cells (REH). The amount of migrated cells was measured by spectrophotometry of crystal violet stained cells. (H) Graph showing the migration of primary MSCs. Migration towards medium was used to calculate the fold-change migration (n=4; one-tailed t-test; paired). (I) Graph showing the migration of primary MSCs toward REH cells (BCP-ALL, black bars) or REH-MSC co-cultures (leukemic niche, blue bars) (n=4; one-tailed t-test, paired). Data are relative to control condition without BCP-ALL cells (medium-only or MSC-only control). Data are expressed as mean±SEM; *P≤0.05, **P≤0.01, ***P≤0.001. See also Online Supplementary Figures S1–S3. N6: NALM6; HPC: hematopoietic progenitor cell.
Next, we hypothesized that BCP-ALL cells alter the chemoattractive properties of their microenvironment. To address this hypothesis, GFP-negative BCP-ALL cells (NALM6) were cultured with MSCs in the bottom compartment of a transwell system as a model of the leukemic niche. Migration of GFP-positive BCP-ALL cells (NALM6) from the upper to the bottom compartment was determined (Online Supplementary Figure S1A–C). GFP-positive BCP-ALL cells migrated significantly more toward ALL-MSC co-cultures than toward MSCs in mono-culture (blue bars vs. gray bars; P<0.01) (Figure 1D and Online Supplementary Figure S1D and E). The leukemic niche induced migration of BCP-ALL cells by 2.2-fold (P<0.001) (Figure 1E). Surprisingly, CXCR4 inhibition by AMD3100 did not affect the preferential migration of BCP-ALL cells towards the leukemic niche (2.2-fold vs. 2.5-fold; P=0.52) (Figure 1E and Online Supplementary Figure S1F). In addition, we compared CXCL12 levels in the supernatant of mono- and co-cultures of primary BCP-ALL cells derived from 9 different BCP-ALL cases and 4 primary MSCs. CXCL12 levels were high in mono-cultures of MSCs, and below detection level in mono-cultures of primary BCP-ALL cells (Online Supplementary Figure S2A and B). Co-culture of primary BCP-ALL cells and MSCs did not increase CXCL12 levels compared to the combined levels produced in both mono-cultures (Figure 1F). These results suggest that BCP-ALL cells create a leukemic niche that attracts leukemic cells in a CXCR4/CXCL12-independent manner.
Next, we assessed the migration of healthy CD34+ hematopoietic progenitor cells (HPCs) toward primary MSCs cultured with or without BCP-ALL cells. As expected, CD34+ HPCs migrated more abundantly toward MSCs than to control medium without MSCs (1.8-fold; P<0.05) (Figure 1G). However, migration of CD34+ HPCs toward ALL-MSC co-cultures was significantly reduced compared to migration toward MSCs in mono-cultures (20% reduction; P<0.05) (Figure 1G). In addition, we assessed the migratory behavior of healthy MSCs toward ALL-MSC co-cultures. We performed a transwell migration assay in which primary MSCs were allowed to migrate toward BCP-ALL cells or other MSCs in the bottom compartment. We observed that MSC migration was induced toward other MSCs compared to control medium (P<0.05) (Figure 1H). Strikingly, MSCs also actively migrated to BCP-ALL cells illustrating that BCP-ALL cells are able to recruit MSCs (P<0.001) (Figure 1H and Online Supplementary Figure S3A). However, MSC migration was significantly inhibited toward the leukemic niche (ALL-MSC co-cultures) compared to a bottom compartment containing only BCP-ALL cells (2–3-fold reduction; P<0.001) (Figure 1I and Online Supplementary Figure S3B and C). Interestingly, increasing the amount of ALL cells (up to 16-fold) in ALL-MSC co-cultures did not re-induce MSC migration (Figure 1I). These results show that ALL-MSC co-cultures efficiently attract leukemic cells, but reduce migration of healthy CD34+ HPCs and primary MSCs, suggesting that ALL-MSC co-cultures produce chemokines that specifically attract BCP-ALL cells.
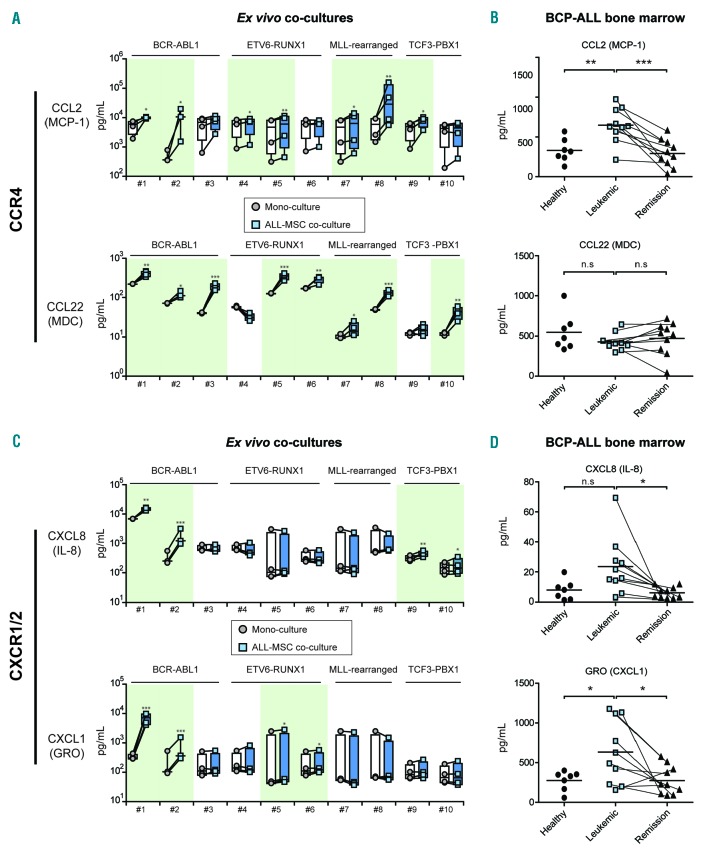
To address which cytokines/chemokines may be altered in the leukemic niche, we quantified the levels of 64 cytokines known to be important in immunology/hematology in the supernatant of ex vivo co-cultures of patient-derived BCP-ALL cells (n=10, representing major BCP-ALL subgroups) and MSCs (n=4) (Online Supplementary Table S1). Cytokine/chemokine profiles of mono-cultures of primary BCP-ALL cells and primary MSCs are shown in Online Supplementary Figure S2. Co-cultures of BCP-ALL cells with MSCs revealed the induction of unique secretion patterns per patient. These patterns were similar for different sources of MSC, i.e. MSCs obtained at the time of leukemia and those obtained from healthy controls. The unique profile per BCP-ALL patients suggests that leukemic cells govern the observed cytokine secretion (Figure 2, Online Supplementary Table S2 and Online Supplementary Figure S4A and B). These unique cytokine/chemokine profiles suggest the need for patient-specific approaches to disrupt the leukemic niche. However, we also found recurrently induced signaling pathways. All ALL-MSC co-cultures showed increased levels of CCL2/MCP-1 and/or CCL22/MDC (Figure 2A), both ligands for the CCR4 receptor. This chemokine receptor is implicated in T-cell leukemia and lymphoma, but has not yet been implicated as an important factor in BCP-ALL.12 In addition, analysis of previously published microarray-based gene expression data showed that CCR4 expression levels in pediatric ALL cells were significantly higher compared to healthy hematopoietic cells (Online Supplementary Figure S4C).
Figure 2.
Chemokines inducing the CCR4 and CXCR1/2 axes are up-regulated in the B-cell precursor (BCP)-acute lymphoblastic leukemia (ALL) niche. (A) Plots showing the secretion of CCL2/MCP-1 and CCL22/MDC by primary mesenchymal stromal cells (MSCs) (n=4) and primary BCP-ALL cells (n=10) in monoculture (circles represent the sum of cytokine secretion in mono-culture of BCP-ALL and mono-culture of MSC) compared to ALL-MSC co-cultures (squares). Data were obtained using a multiplexed fluorescent bead-based immunoassay. Boxes represent p25–p75 intervals. Raw data were logarithmically transformed to obtain a normal distribution of the data and upregulation of cytokines was tested using a one-tailed paired t-test; *P≤0.05, **P≤0.01, ***P≤0.001. (B) Plots showing the serum levels of CCL2/MCP-1 and CCL22/MDC in bone marrow aspirates from healthy controls (n=7, circles), from untreated BCP-ALL patients at diagnosis (n=10, blue squares) and from BCP-ALL patients after induction treatment (n=10, triangles; lines indicate paired samples). Data were obtained using a multiplexed fluorescent bead-based immunoassay in which we measured the levels of 64 cytokines. Healthy (circles) and leukemic samples (blue squares) were compared using a two-tailed unpaired t-test. Leukemic samples before and after induction therapy (blue squares vs. triangles) were compared using a one-tailed paired t-test; *P≤0.05, **P≤0.01, ***P≤0.001. (C) Same as (A) for CXCL8/IL-8 and CXCL1/GRO secretion. (D) Plots showing the serum levels of GRO/CXCL1 and CXCL8/IL-8. See also Online Supplementary Tables S1 and S2, and Online Supplementary Figures S2 and S4.
In addition, we observed recurrent induction of CXCL8/IL-8 and CXCL1/GRO secretion, ligands of the CXCR1 and CXCR2 receptor. CXCR1/2 ligands were induced by BCP-ALL cells from 6 out of 10 patients (Figure 2C). In contrast to CCR4 expression, CXCR1 and CXCR2 expression levels were lower in pediatric ALL compared to healthy hematopoietic cells (Online Supplementary Figure S3C).
We validated the cytokine/chemokine patterns identified in our co-culture model using serum derived from bone marrow aspirates from 10 ALL patients at diagnosis. We compared the cytokine/chemokine patterns in these samples with serum obtained from bone marrow aspirates of the same patients taken at the end of induction chemotherapy. At this stage of treatment, the patients were in remission. Furthermore, we used bone marrow aspirates of 7 healthy controls to analyze cytokine/chemokine patterns of the healthy niche. Levels of the CCR4 ligand CCL2/MCP-1 were significantly higher in the bone marrow of untreated, newly diagnosed BCP-ALL patients compared to healthy controls (2.0-fold increase; P<0.01) (Figure 2B). Interestingly, after induction therapy, the CCL2/MCP-1 levels in the bone marrow of ALL patients decreased to levels observed in healthy controls (2.2-fold decrease; P<0.01) (Figure 2B). This suggests that the induced CCL2/MCP-1 levels at diagnosis are causally linked to the presence of leukemic cells. CCL22/MDC levels were similar between BCP-ALL patients and healthy controls (Figure 2B). Like CCL2/MCP-1, CXCL1/GRO levels were also increased in the bone marrow of BCP-ALL patients (2.3-fold increase; P<0.05) (Figure 2D) and CXCL8/IL8 levels showed a trend towards upregulation (2.5-fold; P=0.052) (Figure 2D). Similar to CCL2/MCP-1 levels, the secreted levels of these CXCR1/2 ligands were significantly reduced after induction chemotherapy (2.3-fold and 3.1-fold, respectively; P<0.05) (Figure 2D). These data implicate that CCR4 and CXCR1/2 signaling are important in the bone marrow of BCP-ALL patients and suggest that CCR4 inhibition (e.g. by mogamulizumab1)13 or CXCR1/2 inhibition might be an alternative or additional way to disrupt the BCP-ALL niche.
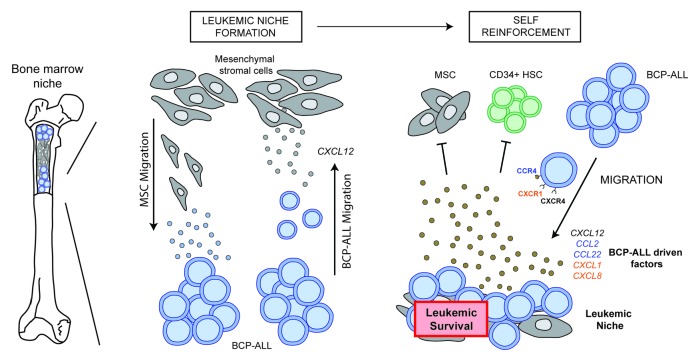
In conclusion, our data indicate that the BCP-ALL niche is highly potent in attracting BCP-ALL cells and repelling the influx of healthy hematopoietic cells and MSCs in a CXCL12-independent manner. Instead, we show the recurrent induction of CCR4 and CXCR1/2 ligands by BCP-ALL cells in the leukemic niche (Figure 3). Our data suggest CCR4 and CXCR1/2 to be valuable future therapeutic targets to interfere with the leukemic niche in ALL.
Figure 3.
Model of the self-reinforcing B-cell precursor (BCP)-acute lymphoblastic leukemia (ALL) niche. Proposed model for the formation of a leukemic niche by BCP-ALL cells. BCP-ALL cells actively migrate toward mesenchymal stromal cells (MSCs) in a CXCL12-dependent manner. Simultaneously, MSC migration is induced by BCP-ALL cells. When BCP-ALL cells and MSCs are in close proximity, a leukemic microenvironment is created that specifically attracts other BCP-ALL cells and inhibits the migration of MSCs and healthy CD34+ cells. This process is CXCR4/CXCL12-independent and is characterized by the secretion of patient-unique cytokines.
Supplementary Material
Acknowledgments
We thank all members of the research laboratory Pediatric Oncology of the Erasmus MC for their help in processing leukemic and mesenchymal stromal cell samples, M. Buitenhuis for critical discussions and reading of the manuscript, the Vlietland Ziekenhuis for collecting and providing cord blood, and the Amsterdam Medical Center for providing the R2 gene genomic analysis and visualization platform.
Footnotes
Funding: the work described in this paper was funded by the KiKa Foundation (Stichting Kinderen Kankervrij – Kika-39), the Dutch Cancer Society (UVA 2008; 4265, EMCR 2010; 4687), the Netherlands Organization for Scientific Research (NWO – VICI M.L. den Boer) and the Pediatric Oncology Foundation Rotterdam.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Colmone A, Amorim M, Pontier AL, et al. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322(5909):1861–1865. [DOI] [PubMed] [Google Scholar]
- 2.Fujisaki J, Wu J, Carlson AL, et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature. 2011;474(7350):216–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakasone ES, Askautrud HA, Kees T, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21(4):488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polak R, de Rooij B, Pieters R, et al. B-cell precursor acute lymphoblastic leukemia cells use tunneling nanotubes to orchestrate their microenvironment. Blood. 2015;126(21):2404–2414. [DOI] [PubMed] [Google Scholar]
- 5.Peled A, Petit I, Kollet O, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283(5403):845–848. [DOI] [PubMed] [Google Scholar]
- 6.van den Berk LC, van der Veer A, Willemse ME, et al. Disturbed CXCR4/CXCL12 axis in paediatric precursor B-cell acute lymphoblastic leukaemia. Br J Haematol. 2014;166(2):240–249. [DOI] [PubMed] [Google Scholar]
- 7.Flomenberg N, Devine SM, Dipersio JF, et al. The use of AMD3100 plus G-CSF for autologous hematopoietic progenitor cell mobilization is superior to G-CSF alone. Blood. 2005;106(5):1867–1874. [DOI] [PubMed] [Google Scholar]
- 8.Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23(1):43–52. [DOI] [PubMed] [Google Scholar]
- 9.Juarez J, Dela Pena A, Baraz R, et al. CXCR4 antagonists mobilize childhood acute lymphoblastic leukemia cells into the peripheral blood and inhibit engraftment. Leukemia. 2007;21(6):1249–1257. [DOI] [PubMed] [Google Scholar]
- 10.Sipkins DA, Wei X, Wu JW, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435(7044):969–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parameswaran R, Yu M, Lim M, et al. Combination of drug therapy in acute lymphoblastic leukemia with a CXCR4 antagonist. Leukemia. 2011;25(8):1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa M, Schmitz R, Xiao W, et al. Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma. J Exp Med. 2014;211(13):2497–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duvic M, Pinter-Brown LC, Foss FM, et al. Phase 1/2 study of mogamulizumab, a defucosylated anti-CCR4 antibody, in previously treated patients with cutaneous T-cell lymphoma. Blood. 2015; 125(12):1883–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.